Abstract
Chromatin states are the key to embryonic stem cell pluripotency and differentiation. Chromatin immunoprecipitation (ChIP) followed by high-throughput sequencing (ChIP-Seq) is increasingly used to map chromatin states and to functionally annotate the genome. Many ChIP-Seq profiles, especially those of histone methylations, are noisy and diffuse. Here we describe SICER (Zang et al., Bioinformatics 25(15):1952–1958, 2009), an algorithm specifically designed to identify disperse ChIP-enriched regions with high sensitivity and specificity. This algorithm has found a lot of applications in epigenomic studies. In this Chapter, we will demonstrate in detail how to run SICER to delineate ChIP-enriched regions and assess their statistical significance, and to identify regions of differential enrichment when two chromatin states are compared.
Keywords: ChIP-Seq, Histone modifications, Epigenetic modifications, Epigenome, SICER
1 Introduction
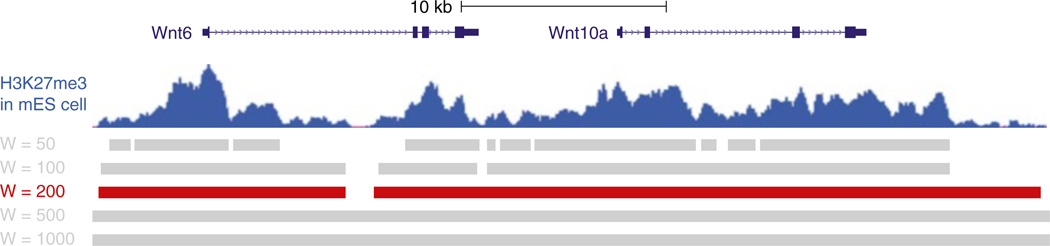
Chromatin structure plays a critical role in embryonic stem cell pluripotency and differentiation. Chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-Seq) is now widely used to quantify chromatin states across genomes. Large amount of ChIP-Seq datasets of genome-wide profiling of epigenetic modifications and chromatin-binding proteins have been generated. The distribution of ChIP-Seq signals has been found to vary widely, ranging from a few nucleosomes to large chromatin domains encompassing multiple genes. For example, H3K4me2 and H3K4me3, which are usually associated with enhancers and promoters, tend to exhibit relatively localized sharp peaks [1, 2]. On the other hand, H3K36me3, a hallmark of elongation, or repressive mark H3K27me3 may span tens or even hundreds of kilo bases (for an example, please see Fig. 1). Diffuse signals can be observed in many libraries. In addition to chromatin modifications, some histone-modifying enzymes [3], chromatin remodeling complexes, and RNA Pol II also exhibit extended domains of enrichment. Because the detection of diffuse signals often suffers from high noise level and lack of saturation in sequencing coverage, it is a challenging task to identify statistically significant ChIP-enriched domains. These generally weak and diffuse signals render approaches seeking strong local enrichment, such as those peak-finding algorithms designed for finding transcription factor (TF) binding sites, inadequate. Toward this end, we developed SICER (Statistical model for Identification of ChIP-Enriched Regions) [4], which achieves high sensitivity and specificity by identifying spatial clusters of ChIP-enriched signals that are unlikely to appear in a background model. As demonstrated in Fig. 1, SICER is able to identify extended domains of ChIP enrichment. Although SICER was designed for analyzing ChIP-Seq data with extended enrichment profile, upon a proper choice of parameters, it could also be applied to ChIP-Seq data with sharp peaks like those for transcription factors.
Fig. 1.
SICER with default parameters identifies extended domain with H3K27me3 enrichment in mouse embryonic stem cell across Wnt6 and Wnt10a gene locus. Top track: H3K27me3 ChIP-Seq data from ENCODE [14]. Middle track: H3K27me3 enrichment peaks identified in ENCODE. Bottom track: H3K27me3 enrichment domains identified by SICER [4] with default parameters
2 Overview of SICER
2.1 Motivation
Classic examples of the mechanism of domain formation of histone modifications include H3K9me3 in yeast. H3K9me3 recruits HP1, which in turn recruits H3K9 methyltransferase Suv39h. Suv39h modifies H3K9 on other nucleosomes in the vicinity, thereby self-propagating the heterochromatin state [5–7]. Another example is H3K27me3. This mark is deposited by the polycomb complex, PRC2, and is believed to recruit the PRC1 complex [8]. In Drosophila, it has been suggested that the looping action of PRC1 and PRC2 that both anchor at the polycomb response elements results in the spreading of H3K27me3 [8]. Inspired by the mechanisms of domain formation illustrated by these examples, SICER [4] regards significant spatial clusters of ChIP enrichment as true signal. The main feature of the SICER algorithm is to pool together signals from nearby nucleosomes that are in the same modification state for identification of statistically significant enrichments. For ChIP-Seq libraries with diffuse profile, this feature alleviates the problem of lack of saturation and markedly improves the signal-to-noise ratio, where at any short scale of one or several nucleosomes, the ChIP enrichment does not appear to be significant enough. This approach also enables a systematic evaluation of statistical significance of identified ChIP-enriched regions against control library when available.
2.2 Algorithm
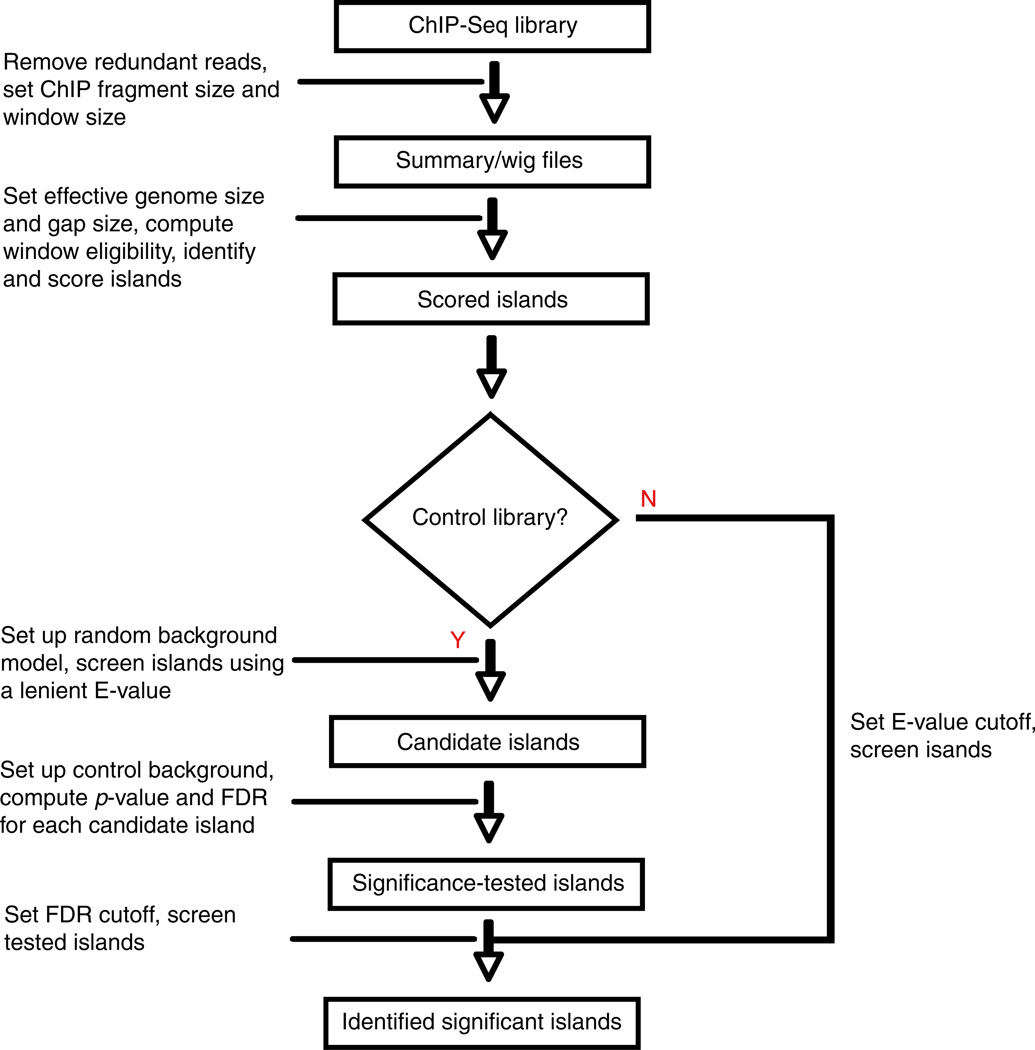
The key concept that SICER uses to capture spatial clustering of reads is island. To delineate the islands and assess the statistical significance of ChIP enrichment on them, SICER [4] carry out the following steps: (1) It partitions the genome into nonoverlapping windows of size w. (2) It identifies windows with enrichment (i.e., “eligible” windows). A window is deemed “eligible” (“ineligible”) if the number of ChIP-Seq reads in this window is equal to or above (below) a read count threshold l0. The threshold l0 is determined based on a Poisson distribution . λ = wN/L is the average number of reads in a window. N is the number of reads in the library and L the effective genome length (further discussion on L can be found in Subheading 2.3.3). The threshold p0 is defaulted to be 0.2 so that all windows with reasonable ChIP enrichment are “eligible.” (3) It identifies islands as clusters of “eligible” windows separated by gaps of size no larger than a predetermined value g. A gap is a contiguous stretch of “ineligible” windows between two neighboring “eligible” windows. When g = 0, islands are uninterrupted clusters of “eligible” windows. See Fig. 2 for an illustration of the definition of islands. (4) It identifies “candidate” islands that exhibit significant clustering of “eligible” windows that are unlikely to appear by chance. SICER assigns a score s(l) for each “eligible” window of read count l as s(l) = − ln P(l,λ). The score S for each island is defined as the aggregated score of all “eligible” windows in the island. Only islands with score S > ST are regarded as “candidate” islands, where ST is an island-score threshold controlling statistical significance of ChIP enrichment on an island against random background. More specifically, ST is determined by requiring that the expected number of islands with scores above ST if reads are randomly distributed be less than an E-value threshold e. (5) If a control library is available, SICER will further filter the “candidate” islands using the control library, retaining only those that exhibit significant enrichment of ChIP signal compared to control on the islands. The statistical significance versus control library is characterized by a p-value based on Poisson distribution. A false discovery rate (FDR) is also reported using p-value adjusted for multiple testing [9]. Because of the presence of systematic biases in a typical ChIP-Seq library, it is highly desirable to have a matching control library.
Fig. 2.
Schematic illustration of definition of islands. Shown is a segment of a genomic landscape of ChIP-Seq reads. The x-axis denotes the genome coordinates, where each interval represents a window. The y-axis denotes the read count. Each black vertical bar represents the read count in the respective window. The regions underlined by the green horizontal bars are the two identified islands under g = 1 and l0 = 2. The two windows underlined by brown boxes are gaps in the first island
The flow chart of SICER is shown in Fig. 3. SICER is essentially a filtering tool. The delineated ChIP-enriched regions can be used to associate with other genomic landmarks. Reads on those ChIP-enriched regions can be identified and used for profiling and other quantitative analysis. Further details of the SICER algorithm can be found in [4].
Fig. 3.
Complete SICER flowchart. For ChIP-Seq library with or without control library, SICER always uses a random background model to identify candidate islands subject to a preset E-value. If a control library is available, SICER further screens islands using a false discovery rate (FDR) or a p-value cutoff against control library
2.3 Considerations for Key Parameters
Choices of key parameters are important for satisfactory results. Of particular importance are window size w, gap size g, effective genome length L, as well as a parameter controlling statistical significance (E-value for random background and p-value or FDR for using a control library as background). Before a discussion of considerations on choices of these parameters, we would like to emphasize that visual examination on the genome browser is indispensable, and positive and negative control cases from known biology would be of tremendous help in making the appropriate choice of the parameters.
2.3.1 Window Size
The choice of the window size, which directly affects the delineation of islands, is an important one. A window too narrow will exaggerate local fluctuation in each window, while a window too large will cause over-smoothing of data and lose resolution. Our experience has been that for transcription factors, a suitable window size choice is around 50–100 bps. On the other hand, for histone modifications and histone variants, a typical choice for window size w is 200 bps, a number approximately the length of a single nucleosome and a linker. As an example, we tested various window sizes (50, 100, 200, 500, and 1,000 bps) with a fixed gap size (3 windows) on the H3K27me3 dataset, and the resulting islands identified were shown in Fig. 4. It is clear that a larger window size results in more extended islands. For this particular dataset on this locus, a window size of 200 bps appears to have a good balance of specificity (that didn’t include too many regions of weak enrichment) and sensitivity (that didn’t produce too many gaps within an extended island) and thus an appropriate choice. In general, we can estimate the window size using the approach developed by Shimazaki and Shinomoto [10, 11], which employed a cost function defined by the mean integrated squared error to find an optimized window size for a histogram. This approach cannot be used blindly. Although the automatically calculated window size results in improved island delineation in many cases, in some other cases it fails to output a reasonable value.
Fig. 4.
The choice of the window size directly affects the delineation of islands. Top track: H3K27me3 ChIP-Seq data from ENCODE. Bottom tracks: islands with significant H3K27me3 enrichment identified by SICER with different choices of window size: (from top to bottom, in bps) 50, 100, 200, 500, 1,000. The gap size is always set to be 3 windows
2.3.2 Gap Size
The adoption of gap reflects the unique strength of SICER in identifying broad ChIP enrichment from poor coverage and/or high background noises. Gap size g by definition must be a multiple of window size chosen. In general the wider the domains are, the larger the gap size should be. For instance, for localized histone modifications like H3K4me3, the gap size can be set to be equal to the window size, g = w, while for a histone modification with an extended profile (e.g., H3K27me3), g = 3w likely works better. For more careful consideration, users can plot the aggregate score of all significant islands as a function of g. If the aggregate score reaches a maximum inside of the range of g explored, the gap size corresponding to the highest aggregate score would be a good choice. On the other hand, the aggregate score may increase monotonically with gap size (see Fig. 5 for an example). If the curve gradually increases toward saturation, we suggest choosing the gap size so that the corresponding aggregate score is sufficiently close to saturation. No sign of saturation suggests poor sequencing coverage (see Fig. 5), and the ultimate solution would be to increase sequencing depth. Another option in the absence of enough sequencing depth is to increase window size, the discussion of which can be found in the previous subsection. In general, we recommend against a gap size beyond 4 windows, for fear of too much spurious clustering. Figure 6 shows the effect of gap size on the H3K27me3 island delineation at the Wnt6 and Wnt10 locus, in which case gap sizes of 2, 3, and 4 windows provide adequate results.
Fig. 5.
Aggregate score on islands as a function of gap size. The window size is fixed at 200 bps. The H3K27me3 ChIP-Seq data exhibits monotonically increasing aggregate score with increasing gap size
Fig. 6.
The choice of the gap size directly affects the delineation of islands. Top track: H3K27me3 ChIP-Seq data from ENCODE. Bottom tracks: islands with significant H3K27me3 enrichment identified by SICER with different choices of gap size: (from top to bottom, in windows) 0, 1, 2, 3, 4. Window size is fixed at 200 bps
2.3.3 Effective Genome Size and Effective Genome Fraction
When short reads are mapped onto the reference genome, normally only those that map to unique genomic loci are retained for further analysis. As a result, genomic regions with degenerate sequences or sequences composed of character “N” are non-mappable. The effective genome length L is defined as the total length of mappable regions in the genome. The effective genome fraction is defined as L divided by the actual genome length. L depends on the species and sequencing protocol (e.g., read length, paired end, or single end). Generally speaking, longer read length and paired-ended sequencing will lead to higher fraction of effective genome. L can be found or computed from Uniqueome [12].
2.3.4 Statistical Significance
In case of random background, an E-value cutoff is used to identify significant islands. E-value is the expected number of islands emerged merely due to local fluctuation from randomly distributed reads along the genome. A smaller E-value means higher stringency. In the case of random background, one can give a rough estimate of error rate empirically by dividing the E-value by the total number of significant islands identified. For example, if E-value is 500, the number of significant islands is 10,000, the empirical error rate is 5 %. If a control library is available, SICER uses a default permissive E-value of 1,000 in identifying candidate islands prior to incorporating control library information. SICER then computes p-value for each candidate island based on a Poisson distribution against read count in the control library and considers multiple-testing correction and uses FDR for statistical significance assessment. An FDR threshold of 0.01 or 0.001 is in general adequate, while an FDR of 10− 8 or less can be used to find the high-confidence ChIP-enriched regions.
3 Material and Method
In this section, we demonstrate how to run SICER and understand SICER output by going through a concrete example, where we apply SICER in the most typical situation: a ChIP-Seq library along with a control library.
3.1 Material
Commands listed in the current manual are executed and time-benchmarked in the following system environment: Mac OS X 10.6.8, dual 2.93 GHz 6-core Intel Xeon processors, 64GB Memory, 64-bit Python 2.7.3 with NumPy, SciPy and PyLab packages, and BEDTools [13] installed (see Note 1).
The dataset analyzed is the ChIP-Seq data of H3K27me3 in mouse embryonic stem cell (ES Bruce4) from the mouse ENCODE project [14], downloaded from the UCSC genome browser [15]. There are 2 replicates of H3K27me3 ChIP-Seq libraries, together with 2 replicates of input control library. For simplicity the replicates are pooled together, yielding the ChIP library ES_H3K27m3. bed with 53 million reads and the control library ES_input.bed with 22 million reads.
The most recent release of SICER (Version 1.1) was downloaded from http://home.gwu.edu/~wpeng/Software.htm.
3.2 Method
3.2.1 Installation of SICER
After downloading SICER software package SICER_V1.1.tgz, the user shall launch Terminal, go to the directory where the downloaded file is, and then execute
$ tar -xvf SICER_V1.1.tgz -C /mydir
Here /mydir represents the directory where the user desires to have SICER installed.
3.2.2 Setting Paths Inside Master Scripts
The master scripts need to be customized to reflect the directory where SICER is located. Use a plain text editor (not rich format editors like Microsoft Word) to open the script SICER.sh, and change the first line right below the shebang (line starts with “#!”) and comment box (lines start with “#”).
PATHTO=/home/data/SICER1.1
to
PATHTO=/mydir/SICER_V1.1
Repeat this for the other main script, SICER-rb.sh. For SICER-df.sh and SICER-df-rb.sh, replace the first line below the comment box to
SICER=/mydir/SICER_V1.1/SICER
3.2.3 Preparation of Data Files
In ENCODE project database, ChIP-Seq data is available in various formats. For simplicity we use the pre-aligned BAM data. After downloading BAM files and accompanying index BAM.BAI files, change working directory to where the files are stored and execute
$ bamToBed -i wgEncodeLicrHistoneEsb4H3k27me3ME0C57bl6StdAlnRep1.bam>ES_H3K27me3_rep1.bed
$ bamToBed -i wgEncodeLicrHistoneEsb4H3k27me3ME0C57bl6StdAlnRep2.bam>ES_H3K27me3_rep2.bed
$ bamToBed -i wgEncodeLicrHistoneEsb4InputME0C57bl6StdAlnRep1.bam>ES_input_rep1.bed
$ bamToBed -i wgEncodeLicrHistoneEsb4InputME0C57bl6StdAlnRep2.bam>ES_input_rep2.bed
Then combine the 2 replicates in each ChIP-Seq sample via
$ cat ES_H3K27me3_rep1.bed ES_H3K27me3_rep2.bed>ES_H3K27me3.bed
$ cat ES_input_rep1.bed ES_input_rep2.bed>ES_input.bed
3.2.4 Execution of SICER
Once we have BED files of the ChIP-Seq library and control library, we can start the SICER analysis. Within the SICER directory, there are four SICER scripts, namely, SICER.sh, SICER-rb.sh, SICER-df.sh, and SICER-df-rb.sh. Among those SICER.sh is the master SICER script that processes ChIP library against a control library. If no control library is available, SICER-rb. sh can be executed in place of SICER.sh. SICER-df.sh and SICER-df-rb.sh are used to compare two epigenomes and will be discussed later (see Notes 2–4).
In Terminal, from where the ChIP and control libraries (ES_H3K27me3.bed and ES_input.bed in this example) are stored, launch SICER with (see Note 2)
$ sh /mydir/SICER_V1.1/SICER/SICER.sh.ES_H3K27me3.bed ES_input.bed.mm9 1 200 150 0.8 600 1e-3
SICER.sh takes 11 ordered command line parameters. The general command structure is
$ sh /mydir/SICER_V1.1/SICER/SICER.sh [Input directory] [ChIP file] [Control file] [Output directory] [Species] [Redundancy threshold] [Window size] [Fragment size] [Effective genome fraction] [Gap size] [FDR]
The detailed description of the command line arguments are listed below:
Input directory: where the ChIP and control libraries data files are stored. In this example, “.” denotes current directory.
ChIP file: the file name of the ChIP library. In this example it is ES_H3K27me3.bed.
Control file: the file name of the control library. In this example it is ES_input.bed.
Output directory: where the output files should be stored. In this example, “.” denotes current directory.
Species: the name of reference genome (see Note 3). In this example it is “mm9.”
Redundancy threshold: number of redundant reads kept for analysis. In this example it is 1. Redundant reads refer to reads with exactly the same genomic location and orientation. For typical ChIP-Seq datasets, this is likely due to PCR amplification artifact. To remove this potential bias, we generally recommend removing the redundancy and retaining only 1 read for each set of redundant reads.
Window size: the width (in bps) of window in comparing ChIP with control library. In this example it is 200 as the default value recommended for histone modification marks.
Fragment size: the average size (in bps) of ChIP fragment. In this example it is 150 as the default recommended value. This parameter is used to assign a ChIP read to the center of the DNA fragment. Typical sonication outputs ChIP fragment of 150–300 bps long.
Effective genome fraction: In this example it is 0.8, which is recommended for single-end ChIP-Seq with 50 bp read length.
Gap size: the gap size (in bps) allowed in SICER filtering. In this example it is 600 bps (or 3 windows), as recommended for extended histone modification marks like H3K27me3. This parameter must be a multiple of window size.
FDR: the desired false discovery rate cutoff in identifying statistically significant islands. In this example FDR = 0.1%.
3.2.5 Analysis of SICER Output
It takes 35 min to finish the SICER.sh run on this data file. Please be advised that the process time increases with increasing ChIP-Seq and control library size.
SICER.sh should generate the following files after the complete workflow. Explanation of each file is as follows:
ES_H3K27me3-1-removed.bed. This file is in BED format and contains all reads in the ChIP library after removal of redundant reads.
ES_H3K27me3-W200-normalized.wig. This file is in WIG format and could be uploaded to the UCSC genome browser for visualization of the ChIP library with redundancy-removed but before island-filtering (ES_H3K27me3-1-removed.bed) with desired window size.
ES_H3K27me3-W200.graph. This file is in bedGraph format and is a summary of the redundancy-removed data file.
ES_H3K27me3-W200.scoreisland. This file stores all identified islands with respective scores and could be used to evaluate the choice of gap size.
ES_H3K27me3-W200-G600-islands-summary. This 8-column file is a summary of all islands identified in the ChIP library. The format is chromosome, start, end, read count in ChIP library, read count in control library, p-value, fold change, FDR).
ES_H3K27me3-W200-G600-islands-summary-FDR1e-3. This is a subset of identified islands whose FDR are less than the given threshold. It is the same 8-column format as the summary.
ES_H3K27me3-W200-G600-FDR1e-3-island.bed. This file has 4 columns and contains information about all identified ChIP-enriched region information under filtering parameters (window size 200, gap size 600, FDR cutoff 1e-3). The 4 columns are chromosome, start, end, and read count in ChIP library.
ES_H3K27me3-W200-G600-FDR1e-3-islandfiltered.bed. This file is in BED format and contains all reads that are within significant islands.
ES_H3K27me3-W200-G600-FDR1e-3-islandfiltered-normalized.wig. This file is in WIG format and could be uploaded directly to UCSC genome browser for visualization of the island-filtered ChIP library.
ES_input-1-removed.bed.
This file is in BED format and contains all reads in the control library after removal of redundant reads to the threshold 1.
Of all these files, the ES_H3K27me3-W200-G600-islands-summary-FDR1e-3 and ES_H3K27me3-W200-G600-FDR1e-3-island.bed are the most important for further analysis. The first one contains the details about each significant island. The second one contains the redundancy-removed raw reads filtered by islands. In addition, the wig files can be used for visual examination of the raw and processed data on the genome browser.
Figure 1 shows the results of SICER on Wnt6 and Wnt10a gene locus. The top data track, showing raw data, suggests that H3K27me3 is enriched in broad domains across the entire Wnt6 and Wnt10a gene locus. SICER under default parameters does a good job in capturing the extended domains of H3K27me3 enrichment.
Fig. 7.

SICER-df identifies regions with reduced H3K9me2 enrichment during adipogenesis. Top track: SICER-filtered H3K9me2 data before differentiation (day 0, red) and after differentiation (day 7, green) overlaid with the same vertical scale. Raw data behaves similarly. Bottom track: Regions with decreased H3K9me2 enrichment as identified by SICER-df. Here window size w = 500 bps (as calculated using Shimazaki and Shinomoto [10]), gap size g = 3w, FDR cutoff is 0.1 %. The profiles were smoothed for illustration purpose (with UCSC genome browser smoothing window set to 10 pixels). There are multiple genes in this locus but only PPARγ is shown
Acknowledgement
This work was supported in part by the Intramural Research Program of the NIDDK, NIH to KG.
Footnotes
SICER prerequisites.
SICER takes advantage of Shell scripting language and therefore runs in a Unix/Linux platform (e.g., Mac OS X and Ubuntu). SICER cannot be run directly under Windows. However, it potentially could work under a Unix/Linux simulator (e.g., Cygwin).
$ python -c 'import struct; print struct.calcsize("P") * 8'
- >>>import numpy
- >>>import scipy
- >>>import pylab
If all packages are installed and functioning correctly, there should be no message displayed.
Avoid running multiple SICER instances under the same directory.
During the execution, SICER generates temporary files with hard-coded names. Therefore, multiple SICER instances running in the same directory will interfere with each other, leading to unpredictable results. This is particularly important if users are using a centralized cluster-computing management system (e.g., Condor).
Adding additional genome.
- Add this entry to dictionary “species_chroms”: “NewGenome”: NewGenome_chroms
- Add this entry to dictionary “species_chrom_lengths”: “NewGenome”: NewGenome_chrom_length
- Add this list to GenomeData.py: NewGenome_chroms=[“chr1,” “chrX”]
- Add this dictionary to GenomeData.py: NewGenome_chrom_lengths={“chr1”: 100, “chrX”: 200}
Compare epigenomes and identify differentially enriched regions.
A frequently encountered case in epigenomic analysis is to identify significant differences between two conditions: wild-type cells versus treated cells, normal cells versus pathological cells, or undifferentiated cells versus differentiated cells. SICER-df.sh and SICER-df-rb.sh are designed to identify domains of differential enrichment. SICER-df.sh shall be used when matching control libraries for the two conditions are available, whereas SICER-df-rb.sh shall be used when random background has to be used. Here we focus on SICER-df.sh. For clarity, we will call the two conditions wild-type (WT) and knockout (KO). SICER-df.sh works as follows: (1) it first identifies significant islands in both WT and KO. This is done by using SICER.sh to process both WT and KO ChIP-Seq libraries against their respective control libraries. (2) It merges the identified ChIP-enriched regions from WT and KO libraries. The merged islands are regarded as the “candidate” islands and constitute the units of comparison. Therefore, the “candidate” islands are required to be significantly ChIP-enriched (compared with the respective control library) in at least one of the two conditions. (3) On each “candidate” island, signal level in KO is compared with that in WT to determine the significance of changes. Regions with increased or decreased enrichment fulfilling a desired FDR requirement are reported. Fold-change values are also reported in the output file.
Figure 7 illustrates the result of SICER-df in identifying regions with differential enrichment of H3K9me2 during adipogenesis [16]. SICER-df finds several domains of decreased H3K9me2 enrichment in a 1.5 Mb region. In particular, an extended region of differential enrichment (~500 kbs) covers the PPARγ gene (see Fig. 7), a master regulator of the adipogenetic process. Interestingly, the specific removal of H3K9me2 correlates well with the induction of PPARγ during adipogenesis and supports the regulatory role of histone methyltransferase G9a-mediated repressive epigenetic mark H3K9me2 [16] on PPARγ expression.
References
- 1.Barski A, Cuddapah S, Cui KR, Roh TY, Schones DE, Wang ZB, Wei G, Chepelev I, Zhao KJ. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Wang ZB, Zang CZ, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui KR, Roh TY, Peng WQ, Zhang MQ, Zhao KJ. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18(7):1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 7.Lachner M, O’Carroll N, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8(1):9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 10.Shimazaki H, Shinomoto S. A method for selecting the bin size of a time histogram. Neural Comput. 2007;19:1503–1527. doi: 10.1162/neco.2007.19.6.1503. [DOI] [PubMed] [Google Scholar]
- 11.Song Q, Smith AD. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011;27(6):870–871. doi: 10.1093/bioinformatics/btr030. doi:10.1093/bioinformatics/btr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koehler R, Issac H, Cloonan N, Grimmond SM. The uniqueome: a mappability resource for short-tag sequencing. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq640. doi:10.1093/bioinformatics/btq640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. doi:10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The EPC. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. doi:10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1172. doi:10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Xu S, Lee J-E, Baldridge A, Grullon S, Peng W, Ge K. Histone H3K9 methyltransferase G9a represses PPAR[gamma] expression and adipogenesis. EMBO J. 2013;32(1):45–59. doi: 10.1038/emboj.2012.306. http://www.nature.com/emboj/journal/v32/n1/suppinfo/emboj2012306a_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]