Abstract
The peanut is a special plant for its aerial flowering but subterranean fructification. The failure of peg penetration into the soil leads to form aerial pod and finally seed abortion. However, the mechanism of seed abortion during aerial pod development remains obscure. Here, a comparative transcriptome analysis between aerial and subterranean pods at different developmental stages was produced using a customized NimbleGen microarray representing 36,158 unigenes. By comparing 4 consecutive time-points, totally 6,203 differentially expressed genes, 4,732 stage-specific expressed genes and 2,401 specific expressed genes only in aerial or subterranean pods were identified in this study. Functional annotation showed their mainly involvement in biosynthesis, metabolism, transcription regulation, transporting, stress response, photosynthesis, signal transduction, cell division, apoptosis, embryonic development, hormone response and light signaling, etc. Emphasis was focused on hormone response, cell apoptosis, embryonic development and light signaling relative genes. These genes might function as potential candidates to provide insights into seed abortion during aerial pod development. Ten candidate genes were validated by Real-time RT-PCR. Additionally, consistent with up-regulation of auxin response relative genes in aerial pods, endogenous IAA content was also significantly increased by HPLC analysis. This study will further provide new molecular insight that auxin and auxin response genes potentially contribute to peanut seed and pod development.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-014-0193-x) contains supplementary material, which is available to authorized users.
Keywords: Aerial pod, Subterranean pod, Transcriptome, Peanut, Seed abortion, Development
Introduction
Peanut (Arachis hypogaea L.) is an important oilseed and economic crop cultivated in worldwide for providing human nutrition and oil production. Different to other plant, the peanut plant produces flowers aerially, while develops fruit and seeds underground with fascinating gravitropic growth habits (Zhu et al. 2013). In the reproduction cycle, when the fertilization is succeeded after flowering, the ovule-carrying peg (gynophore) starts to form and then down elongation to bury the fertilized ovule into the soil. However, only until the peg carries the ovule into the soil where can the pod normally swell to allow room for the embryo to grow and eventually become subterranean pod (Feng et al. 1995; Moctezuma and Feldman 1999, 2003). The failure of peg penetration into the soil leads to suppression of pod swelling initiation and form aerial pod, finally causing seed abortion and seriously impacting on the peanut production (Chen et al. 2013). For instance, when gynophore penetration into the soil is prevented by any means of a physical barrier but still under a light treatment, the pod will not form normally (Zamski and Ziv 1976; Thompson et al. 1985; Moctezuma 2003). Therefore, it is essential to gain a clearer understanding of these occurring mechanisms during peanut pod development.
Seed formation in peanut is a central stage of pod development. This complex process is initiated by a successful double fertilization that not only results in a diploid embryo and a triploid endosperm, but also triggers development of seed coat by tissue differentiation and cell expansion (Sin et al. 2006; Capron et al. 2012). Accumulating evidence illustrates that seed development is highly coordinated by both endogenous signal and environment stimuli. For instance, several plant hormones have long been known to play a significant role in peanut gynophore elongation and embryo differentiation, such as auxin (Jacobs 1951; Moctezuma and Feldman 1996), the ration of NAA and kinetin (Ziv and Zamskj 1975), ABA (Ziv and Kahana 1988), ethylene (Shlamovitz et al. 1995). In addition, mechanical stimulus and alternation of light and dark conditions also controlled the cessation of embryo differentiation during peg elongation phase, and the resumption of embryo development following quiescence in underground phase (Zamski and Ziv 1976; Stalker and Wynne 1983; Thompson et al. 1985; Shlamovitz et al. 1995; Nigam et al. 1997). At present, despite a comprehensive understanding of physiological and environmental factors that influence seed and pod development, isolation and characterization of candidate genes is of vital importance for improving peanut seed quality and yield.
Over the past decade, with the advent of rapid and high-throughput technology for quantification of the transcriptome (Malone and Oliver 2011), progress on seed development (Guo et al. 2008; Zhang et al. 2012) and tissue expression (Payton et al. 2009; Wang et al. 2012) in peanut (Haegeman et al. 2009; Tirumalaraju et al. 2011; Chen et al. 2012) has been studied intensely using DNA microarrays or RNA sequencing. For instance, they are explored to investigate how the transcriptome is deployed in aerial and subterranean pods (Chen et al. 2013), and how gene expression varies in response to disease infection (Guo et al. 2008; Wang et al. 2012). Furthermore, in our previous studies (Chen et al. 2013; Zhu et al. 2013), both RNA-seq and proteomics analysis shed light on the potential candidate genes and proteins that regulated aerial and subterranean pods development. These studies not only revealed that the embryo of the aerial pod ceased growth at early stages and finally aborted, but also underlined two senescence associated genes and one late embryogenesis-abundant gene as candidates to embryo abortion of aerial pod; additionally, proteins involved in lignin synthesis and ubiquitin proteasome system might regulate pods swelling to allow room for embryo development.
However, in previous RNA-seq analysis, limited knowledge is available in candidate genes that potentially contribute to seed abortion during aerial pods development. Little is known about stage-specific genes expression alternation during aerial and subterranean pods development due to pooling many samples from aerial and subterranean pods for one aerial library, and two subterranean libraries, respectively. Recently, many studies proved that microarrays remained useful and accurate tools for measuring gene expression alternation across development stages (Bloom et al. 2009; Willenbrock et al. 2009; Bradford et al. 2010), which not only achieved mature and stable analytic strategies, but also developed appropriate standards for this tools (Stears et al. 2003; Malone and Oliver 2011). Therefore, both DNA microarrays and RNA sequencing could complement with each other to profile the transcriptome for addressing problems during peanut aerial and subterranean pods development. In this study, to better understand the mechanism of seed abortion and pod development, we compared the transcriptome profiles of peanut aerial and subterranean pods at different developmental stages by microarrays approaches combined with previous RNA-Seq and proteomics analysis. The objectives of the present study were to: (1) compare the differentially expression of genes between developing aerial and subterranean pods; (2) identify potential candidate genes related to seed abortion; and (3) highlight stage-specific genes expression alternation during aerial and subterranean pods development.
Materials and methods
Plant materials and treatment
A peanut cultivar, ‘Yueyou-7’, was provided by Crops Research Institute, Guangdong Academy of Agricultural Sciences (GAAS, China). We identified selfed flowers with colored plastic thread, and marked elongating aerial pegs by tying with colored tags at the eighth day after flowering (DAF). After these treatments, one-third of marked pegs were artificially covered with soil, while the other two-third were put thick plastic membrane to prevent them from penetrating into the soil. Both of them were of the same age and grown in experimental flowerpot with normal management. We collected aerial pods and subterranean pods at 1, 2, 4, 8 days after marked (DAM), corresponding to 9, 10, 12, 16 DAF. In order to get the same parts of the pods, all important components such as the ovules and meristem from aerial and subterranean pods were excised. Especially on the early development stages, the same length (about 10 mm) from the tips of aerial and subterranean pods was excised. At the 4 and 8 DAM, aerial pods were excised around 10 mm from the apex, while subterranean pods were collected the swelling part for total RNA isolation. The materials were collected and immediately frozen in liquid nitrogen, and then stored in a freezer at −80 °C.
Fixation, sectioning and staining
Material was fixed in FAA (50 % alcohol:acetic acid:formaldehyde solution = 89:6:5) at room temperature. The samples were then vacuum infiltrated to remove trapped air. Samples were washed by 50 % alcohol, dehydrated using an ethyl alcohol series, cleared in xylene and embedded in paraffin wax. The specimens were sectioned to a thickness of 8 μm. Sections were stained with toluidine blue, observed and photographed using a Leica DMLB light microscope (Leica Microsystems GmbH, Wetzlar, Germany).
RNA extraction and microarrays procedure
Total RNA for microarray and quantitative real-time RT-PCR analysis were isolated from the same aerial and subterranean pods samples using a modified CTAB-based protocol (Chang et al. 1993) with high salt and further purified with the RNeasy Plant Mini Kit (Qiagen, Shanghai, China). A NanoDrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE) and agarose gel electrophoresis were used to test RNA quality and quantity. Gene expression profiles were generated using a high-density peanut microarray with pooling three biological replicates together for each development stage. Each microarray used a customized NimbleGen oligonucleotide microarray (4X 72 k) representing 36,158 unigenes.
The microarray hybridization procedure in this study was performed same as our lab Chen’ methods (Chen et al. 2012). Probes on the microarray ranging from 60 to 70 mer were synthesized by Sigma-Aldrich (Saint Louis, MO, USA) and then spotted to Corning ultraGAPs glass slides with three replications of each oligonucleotide at different locations on the slide to accommodate bioinformatics statistic analysis. Microarray procedure was performed according to the methods described previously by our lab Wang et al. (2012). Double-stranded cDNA synthesis, cleaned, fluorescently labeling, microarrays hybridization, washing and scanning were conducted at CapitalBio Corporation (Beijing, China) using Roche (Shanghai, China) NimbleGen Systems.
Data analysis and functional annotation
All microarrays were scanned with a LuxScan 10 KA scanner using LUXSCAN 4.0 software (CapitalBio, Beijing, China) to generate the raw data files. Fluorescence data were processed with SpotData software to quantify signal at CapitalBio Corporation as described previously (Graubert et al. 2007). For statistical analysis, the data normalization was performed by rank-consistency-filtering with Lowess intensity normalization method based on a robust multichip analysis (RMA, CapitalBio). And expression ratios were collected only on those spots with signal intensity (Cy3) ≥400 in at least one dye channel on the microarray slides after subtraction of the background in all experiments. Statistical analysis involved unpaired t test with using of GeneSpringGX11 (Agilent Technologies). The Benjamini-Hochberg FDR method was used to obtain corrected P values (false discovery rate, FDR) for multiple testing (p ≤ 0.05). The microarray analysis was employed to measure global gene expression in aerial and subterranean pods across four different developmental days. Comparative transcriptomics analysis was conducted between aerial pods and subterranean pods at the same developmental days. And we set the RNA sample from aerial pods as the reference. The intensity values of each sample were further transformed on log2-scale and used for performing differential expression analysis. The probe sets had a P value < 0.01 and >twofold changes in at least one of the comparisons were considered as differentially expressed genes (DEGs) for further analysis. To show the alternation of each DEGs during the whole development stages, their fold-change data were imported into MultiExperiment Viewer (MeV v4.8, http://www.tm4.org/mev/) for hierarchical clustering analysis with the average linkage method. Venn diagram was also constructed with the Venny tool (Oliveros 2007) to show the stage-specific expressed genes in each comparison and the overlaps between four binary comparison groups. The normalized data, Gene ID, gene sequence, Probes ID and probes sequence from peanut aerial and subterranean pods development microarray expression analysis were listed in Supplemental Table 7, 8 and 9.
The gene annotation was performed according to the method described by Shi et al. (2006). Descriptions of each DEGs were BLAST searched against the NCBI protein database (http://www.ncbi.nlm.nih.gov) and UniProt database (http://www.uniprot.org) for retrieval of updated annotation of homologous proteins with a cutoff of 1E−5. The gene annotation was based on the similarity (>80 % identity) of the homologous proteins and the evolutionary relationship between species (mainly the legume family and Arabidopsis thaliana). The DEGs of each comparison and their annotation were listed in Supplemental Table 3. Furthermore, the UniProtKB accession numbers assigned to the DEGs were submitted to gene ontology (GO) terms using MAS (molecule function annotation system, http://bioinfo.capitalbio.com/mas) to organize genes into hierarchical categories on the basis of biological process and molecular function. GO terms with false discovery rate (FDR) corrected P values < 0.05 were considered statistically significant.
Real-time PCR quantification
First-strand cDNA was synthesized from 1 μg total RNA using the ReverTra Ace-α-First Strand cDNA Synthesis kit (TOYOBO). Gene-specific primers were designed with the Primer Premier 5.0 (PREMIER Biosoft International, USA). Quantitative real-time RT-PCR was performed with Realtime PCR Master Mix (TOYOBO) and a LightCycler 480 instrument (Roche) equipped with LightCycler Software Version 1.5 (Roche) based on the manufacturer’s instructions. The actin gene was amplified along with the target gene as reference to normalize expression between different samples. All assays for a target gene were performed in triplicate synchronously under identical conditions.
Plant hormone extraction and measurement
The method for the extraction and purification of endogenous plant hormones GA3 and IAA were modified from those described by Kojima (1995) and Yang et al. (2000). One gram plant tissue were ground into fine powder in liquid nitrogen, then added 5 ml 80 % (v/v) methanol containing 10 mg/l butylated hydroxytoluene (BHT) as an antioxidant and transferred this mixture into a conical tube. The methanolic extracts were kept for continuous stirring at 4 °C in the dark for about 12 h. Then centrifuged with 5,000 rpm for 30 min at 4 °C. This procedure was repeated for twice and put the supernatant together, then concentrated to a water residue in vacuum at 35 °C by rotatory evaporation. The volume was adjusted to 2 ml with distilled water and added two volumes of cold chloroform to wash them. Centrifuged at 5,000 rpm for 5 min at 4 °C and adjusted the aqueous phase to pH 2.7 with HCl. The ethyl acetate was layered to the aqueous phase and the two-phase system was gently stirred for 3 min. Repeated for thrice and put the ethyl acetate phase together. The combined ethyl acetate phases were reduced to dryness in vacuum at 35 °C by rotatory evaporation. The solid residue was dissolved in 500 μl 100 % (v/v) methanol, and further filtered to measure GA3 and IAA by high performance liquid chromatograph (HPLC) according to Ross’ methods (Ross et al. 2004). All the measurements were performed with three biological replicates together for each development stage.
Results
Anatomical analysis of aerial pod
It is still obscure why the aerial pod always remains small pod and can’t develop normally. To better understand the cellular structure of aerial pod, we conducted anatomical analysis by toluidine blue staining of tissue sections. The results revealed that the aerial pod was a special reproductive organ, which showed unique anatomical and morphological characteristics (Fig. 1a–c). The shape of aerial pod looked like a needle, and the fertilized ovary was located at nearly 3–5 mm after the tip (Fig. 1a, b). While at the late development stage, the embryo was abortion due to the failure of aerial pod penetration into the soil (Fig. 1c).
Fig. 1.

Anatomical analysis of the peanut aerial pod. The embryo of aerial pod was abortion at the late development stage. a the aerial pod; b Longitudinal section of early aerial pod; c Longitudinal section of old aerial pod. Embryos are indicated with red arrows. Bars = 200 μm
Differential expression in peanut pod development
As shown in the previous study, the seed in aerial pods aborted at the 6 days after marked (DAM) (Chen et al. 2013). To identify differentially expressed genes (DEGs) relating to seed abortion during peanut aerial and subterranean pods development, we used a customized NimblerGen onligonucleotide microarrays at the 1, 2, 4, 8 DAM. Results showed that a total of 18,366 genes, accounting for 50.79 % of all transcripts in the microarrays, were expressed in all the eight samples and 7,835 genes (21.67 %) were not expressed in all the eight samples. In addition, many specific expressed genes which detected only in aerial or subterranean pods across four time-points were identified. Among them, 1,698 genes were detected only in aerial pods (Supplemental Table 1), while 703 genes only in subterranean pods (Supplemental Table 2). We set the sample of aerial pods as control, and totally 6,203 genes were differentially expressed in aerial and subterranean pods development across 4 consecutive time-points (Supplemental Table 3). All of these DEGs showed up-regulation or down-regulation with at least twofold changes. Clustering analysis showed that the expression profiles of DEGs varied significantly in developing peanut aerial and subterranean pods at different DAM (Fig. 2). Based on the expression pattern of subterranean pods versus aerial pods at 1, 2, 4 and 8 DAM, we classified these DEGs into the following 4 major groups, with a few exceptions: group I, a large group of genes whose expression significantly increased at 1 DAM but down-regulated at other DAM; group II, a group of genes whose expression significantly increased at 2 DAM but down-regulated at other DAM; group III, a large group of genes whose expression significantly increased at both 2 and 4 DAM, together with a large group of genes whose expression significantly increased at both 4 and 8 DAM; group IV, a group of genes whose expression significantly increased at 8 DAM but down-regulated at other DAM. The clustering result indicated that seed aborted in aerial pods was a complex process and coordinated by a large group of DEGs at different development stages.
Fig. 2.
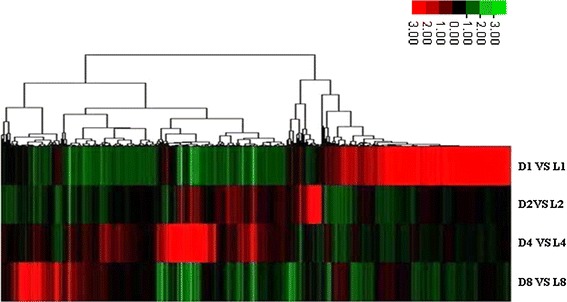
The result of clustering analysis on the differentially expressed genes of subterranean pods versus aerial pods at the different development days. Probe sets with P < 0.01 and fold changes (FC) >2 in at least one of the comparison are included. The columns are sorted by hierarchical clustering using the average linkage methods. The ratios are shown in a red-green color scale, where red indicates up-regulation and green indicates down-regulation. Each row represents a sample of subterranean pods versus aerial pods obtained from three biological replicates and each column represents a differentially expressed probe set. D1 versus L1, D2 versus L2, D4 versus L4 and D8 versus L8: the comparison of subterranean pods versus aerial pods at 1, 2, 4 and 8 days after marked
Comparative analysis of aerial and subterranean pods development
To investigate expression alternation of stage-specific genes and seed abortion candidate genes during aerial and subterranean pods development, we conducted comparative analysis of the transcriptome profiles between aerial and subterranean pods development at 1, 2, 4, 8 DAM. As shown in Fig. 3, comparing subterranean pods with aerial pods, considerable DEGs were identified at 1, 2, 4, 8 DAM. Totally 3,609 differentially expressed genes were detected at 1 DAM (D1 vs L1), including 1,781 up-regulated and 1,828 down-regulated genes. Similarly, 665 DEGs (including 371 up-regulated and 294 down-regulated genes), 1,612 DEGs (including 1,055 up-regulated and 557 down-regulated genes), 2,165 DEGs (including 663 up-regulated and 1,502 down-regulated genes) were identified at 2, 4, 8 DAM (D2 vs. L2, D4 vs. L4, D8 vs. L8), respectively (Supplemental Table 3). These results indicated that dramatical changes in transcriptome profiles of peanut pod development began to occur at the 1 DAM when aerial pegs penetrated into the soil. In addition, many stage-specific DEGs were also analyzed across 4 consecutive time-points using a Venn diagram (Fig. 4). Approximately 2,724, 230, 635 and 1,143 stage-specific DEGs were identified at 1, 2, 4, 8 DAM, respectively. And 50 DEGs were shared among all 4 consecutive time-points. These DEGs could be contributed to seed and pod development under disparate conditions, especially light conditions.
Fig. 3.

Comparative analysis of differentially expressed genes (DEGs) between aerial and subterranean pods at 1, 2, 4, 8 days after marked. The number of up-regulated and down-regulated genes between aerial and subterranean pods at different DAM are indicated. D1, D2, D4 and D8: the development of subterranean pods after marked at 1, 2, 4 and 8 days; L1, L2, L4 and L8: the development of aerial pods after marked at 1, 2, 4 and 8days
Fig. 4.

Venn diagram depicting the number of differentially expressed genes between aerial and subterranean pods development at 1, 2, 4 and 8 days after marked in each comparison and the overlaps between four binary comparison groups. For each developmental stage, the total number of differentially expressed genes and the number of stage-specific expressed genes are indicated. D1 versus L1, D2 versus L2, D4 versus L4 and D8 versus L8: the comparison of subterranean pods versus aerial pods at 1, 2, 4 and 8 days after marked
Gene ontology enrichment analysis for differentially expressed genes
To identify biological process of differentially expressed genes (DEGs), gene ontology (GO) analysis was conducted using Molecule Annotation System (MAS, http://bioinfo.capitalbio.com/mas3). The GO analysis obtained using the annotation procedure through homology analysis to generate a concise functional annotation. As shown in Fig. 5, the known DEGs were mainly classified into 29 functional categories and involved in 34 biological processes. The results showed that these DEGs mainly distributed in plasma membranes and nucleus after genes expression, and participated in the biological process of biosynthesis (2.1 %), metabolism (12.2 %), regulation of transcription (8.9 %), transporting (7.6 %), stress response (4.9 %), cell division and differentiation (1.1 %), cell apoptosis (1.3 %), hormone response (1.1 %), embryonic development (0.7 %), photomorphogenesis (0.5 %), photoperiodism (0.7 %), photosynthesis (2.1 %), lignin synthesis (1.6 %), and so on. All these results indicated the biological process of DEGs varying in a broad range. Through comparative analysis, the two most abundant sub-classes were biosynthesis processes and metabolic processes. Six other subclasses, including regulation of transcription, transport, oxidation and reduction processes, defense and stress response were also enriched. However, there were several important subcategories for pod development which were represented by genes for hormone response, signal transduction and embryonic development.
Fig. 5.

Statistics of differentially expressed genes assigned to GO functional categories based on biological process. Some genes are assigned to more than one GO functional category for participating in multiple biological processes. The percentages for GO terms are calculated by the number of DEGs in one GO term dividing to the total number of DEGs in all GO term
Furthermore, to determine the biological significance of the differentially expressed genes, GO terms enrichment analysis of the total DEGs was also performed (P ≤ 0.05) using the agriGO tools (http://bioinfo.cau.edu.cn/agriGO/). As shown in Supplemental Table 4, the biological processes of response to stimulus (GO:0050896), response to stress (GO:0006950), response to hormone stimulus (GO:0009725) and developmental process (GO:0032502) were enriched GO terms, indicating that hormone and environment stimuli played a vital role in peanut pods development. In the tree traversing of enriched GO terms such as response to hormone stimulus (GO:0009725), 19 DEGs were involved in response to auxin stimulus (GO:0009733); 8 DEGs were involved in response to gibberellin stimulus (GO:0009739); 7 DEGs were related to abscisic acid stimulus (GO:0009737); 5 DEGs were associated to ethylene stimulus (GO:0009723); 7 DEGs were participated in hormone-mediated signaling pathway (GO:0009755). Moreover, GO terms enrichment analysis of the stage-specific DEGs at 1, 2, 4 and 8 DAM were conducted. During aerial and subterranean pods development, the significant GO for stage-specific expressed genes at 1 DAM were mainly classified into the following categories: ion binding (GO:0005506), transferase activity(GO:0016740), catalytic activity (GO:0003824), DNA binding (GO:0003677), oxidoreductase activity (GO:0016491), hydrolase activity (GO:0016787), enzyme activity (GO:0004857), transporter activity (GO:0005215), ATP binding (GO:0005524) and peptidase activity (GO:0004176). Similarly, stage-specific expressed genes at 2 and 8 DAM were significantly enriched with ion binding (GO:0005506). The enrichment GO term for stage-specific expressed genes at 4 DAM mainly belonged to ion binding (GO:0005506), transferase activity(GO:0016740), catalytic activity (GO:0003824), enzyme activity (GO:0004857), ATP binding (GO:0005524) and cofactor binding (GO:0048037).
Candidate genes related to seed abortion during aerial pods development
Many physiological studies revealed that seed development in peanut was highly coordinated by plant hormones, and also mainly controlled by the alternation of light and dark conditions. Based on GO analysis, some differentially expressed genes participated in biological process of hormone response, cell apoptosis, embryonic development and light signaling. We speculated that these differentially expressed genes might function as candidate genes to provide insights into seed abortion during aerial pods development. These potential candidate genes contained 39 hormone response relative genes, 16 cell apoptosis relative genes, 17 embryonic development relative genes and 10 light signaling relative genes, respectively (Table 1). In the identified hormone relative genes, many shared homology with genes coding for proteins well known to be involved in cell apoptosis, development and light signaling pathways. It indicated that hormone relative genes were in a central position of signaling pathway to regulate seed abortion during aerial pods development. Especially, 19 auxin-related genes, accounting for 48.7 % of all hormone relative genes, were significant differentially expressed during seed and pod development, suggesting that auxin response factor and auxin-induced protein might be involved in seed abortion. However, these genes that might lead to seed abortion need to be confirmed in further functional studies.
Table 1.
The annotation of candidate genes related to seed abortion during aerial pods development
| Gene ID | Uniprot NO. | Species | Protein name | E value |
|---|---|---|---|---|
| Hormone response relative genes | ||||
| AHTC1008761 | Q2HRH3 | Medicago truncatula | Gibberellin regulated protein | 1.00E−46 |
| AHTC1031559 | A9P6A4 | Medicago truncatula | Ethylene-responsive transcription factor 1A | 8.00E−21 |
| AHTC1009678 | O04280 | Phaseolus vulgaris | Gibberellin 20-oxidase | 1.00E−141 |
| AHTC1027127 | B9R824 | Ricinus communis | Auxin-induced protein 5NG4 | 4.00E−39 |
| AHTC1021447 | Q8GV76 | Medicago truncatula | Auxin efflux carrier protein | 3.00E−72 |
| AHTC1035475 | A2Q374 | Medicago truncatula | Gibberellin regulated protein | 2.00E−41 |
| AHTC1012926 | B9SHD1 | Ricinus communis | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | 4.00E−66 |
| AHTC1013072 | Q76FZ8 | Pisum sativum | Brassinosteroid receptor | 2.00E−75 |
| AHTC1007964 | B9R7Q4 | Ricinus communis | BRASSINAZOLE-RESISTANT 1 protein | 1.00E−37 |
| AHTC1015327 | B9RWA6 | Ricinus communis | Gibberellin receptor GID1 | 2.00E−31 |
| AHTC1026621 | Q6L8U0 | Cucumis sativu | Auxin response factor 4 | 2.00E−33 |
| AHTC1021732 | B9N158 | Populus trichocarpa | Auxin efflux carrier component | 1.00E−58 |
| AHTC1028384 | Q8GV76 | Medicago truncatula | Auxin efflux carrier component | 2.00E−83 |
| AHTC1008463 | B9S5C3 | Ricinus communis | Ethylene-overproduction protein | 6.00E−46 |
| AHTC1013682 | Q9ATR0 | Pisum sativum | Brassinosteroid biosynthetic protein LKB | 2.00E−92 |
| AHTC1004450 | Q05G09 | Lupinus albus | Auxin efflux carrier | 7.00E−96 |
| AHTC1016323 | C6ZJZ5 | Glycine max | Auxin efflux carrier protein 2 | 2.00E−76 |
| AHTC1003763 | B9I0L4 | Populus trichocarpa | Auxin efflux carrier family protein | 3.00E−124 |
| AHTC1020717 | Q05680 | Glycine max | Auxin-responsive GH3 product | 1.00E−54 |
| AHTC1000327 | Q45W71 | Arachis hypogaea | Auxin-repressed protein | 2.00E−43 |
| AHTC1017738 | Q8S4Q2 | Arachis ipaensis | Ethylene-responsive transciptional coactivator-like protein | 6.00E−58 |
| AHTC1014115 | A5HSG1 | Arachis ipaensis | Ethylene-responsive transcription factor | 3.00E−36 |
| AHTC1034938 | Q76FZ8 | Pisum sativum | Brassinosteroid receptor | 5.00E−26 |
| AHTC1000612 | Q4W8C3 | Phaseolus angularis | Gibberellin 2-oxidase | 3.00E−125 |
| AHTC1010480 | Q9FNV7 | Robinia pseudoacacia | Auxin-repressed protein | 7.00E−39 |
| AHTC1026648 | B0L633 | Cicer arietinum | GA-like protein | 5.00E−19 |
| AHTC1008511 | B9SWW7 | Ricinus communis | Auxin response factor GTPase activator | 1.00E−73 |
| AHTC1023667 | A9QNE7 | Solanum lycopersicum | ABA 8’-hydroxylase | 7.00E−36 |
| AHTC1005584 | B6VB01 | Arachis hypogaea | Auxin binding protein 1 | 4.00E−99 |
| AHTC1003543 | Q94F62 | Arabidopsis thaliana | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | 2.00E−12 |
| AHTC1006456 | Q0GXX3 | Medicago truncatula | Auxin conjugate hydrolase | 0 |
| AHTC1004284 | Q8S4Q2 | Ammopiptanthus mongolicus | Ethylene-responsive transciptional coactivator-like protein | 6.00E−28 |
| AHTC1025214 | B2BA73 | Pisum sativum | Gibberellin 3-oxidase | 1.00E−28 |
| AHTC1009513 | B9STH7 | Ricinus communis | Auxin-induced protein 5NG4 | 4.00E−64 |
| AHTC1032952 | Q45W71 | Arachis hypogaea | Auxin-repressed protein | 2.00E−18 |
| AHTC1034412 | Q8W3P8 | Phaseolus angularis | ABA-glucosyltransferase | 6.00E−60 |
| AHTC1022224 | P33081 | Glycine max | Auxin-induced protein 15A | 5.00E−21 |
| AHTC1019083 | P33079 | Glycine max | Auxin-induced protein 10A5 | 4.00E−31 |
| AHTC1030208 | P33080 | Glycine max | Auxin-induced protein X10A | 8.00E−34 |
| Cell apoptosis relative genes | ||||
| AHTC1006425 | B9RDP2 | Ricinus communis | Dead box ATP-dependent RNA helicase | 2.00E−72 |
| AHTC1016793 | B9T0X5 | Ricinus communis | Dead box ATP-dependent RNA helicase | 6.00E−63 |
| AHTC1010861 | B9RWT5 | Ricinus communis | Dead box ATP-dependent RNA helicase | 7.00E−85 |
| AHTC1020385 | Q0H950 | Glycine max | Lethal leaf spot 1-like protein | 2.00E−95 |
| AHTC1004442 | A3QRM3 | Glycine max | Senescence-associated nodulin 1A | 6.00E−76 |
| AHTC1008715 | Q0H950 | Glycine max | Lethal leaf spot 1-like protein | 1.00E−62 |
| AHTC1010962 | D3G9M3 | Glycine max | Vascular associated death 1 | 2.00E−52 |
| AHTC1008318 | B5TV63 | Camellia sinensis | Senescence-related protein | 7.00E−62 |
| AHTC1004846 | B9RDP2 | Ricinus communis | Dead box ATP-dependent RNA helicase | 4.00E−138 |
| AHTC1027355 | Q2HVE0 | Medicago truncatula | Leucine-rich repeat | 2.00E−17 |
| AHTC1025672 | Q2YE88 | Glycine max | NB-LRR type disease resistance protein Rps1-k-1 | 5.00E−15 |
| AHTC1014344 | Q2YE88 | Glycine max | NB-LRR type disease resistance protein Rps1-k-1 | 6.00E−35 |
| AHTC1028456 | Q84ZU8 | Glycine max | R 10 protein | 3.00E−53 |
| AHTC1033023 | Q84ZU5 | Glycine max | R 8 protein | 8.00E−31 |
| AHTC1035719 | Q8W2C0 | Glycine max | candidate resistance protein KR1 | 4.00E−21 |
| AHTC1003543 | Q94F62 | Arabidopsis thaliana | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | 2.00E−12 |
| Embryonic development relative genes | ||||
| AHTC1032586 | B4UW62 | Arachis hypogaea | Embryo-abundant protein EMB | 2.00E−22 |
| AHTC1010653 | Q9SWB3 | Glycine max | Seed maturation protein PM39 | 2.00E−22 |
| AHTC1001743 | Q39871 | Glycine max | Late embryongenesis abundant protein | 2.00E−56 |
| AHTC1000013 | O49817 | Cicer arietinum | Late embryogenesis abundant protein 2 | 1.00E−36 |
| AHTC1001476 | Q39801 | Glycine max | 51 kDa seed maturation protein | 6.00E−26 |
| AHTC1014629 | Q39871 | Glycine max | Late embryongenesis abundant protein | 1.00E−58 |
| AHTC1001477 | Q39801 | Glycine max | 51 kDa seed maturation protein | 7.00E−49 |
| AHTC1014265 | O49817 | Cicer arietinum | Late embryogenesis abundant protein 2 | 7.00E−19 |
| AHTC1014482 | O49817 | Cicer arietinum | Late embryogenesis abundant protein 2 | 3.00E−38 |
| AHTC1000135 | O49817 | Cicer arietinum | Late embryogenesis abundant protein 2 | 3.00E−39 |
| AHTC1005600 | Q2XSI1 | Glycine latifolia | Seed maturation protein | 1.00E−23 |
| AHTC1006589 | Q9ZTZ3 | Glycine max | 24 kDa seed maturation protein | 3.00E−54 |
| AHTC1011324 | Q9SWS4 | Glycine max | Ripening related protein | 1.00E−43 |
| AHTC1003523 | Q9SWS4 | Glycine max | Ripening related protein | 1.00E−36 |
| AHTC1014148 | O49817 | Cicer arietinum | Late embryogenesis abundant protein 2 | 4.00E−7 |
| AHTC1011166 | Q9SYM4 | Arabidopsis thaliana | alpha-trehalose-phosphate synthase | 2.00E−136 |
| AHTC1022204 | O49552 | Arabidopsis thaliana | DNA damage-binding protein 1b | 5.00E−30 |
| Light signaling relative genes | ||||
| AHTC1014391 | Q8LEA8 | Arabidopsis thaliana | Phytochrome A-associated F-box protein | 7.00E−44 |
| AHTC1029353 | B9MST1 | Glycine max | Circadian clock-associated FKF1 | 2.00E−92 |
| AHTC1003429 | B9MST1 | Glycine max | Circadian clock-associated FKF1 | 0 |
| AHTC1021859 | B9MST1 | Glycine max | Circadian clock-associated FKF1 | 5.00E−49 |
| AHTC1000126 | Q850G4 | Arachis hypogaea | Putative early light induced protein | 6.00E−98 |
| AHTC1000086 | Q850G4 | Arachis hypogaea | Putative early light induced protein | 9.00E−22 |
| AHTC1024152 | Q8GWZ0 | Arabidopsis thaliana | uncharacterized protein | 3.00E−06 |
| AHTC1013035 | Q8GWZ0 | Arabidopsis thaliana | uncharacterized protein | 2.00E−26 |
| AHTC1022204 | O49552 | Arabidopsis thaliana | DNA damage-binding protein 1b | 5.00E−30 |
| AHTC1024768 | Q5XEU1 | Arabidopsis thaliana | At2g21070 | 5.00E−16 |
The eighty-two candidate genes identified in this study are shown. Gene ID is provided on the left side of the table. Based on the functional annotation and GO analysis as described in the section of materials and methods, they are mainly hormone response, cell apoptosis, embryonic development and light signaling relative genes. Their hits of Uniprot accession number, species, protein name and E value are shown in the table
Validation of microarrays data by real-time RT-PCR
In order to confirm the microarrays results, ten genes were randomly selected from seed abortion relative genes based on the GO analysis and subjected to real-time RT-PCR analysis. These genes (as shown in Table 2) were involved in hormone response or cell apoptosis. Primers for these genes were listed in Supplemental Table 5, and the actin gene was used as an internal control. As shown in Fig. 6, the expression pattern of 10 selected DEGs analyzed by real-time RT-PCR were consistent with their respective microarrays data. Compared with subterranean pods, all the selected genes were significantly up-regulated in aerial pods at 8 DAM, suggesting that they might function in seed abortion during aerial pods development. In 3 selected genes (AHTC1026322, AHTC1028456, AHTC1035719), their expression levels were higher in subterranean pods at 1–4 DAM, while rapidly decreased on 8 DAM. The expression levels of other 7 selected genes were similarly with each other at 1 DAM to 4 DAM, while significantly up-regulated in aerial pods at 8 DAM.
Table 2.
The selected differentially expressed genes for real time RT-PCR analysis
| Gene ID | Uniprot no. | Gene function | Protein name | Species | E value |
|---|---|---|---|---|---|
| AHTC1025948 | Q0WQQ1 | Hormone response | ADP-ribosylation factor GTPase-activating protein AGD15 | Arabidopsis thaliana | 7.00E−10 |
| AHTC1022224 | P33081 | Hormone response | Auxin-induced protein 15A | Glycine max | 5.00E−21 |
| AHTC1019083 | P33079 | Hormone response | Auxin-induced protein 10A5 | Glycine max | 4.00E−31 |
| AHTC1030208 | P33080 | Hormone response | Auxin-induced protein X10A | Glycine max | 8.00E−34 |
| AHTC1026322 | Q2HSV9 | Hormone response | Transcriptional factor B3; Auxin response factor | Medicago truncatula | 1.00E−48 |
| AHTC1027355 | Q2HVE0 | Apoptosis | Leucine-rich repeat | Medicago truncatula | 2.00E−17 |
| AHTC1025672 | Q2YE88 | Apoptosis | NB-LRR type disease resistance protein Rps1-k-1 | Glycine max | 5.00E−15 |
| AHTC1028456 | Q84ZU8 | Apoptosis | R 10 protein | Glycine max | 3.00E−53 |
| AHTC1033023 | Q84ZU5 | Apoptosis | R 8 protein | Glycine max | 8.00E−31 |
| AHTC1035719 | Q8W2C0 | Apoptosis | candidate resistance protein KR1 | Glycine max | 4.00E−21 |
Ten DEGs were randomly selected from seed abortion candidate genes to validate the microarrays data by real-time RT-PCR analysis. Based on GO functional categories, they were involved in the biological process of hormone response and cell apoptosis. Gene ID is provided on the left side of the table. Their hits of Uniprot accession number, gene function, protein name, species and E-value are shown in the table
Fig. 6.
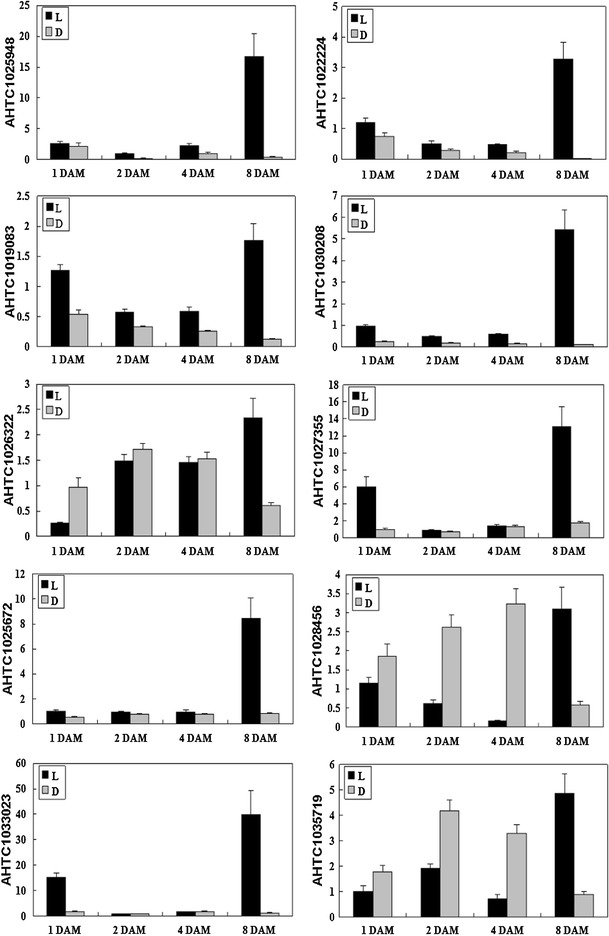
Real time RT-PCR analysis on mRNA transcription of the selected differentially expressed genes. L: aerial pods; D: subterranean pods; 1–8 DAM: the development days of aerial and subterranean pods after marked
Measurement of endogenous plant hormone
With the significant up-regulation of hormone response relative genes in aerial pods, we examined the changes of plant hormone in order to clarify its possible roles in aerial and subterranean pods development. Levels of GA3 and IAA were investigated during the whole process by HPLC (Fig. 7). The results showed that in the development from 2 DAM to 8 DAM, the GA3 content in subterranean pods was higher than aerial pods, while lower in the other development days. The IAA contents of aerial pods were similarly with each other at 1 DAM to 8 DAM, while significantly increased with six times more than subterranean pods during the development from 12 DAM to 20 DAM. These results well agreed with the microarrays data and real time RT-PCR result of hormone response relative genes, indicating that GA3 and IAA might be involved in seed abortion of aerial pods.
Fig. 7.

Change of GA3 (a) and IAA (b) content in peanut aerial and subterranean pods development at different days after marked. L: aerial pods; D: subterranean pods; 1–20 days: at the development days after marked. FW fresh weight. Vertical bars represent standard error of means
Discussions
In recent years, the powerful tool of microarrays was broadly applied to investigate fruit development, as reported for strawberry (Aharoni and O’Connell 2002), tomato (Alba et al. 2005), pear (Fonseca et al. 2004), and apple (Lee et al. 2007). In this study, we identified 6,203 differentially expressed genes via microarrays analysis, and also detected 4,732 stage-specific expressed genes and 2,401 specific expressed genes only in aerial or subterranean pods across various stages. Additionally, through Gene Ontology analysis, many differentially expressed genes participated in the biological process of cell apoptosis, hormone response, embryonic development and light signaling, which were identified as potential candidate genes responsible for the normal development of subterranean pods and also seed abortion in aerial pods. In a manner consistent with significant up-regulation of auxin response relative genes in aerial pods, the changes of plant hormone IAA potentially contributed to seed abortion during aerial pods development, providing new insight that auxin response factors might be involved in seed abortion. Real time RT-PCR analysis also validated the expression alternation of candidate genes occurring at transcriptional level. Interestingly, seed development is a complex process with fascinating characteristic in seed biology, while several distinctive features can make peanut as an excellent model to study seed and pod development, especially for aerial flowering, gravitropic peg elongation and subterranean fructification.
DEGs involved in UPS and photosynthesis
Comparison of gene expression profiling between the aerial and subterranean pods development is essential for the elucidation of molecular networks in peanut pod development. Combined with previous RNA-Seq and proteomics analysis (Chen et al. 2013; Zhu et al. 2013), this study further facilitated that the differentially expressed genes and proteins involved in ubiquitin proteasome system (UPS) and cell wall modification, might function as candidates to regulate peanut pod development. As shown in Supplemental Table 6, 21 UPS relative genes were identified as DEGs during aerial and subterranean pods development. These genes might play a critical role in pod swelling process to allow room for embryo development. Gene probesets matching to identified proteins indicated a good consistence of expression alternations between protein and mRNA data. Lots of UPS relative genes were up-regulated in subterranean pods during the early development stages, while significantly decreased at the late stages. However, the expression levels of cell wall modification relative genes were up-regulated in aerial pods. In addition, similar to proteins expression, a great number of photosynthesis relative genes were also significantly up-regulated and enriched in aerial green pods. All these well agreed with the previous RNA-seq analysis, validating the significance of the large number of DEGs changes found in this study.
Potential functions of auxin response genes in seed and pod development
In recent years, the prominent role of auxin signaling in patterning the early embryo was becoming increasingly clear. When auxin was perceived by its receptor, the auxin response factors (ARFs) would be released to exert their function as activators or repressors of transcription (Möller and Weijers 2009). Studies in Arabidopsis had led to an understanding of embryo development processes that were controlled by auxin response factors to coordinate several cell specification and pattern formation (Abel and Theologis 1996; Guilfoyle et al. 1998; Jenik and Barton 2005). For instance, many auxin response factors linked auxin signaling to control the seed development, seed size and cotyledons transition of embryogenesis by regulating cell division and organ growth, such as ARF5 (Okushima et al. 2005; Xing et al. 2011), DR5 (Benkova et al. 2003), ARF7 (Harper et al. 2000) and ARF2 (Schruff et al. 2006). Additionally, the PIN family of auxin efflux facilitators such as PIN1, PIN3, PIN4 and PIN7 (Friml et al. 2002a, b, 2003, 2004), were responsible for the dynamic and shifting pattern of auxin accumulation in the embryo by expressing at stage-specific and tissue-specific during embryonic development.
GAs, auxin, ABA, and ethylene have been implicated in regulating the peanut seed development and pod maturation (Jacobs 1951; Ziv and Kahana 1988; Shlamovitz et al. 1995; Moctezuma and Feldman 1996; Ozga and Reinecke 2003). In this study, a number of differentially expressed genes related to hormone response were identified during aerial and subterranean pods development, while in our previous RNA-seq and proteomics study we could not detect significantly up-regulated genes involved in this pathways. Among them, auxin-related genes accounted for 48.7 % of all hormone response relative genes. Together with IAA content was significantly increased during aerial pods development, many auxin response relative genes (Table 1) were identified as candidates to seed abortion, such as auxin response factor, auxin-induced protein, auxin-repressed protein, auxin response factor GTPase activator, auxin efflux carrier and auxin binding protein. Moreover, the expression pattern of most auxin-related genes were up-regulated during aerial pods development. All these revealed that auxin and auxin response genes potentially played a crucial role in peanut seed and pod development.
Transcriptional regulation of cell apoptosis and embryonic development
Embryonic development was the main biological process that determined the size and ultimate fate of the seed by cell division and enlargement (Ohto et al. 2009). Based on the embryo abortion during aerial pods development, previous studies underlined the importance of three candidate genes such as two senescence associated genes and one late embryogenesis-abundant gene (Chen et al. 2013). In this study, we identified 16 cell apoptosis relative genes and 17 embryonic development relative genes as candidate genes to seed abortion in aerial pod. We also detected two senescence associated genes and seven late embryogenesis-abundant genes which were specially and significantly up-regulated in the aerial pod. Several reports of senescence-associated genes appeared to trigger senescence program preceding death in response to multiple developmental and environmental signals (Quirino et al. 1999; Gepstein et al. 2003; Lim and Nam 2005; Espinoza et al. 2007). Late embryogenesis abundant proteins accumulated late in plant seed development and played crucial roles in varying stressful environmental conditions (Xu et al. 1996; NDong et al. 2002; Hundertmark and Hincha 2008).
Transcriptional regulation of light signaling pathway
As Thompson et al. (1985) reported, light leaded to the cessation of embryo differentiation during peg elongation phase, and dark stimulated the resumption of embryo development following quiescence in underground phase. Ten differentially expressed genes involved in light signaling pathway were identified in this study. Phytochrome A-associated F-box protein (AHTC1014391) were identified and significantly up-regulated in subterranean pods at late stages. Some studies suggested that phytochrome was localized in tissue-specific of the developing embryo and integument, which might play an important role in the underground phase, but not in the peg elongation phase (Thompson et al. 1992; Moctezuma 2003). The discovery of these genes indicated that regulation of seed and pod development were important in the acclimation to disparate growth conditions, especially under dark conditions.
New insights into peanut seed and pod development
Seed formation and pod swelling are of two vital important processes of peanut pod development. However, seed and pod development programs in peanut are highly complex and need to be finely controlled and coordinated by the intervention of several cross talks. Based on the data presented in this work and in our previous investigations, we propose a preliminary overview of the important biological processes occurring during peanut pod development, which are in part schematically represented in Supplemental Fig. 1. The plant hormone auxin significantly increases in aerial pods under light conditions, which in turn directly or indirectly activates auxin response factors to trigger the auxin signal transduction. When the signals transduce to UPS or lignin synthesis pathways, the initiation of pod swelling in peg tips will be suppressed. Alternatively, when the signals transduce to cell apoptosis or embryonic development pathways, it may lead to seed abortion. All these pathways would work together, leading to the seed and pod normally developing. In addition, it is clear that the UPS not only plays an essential role in hormone perception and responses, but also contributes to plant cell division and PCD (Dharmasiri and Estelle 2002; Yanagawa et al. 2002; Kim et al. 2003; Santner and Estelle 2010; Kepinski and Leyser 2002), indicating that UPS may act as a dual coordinator between pod swelling and seed abortion.
Concluding remarks
In conclusion, seed abortion within aerial pods caused by peg penetration failure is a major limitation of seed yield, seriously impacting on peanut production. In this study, we have performed a comparative investigation of transcriptome profile between aerial and subterranean pods development. Simultaneously, together with endogenous IAA significantly increased in aerial pods, many candidate genes to seed abortion were identified, providing new molecular view that auxin response genes potentially played vital roles in seed and pod development. Although development of peanut aerial and subterranean pods have been studied intensely by DNA microarrays combined with previous RNA sequencing and proteomics analysis, many questions still wait to be answered. It is a key gap in our understanding what and how distinct genes in different seed tissue play important roles in cell division, differentiation and morphogenesis during early seed and embryo development. Our researches of identification and characterization of potential candidate genes and proteins can initiate the long way to unravel regulatory networks that program and coordinate the developmental and physiological events occurring. More detailed analysis by reverse genetic approaches is ongoing to further characterize their possible functional roles in seed and pod development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 9 (FASTA_ONELINESEQ 26120 kb)
Supplementary material 10 (FASTA 5785 kb)
Acknowledgments
This research was funded by grants from National Natural Science Foundation of China (No. 30971819 and 30900907), Natural Science Foundation of Guangdong Province (No.10151064001000002), Science and Technology Planning Project of Guangdong Province (2011B010500019), Pearl River Science and Technology Nova of Guangzhou (No. 2011J2200035) and supported by the earmarked fund for Modern Agro-industry Technology Research System (No. nycycx-19, CARS-14). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We declare no conflict of interests.
Abbreviations
- DAF
Day after flowering
- DAM
Days after marked
- UPS
Ubiquitin proteasome system
- DEGs
Differentially expressed genes
- GO
Gene ontology
- PCD
Programmed cell death
Contributor Information
Wei Zhu, Email: zhuwei0501@163.com.
Xiaoping Chen, Email: xpchen1011@gmail.com.
Haifen Li, Email: 565340390@163.com.
Fanghe Zhu, Email: gxzhufanghe@163.com.
Yanbin Hong, Email: hongyanbin1979@yahoo.com.cn.
Rajeev K. Varshney, Email: varshney.raj@gmail.com
Xuanqiang Liang, FAX: +862085514269, Email: liang-804@163.com.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, O’Connell A. Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot. 2002;53:2073–2087. doi: 10.1093/jxb/erf026. [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genom. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genom. 2010;11:282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron D, Mouzeyar S, Boulaflous A, Girousse C, Rustenholz C, Laugier C, Paux E, Bouzidi MF. Transcriptional profile analysis of E3 ligase and hormone-related genes expressed during wheat grain development. BMC Plant Biol. 2012;12:35. doi: 10.1186/1471-2229-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- Chen XP, Hong YB, Zhang EH, Liu HY, Zhou GY, Li S, Zhu F, Guo B, Yu J, Liang X. Comparison of gene expression profiles in cultivated peanut (Arachis hypogaea) under strong artificial selection. Plant Breeding. 2012;131:620–630. doi: 10.1111/j.1439-0523.2012.01997.x. [DOI] [Google Scholar]
- Chen XP, Zhu W, Azam S, et al. Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant Biotechnol J. 2013;11:115–127. doi: 10.1111/pbi.12018. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Estelle M. The role of regulated protein degradation in auxin response. Plant Mol Biol. 2002;49:401–409. doi: 10.1023/A:1015203013208. [DOI] [PubMed] [Google Scholar]
- Espinoza C, Medina C, Somerville S, Johnson PA. Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J Exp Bot. 2007;58:3197–3212. doi: 10.1093/jxb/erm165. [DOI] [PubMed] [Google Scholar]
- Feng QL, Stalker HT, Pattee HE, Isleib TG. Arachis hypogaea plant recovery through in vitro culture of peg tips. Peanut Sci. 1995;22:129–135. doi: 10.3146/i0095-3679-22-2-11. [DOI] [Google Scholar]
- Fonseca S, Hackler L, Zvara A, Ferreira S, Baldé A, Dudits D, Pais MS, Puskas LG. Monitoring gene expression along pear fruit development, ripening and senescence using cDNA microarrays. Plant Sci. 2004;167:457–469. doi: 10.1016/j.plantsci.2004.03.033. [DOI] [Google Scholar]
- Friml J, Benkova E, Blilou I, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/S0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen I, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, et al. A PINOID-dependent binary switch in apical–apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariy I, Dor C, Bassani M. Large-scale identification of leaf senescence-associated genes. Plant J. 2003;36:629–642. doi: 10.1046/j.1365-313X.2003.01908.x. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Cahan P, Edwin D, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3(1):e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes? Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BZ, Chen XP, Dang P, Scully BT, Liang XQ, Holbrook CC, Yu J, Culbreath AK. Peanut gene expression profiling in developing seeds at different reproduction stages during Aspergillus parasiticus infection. BMC Dev Biol. 2008;8:12–28. doi: 10.1186/1471-213X-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Jacob J, Vanholme B, Kyndt T, Mitreva M, Gheysen G. Expressed sequence tags of the peanut pod nematode Ditylenchus africanus: the first transcriptome analysis of an Anguinid nematode. Mol Biochem Parasit. 2009;167:32–40. doi: 10.1016/j.molbiopara.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, Evans ELS, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs W. Auxin relationships in an intercalary meristem: further studies on the gynophore of Arachis hypogaea L. Am J Bot. 1951;38:307–310. doi: 10.2307/2438005. [DOI] [Google Scholar]
- Jenik PD, Barton MK. Surge and destroy: the role of auxin in plant embryogenesis. Development. 2005;132:3577–3585. doi: 10.1242/dev.01952. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. Ubiquitination and auxin signaling: a degrading story. Plant Cell. 2002;14:S81–S95. doi: 10.1105/tpc.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn JW, Jin UH, Choi D, Paek KH, Pai HS. Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem. 2003;278:19406–19415. doi: 10.1074/jbc.M210539200. [DOI] [PubMed] [Google Scholar]
- Kojima K. Simultaneous measurement of ABA, IAA and GA’s in citrus- Role of BA in relation to sink ability. J Agric Res Quart. 1995;29:179–185. [Google Scholar]
- Lee YP, Yu GH, Seo YS, Han SE, Choi YO, Daeil K, Mok IG, Kim WT, Sung SK. Microarray analysis of apple gene expression engaged in early fruit development. Plant Cell Rep. 2007;26:917–926. doi: 10.1007/s00299-007-0308-9. [DOI] [PubMed] [Google Scholar]
- Lim PO, Nam HG. The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol. 2005;67:49–83. doi: 10.1016/S0070-2153(05)67002-0. [DOI] [PubMed] [Google Scholar]
- Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34–43. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moctezuma E. The peanut gynophore: a developmental and physiological perspective. Can J Bot. 2003;81:183–190. doi: 10.1139/b03-024. [DOI] [Google Scholar]
- Moctezuma E, Feldman LJ. IAA redistributes to the upper side of gravistimulated peanut (Arachis hypogaea) gynophores. Plant Physiol. 1996;111:S73. [Google Scholar]
- Moctezuma E, Feldman LJ. The role of amyloplasts during gravity perception in gynophores of the peanut plant. Ann Bot. 1999;84:709–714. doi: 10.1006/anbo.1999.0963. [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D. Auxin control of embryo patterning. CSH Perspect Biol. 2009 doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NDong C, Danyluk J, Wilson KE, Pocock T, Huner NP, Sarhan F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analysis. Plant Physiol. 2002;129:1368–1381. doi: 10.1104/pp.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam SN, Dwivedi SL, Ramraj VM, Chandra S. Combining ability of response to photoperiod in peanut. Crop Sci. 1997;37:1159–1162. doi: 10.2135/cropsci1997.0011183X003700040022x. [DOI] [Google Scholar]
- Ohto M, Floyd SK, Fischer RL, Goldberg RB, Harada JJ. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 2009;22:277–289. doi: 10.1007/s00497-009-0116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC (2007) Venny. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html Verified 24 October 2012
- Ozga JA, Reinecke DM. Hormonal interactions in fruit development. J Plant Growth Regul. 2003;22:73–81. doi: 10.1007/s00344-003-0024-9. [DOI] [Google Scholar]
- Payton P, Kottapalli KR, Rowland D, Faircloth W, Guo BZ, Burow M, Puppala N, Gallo M. Gene expression profiling in peanut using high density oligonucleotide microarrays. BMC Genom. 2009;10:265. doi: 10.1186/1471-2164-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol. 1999;40:267–278. doi: 10.1023/A:1006199932265. [DOI] [PubMed] [Google Scholar]
- Ross ARS, Ambrose SJ, Cutler AJ, Feurtado JA, Kermode AR, Nelson K, Zhou R, Abrams SR. Determination of endogenous and supplied deuterated abscisic acid in plant tissues by high-performance liquid chromatography electrospray ionization tandem mass spectrometry with multiple reaction monitoring. Anal Biochem. 2004;329:324–333. doi: 10.1016/j.ab.2004.02.026. [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010;61:1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signaling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlamovitz N, Ziv M, Zamski E. Light, dark and growth regulator involvement in groundnut (Arachis hypogaea L.) pod development. Plant Growth Regul. 1995;16:37–42. doi: 10.1007/BF00040505. [DOI] [Google Scholar]
- Sin SF, Yeung EC, Chye ML. Down regulation of Solanum americanum genes encoding proteinase inhibitor II causes defective seed development. Plant J. 2006;45:58–70. doi: 10.1111/j.1365-313X.2005.02597.x. [DOI] [PubMed] [Google Scholar]
- Stalker HT, Wynne JC. Photoperiodic response of peanut species. Peanut Sci. 1983;10:59–62. doi: 10.3146/i0095-3679-10-2-4. [DOI] [Google Scholar]
- Stears RL, Martinsky T, Schena M. Trends in microarray analysis. Nat med. 2003;9:140–146. doi: 10.1038/nm0103-140. [DOI] [PubMed] [Google Scholar]
- Thompson LK, Ziv M, Deitzer GF. Photocontrol of peanut (Arachis hypogaea L.) embryo and ovule development in vitro. Plant Physiol. 1985;78:370–373. doi: 10.1104/pp.78.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LK, Burgess CL, Skinner EN. Localization of phytochrome during peanut (Arachis hypogaea) gynophore and ovule development. Am J Bot. 1992;79:828–832. doi: 10.2307/2444950. [DOI] [Google Scholar]
- Tirumalaraju SV, Jain M, Gallo M. Differential gene expression in roots of nematode-resistant and -susceptible peanut (Arachis hypogaea L.) cultivars in response to early stages of peanut root-knot nematode (Meloidogyne arenaria) parasitization. Plant Physiol. 2011;168:481–492. doi: 10.1016/j.jplph.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen XP, Li HF, et al. Transcriptome identification of the resistance-associated genes (RAGs) to Aspergillus flavus infection in pre-harvested peanut (Arachis hypogaea) Funct Plant Biol. 2012 doi: 10.1071/FP12143. [DOI] [PubMed] [Google Scholar]
- Willenbrock H, Salomon J, Sokilde R, Braken KB, Hansen TN, Nielsen FC, Moller S, Litman T. Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. RNA. 2009;15:2028–2034. doi: 10.1261/rna.1699809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Pudake RN, Guo G, Xing GF, Hu ZR, Zhang Y, Sun Q, Ni Z. Genome-wide identification and expression profiling of auxin response factor gene family in maize. BMC Genom. 2011;12:178. doi: 10.1186/1471-2164-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T-HD, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y, Hasezawa S, Kumagai F, et al. Cell cycle dependent dynamic change of 26S proteasome distribution in tobacco BY-2 cells. Plant Cell Physiol. 2002;43:604–613. doi: 10.1093/pcp/pcf072. [DOI] [PubMed] [Google Scholar]
- Yang JC, Peng S, Zhu QS, Gu SL. Grain filling pattern and cytokinin content in the grains and roots of rice plants. Plant Growth Regul. 2000;30:261–270. doi: 10.1023/A:1006356125418. [DOI] [Google Scholar]
- Zamski E, Ziv M. Pod formation and its geotropic orientation in the peanut, Aruchis hypogaea L., in relation to light and mechanical stimulus. Ann Bot. 1976;40:63l–636. [Google Scholar]
- Zhang J, Liang S, Duan J, Wang J, Chen S, Cheng Z, Zhang Q, Liang X, Li Y. De novo assembly and characterization of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.) BMC Genom. 2012;13:90. doi: 10.1186/1471-2164-13-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhang E, Li H, Chen X, Zhu F, Hong Y, Liao B, Liu S, Liang X. Comparative proteomics analysis of developing peanut aerial and subterranean pods identifies pod swelling related proteins. J Proteomics. 2013;91:172–187. doi: 10.1016/j.jprot.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Ziv M, Kahana O. The role of the peanut (Araschis hypogaea) ovular tissue in the photo-morphogenetic response of the embryo. Plant Sci. 1988;57:159–164. doi: 10.1016/0168-9452(88)90082-9. [DOI] [Google Scholar]
- Ziv M, Zamskj E. Geotropic responses and pod development in gynophore explants of peanut (Arachis hypogaea L.) cultured in vitro. Ann Bot. 1975;39:579–583. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 9 (FASTA_ONELINESEQ 26120 kb)
Supplementary material 10 (FASTA 5785 kb)


