Abstract
Tamoxifen (Tam) is a selective estrogen receptor modulator used to inhibit breast tumor growth. Tam can be directly N-glucuronidated via the tertiary amine group or O-glucuronidated after cytochrome P450–mediated hydroxylation. In this study, the glucuronidation of Tam and its hydroxylated and/or chlorinated derivatives [4-hydroxytamoxifen (4OHTam), toremifene (Tor), and 4-hydroxytoremifene (4OHTor)] was examined using recombinant human UDP-glucuronosyltransferases (UGTs) from the 1A subfamily and human hepatic microsomes. Recombinant UGT1A4 catalyzed the formation of N-glucuronides of Tam and its derivatives and was the most active UGT enzyme toward these compounds. Therefore, it was hypothesized that single nucleotide polymorphisms (SNPs) in the promoter region of UGT1A4 have the ability to significantly decrease the glucuronidation rates of Tam metabolites in the human liver. In vitro activity of 64 genotyped human liver microsomes was used to determine the association between the UGT1A4 promoter and coding region SNPs and the glucuronidation rates of Tam, 4OHTam, Tor, and 4OHTor. Significant decreases in enzymatic activity were observed in microsomes for individuals heterozygous for −163G/A and −217T/G. These alterations in glucuronidation may lead to prolonged circulating half-lives and may potentially modify the effectiveness of these drugs in the treatment of breast cancer.
Introduction
Tamoxifen [Tam; (Z)-2-[4-(1,2-diphenylbut-1-enyl)phenoxy]-N,N-dimethylethanamine] is prescribed within the United States as a component of adjuvant therapy to prevent estrogen receptor (ER)–positive breast cancer recurrence, as well as to treat metastatic breast cancer and to prevent disease in high-risk populations and in women with ductal carcinoma in situ (Sakorafas et al., 2008). Phase I metabolism of Tam is complex; it occurs via hepatic cytochrome P450 (P450) and involves many enzymes of this superfamily. The major metabolite of P450 metabolism of Tam is N-desmethyltamoxifen (N-desTam), which is produced by several P450s. This compound can then be converted to endoxifen, and CYP2D6 is the major enzyme involved in this process. In addition, Tam can be metabolized into its active metabolite 4-hydroxytamoxifen (4OHTam) by several P450s, including CYP2D6 (Hoskins et al., 2009). It has also been demonstrated that human flavin-containing monooxygenases FMO1 and FMO3 catalyze Tam N-oxidation to form Tam-N-oxide and that many P450s, but not FMOs, reduce Tam-N-oxide back to Tam, with CYP2A6, CYP1A1, and CYP3A4 producing the greatest reduction (Parte and Kupfer, 2005).
Glucuronidation, a phase II metabolism reaction, is one of the major means of biotransformation and elimination of Tam and its major active metabolites (Ogura et al., 2006; Sun et al., 2006, 2007; Mazzarino et al., 2013). Glucuronidation, catalyzed by UDP-glucuronosyltransferases (UGTs), is an important biochemical process that leads to the conjugation of a number of hydrophobic endogenous and exogenous substrates with UDP-glucuronic acid (UDP-GlcUA; the cosubstrate in the UGT-catalyzed reactions) to generally inactive, more hydrophilic compounds (Dutton, 1980; Tukey and Strassburg, 2000). UGT1A4 exhibits the highest enzymatic activity toward Tam catalyzing the formation of the N-linked glucuronide; however, it was previously demonstrated that UGT1A3 and UGT2B10 can also form Tam-N-glucuronides (Sun et al., 2007; Kato et al., 2013). Therefore, interindividual variability in UGT1A4 genes and expression levels may impact the clinical efficacy of this valuable drug.
The single nucleotide polymorphism (SNP) is most common type of genetic polymorphism, with the total number of variant positions among humans estimated to be more than 10,000,000 (Nussbaum et al., 2007). SNPs identified in UGT genes are associated with subpopulations of individuals that are more susceptible to various diseases and may play an important role in individual pharmacological responses to various xenobiotics and drugs (Tukey and Strassburg, 2001; Burchell, 2003; Crettol et al., 2010; Ghotbi et al., 2010; Gulcebi et al., 2011; López et al., 2011; Zhou et al., 2011). Specifically, studies of recombinant UGT1A4 variants showed that these functional polymorphisms can significantly alter the glucuronidation rates of Tam and its metabolites (Sun et al., 2006; Benoit-Biancamano et al., 2009).
This study focuses on SNPs found in the promoter region of UGT1A4, and the 3′ untranslated region (UTR) that is common to all UGT1A enzymes. Since these SNPs are not in the coding region, they are not thought to affect the structure or activity of the expressed UGT1A4 protein. However, these promoter SNPs are shown to affect UGT transcription rates leading to altered mRNA levels (Erichsen et al., 2008; Edavana et al., 2013). We previously showed that these altered mRNA levels are correlated with glucuronidation activity, implying that UGT1A4 protein expression levels are also affected (Edavana et al., 2013). We also previously showed that none of these UGT1A4 promoter SNPs are in linkage disequilibrium either with each other or with coding region SNPs (Edavana et al., 2013). The rs8330 UGT1A SNP studied here is located within the 3′UTR of UGT1A isoforms and is shared among all UGT1A isoforms. SNPs in the 3′UTR may impact mRNA stability and translation efficiency leading to impaired UGT1A expression and activity (Court et al., 2013).
In this work, we carried out extensive studies on the glucuronidation of Tam, its chlorinated derivative toremifene [Tor; 2-{4-[(1Z)-4-chloro-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine], and their hydroxylated metabolites 4OHTam and 4-hydroxytoremifene (4OHTor), using recombinant human UGTs from the 1A subfamily and a set of human liver microsomes from patients that were previously genotyped to identify SNPs located in the UGT1A4 promoter (Edavana et al., 2013). These studies were driven by the hypothesis that these SNPs in the promoter region of UGT1A4 have the ability to significantly decrease the glucuronidation rates of Tam and its derivatives in the human liver. Because of the importance of these drugs in cancer treatment and prevention, the information presented here could serve as a tool for better understanding variations in Tam metabolism and could lead to improved breast cancer treatment.
Materials and Methods
All chemicals used for this study were of at least reagent grade. Tam and 4OHTam were purchased from Sigma-Aldrich (St. Louis, MO). Tor and 4OHTor were procured from Orion Pharma (Helsinki, Finland). Ethyl alcohol (95%) was purchased from AAPER (Shelbyville, KY). Unless otherwise specified, all other chemicals and reagents were of reagent grade and were purchased from Sigma-Aldrich. Recombinant human UGTs were produced in baculovirus-infected insect cells as previously described (Kurkela et al., 2003; Kuuranne et al., 2003).
Human Liver Microsomes.
The human liver microsomes used for this study were previously described (Edavana et al., 2012). In brief, human normal liver specimens (n = 65; from noncancerous subjects, collected and frozen 0–3 hours after surgical excision) were obtained from the Cooperative Human Tissue Network (Birmingham, AL). All specimens assayed in this study were from donors of white or unknown race. Donors ranged in age from 26 to 102 years and comprised 54 men, 36 women, and 6 individuals of unknown sex. Microsomes were prepared from human liver tissue as previously described (Turesky et al., 1991) and were stored frozen at −80°C until assayed. Liver microsomal protein levels were determined using the Bradford (1976) method with bovine serum albumin as a standard.
Human Liver Microsome and Recombinant UGT Screening Incubations.
Human liver microsomes (15 µg) or recombinant UGT membrane protein (5 µg) were assayed for activity toward Tam, 4OHTam, Tor, and 4OHTor. The cloning and expression and characterization of UGT1A1, UGT1A3, UGT1A4, and UGT1A6–UGT1A10 in baculovirus-infected Sf9 insect cells as His-tagged proteins and the preparation of enriched membrane fractions were previously reported (Kurkela et al., 2003; Kuuranne et al., 2003). These preparations were shown to contain similar amounts of protein by Western blot analysis using an anti-His antibody directed at the His tag on each of these recombinant enzymes. Each enzyme tested in this study is known to be active toward substrates specific for that enzyme, and metabolite formation was previously shown to be linear with respect to both protein concentration and incubation time up to 2 mg protein/ml and 180 minutes, respectively (Kaivosaari et al., 2008). UGT2B10 activity was assayed using cells homogenates, not enriched membranes, as was previously reported (Kato et al., 2013).
Protein was incubated in 100 µM Tris-HCl (pH 7.4)/5 mM MgCl2/5 mM saccharolactone with 100 or 200 µM substrate in a total final volume of 30 μl. Substrates were dissolved in H2O/EtOH/dimethylsulfoxide at a ratio of 25:50:25 with a final reaction concentration of 2% dimethylsulfoxide and 1% ethanol, and controls omitting substrate/cosubstrate were run with each assay. No additional detergents or other activators were used with recombinant proteins; however, human liver microsomes were activated with the addition of alamethicin (2.5 µg). Reactions were started by the addition of UDP-GlcUA (2 mM) and incubated for 180 minutes at 37°C. The reactions were stopped by addition of 30 μl ethanol, followed by centrifugation at 14,000 rpm for 8 minutes to pellet the protein.
High-pressure liquid chromatography (HPLC) analyses of the supernatants were performed using an HP1050 HPLC system equipped with the Agilent ChemStation software package (Agilent Technologies, Santa Clara, CA). Samples were separated using an Agilent Eclipse XDB-C18 (4.6 × 150 mm, 5 μm) with an Eclipse XDB-CN Analytical Guard Column (4.6 × 12.5 mm, 5 μm) at 37°C. The solvent system consisted of 10 mM ammonium acetate (A), and acetonitrile (B). For assays with Tam and Tor, products were separated at a flow rate of 1.25 ml/min, with the following elution gradient: 75% A (5 minutes), linear gradient from 75% A to 75% B (5–15 minutes), 75% B (15–23 minutes), and 75% A (23.01–24 minutes). The column was re-equilibrated at initial conditions for 10 minutes between runs. For assays using the hydroxylated derivatives as substrates, the reaction products were separated using a flow rate of 1.00 ml/min, with the following elution gradient: 75% A (5 minutes), linear gradient from 75% A to 75% B (5–20 minutes), and 75% B (20–23 minutes). The elution of each metabolite was monitored with a UV-Vis Diode Array Detector (Agilent Technologies, Santa Clara, CA) at 280 nm. Primary standards for the glucuronidated metabolites are not available; however, it was previously shown that the addition of the glucuronic acid moiety does not alter the extinction coefficient from that of the unreacted substrate (Doerge et al., 2000). Therefore, product concentrations were calculated using the external standard response for each substrate under identical conditions. Concentration curves were established for each of the substrates based on peak areas measured from increasing concentrations of substrate dissolved in reaction mix, incubated at identical conditions as the reactions themselves, and separated using the appropriate HPLC methods. Each concentration was assessed in triplicate and the data were fit to a linear equation.
Steady-State Enzyme Kinetics Assays.
Kinetic parameters were determined by incubating recombinant UGT1A4 (5 µg total membrane protein) in the presence of varying concentrations of substrate (5–500 µM) at a fixed concentration of UDP-GlcUA (3 mM) for 180 minutes. All other conditions were identical to those of the screening experiments. Curve fitting was conducted utilizing GraphPad Prism v4.0b software (GraphPad Software Inc., San Diego, CA). Kinetic constants were obtained by fitting experimental data to the uncompetitive substrate inhibition (USI) model, where Ks is the inhibition constant describing the reduction in rate as follows:
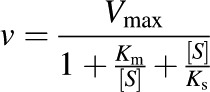 |
The fit of the data for each model was assessed from the standard error, 95% confidence intervals, and R2 values. Kinetic curves were also analyzed as Eadie–Hofstee plots to support kinetic models. Kinetic constants were reported as the mean ± standard error.
Genotyping.
Genotyping for UGT1A4 5′ sequence SNPs (−36G/A, −163G/A, −217T/G, and −219C/T) and 3′UTR SNPs were performed as previously described (Saeki et al., 2005; Benoit-Biancamano et al., 2009) using an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA). The UGT1A-3′UTR SNP (rs8330) was genotyped by sequencing DNA obtained from this same group of human liver samples (n = 36). Briefly, PCR amplification was performed using forward (5′-GAA ACA TGG CCT GTT TG-3′) and reverse (5′-CAA AGC AAG AAA TCA TAT GC-3′) primers using Jumpstart PCR Master mix (Invitrogen, Carlsbad, CA) with the following PCR conditions: 95°C for 5 minutes, followed by 40 cycles at 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute, followed by a final extension at 72°C for 7 minutes. PCR products were purified using the DNA Clean & Concentrator Kit (Zymoresearch, Irvine, CA), followed by dye terminator incorporation (DTCS Quick Start Kit; GenomeLab, Fullerton, CA) and ethanol precipitation. The resultant product was sequenced on a Beckman Coulter Genome Laboratory GeXP Genetic Analysis System (Beckman Coulter, Fullerton, CA).
Statistical Analysis.
Genotyping data were examined for possible genotyping errors by testing deviation from the Hardy–Weinberg equilibrium. Heterozygous and homozygous variant genotypes were combined as one genotype group in analysis for SNPs with < 5 subjects per genotype group. Median enzymatic activity levels and standard deviations were summarized by genotype and activity and were compared using the Wilcoxon rank sum test and the Kruskal–Wallis test for differences between genotypes (Table 1). Statistical analyses were performed with SAS software (version 9.2; SAS Institute Inc., Cary, NC). A P value < 0.05 (two-sided) was considered statistically significant.
TABLE 1.
Correlations between genotypes and enzymatic activity
Data are presented as the median (standard deviation) of specific enzymatic activity. P values were calculated using nonparametric analysis of variance (Kruskal–Wallis test or Wilcoxon rank sum test).
| SNP | Tamox-N-Gluc |
4OHTam-N-Gluc |
4OHTam-O-Gluc |
Tor-N-Gluc |
4OHTor-N-Gluc |
4OHTor-O-Gluc |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | P | Median | P | Median | P | Median | P | Median | P | Median | P | |
| nmol/mg protein per min | ||||||||||||
| −36G/A | ||||||||||||
| AA + AG | 0.11 (0.06) | 0.12 (0.07) | 0.13 (0.06) | 0.17 (0.07) | 0.09 (0.03) | 0.25 (0.16) | ||||||
| GG | 0.14 (0.07) | 0.45 | 0.15 (0.07) | 0.21 | 0.15 (0.11) | 0.11 | 0.18 (0.09) | 0.50 | 0.09 (0.04) | 0.96 | 0.27 (0.15) | 0.85 |
| −163G/A | ||||||||||||
| AA | 0.16 (0.05) | 0.21 (0.06) | 0.12 (0.15) | 0.21 (0.08) | 0.13 (0.05) | 0.14 (0.26) | ||||||
| AG | 0.08 (0.08) | 0.10 (0.07) | 0.12 (0.12) | 0.11 (0.11) | 0.07 (0.03) | 0.25 (0.14) | ||||||
| GG | 0.16 (0.05) | <0.0001a | 0.17 (0.04) | 0.0002a | 0.18 (0.09) | 0.052 | 0.19 (0.06) | 0.007a | 0.10 (0.02) | <0.0001a | 0.29 (0.13) | 0.29 |
| −217T/G | ||||||||||||
| GG + TG | 0.07 (0.05) | 0.09 (0.06) | 0.10 (0.05) | 0.09 (0.07) | 0.06 (0.04) | 0.21 (0.13) | ||||||
| AA | 0.16 (0.06) | <0.0001a | 0.17 (0.05) | <0.0001a | 0.19 (0.11) | <0.0001a | 0.19 (0.08) | <0.0001a | 0.10 (0.03) | 0.001a | 0.29 (0.15) | 0.022a |
| −219C/T | ||||||||||||
| CC | 0.16 (0.06) | 0.16 (0.05) | 0.16 (0.09) | 0.18 (0.06) | 0.10 (0.03) | 0.29 (0.12) | ||||||
| CT | 0.13 (0.08) | 0.15 (0.08) | 0.13 (0.14) | 0.16 (0.11) | 0.08 (0.03) | 0.24 (0.18) | ||||||
| TT | 0.12 (0.07) | 0.13 | 0.14 (0.07) | 0.53 | 0.12 (0.06) | 0.30 | 0.17 (0.10) | 0.56 | 0.08 (0.07) | 0.047a | 0.19 (0.11) | 0.23 |
| 3′UTR | ||||||||||||
| CC | 0.12 (0.06) | 0.12 (0.07) | 0.13 (0.07) | 0.17 (0.09) | 0.09 (0.03) | 0.26 (0.11) | ||||||
| GG + CG | 0.14 (0.05) | 0.79 | 0.15 (0.05 | 0.67 | 0.22 (0.10) | 0.019a | 0.18 (0.07) | 0.47 | 0.08 (0.02) | 0.49 | 0.31 (0.14) | 0.053 |
Significant correlation.
Results
Glucuronidation by Recombinant UGTs.
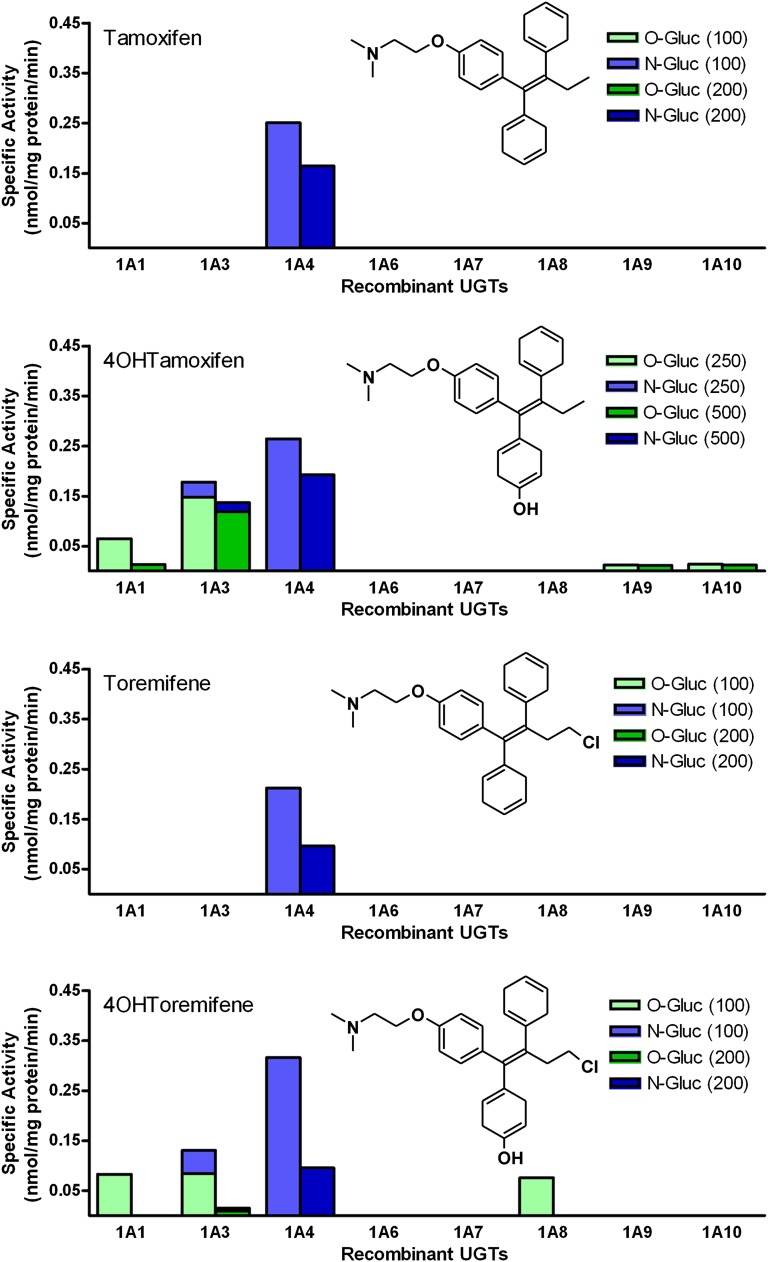
As an initial screen for glucuronidation activity toward Tam, eight His-tagged human recombinant UGT1A subfamily enzymes were evaluated for their ability to glucuronidate 100 and 200 µM Tam, 4OHTam, Tor, and 4OHTor (Fig. 1). The results revealed that UGT1A4 was the most active enzyme and catalyzed direct N-glucuronidation of each compound. Glucuronidation by UGT1A3 was specific for the hydroxylated derivatives, and resulted predominantly in O-glucuronide formation with very little N-glucuronide formed. UGT1A1, UGT1A8, UGT1A9, and UGT1A10 also catalyzed O-glucuronidation in at least one of the hydroxylated derivatives, but UGT1A3 was the most active enzyme toward both compounds. The relative activity of UGT1A3 varied between the hydroxylated compounds, but the UGT1A4-catalyzed glucuronidation rates were very consistent among all of the compounds tested. Of the two concentrations tested, higher glucuronidation rates were observed with the lower substrate concentration, suggesting substrate inhibition at higher concentrations. This information helped us choose suitable concentrations for more detailed steady-state kinetics experiments. UGT2B10 (cell homogenate) was screened for activity but none was seen using our HPLC–diode array detector method. This is reasonable based on the small amount of activity (approximately 15- and 200-fold less N-glucuronide formed than with UGT1A3 and UGT1A4, respectively) measured by Kato et al. (2013), in which product formation was assessed using liquid chromatography coupled to tandem mass spectrometry, which is known to be much more sensitive. We also screened these UGTs for activity toward N-desTam, but no glucuronidation activity was seen (data not shown).
Fig. 1.
Glucuronidation activity screening. Selected recombinant UGT1A subfamily enzymes were screened for activity toward Tam, 4OHTam, Tor, and 4OHTor. Glucuronidation activities were measured by incubating recombinant protein (5 µg) with substrate (100 or 200 µM) and UDP-GlcUA (2 mM).
Steady-State Kinetics for Glucuronidation by Recombinant UGTs.
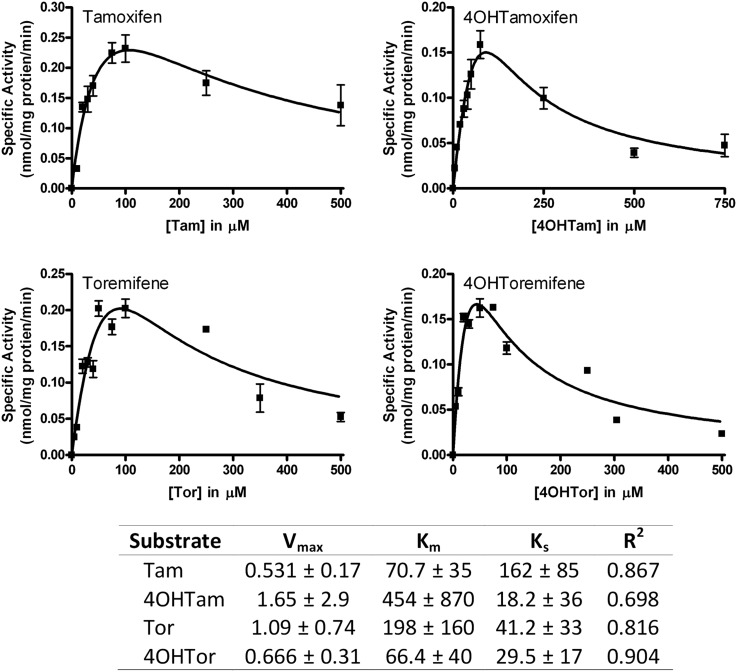
Based on the specific activity screening results, we selected UGT1A4 for further catalytic studies to determine the respective steady-state parameters for glucuronidation of these compounds (Fig. 2). Each of the four experiments resulted in data that best fit the USI model, with Ks values of 162, 18.2, 41.2, and 29.5 μM for Tam, 4OHTam, Tor, and 4OHTor, respectively. The USI model suggests the existence of multiple binding sites within UGT1A4, which fits well with previously published research, which analyzed the steady-state kinetics of UGT1A4 toward dihydrotestosterone and trans-androsterone and compared them with those for Tam to support the hypothesis that UGT1A4 has multiple substrate binding sites (Zhou et al., 2010), and suggests that hydroxylation and/or chlorination of Tam does not affect this phenomenon.
Fig. 2.
Steady-state glucuronidation kinetics of Tam, 4OHTam, Tor, and 4OHTor with UGT1A4. Glucuronidation activities of recombinant proteins were measured by incubating membrane fractions containing recombinant UGT1A4 (5 µg) with increasing concentrations (shown in the figure) of substrate at a constant concentration of UDP-GlcUA (3 mM). Curve fits and kinetic constants were determined using GraphPad Prism 4 software and the resulting parameters are shown.
Effects of UGT1A4 Genotypes on Glucuronidation in Human Liver Microsomes.
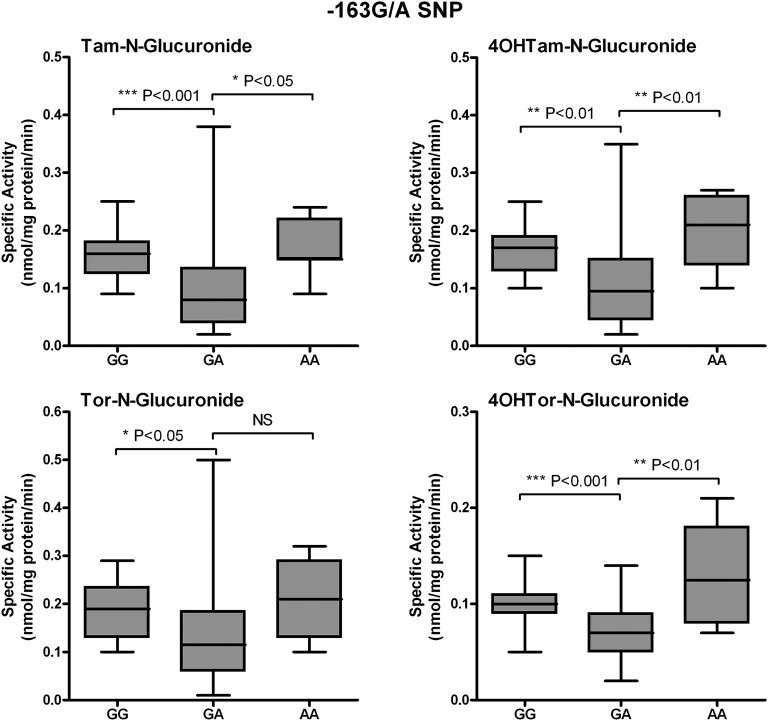
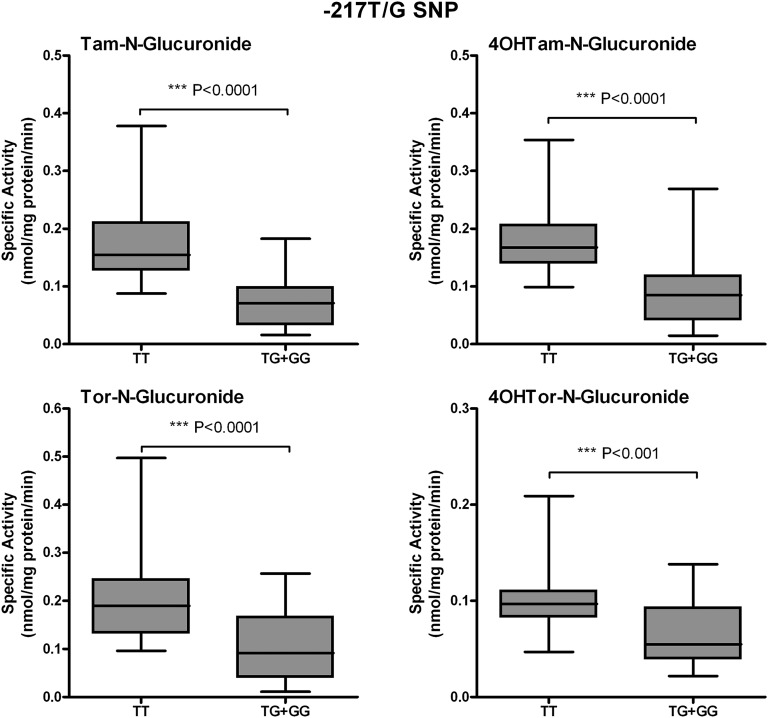
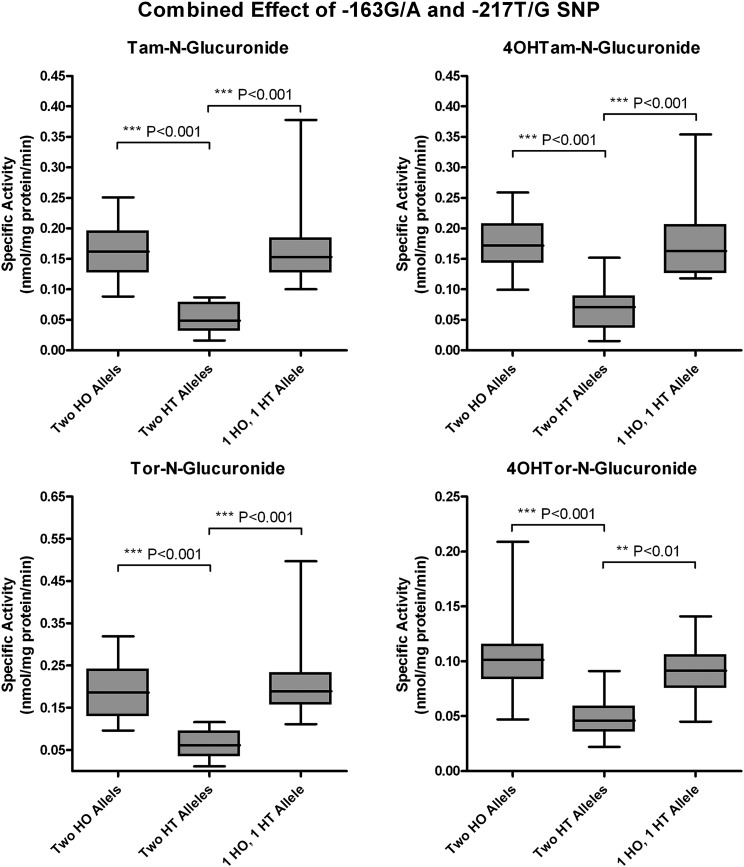
To determine the contribution of the UGT1A-3′UTR SNP (rs8330) and promoter SNPs to Tam, 4OHTam, Tor, and 4OHTor glucuronidation, activity assays were performed using human liver microsomes (64 human subjects), which had undergone dideoxy sequencing of their genomic DNA (Table 1). We previously examined SNPs in the promoter region of UGT1A4, and sequencing analysis revealed the existence of four different SNPs of allele frequencies: −36G/A (0.08), −163G/A (0.32), −217T/G (0.18), and −219C/T (0.32) (Edavana et al., 2013). Here, SNP −163G/A was significantly associated with Tam-N-gluc (P < 0.0001), 4OHTam-N-gluc (P = 0.002), 4OHTam-O-gluc (0.052), Tor-N-gluc (0.007), and 4OHTor-N-gluc (P < 0.0001). SNP −217T/G was also significantly correlated with Tam-N-gluc (P < 0.0001), 4OHTam-N-gluc (P < 0.0001), 4OHTam-O-gluc (P < 0.0001), Tor-N-gluc (P < 0.0001), 4OHTor-N-gluc (P = 0.001), and 4OHTor-O-gluc (P < 0.022). Microsomes from individuals with the homozygous GG or AA alleles at position −163 show significantly higher N-glucuronidation activity toward all four compounds tested than those with the heterozygous GA allele at this position (Fig. 3). In addition, microsomes from samples with the common homozygous TT allele at position −217 exhibited significantly greater glucuronidation activity compared with microsomes from samples with either the TG or GG allele at this position (Fig. 4). Further analysis of the data revealed that glucuronidation activity was significantly decreased only in samples that were heterozygous at both the −163 and −217 positions (Fig. 5). There was no significant difference in activity between samples from individuals who were homozygous at both positions (whether common or uncommon alleles) or from those that were heterozygous at only one of these two alleles.
Fig. 3.
Correlation between −163G/A promoter SNP genotypes and phenotypes. Glucuronidation activity toward Tam, 4OHTam, Tor, and 4OHTor among human liver microsomes was compared among livers of known genotypes at the −163G/A position. Data are displayed as box and whisker plots and were analyzed using nonparametric analysis of variance (Kruskal–Wallis test) to determine overall significance of the effect of this SNP with a Dunn’s multiple comparison post test to determine the significance of the activity difference between each genotype. No significant difference was seen between the activities of samples with either of the two homozygous alleles.
Fig. 4.
Correlation between −217T/G promoter SNP genotypes and phenotypes. Glucuronidation activity toward Tam, 4OHTam, Tor, and 4OHTor among human liver microsomes was compared among livers of known genotypes at the −217T/G position. Because of the limited number of samples with the GG genotype, data were grouped as TT versus TG + GG. Data are displayed as box and whisker plots and were analyzed using a nonparametric t test (Wilcoxon rank sum test) to determine the overall significance of the effect of this SNP.
Fig. 5.
Combined effect of −163G/A and −217T/G promoter SNP genotypes on phenotype. N-Glucuronidation activity of human liver microsomes toward Tam, 4OHTam, Tor, and 4OHTor was compared among livers of known genotypes at both the −163G/A and −217T/G positions. Samples were grouped as follows: homozygous (HO) at both positions (two HO alleles), heterozygous (HT) at both positions (two HT alleles), or one homozygous and one heterozygous allele (one HO allele and one HT allele). Data are displayed as box and whisker plots and were analyzed using nonparametric analysis of variance (Kruskal–Wallis test) and a Dunn’s multiple comparison post test to determine the significance of the activity difference between each genotype group. No significant difference was seen between the activities of the group with two HO alleles and the groups with one HO allele and one HT allele.
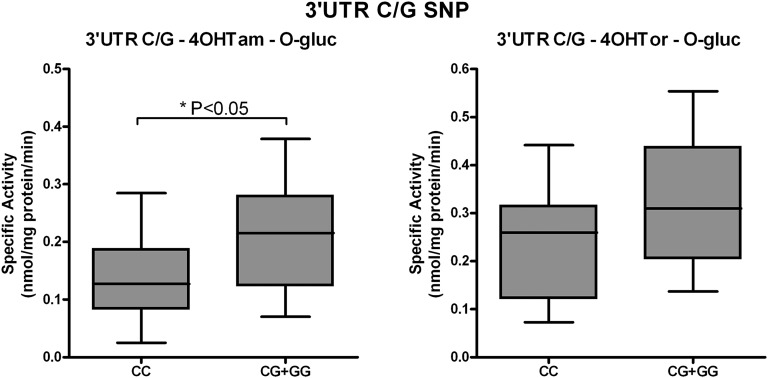
In addition to the UGT1A4 promoter SNPs investigated, the UGT1A-3′UTR SNP (rs8330) genotypes from human liver DNA specimens were correlated with 4OHTam and 4OHTor glucuronidation activities from 36 microsomal samples (Fig. 6). Microsomes from individuals homozygous for the common allele (CC) had significantly lower O-glucuronidation of 4OHTam than individuals with either of the variant alleles (GG or CG) (P < 0.05). A similar increase in 4OHTor-O-glucuronidation was seen in individuals with variant 3′UTR alleles; however, this increase did not quite reach statistically significant levels based on our data set (P = 0.053).
Fig. 6.
Correlation between 3′UTR SNP genotypes and phenotypes. O-Glucuronidation activity toward 4OHTam and 4OHTor by human liver microsomes was compared among livers of known 3′UTR SNP genotypes. Because of the limited number of samples with the GG genotype, data were grouped as CC versus CG + GG. Data are displayed as box and whisker plots and were analyzed using a nonparametric t test (Wilcoxon rank sum test) to determine the overall significance of the effect of this SNP. No significant differences were seen unless otherwise indicated.
Little or no significant effect of SNP −36G/A, −219C/T, or UGT1A-3′UTR was seen on the N-glucuronidation of these compounds (Supplemental Fig. 1), and O-glucuronidation of 4OHTam and 4OHTor was only affected by the SNP at position −217 (Supplemental Fig. 2). Of the UGT1A4 SNPs studied, only −163G/A conformed to the Hardy–Weinberg equilibrium, which is likely due to unknown population stratification. Furthermore, there was no statistical evidence of linkage of the UGT1A4 promoter SNPs with each other or with the 3′UTR SNPs (Table 1).
Discussion
Breast cancer is the most common form of cancer in women. In two-thirds of patients with breast cancer, endocrine therapy is the standard treatment. There are two classes of endocrine therapy: selective ER modulators, which act on the ER, and aromatase inhibitors, which either block the production of estrogens or block the action of estrogens on the ER (Ingle, 2013). Tam (a selective ER modulator) significantly decreases relapse and mortality in steroid hormone receptor–positive breast cancer and is currently prescribed in >120 countries worldwide (Brauch et al., 2009). Tam is also used as a chemotherapeutic agent in healthy women who are at high risk of developing ER-positive breast cancer (Ogura et al., 2006).
Variants of the CYP2D6 gene result in a protein with reduced or absent enzyme activity that may be associated with lower plasma levels of active Tam metabolites (de Leon et al., 2006; Lim et al.. 2007). Given the integral role of this gene in the production of active Tam metabolites, many studies examining the relationship between CYP2D6 genotype and tamoxifen response have been performed, but have generated controversial results (Kiyotani et al., 2013). N-desTam is primarily a product of CYP3A4 metabolism; although it is the major metabolite of Tam, like Tam itself, N-desTam has little affinity for the ER. 4OHTan is produced by multiple P450s, and has 100 times greater affinity for the ER than Tam or N-desTam. Endoxifen is produced primarily by CYP2D6 and has similar affinity for the ER as 4OHTam, and its circulating levels are approximately eight times higher than 4OHTam; thus, it is considered by some to be the major active metabolite of Tam.
Recent studies from the Breast International 1-98 Collaborative Group as well as the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial investigators (Rae et al., 2012; Regan et al., 2012), however, did not support the hypothesis that CYP2D6 genotype is a biomarker of Tam response. Other drug-metabolizing enzymes, including UGTs, play a major role in metabolizing Tam (Kaku et al., 2004; Ogura et al., 2006; Sun et al., 2006, 2007). Thus, genetic variation in these enzymes may merit investigation as additional biomarkers of Tam efficacy and toxicity.
In this work, we investigated the glucuronidation of Tam and three of its derivatives, with a set of well defined recombinant human UGT enzymes from the UGT1A subfamily. We confirmed that UGT1A4 is the major enzyme involved in the glucuronidation of all four of the selected compounds. We recently identified that the −163G/A, −219T/G, and −217C/T SNPs located in the UGT1A4 promoter affect the glucuronidation of another breast cancer drug, anastrozole (Edavana et al., 2013). We observed that microsomes isolated from heterozygous individuals had significantly lower glucuronidation activity toward this drug compared with those individuals who were homozygous for the common allele, and these changes correlated with altered UGT1A4 mRNA expression (Edavana et al., 2013). Therefore, we focused our attention of the role of UGT1A4 promoter and 3′UTR SNPs in the glucuronidation of these compounds.
Here we used this same well characterized set of human liver microsomes to assess the glucuronidation of our selected Tam derivatives. We have examined the association between UGT1A4 promoter and 3′UTR SNPs and the N-glucuroindation of Tam, 4OHTam, Tor, and 4OHTor as well as the O-glucuronidation of 4OHTam and 4OHTor. We found similar decreases in the glucuronidation of Tam and its derivatives in heterozygous individuals. In addition, we observed that microsomes isolated from individuals who were heterozygous at both the −163G/A and −217T/G positions had significantly lower glucuronidation activity toward Tam and its derivatives compared with those who were homozygous at these two positions or who were heterozygous at only the −163 or −217 allele. These data suggest that there is some redundancy in the UGT1A4 promoter region to ensure adequate levels of UGT1A4 activity. However, if both of these positions are altered, it has a significant effect on the activity of this enzyme. This variability in glucuronidation of various xenobiotics in human subjects suggests that the pharmacogenomics of different breast cancer therapy xenobiotics could be greatly affected by genetic UGT1A4 variants that affect the levels of this enzyme expressed in hepatic tissue.
In addition to examining SNPs located in the promoter region of UGT1A4, we determined the association of the UGT1A-3′UTR SNP (rs8330) with glucuronidation activity toward Tam and its derivatives. The importance of SNPs located within the common 3′UTR of UGT1A subfamily enzymes lies in their potential to significantly impact mRNA stability and translation efficiency, thus affecting transcript levels and enzyme activity. A recent study by Court et al. (2013) determined the association of rs8330 genotypes with glucuronidation of acetaminophen, which has both an amide and an alcohol linkage, and demonstrated that the rs8330 SNP was associated with acetaminophen glucuronidation activities in human liver bank samples (n = 48). In that study, individuals who were heterozygous (CG) for the rs8330 had mean acetaminophen glucuronidation activities that were significantly higher than the common rs8330 CC homozygotes, but significantly lower than the variant rs8330 GG homozygotes (CC < CG < GG). Our findings with O-4OHTam glucuronidation are similar. We demonstrate that individuals homozygous for the rs8330 common allele (CC) had significantly lower glucuronidation activity toward our selected compounds than individuals with either the CG or GG allele. Importantly, the 3′UTR SNP was found to significantly affect only O-4OHTam glucuronidation. The fact that this SNP is in the 3′UTR, which is common to all UGT1A subfamily enzymes, and that only O-glucuronidation was altered by changes at this position indicates that this effect was most likely due to alterations in another UGT1A enzyme, such as UGT1A1 or UGT1A3 (Fig. 1), and not by UGT1A4.
Although the effects of functional UGT polymorphisms on the glucuronidation of Tam were the focus of previous research (Sun et al., 2006; Blevins-Primeau et al., 2009; Lazarus et al., 2009; Lazarus and Sun, 2010), to our knowledge, this study is the first to describe the association of UGT1A4 promoter SNPs and glucuronidation of N-Tam, N-4OHTam, O-4OHTam N-Tor, N-4OHTor, and O-4OHTor. Here we demonstrated a significant impact of genetic variability in the UGT1A4 promoter region on xenobiotic glucuronidation, which highlights the importance of UGT1A4 genotype in breast cancer pharmacogenomic studies. The novel data presented in this work indicate that genetic variation in UGT1A4, which affects the expression levels of this enzyme, could potentially contribute to the bioavailability and disposition of Tam and its derivatives in patients, could be a potential biomarker of Tam response, and should be further investigated as a phase II biomarker of Tam efficacy and toxicity.
Supplementary Material
Abbreviations
- 4OHTam
4-hydroxytamoxifen
- 4OHTor
4-hydroxytoremifene
- ER
estrogen receptor
- FMO
flavin-containing monooxygenase
- GlcUA
glucuronic acid
- HPLC
high-pressure liquid chromatography
- N-desTam
N-desmethyltamoxifen
- P450
cytochrome P450
- SNP
single nucleotide polymorphism
- Tam
tamoxifen [(Z)-2-[4-(1,2-diphenylbut-1-enyl)phenoxy]-N,N-dimethylethanamine]
- Tor
toremifene [2-{4-[(1Z)-4-chloro-1,2-diphenylbut-1-en-1-yl]phenoxy}-N,N-dimethylethanamine]
- UGT
UDP-glucuronosyltransferase
- USI
uncompetitive substrate inhibition
- UTR
untranslated region
Authorship Contributions
Participated in research design: Kadlubar, Radominska-Pandya.
Conducted experiments: Greer, Dates, Starlard-Davenport, Edavana.
Contributed new reagents or analytic tools: Finel.
Performed data analysis: Bratton, Dhakal.
Wrote or contributed to the writing of the manuscript: Greer, Dates, Starlard-Davenport, Edavana, Bratton, Dhakal, Finel, Kadlubar, Radominska-Pandya.
Footnotes
This research was supported by the Department of Defense [Grant W81XWH1110795]; the National Institutes of Health National Center for Research Resources and the National Center for Advancing Translational Sciences [UL1TR000039] through the University of Arkansas for Medical Sciences Translational Research Institute Arkansas Breast Cancer Research Project; the National Institutes of Health National Cancer Institute [Grant R01-CA118981]; and the Academy of Finland [Grant 1260010].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Benoit-Biancamano MO, Adam JP, Bernard O, Court MH, Leblanc MH, Caron P, Guillemette C. (2009) A pharmacogenetics study of the human glucuronosyltransferase UGT1A4. Pharmacogenet Genomics 19:945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins-Primeau AS, Sun D, Chen G, Sharma AK, Gallagher CJ, Amin S, Lazarus P. (2009) Functional significance of UDP-glucuronosyltransferase variants in the metabolism of active tamoxifen metabolites. Cancer Res 69:1892–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Brauch H, Mürdter TE, Eichelbaum M, Schwab M. (2009) Pharmacogenomics of tamoxifen therapy. Clin Chem 55:1770–1782 [DOI] [PubMed] [Google Scholar]
- Burchell B. (2003) Genetic variation of human UDP-glucuronosyltransferase: implications in disease and drug glucuronidation. Am J Pharmacogenomics 3:37–52 [DOI] [PubMed] [Google Scholar]
- Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, Duan SX, Greenblatt DJ, Lee WM, Acute Liver Failure Study Group (2013) The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther 345:297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crettol S, Petrovic N, Murray M. (2010) Pharmacogenetics of phase I and phase II drug metabolism. Curr Pharm Des 16:204–219 [DOI] [PubMed] [Google Scholar]
- de Leon J, Susce MT, Murray-Carmichael E. (2006) The AmpliChip CYP450 genotyping test: Integrating a new clinical tool. Mol Diagn Ther 10:135–151 [DOI] [PubMed] [Google Scholar]
- Doerge DR, Chang HC, Churchwell MI, Holder CL. (2000) Analysis of soy isoflavone conjugation in vitro and in human blood using liquid chromatography-mass spectrometry. Drug Metab Dispos 28:298–307 [PubMed] [Google Scholar]
- Dutton GJ. (1980) Glucuronidation of Drugs and Other Compounds, CRC Press Inc., Boca Raton, FL. [Google Scholar]
- Edavana VK, Dhakal IB, Williams S, Penney R, Boysen G, Yao-Borengasser A, Kadlubar S. (2013) Potential role of UGT1A4 promoter SNPs in anastrozole pharmacogenomics. Drug Metab Dispos 41:870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edavana VK, Dhakal IB, Yu X, Williams S, Kadlubar S. (2012) Sulfation of 4-hydroxy toremifene: individual variability, isoform specificity, and contribution to toremifene pharmacogenomics. Drug Metab Dispos 40:1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erichsen TJ, Ehmer U, Kalthoff S, Lankisch TO, Müller TM, Munzel PA, Manns MP, Strassburg CP. (2008) Genetic variability of aryl hydrocarbon receptor (AhR)-mediated regulation of the human UDP glucuronosyltransferase (UGT) 1A4 gene. Toxicol Appl Pharmacol 230:252–260 [DOI] [PubMed] [Google Scholar]
- Ghotbi R, Mannheimer B, Aklillu E, Suda A, Bertilsson L, Eliasson E, Osby U. (2010) Carriers of the UGT1A4 142T>G gene variant are predisposed to reduced olanzapine exposure—an impact similar to male gender or smoking in schizophrenic patients. Eur J Clin Pharmacol 66:465–474 [DOI] [PubMed] [Google Scholar]
- Gulcebi MI, Ozkaynakcı A, Goren MZ, Aker RG, Ozkara C, Onat FY. (2011) The relationship between UGT1A4 polymorphism and serum concentration of lamotrigine in patients with epilepsy. Epilepsy Res 95:1–8 [DOI] [PubMed] [Google Scholar]
- Hoskins JM, Carey LA, McLeod HL. (2009) CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat Rev Cancer 9:576–586 [DOI] [PubMed] [Google Scholar]
- Ingle JN. (2013) Pharmacogenomics of endocrine therapy in breast cancer. J Hum Genet 58:306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaivosaari S, Toivonen P, Aitio O, Sipilä J, Koskinen M, Salonen JS, Finel M. (2008) Regio- and stereospecific N-glucuronidation of medetomidine: the differences between UDP glucuronosyltransferase (UGT) 1A4 and UGT2B10 account for the complex kinetics of human liver microsomes. Drug Metab Dispos 36:1529–1537 [DOI] [PubMed] [Google Scholar]
- Kaku T, Ogura K, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. (2004) Quaternary ammonium-linked glucuronidation of tamoxifen by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol 67:2093–2102 [DOI] [PubMed] [Google Scholar]
- Kato Y, Izukawa T, Oda S, Fukami T, Finel M, Yokoi T, Nakajima M. (2013) Human UDP-glucuronosyltransferase (UGT) 2B10 in drug N-glucuronidation: substrate screening and comparison with UGT1A3 and UGT1A4. Drug Metab Dispos 41:1389–1397 [DOI] [PubMed] [Google Scholar]
- Kiyotani K, Mushiroda T, Zembutsu H, Nakamura Y. (2013) Important and critical scientific aspects in pharmacogenomics analysis: lessons from controversial results of tamoxifen and CYP2D6 studies. J Hum Genet 58:327–333 [DOI] [PubMed] [Google Scholar]
- Kurkela M, García-Horsman JA, Luukkanen L, Mörsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. (2003) Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem 278:3536–3544 [DOI] [PubMed] [Google Scholar]
- Kuuranne T, Kurkela M, Thevis M, Schänzer W, Finel M, Kostiainen R. (2003) Glucuronidation of anabolic androgenic steroids by recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos 31:1117–1124 [DOI] [PubMed] [Google Scholar]
- Lazarus P, Blevins-Primeau AS, Zheng Y, Sun D. (2009) Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen. Ann N Y Acad Sci 1155:99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus P, Sun D. (2010) Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev 42:182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HS, Ju Lee H, Seok Lee K, Sook Lee E, Jang IJ, Ro J. (2007) Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol 25:3837–3845 [DOI] [PubMed] [Google Scholar]
- López M, Dorado P, Monroy N, Alonso ME, Jung-Cook H, Machín E, Peñas-Lledó E, Llerena A. (2011) Pharmacogenetics of the antiepileptic drugs phenytoin and lamotrigine. Drug Metabol Drug Interact 26:5–12 [DOI] [PubMed] [Google Scholar]
- Mazzarino M, Biava M, de la Torre X, Fiacco I, Botrè F. (2013) Characterization of the biotransformation pathways of clomiphene, tamoxifen and toremifene as assessed by LC-MS/(MS) following in vitro and excretion studies. Anal Bioanal Chem 405:5467–5487 [DOI] [PubMed] [Google Scholar]
- Nussbaum R, McInnes RR, Willard HF. (2007) The human genome and the chromosomal basis of heredity, in Thompson and Thompson Genetics in Medicine (Nussbaum R, McInnes RR, Willard HF. eds) pp 5–25, Elsevier Saunders, Philadelphia [Google Scholar]
- Ogura K, Ishikawa Y, Kaku T, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. (2006) Quaternary ammonium-linked glucuronidation of trans-4-hydroxytamoxifen, an active metabolite of tamoxifen, by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol 71:1358–1369 [DOI] [PubMed] [Google Scholar]
- Parte P, Kupfer D. (2005) Oxidation of tamoxifen by human flavin-containing monooxygenase (FMO) 1 and FMO3 to tamoxifen-N-oxide and its novel reduction back to tamoxifen by human cytochromes P450 and hemoglobin. Drug Metab Dispos 33:1446–1452 [DOI] [PubMed] [Google Scholar]
- Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, Salter J, Sestak I, Cuzick J, Dowsett M, ATAC trialists (2012) CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 104:452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, Kammler R, Dell’orto P, Biasi MO, Thürlimann B, Lyng MB, et al. Breast International Group (BIG) 1-98 Collaborative Group (2012) CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 trial. J Natl Cancer Inst 104:441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki M, Saito Y, Jinno H, Sai K, Hachisuka A, Kaniwa N, Ozawa S, Kawamoto M, Kamatani N, Shirao K, et al. (2005) Genetic variations and haplotypes of UGT1A4 in a Japanese population. Drug Metab Pharmacokinet 20:144–151 [DOI] [PubMed] [Google Scholar]
- Sakorafas GH, Farley DR, Peros G. (2008) Recent advances and current controversies in the management of DCIS of the breast. Cancer Treat Rev 34:483–497 [DOI] [PubMed] [Google Scholar]
- Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, Lazarus P. (2006) Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res 8:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Sharma AK, Dellinger RW, Blevins-Primeau AS, Balliet RM, Chen G, Boyiri T, Amin S, Lazarus P. (2007) Glucuronidation of active tamoxifen metabolites by the human UDP glucuronosyltransferases. Drug Metab Dispos 35:2006–2014 [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. (2000) Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol 40:581–616 [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. (2001) Genetic multiplicity of the human UDP-glucuronosyltransferases and regulation in the gastrointestinal tract. Mol Pharmacol 59:405–414 [DOI] [PubMed] [Google Scholar]
- Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. (1991) Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis 12:1839–1845 [DOI] [PubMed] [Google Scholar]
- Zhou J, Argikar UA, Remmel RP. (2011) Functional analysis of UGT1A4(P24T) and UGT1A4(L48V) variant enzymes. Pharmacogenomics 12:1671–1679 [DOI] [PubMed] [Google Scholar]
- Zhou J, Tracy TS, Remmel RP. (2010) Glucuronidation of dihydrotestosterone and trans-androsterone by recombinant UDP-glucuronosyltransferase (UGT) 1A4: evidence for multiple UGT1A4 aglycone binding sites. Drug Metab Dispos 38:431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.