Abstract
Biological processes frequently require the formation of multi-protein or nucleoprotein complexes. Some of these complexes have been produced in homogeneous form, crystallized, and analysed at high resolution by X-ray crystallography (for example, see refs 1–3). Most, however, are too large or too unstable to crystallize. Individual components of such complexes can often be purified and analysed by crystallography. Here we report how the coordinated application of cryoelectron microscopy, three-dimensional image reconstruction, and X-ray crystallography provides a powerful approach to study large, unstable macromolecular complexes. Three-dimensional reconstructions of native cowpea mosaic virus (CMPV) and a complex of CPMV saturated with a Fab fragment of a monoclonal antibody against the virus have been determined at 23 Å resolution from low-irradiation images of unstained, frozen-hydrated samples. Despite the nominal resolution of the complex, the physical footprint of the Fab on the capsid surface and the orientation and position of the Fab have been determined to within a few angstroms by fitting atomic models of CPMV4 and Fab (Kol)5 to reconstructed density maps.
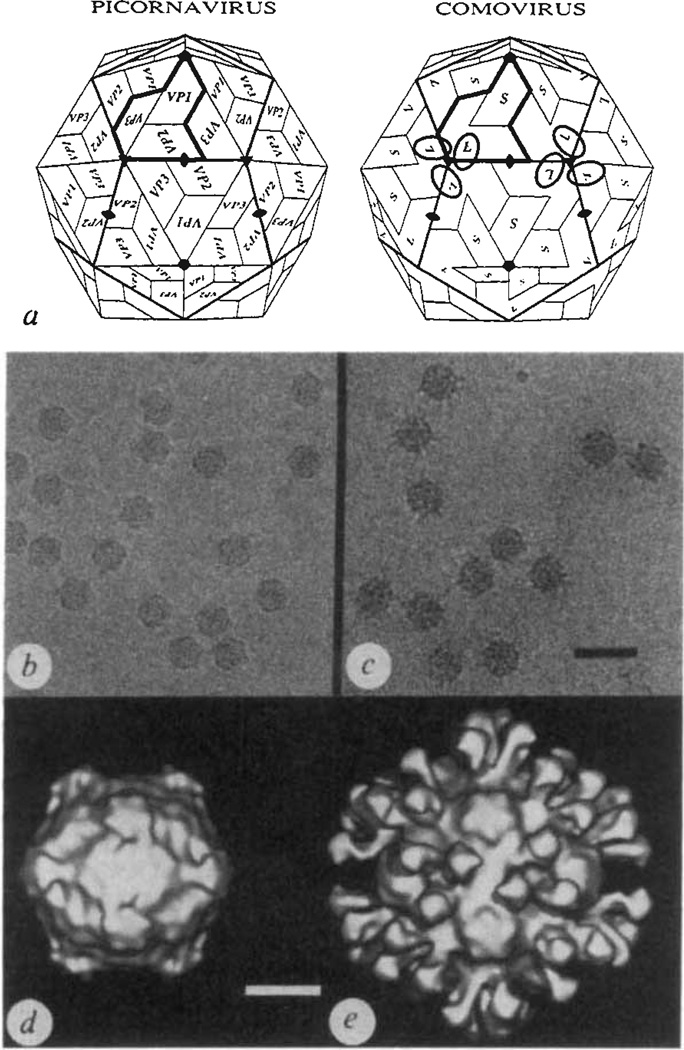
Comoviruses are plant viruses whose structural and biological properties are strikingly similar to the animal picornaviruses6,7. For example, the protein capsids of CPMV, the type member of the comovirus group, and poliovirus, the most extensively studied picornavirus, are comparable (Fig. 1a). The CPMV coat contains 60 copies each of large (relative molecular mass 42,000 (Mr 42K) and small (24K) protein subunits arranged with icosahedral symmetry. Each large subunit (L) is composed of two antiparallel β-barrel domains which are positioned close to the icosahedral 3-fold axes. The β-barrel domain at the amino-terminal end of L corresponds to the VP2 subunit in poliovirus and the β-barrel at the carboxy-terminal end of L corresponds to VP3 in poliovirus. The small subunits (S), packed near the icosahedral 5-fold axes, correspond to VP1 in poliovirus.
FIG. 1.
The capsid architectures of picorna and comoviruses displayed with electron micrographs and three-dimensional image reconstructions of frozen-hydrated native CPMV and CPMV–Fab complexes, a, Schematic comparison of picornavirus and comovirus capsids. In each case one trapezoid represents an eight-stranded antiparallel β-barrel (see Fig. 2d). The icosahedral asymmetric unit of the picornavirus capsid (shaded in the heavy outline) contains three β-barrels, labelled VP1, VP2 and VP3, each with a characteristic amino-acid sequence. The comovirus capsid is similar to the picornavirus capsid except that two of the β-barrels, corresponding to VP2 (N terminus) and VP3 (C terminus), are covalently linked to form a single polypeptide, the large protein subunit (L), and the small protein subunit (S) corresponds to VP1. The ellipsoids on the comovirus capsid show the rough location of 6 of the 60 equivalent Fab binding sites (compare with e). b and c, Electron micrographs of frozen-hydrated samples of CPMV (b) and CPMV complexed with monoclonal antibody (5B2) Fab fragments (c). Scale bar, 500 Å. Both samples were prepared and photographed with established cryomicroscopy procedures14–17. Roughly 4 ml each of a 2.0 mg ml−1 CPMV (b) or 0.5 mg ml−1 CPMV–Fab (c; stoichiometry of Fabs to virus in solution was 600:1) sample at pH 7.4 in PBS was applied to holey-carbon films, blotted with filter paper and rapidly plunged into liquid ethane at about −170 °C. Images were recorded at minimal electron dose (~16 e− Å−2) at a nominal magnification of ×49,000 and at 80 kV in a Philips EM420 electron microscope. The images in b and c were recorded at about 0.8 and 0.9 µm underfocus, respectively, d, e, Surface-shaded representations of three-dimensional reconstructions of native virus (d) and virus–Fab complex (e), both viewed in the standard crystallographic 2-fold orientation, were computed with established methods18–20. A total of 17 (d) or 22 (e) particle images were combined to reconstruct both structures to a resolution limit of 23 Å. Scale bar, 100 Å.
CPMV is strongly immunogenic when injected into rabbits or mice. We prepared nine monoclonal antibodies against CPMV using standard methods8. The binding properties of these antibodies were analysed by enzyme-linked immunosorbent assay (ELISA) and all nine would only bind to virus particles presented in a double antibody sandwich-type ELISA in which the virus is bound to a polyclonal antibody attached to the plastic plate. Denaturation of the virus by its direct adsorption onto a plastic surface destroys the antigenic sites for all nine monoclonal antibodies. Only monoclonal antibodies 5B2 and 10B7 reacted strongly with CPMV that was free in solution (antibody binding was assayed by electron microscopy analysis of negatively stained complexes marked with gold-labelled anti-mouse immunoglobulin). Fab fragments from monoclonal antibody 5B2 were prepared by digestion of IgGs with insoluble papain (Sigma). The papain was removed by low-speed centrifugation and the digestion products were incubated with CPMV for 2 h at 37 °C before centrifugation of the reaction mixture on a 10–40% sucrose gradient to separate Fab and virus/Fab complexes. A single peak containing virus and virus complexed with varying amounts of Fab was fractionated and visualized by electron microscopy. Unstained aliquots of native CPMV and of virus saturated with Fab were quick-frozen and low-irradiation images (Fig. 1b, c) were recorded to compute three-dimensional reconstructions (Fig. 1d, e).
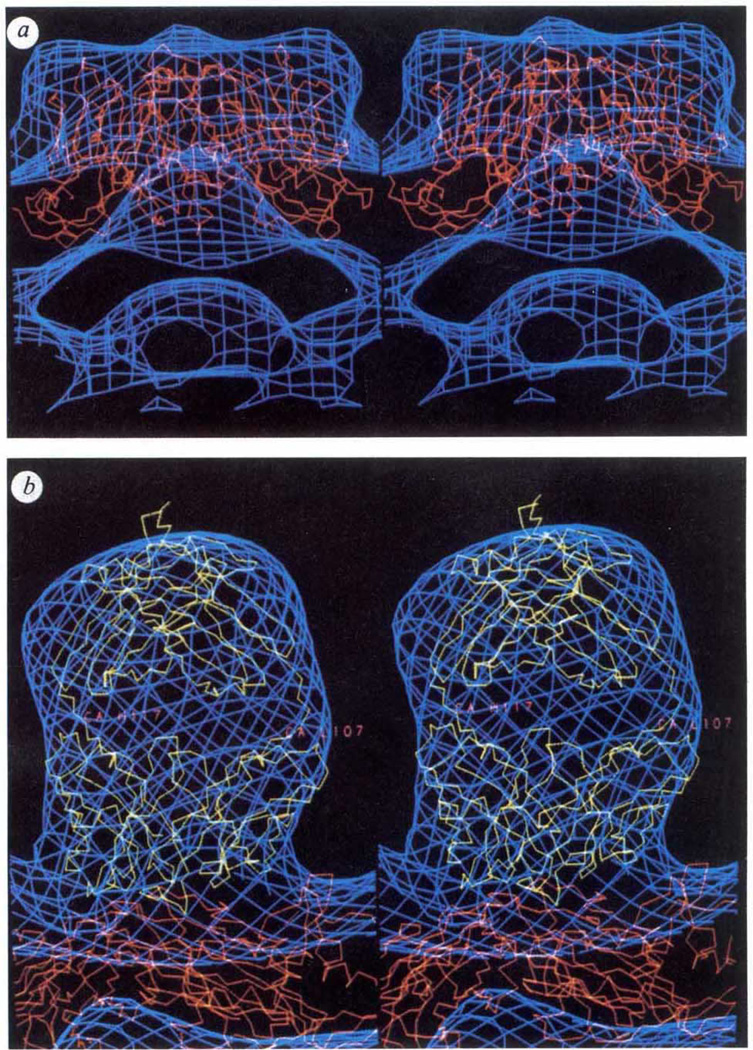
The Cα-backbone model of CPMV, derived from the 3.0 Å-resolution X-ray structure4, was placed in both enantiomorphs of the native (Fig. 1d) and the virus–Fab-complex (Fig. 1e) reconstructed densities (the correct enantiomorph of the electron microscope density cannot be determined from a single micrograph). The program FRODO9 was used to align the symmetry axes of the X-ray and electron microscope structures and to compute an overall scale factor to adjust for small errors in the microscope magnification. The exceptionally good fits of the X-ray model with both reconstructions allowed the correct enantiomorph of the density to be unambiguously identified and suggests that Fab binding induces no major change in the capsid structure.
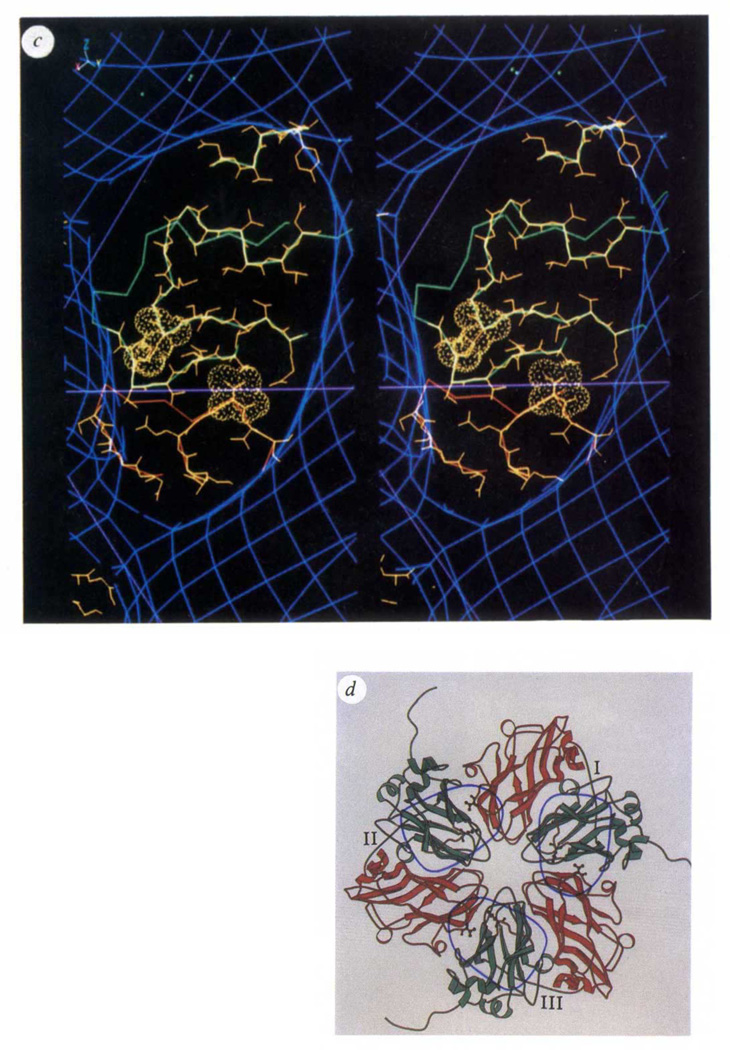
The Fab density, centred 25 Å from the icosahedral 3-fold symmetry axes, covers an ellipsoidal area roughly 40 × 20 Å on the viral surface and extends radially about 70 Å from it (Figs 1e and 2b). The overall shape and extent of the virus–Fab interaction agrees very well with protein–Fab interactions that have been analysed at high resolution by X-ray crystallography1. The roughly 30 amino-acid residues of the virus that lie under the ‘footprint’ of the Fab were identified from inspection of the high-resolution X-ray model of CPMV (Fig. 2c). The Fab footprint covers regions of the CPMV surface that correspond to subunits VP2 and VP3 in poliovirus. This antigenic region is composed of a quaternary interaction between the ‘VP2’ β-barrel domain of one L subunit and the ‘VP3’ β-barrel domain in a neighbouring 3-fold-related L subunit. Exposed loops from each of these domains are in favourable positions to interact with the Fab and each loop contains a residue spatially equivalent to the corresponding two amino acids that define the 3B antigenic site of poliovirus10 (mutation of either residue in poliovirus prevents neutralization by a specific monoclonal antibody to poliovirus) (Fig. 2d). The common immunogenic sites on CPMV and poliovirus probably arise because of the overall similarity in the capsids of the two viruses. Some immunogenic regions on mammalian viruses are readily mutable as a result of evolutionary pressure exerted by the circulating immune system on exposed elements of the structure. Other sites may be surface regions that are good immunogens, but lack the structural flexibility to readily mutate. Page et al.10 point out that immunogenic site 3B in type I poliovirus is likely to be such an epitope because of the small number of viable mutants observed. This prediction is consistent with the discovery of a similar epitope on CPMV. By contrast, site 2 on type I poliovirus displays a large number of viable mutations positioned on exposed loops that are not present in CPMV.
FIG 2.
Comparisons of CPMV and Fab atomic models with reconstructed electron density derived from electron micrographs of frozen-hydrated specimens, a, b, Stereoviews comparing Cα backbone models of CPMV (red) and Fab (Kol) (yellow) with electron density (blue) derived from the electron-microscopy reconstruction described in Fig. 1. a, Native virions viewed in a direction perpendicular to a vertical fivefold icosahedral symmetry axis. The surface bulge in the density corresponds to amino acids in a β-turn between the outermost strands of the S subunit. b, A close-up view showing the Fab (Kol) and CPMV Cα backbones fitted to the CPMV–Fab reconstruction. The small portions of Kol that protrude outside the Fab electron microscope envelope at the top and into the surface of the CPMV L subunit are regions of the polypeptide that are not conserved in sequences of other IgG molecules, and are therefore likely to be different in the 5B2 Fab structure. When contoured at a higher density level the Fab envelope divides into well-defined globular regions which correspond closely with the Fab two-domain structure. Note that the electron microscope envelope, contoured at a single density level, fits the CPMV and Fab X-ray models equally well. This suggests that the frozen-hydrated CPMV samples imaged were nearly fully saturated with the Fabs and most of the Fab molecules must be fairly rigidly attached in equivalent orientations with respect to the viral surface.
c, Stereoview of the Fab footprint on the virion surface showing regions of the CPMV polypeptide chain that lie under the Fab. The region in red corresponds to the ‘VP3’ domain of one L subunit; green denotes the ‘VP2’ domain of a 3-fold-related L subunit. Main chain and side chains of residues that may interact with the Fab are shown in yellow. The amino acids displayed with surface rendering (Lys 34 in the ‘VP2’ domain and Thr 24 in the 3-fold-related ‘VP3’ domain) are spatially equivalent to residues 72 and 76 in the poliovirus VP2 and VP3 domains. Mutations of either of these residues in poliovirus prevents neutralization by poliovirus monoclonal antibodies 10 and 13 (ref. 10), thus defining the 3B antigenic site on serotype I poliovirus. Lines in magenta correspond to the boundaries in Fig. 1a (picornavirus) that separate VP2 from VP3 near the 3-fold axis on the left side of the figure, d, A schematic representation21 of 3-fold-related L subunits, each numbered near the peptide that connects ‘VP2’ (green) and ‘VP3’ (red) domains. Ellipsoids identify the viral surface in contact with each Fab. The side chains shown correspond to the residues with surface rendering in Fig. 2c. The ellipsoid on the right (covering portions of L subunits I and III) is in the same orientation as the ellipsoid in c.
An atomic structure for Fab 5B2 is unknown, although atomic coordinates for other Fab fragments are available in the Brookhaven protein data bank. The Cα coordinates for Fab (Kol)5 were used for initial modelling of the relevant density in the CPMV–Fab reconstruction because Fab (Kol) has a 165° elbow angle (the angle between pseudo 2-fold axes relating heavy and light chains in the variable domain and heavy and light chains in the constant domain), which is consistent with the extended shape of the density ascribed to the Fab in the reconstruction. The variable domain of the Fab (Kol) model was positioned in contact with the virus surface and adjusted as a rigid body to fit the electron microscope density. Only two orientations of the Fab, which were related by a 180° rotation about the axis perpendicular to the particle surface, gave excellent agreement between the model and the ellipsoidal-shaped density.
The factors determining virus neutralization by antibodies are not fully understood. But for some antibodies, bivalent binding of the IgG was determined to be important to its ability to neutralize the virion11. The centres of pairs of 5B2 Fab fragments, related by an adjacent icosahedral 2-fold symmetry axis, are 60 Å apart. This is near the minimum distance thought to be reasonable for bidentate binding of an IgG12. Model building shows that bivalent binding of the 5B2 IgG is possible if its Fab portions can adopt a smaller elbow angle.
The excellent correspondence of the X-ray and electron microscope structures for both the Fab and CPMV density distributions demonstrate the high fidelity of reconstructed density maps computed from images of frozen-hydrated specimens. Even though it is impossible to accurately model the detailed interactions between the Fab and the virus with these data, our results clearly suggest candidates for site-directed mutagenesis of an infectious clone13 to explore the role of individual amino acids in complex formation. The methods used for the analysis of the CPMV–Fab complex can be extended to study other viruses or macromolecules and, for example, reveal details of critical molecular interactions that occur between them and antibodies or receptor molecules.
ACKNOWLEDGEMENTS
We thank Y. Li for virus preparations, M. H. V. Van Regenmortel for help with preparation of the monoclonal antibodies and for support. R. Holland Cheng for help in converting electron density maps for different display systems, S. Fateley for help in preparing the manuscript and D. L. D. Caspar for critically reading the manuscript. This work was supported by NIH (T.S.B. and J.E.J.), a National Science Foundation Grant (T.S.B.), and The Lucille P. Markey Trust.
References
- 1.Davies DR, Padlan EA, Sheriff S. A. Rev. Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 2.Ruff M, et al. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 3.Rossmann MG, Johnson JE. A. Rev. Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Stauffacher CV, Johnson JE. Semin. Virol. 1990;1:453–466. [Google Scholar]
- 5.Marquart M, Deisenhofer J, Huber R, Palm W. J. molec. Biol. 1980;141:369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- 6.Goldbach R, van Kammen A. In: Molecular Plant Virology. Davies JW, editor. Vol. 2. Boca Raton, Florida: CRC; 1985. pp. 83–120. [Google Scholar]
- 7.Chen Z, et al. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 8.Fazekas de St. Groth S, Scheidegger D. J. immunol. Meth. 1980;35:1–121. [Google Scholar]
- 9.Jones TA. In: Computational Crystallography. Sayre D, editor. Clarendon, Oxford: 1982. pp. 303–317. [Google Scholar]
- 10.Page GS, et al. J. Virol. 1988;62:1781–1794. doi: 10.1128/jvi.62.5.1781-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Icenogle JH, et al. Virology. 1983;127:412–425. doi: 10.1016/0042-6822(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 12.Mosser AG, Leippe DM, Rueckert RR. In: Molecular Aspects of Picornavirus Infection and Detection. Semler BL, Ehrenfeld E, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 155–167. [Google Scholar]
- 13.Holness CL, Lomonossoff GP, Evans D, Maule AJ. Virology. 1989;172:311–320. doi: 10.1016/0042-6822(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 14.Adrian MJ, Dubochet J, Lepault J, McDowall AW. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 15.Milligan RA, Brisson A, Unwin PNT. Ultramicroscopy. 1984;13:1–10. doi: 10.1016/0304-3991(84)90051-2. [DOI] [PubMed] [Google Scholar]
- 16.Dubochet J, et al. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 17.Prasad BVK, Burns JW, Marietta E, Estes MK, Chiu W. Nature. 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 18.Baker TS, Drak J, Bina M. Proc. natn. Acad. Sci. U.S.A. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker TS, Newcomb WW, Booy FP, Brown JC, Steven AC. J. Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeager M, Dryden KA, Olson NH, Greenberg HB, Baker TS. J. Cell. Biol. 1990;110:2133–2144. doi: 10.1083/jcb.110.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraulis PJ. J. Appl. Crystallogr. (in the press). [Google Scholar]