Abstract
Glucocorticoids regulate many crucial biologic functions through their cytoplasmic/nuclear glucocorticoid receptors (GR). Excess, deficiency, or alteration in tissue sensitivity to glucocorticoids has been associated with major causes of human morbidity and mortality. Brx, a cytoplasmic Rho family guanine nucleotide exchange factor, binds to and influences the activity of several nuclear hormone receptors. We examined the functional and molecular interactions between GR and Brx. The glucocorticoid sensitivity of lymphocytes obtained from mice haplo-insufficient for Brx was significantly decreased. Conversely, GR-mediated transcriptional activity of a glucocorticoid response element (GRE)-mediated glucocorticoid-responsive promoter was enhanced by Brx in a guanine nucleotide exchange factor domain-dependent fashion. Brx interacted with GR, forming a ternary complex with RhoA. In a chromatin immunoprecipitation assay, Brx and RhoA were co-precipitated with GREs only in the presence of ligand-activated GR. Extracellularly administered lyso-phosphatidic acid, which activates its signaling cascade through a specific membrane GTP-binding protein (G-protein)-coupled receptor in a G-protein α13-, Brx-, and RhoA-dependent fashion, enhanced GR transcriptional activity, whereas depletion of endogenous Brx attenuated this effect. These findings suggest that glucocorticoid signaling and, hence, the tissue sensitivity to glucocorticoids, may be coupled to extracellular signals via Brx and small G-proteins. Nuclear Brx might act as a local GRE-GR-transcripto-some activator by mediating the effect of small G-proteins on glucocorticoid-regulated genes.
Glucocorticoids exert profound influences on many physiologic functions by virtue of their extremely diverse roles in growth, development, and maintenance of cardiovascular, metabolic, and immune homeostasis (1, 2). At pharmacologic doses, glucocorticoids also act as potent immunosuppressive and anti-inflammatory agents that make them irreplaceable therapeutic means for many inflammatory, autoimmune, and lymphoproliferative diseases (3). The actions of glucocorticoids are mediated by an intracellular receptor protein, the glucocorticoid receptor (GR),2 which belongs to the steroid/sterol/thyroid/ retinoid/orphan receptor superfamily of nuclear transcription factors (4–6). GR is ubiquitously expressed in almost all human tissues and organs (5). In its unliganded state, GR is located primarily in the cytoplasm, as part of hetero-oligomeric complexes containing heat shock proteins 90, 70, and 50, and possibly other proteins (7). After ligand binding GR undergoes conformational changes, dissociates from heat shock proteins, homodimerizes, and translocates into the nucleus. There, the ligand-activated GR directly interacts with DNA sequences, the glucocorticoid response elements (GREs), in the promoter regions of target genes and regulates their transcriptional activity (7).
Accumulating evidence indicates that many extracellular molecules including hormones, growth factors, and cytokines, influence biologic activity of glucocorticoids at several GR activation steps. Resultant changes in GR transcriptional activity play a role in the physiologic regulation of glucocorticoid actions and the development of pathologic conditions, such as glucocorticoid-resistant asthma and “dysmetabolic syndrome,” which is associated with visceral-type obesity, hyperlipidemia, and insulin resistance/overt diabetes mellitus (5, 7–9). Such extracellular factors convey biologic information to cells by binding to specific cell surface receptors and by activating downstream intermediate signaling effector molecules (10–12). The small guanine nucleotide-binding proteins (G-proteins) are examples of intracellular signal mediators that influence diverse biologic processes, such as cell growth, differentiation, apoptosis, and subcellular compartment shuttling of intracellular molecules (13).
The small G-proteins are classified into five subgroups, the Ras, Ran, Rab, Sar/Arf, and Rho families (13). The Rho family proteins, which include RhoA, Cdc42, and Rac1, play an important role in the reorganization of cytoskeleton, embryonic development, and regulation of gene expression after their activation by numerous extracellular stimuli (13–15). For example, lysophosphatidic acid (1-acyl-glycerol-3-phosphate (LPA)), a lipid compound produced locally from activated platelets, binds to specific cell membrane receptors, activates classic G-proteins Gα12/13, subsequently stimulates RhoA, and induces morphologic changes of responsive cells via modulation of stress fiber formation and cell-cell interactions (14, 16–19). LPA has growth factor-like actions, such as stimulation of cellular proliferation, migration, and survival (20). It increases endothelial permeability and inhibits gap junction-mediated communication between adjacent cells as well as promotes wound healing and suppresses intestinal damage after irradiation (20). LPA also functions as an inflammatory mediator exerting many biologic activities on the immune system (21). Pathologically, LPA produced from locally activated platelets may play a role in the development of atherosclerotic regions in the vascular walls (22).
The small G-proteins exist in active GTP-bound and inactive GDP-bound forms (13, 14), turning on and off the activity of its target molecules via physical interactions (13). The guanine nucleotide exchange factors (GEFs), which contain specific protein motifs characterized by the Dbl homology (DH) domain followed by the pleckstrin homology domain, catalyze the conversion of the GDP-bound to the GTP-bound form, indicating that this class of proteins plays an essential role in the activation of small G-proteins (23, 24). After regulating the activity of their target molecules, GTP-bound small G-proteins are immediately returned to the GDP-associated form via their intrinsic GTPase activity (13).
Several Rho family small G-protein-specific GEFs have been recently cloned (23, 24). We first cloned a 1429-residue GEF protein called Brx by using the ligand binding domain of the retinoic X receptor β as bait in expression cloning (25); interestingly, Brx not only acts as a Rho family GEF but also binds to nuclear hormone receptors via its C-terminal portion (nuclear receptor-interacting domain (NRID)) and enhances transcriptional activity of the estrogen receptors α (25) and β (26). AKAP-Brx (Lbc), a larger splicing variant of Brx with an additional 1389-amino acid residues, was subsequently reported (27). This transcript possesses a protein kinase A docking domain in its N-terminal portion and a full Brx sequence-containing GEF domain and NRID at its C-terminal half (27). Inclusion of NRID and/or a protein kinase A docking domain in Brx and AKAP-Brx indicates that this class of GEFs acts as an integrator of signal transduction pathways to tune their independent signals toward orchestrated final biologic actions.
In this report we show that Brx modifies the actions of glucocorticoids, enhancing the transcriptional activity of GR by interacting with GR and by attracting Rho family G-proteins to the GR-induced transcriptosome. It appears that Brx, via its two functionally distinct domains GEF and NRID, acts as a bridging factor between several intracellular signaling systems, such as those induced through transmembrane receptors and small G-proteins and those of the GR and other nuclear hormone receptors. These results indicate that Brx acts as a determinant of target tissue sensitivity to glucocorticoids and, thus, may play a role in the development of highly prevalent human disorders with major metabolic and cardiovascular morbidity and mortality (7, 8).
MATERIALS AND METHODS
Plasmids
pBK/RSV-FLAG-Brx, Brx-(1042–1429) and Brx-(527–950), which respectively express FLAG-tagged full-length Brx, Brx-(1042–1429), or Brx-(527–950), were described previously (25). pBK/RSV-FLAG-BrxY769F was constructed with the PCR-assisted mutagenesis reaction by using pBK/RSV-FLAG-Brx as a template. This plasmid expresses a FLAG-Brx mutant, which has a replacement of tyrosine at amino acid 769 with phenylalanine that causes the inactivation of the GEF activity in the wild type Brx (23). pCDNA3-RhoA wild type (WT) and constitutive active form (QL) and pCDNA3-Cdc42 WT and QL, which, respectively, express the WT and QL of RhoA and Cdc42, were kindly gifted by Dr. J. S. Gutkind (National Institutes of Health, Bethesda, MD). pCEV-C3 and pCDNA3-N17Cdc42, which express the botulinum toxin C3 and a dominant negative form of Cdc42, and, thus, inhibit the action of RhoA and Cdc42, respectively, were also provided by Dr. J. S. Gutkind. pCDNA3-Gα13G225A, which expresses a transdominant negative form of Gα13 (27), was created by introducing a point mutation that replaced glycine at amino acid 225 with alanine, with the PCR-assisted mutagenesis reaction using pCDNA3-Gα13 as a template, which was provided by Dr. J. S. Gutkind. pRShGRα, which expresses the full-length human GRα, was a generous gift from Dr. R. M. Evans (Salk Institute, La Jolla, CA). pMMTV-Luc, which has the luciferase gene under the control of the GREs-containing and glucocorticoid-responsive murine mammary tumor virus (MMTV) promoter, was a kind gift from Dr. G. L. Hager (NCI, National Institutes of Health, Bethesda, MD). pBK/RSV, pCDNA3, and pSV40-β-Gal were purchased from Stratagene (La Jolla, CA), Invitrogen, and Promega (Madison, WI), respectively.
Brx-haplo-insufficient Mice and Separation of Splenic Lymphocytes
Briefly, targeted disruption of the brx gene was performed using standard homologous recombination by insertion of a Neor cassette, resulting in elimination of an exon encoding amino acids 752–1044 of Brx transcript (25). These residues are required for GEF activity and are highly conserved throughout Rho GEF family members, as described (see Ref. 28). Mice homozygous for the null brx allele died in utero (29). Mice haplo-insufficient (+/−) for the null brx allele appeared grossly normal. Genotypes of adult, embryos, and neonate mice were confirmed using Southern analysis and PCR Details of the construction and phenotype of the brx null mice will be presented elsewhere.3 Spleens were dissected from mice haplo-insufficient for the brx gene and wild type mice, minced, and passed through nylon cell strainers (BD Biosciences). The obtained extracts were further purified with the gradient separation using the Ficoll-Paque PLUS (Amersham Biosciences) to obtain the splenic lymphocytes.
Thymidine Incorporation Assay in Splenic Lymphocytes
1 × 105 of the splenic lymphocytes were plated in 24-well plates, 5 µg/well of concanavalin A (Sigma) and increasing concentrations of dexamethasone (Sigma) were added, and the cells were cultured for 72 h. 0.25 µCi/well of [3H]thymidine (Amersham Biosciences) was then added into the wells. After 4 h of additional culture, the cells were harvested and treated with 10% trichloroacetic acid (MG Scientific Inc., Pleasant Prairie, WI). Cell pellets were then dissolved in 1 M NaOH, and their radioactivity was determined using a LS 6000IC β-counter (Beckman Coulter, Inc.). Results were demonstrated as percentages of radioactivity obtained in each concentration of dexamethasone to that observed without dexamethasone. The experiments were performed with hexaplicate and repeated three times.
Cell Culture and Transfection
Human cervical carcinoma HeLa and African green monkey kidney CV-1 cells were obtained from American Type Culture Collection (ATCC) (Manassas. VA). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 100 units of penicillin, 1 µg/ml of streptomycin sulfate, 10% of fetal bovine serum, and 25 mM of HEPES. Human colon carcinoma HCT116 cells were also purchased from ATCC and were maintained in McCoy’s 5A medium with the same supplements. Twenty-four hours before transfection, 1 × 105 cells were seeded in 12-well plates. Transfection was performed with FuGENE 6 (Roche Applied Science) or Lipofectin™ (Invitrogen). The cells were co-transfected with 0.5 µg/well of pMMTV-Luc, 0.2 µg/well of pSV40-β-Gal, 0.1–0.5 µg/well of FLAG-Brx-expressing plasmids, and/or 0.1–0.5 µg/well of RhoA-, Cdc42-, Gα13G225A-, C3-, or N17Cdc42-expressing plasmids. Because HeLa cells express endogenous GR, whereas CV-1 and HCT116 cells are devoid of the receptor, 0.1 µg/well of pRShGRα was included in the transfection medium for the latter two cell types (30). Twenty-four hours after the transfection 10−6 m dexamethasone was added to the medium. After an additional 24 h the cells were lysed with a reporter lysis buffer (Promega), and the luciferase and β-galactosidase activities were determined using a Monolight 2010 luminometer (BD PharMingen), as previously described (31, 32). Luciferase activity was normalized for β-galactosidase activity to correct for transfection efficiency.
Western Blot and Co-immunoprecipitation Assay
2 × 106 of HeLa cells were plated in 75-cm2 flasks 24 h before transfection. The cells were transfected with 15 µg of pBK/RSV-FLAG-Brx or -BrxY769F using FuGENE 6. Forty-eight hours after the transfection the cells were treated with 10−6 m dexamethasone or vehicle for 5 h. Cell lysis and co-immunoprecipitation reactions were carried out in the lysis buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 0.2% Nonidet P-40, Complete™ tablets 1 tablet/50 ml, as reported previously (31). Proteins were precipitated with anti-Brx, -FLAG (M2), or -GRα (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies or control serum bound to protein A/G-agarose PLUS (Santa Cruz Biotechnology). Precipitated proteins were run on 6% or 8% SDS-PAGE gels and subsequently blotted on nitrocellulose membranes. Membrane-attached FLAG-Brx, RhoA, or GR were visualized with anti-Brx, -RhoA, and -GRα antibodies (Santa Cruz Biotechnology) and subsequent treatment with peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology). Endogenous or expressed RhoA, GR, and/or FLAG-Brx were determined with Western blots in 10% of cell lysates, which were used in the co-immunoprecipitation reactions. To compare expression levels of wild type FLAG-Brx, BrxY769, and Brx-(1042–1429), HeLa cells were transfected with their expression plasmids or a control plasmid together with pEGFP-C1 (Clontech), expressing enhanced green fluorescence protein. Cells were harvested and lysed in lysis buffer, and whole cell homogenates were run on 8–16% SDS-PAGE gels. After blotting, bands were visualized with anti-FLAG(M2) antibody for Brx-related molecules or anti-GFP antibody (Clontech). Band density in three different experiments was quantified with the Science Lab 2003 Image Gauge Version 4.1 software (Fuji Photo Film USA, Inc., Valhalla, NY), and relative expression levels of Brx-related molecules were shown as a ratio of their band density divided by that of enhanced green fluorescence protein.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed in HCT116/MMTV cells, which have the genomic integrated MMTV-luciferase gene (33), using a chromatin immunoprecipitation kit (Upstate Biotechnology, Charlottesville, VA) according to the manufacturer’s instructions with minor modifications. They were transfected with pRShGRα to express functional GR. In some experiments the cells were also transfected with the Brx siRNA (5′-AAAGCCA-GAGGAAGAGCAUUUdTdT-3′), which targets nucleotides 219–239 of the Brx coding region, that were produced by Qiagen (Valencia, CA), or control lamin A/C siRNA (Qiagen) using the x-tremeGENE siRNA transfection reagent (Roche Applied Science). After 24 h of transfection, the cells were exposed to either 10−6 m dexamethasone or vehicle for 5 h. The cells were then fixed, DNA and bound proteins were cross-linked, and ChIP assays were performed by co-precipitating the DNA-protein complexes with anti-Brx, anti-RhoA, anti-FLAG, anti-GRα antibodies, or rabbit control IgG (Santa Cruz Biotechnology), as previously reported (34). The promoter region −219 to −47 of the MMTV long terminal repeat that contains two functional GREs was amplified from the prepared DNA samples using a primer pair 5′-AACCTTGCG-GTTCCCAG-3′ and 5′-GCATTTACATAAGATTTGG-3′ (fragment size, 173 bp). A distal region −1158 to −935 of the MMTV promoter, which does not contain GREs, was amplified using a primer pair 5′-GAGAGTGTCCTACACCTAGG-3′ and 5′-GTAATCTTGCA-CAGAAGAGC-3′ (fragment size, 224 bp). The promoter region −777 to −516 of the glucocorticoid-unresponsive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter, which does not contain GREs, was also amplified using a primer pair 5′-GATTGTCTGC-CCTAATTATC-3′ and 5′-CAGGCAAAGGCCTAGGAG-3′ (fragment size, 261 bp). Amplified products were then run on a 2–3% agarose gel, and the visualized DNA bands were photographed.
Inhibition of Brx Expression by siRNA and Evaluation of the FKBP51 and Brx/AKAP-Brx mRNA Levels in the Quantitative Realtime PCR
8 × 104 of HCT116 cells were plated on 24-well plates. After 24 h, 1 µg of the siRNA for Brx or lamin A/C and 0.5 µg of pRShGRα were transfected with x-tremeGENE siRNA transfection reagent. Twenty-four hours after transfection, cells were treated with 1–10 µg/ml LPA (Avanti Polar Lipids, Inc., Alabaster, AL) and/or 10−6 m dexamethasone and were cultured for an additional 24 h. The cells were then lysed with TRIzol (Invitrogen), and total RNA was isolated according to the manufacturer’s instructions. The reverse transcription reaction was carried out using the random hex-amers with TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA). To detect mRNA levels of Brx/AKAP-Brx, 51-kDa FK506-binding protein (FKBP51) and control human acidic ribosomal phosphoprotein P0 (RPLP0), primer pairs (Brx/AKAP-Brx: forward primer, 5′-CAGTGATGACATGGACAG-3′, reverse primer, 5′-TCGGTGGATGAACTGGATC-3′; FKBP51: forward primer, 5′-AAAAGGCCAAGGAGCACAAC-3′, reverse primer, 5′-TTGAGGAGGGGCCGAGTTC-3′; RPLP0: forward primer, 5′-CGCGACCTGGAAGTCCAACT-3′, reverse primer, 5′-CCAT-CAGCACCACAGCCTTC-3′) were used (35). The real-time PCR reaction, consisting of the heat activation of the Taq polymerase (10 min at 95 °C) and the subsequent 60 PCR cycles (denaturing, 15 s at 95 °C; annealing/extension, 1 min at 60 °C) was performed in quadruplicate using the SYBR Green PCR Master Mix (Applied Biosystems) in an ABI PRIZM 7700 SDS lightcycler (Applied Biosystems). Obtained CT (threshold cycle) values of FKBP51 and Brx/AKAP-Brx were normalized for those of RPLP0, and their relative mRNA expressions were demonstrated as -fold induction to the base line. The dissociation curves of the primer pairs used showed a single peak, and samples after PCR reactions had a single expected DNA band in an agarose gel analysis (data not shown).
Statistical Analysis
Statistical analysis was carried out by analysis of variance followed by Student’s t test with Bonferroni correction for multiple comparisons or unpaired t test with the two-tailed p value. All experiments were repeated at least three times, and representative results are shown in the figures.
RESULTS
Brx Is Necessary for Glucocorticoid Action in Mouse Splenic Lymphocytes
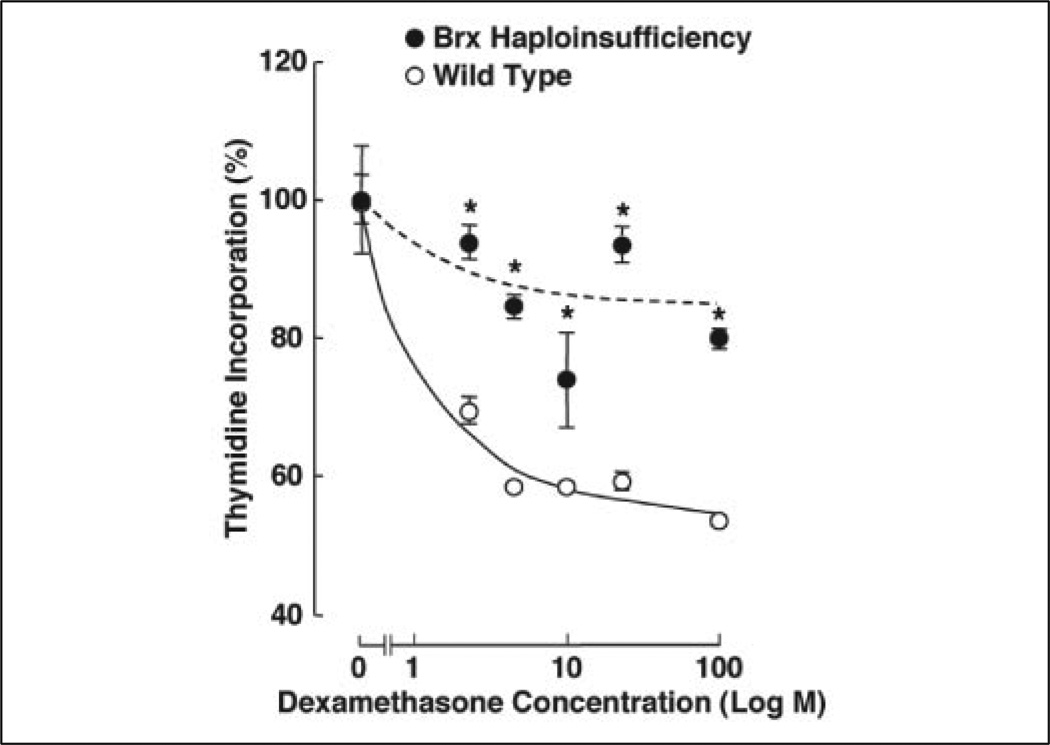
In previous studies we noted that Brx was expressed in immune organs, such as peripheral and splenic lymphocytes (25), suggesting that Brx may play an important role in the regulation of their functions. Because glucocorticoids also exert strong effects on the activity of the immune system, we examined the effect of dexamethasone on thymidine incorporation by splenic lymphocytes obtained from Brx haplo-insufficient mice to examine physiologic relevance of Brx in glucocorticoid action (Fig. 1). Glucocorticoid-induced suppression of thymidine incorporation by lymphocytes is a classic ex vivo indicator of the effectiveness of glucocorticoid hormones, positively correlating with the transactivation activity of the GR (36). Administration of increasing concentrations of dexamethasone effectively suppressed the incorporation of [3H]thymidine into splenic lymphocytes purified from wild type mice in a dose-dependent fashion, whereas this effect was blunted in the cells harvested from Brx haplo-insufficient mice. These results clearly indicate that Brx functions as a physiologic and positive regulator of glucocorticoid action in these cells.
FIGURE 1. Haplo-insufficiency of the brx gene partially abrogates the suppressive effect of dexamethasone on thymidine incorporation in splenic lymphocytes.
Splenic lymphocytes were isolated from the wild type and brx-haplo-insufficient mice and exposed to increasing concentrations of dexamethasone, and uptake of 3H-labeled thymidine was examined. Closed (Brx haplo-insufficient) and open (wild type) circles indicate the mean ± S.E. values of percentages of the incorporated [3H]thymidine obtained in the cells treated with indicated concentrations of dexamethasone to untreated cells. *, p < 0.01, compared with the wild type treated with the same concentrations of dexamethasone.
Brx Enhances GR-induced Transactivation in HeLa and CV-1 Cells
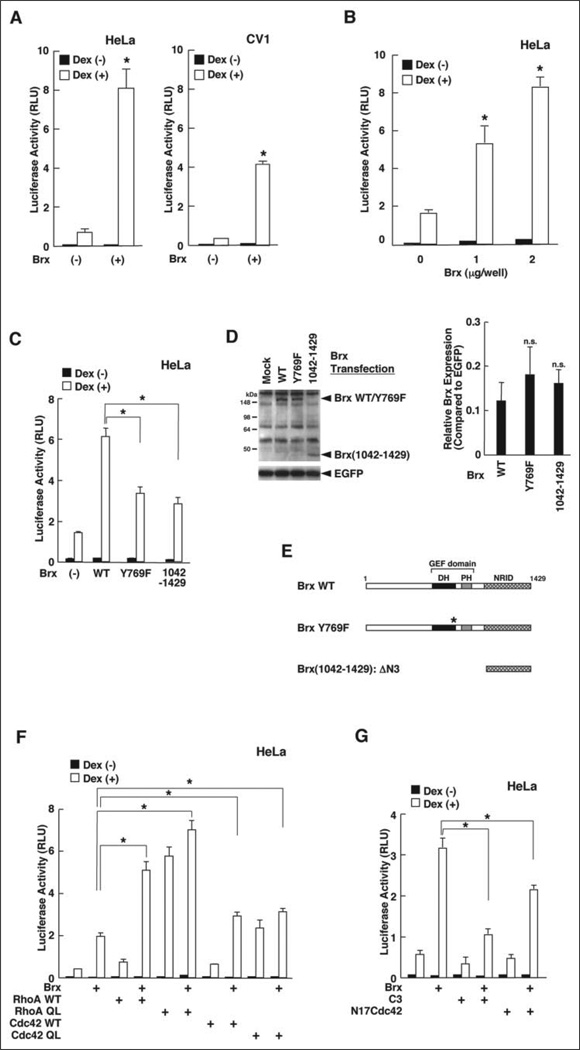
We next examined molecular mechanisms of Brx action on the regulation of glucocorticoid activity. We employed the glucocorticoid-responsive MMTV promoter in transfection-based reporter assays to examine the transcriptional activity of the GR in two different cell lines, HeLa and CV-1 cells. As observed with ERα, overexpression of Brx strongly potentiated GR-induced transcriptional activity of this promoter in a dexamethasone-dependent fashion in these cells (Fig. 2A). Brx enhanced GR-induced transactivation of this promoter in HeLa cells in a dose-dependent fashion (Fig. 2B).
FIGURE 2. Brx enhances the transcriptional activity of the GR by activating the small G-protein via its GEF domain.
A, Brx enhances GR-induced trans-activation of the MMTV promoter in HeLa (left panel) and CV-1 (right panel) cells. HeLa (left panel) or CV-1 (right panel) cells were transfected with FLAG-Brx-expressing plasmids together with pMMTV-Luc and pSV40-β-Gal. Bars represent the mean ± S.E. values of the luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone (Dex). *, p < 0.01, compared with the base line. B, Brx enhances GR-induced transactivation of the MMTV promoter in a dose-dependent fashion. HeLa cells were transfected with the indicated amounts of FLAG-Brx-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal. After adding dexamethasone, the cells were incubated for 24 h. Bars represent the mean ± S.E. values of the luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01, compared with the base line. RLU, relative luminescence units. C, inactivation of Brx GEF domain attenuated Brx-in-duced enhancement of GR transactivation. HeLa cells were transfected with wild type FLAG-Brx- or the indicated mutant FLAG-Brx-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal. Bars represent the mean ± S.E. values of the luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01. D, exogenous wild type Brx, mutant BrxY769F, and fragment Brx-(1042–1429) are expressed in similar quantities in HeLa cells. HeLa cells were transfected with the same amounts of the indicated FLAG-Brx-expressing plasmids. Whole cell homogenates were run on 8–16% SDS-PAGE gels, and blotted proteins were visualized with anti-FLAG (M2) or anti-GFP antibody. A representative gel image is shown in the left panel. The band densities from three independent experiments were measured, and the results were corrected by enhanced green fluorescence protein (EGFP) expression and shown as the mean ± S.E. in the right panel. n.s., statistically non-significant, compared with the wildtype Brx. E, linearized molecules of used Brx fragments and their functional domains. DH, Dbl homology; PH, pleckstrin homology; NRID, nuclear receptor-interacting domain. F, RhoA and Cdc42 cooperatively enhance Brx-potentiated GR transactivation. HeLa cells were transfected with FLAG-Brx-expressing plasmid and/or the indicated RhoA-or Cdc42-related molecule-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal. Bars represent the mean ± S.E.values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01, compared with the base line. QL, constitutively active mutant. G, neutralization of RhoA or Cdc42 activity diminishes Brx-induced potentiation of GR transactivation. HeLa cells were transfected with FLAG-Brx-expressing plasmid and/or C3-or N17Cdc42-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal. Bars represent the mean ± S.E. values of the luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01.
The GEF Domain of Brx Plays a Significant Role in Brx-induced Enhancement of GR Transactivation by Co-operating with RhoA rather than Cdc42
Brx and its longer splice form AKAP-Brx have a NRID at the C terminus, whereas they possess a central GEF domain (25, 27). The former plays an important role in Brx-induced enhancement of ERα transcriptional activity (25), whereas the latter mediates their physical/functional association with several small GTP-binding proteins and subsequent conversion of these proteins from the inactive, GDP-bound form to the active, GTP-bound form (23, 27). We, therefore, examined the importance of the Brx GEF domain in enhancement of GR-induced transactivation in HeLa cells to elucidate the molecular mechanism of Brx-induced potentiation of GR transactivation. We tested a GEF inactive point mutant BrxY769F and Brx-(1042–1429), which lacked the GEF domain, but contained the NRID, and compared their activity on GR transactivation to that of wild type Brx (Fig. 2, C and D). Simplified diagrams of these Brx constructs are shown in Fig. 2E. Wild type Brx strongly potentiated GR-induced transactivation of the MMTV promoter in a dexamethasone-dependent fashion, whereas BrxY769F weakly enhanced GR-induced transactivation, and its effect was significantly blunted compared with that of wild type Brx (Fig. 2C). Brx-(1042–1429) demonstrated similar activity as BrxY769F. Wild type Brx, BrxY769F, and Brx-(1042–1429) were expressed at similar levels (Fig. 2D). These results indicate that the GEF domain of Brx acts to potentiate GR transcriptional activity, whereas its NRID domain retains a weak effect.
Because the activity of the former domain is significant, we further examined the effects of Brx on the GR transcriptional activity using wild type and constitutively active form (GTP-bound equivalent) of RhoA and Cdc42 (Fig. 2F). Co-expression of wild type RhoA and Cdc42 weakly enhanced GR-mediated transactivation of the MMTV promoter, whereas they strongly potentiated Brx-induced enhancement of GR transactivation in HeLa cells. RhoA acted more potently than Cdc42. Expression of the constitutive active forms of RhoA and Cdc42 strongly potentiated GR-induced transactivation and weakly potentiated Brx activity. Again, the RhoA mutant was stronger than that of Cdc42. We interpret these results to suggest that potentiation of GR-induced transactivation by constitutively active forms of RhoA and Cdc42 was near maximal, and therefore, co-expression of Brx did not enhance activation further. Taken together, these results indicate that Brx enhances GR-induced transcriptional activity through small G-proteins such as RhoA and Cdc42 by converting them from inactive GDP-bound to active GTP-bound forms.
To confirm these results using another approach, we tested inhibitory molecules directed against either RhoA or Cdc42 for an effect on Brx-induced enhancement of GR transactivation (Fig. 2G). Expression of the botulinum toxin C3, a well known RhoA inhibitor, strongly suppressed Brx-induced enhancement, whereas the effect of the Cdc42 inhibitor N17Cdc42 was relatively weak, further supporting the hypothesis that Brx enhances GR-induced transcriptional activity by converting small G-proteins from their inactive to active forms via its GEF domain. Enhancement of GR transactivation by Brx was more dependent upon RhoA than Cdc42.
Brx Forms a Ternary Complex with GR and RhoA in Vivo
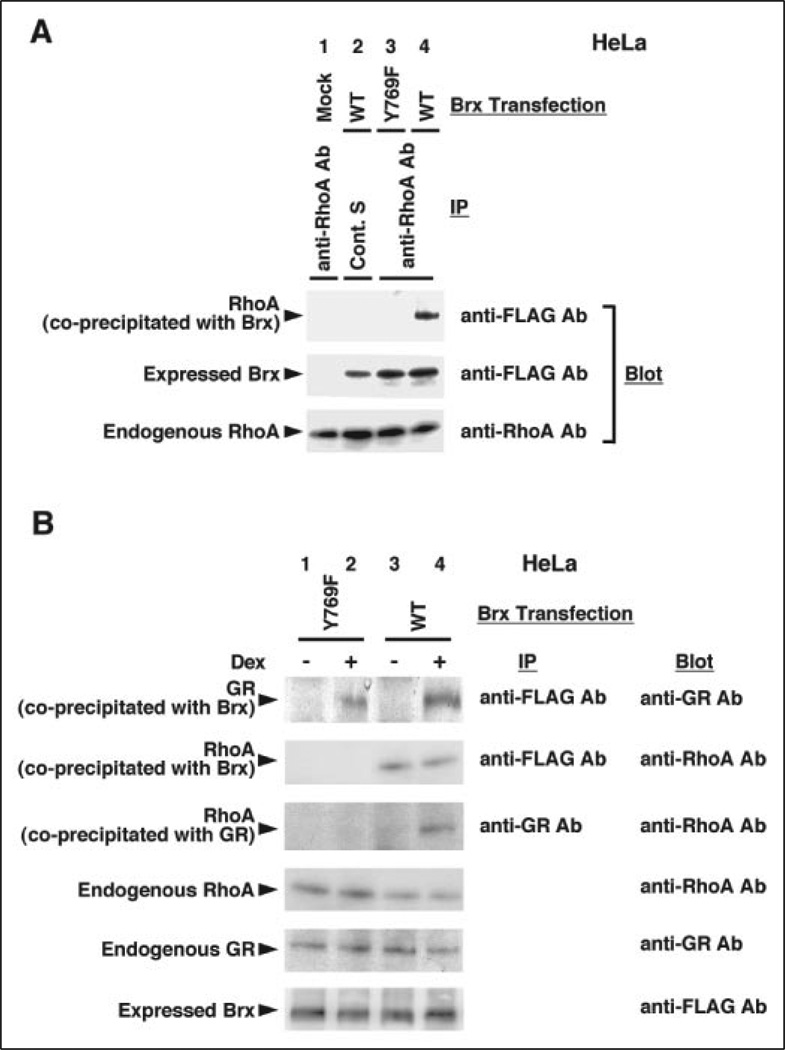
Brx and AKAP-Brx physically interacted with the small G-proteins RhoA and Cdc42 both in vitro and in vivo (27), and it was shown to interact with one of the nuclear receptors ERα through its C-terminal NRID in vitro (25). Because we observed dependence upon the small G-proteins in Brx-induced GR transactivation, we suspected that small G-proteins and nuclear receptors might form a ternary complex with Brx in vivo. Thus, we examined the interaction of Brx, RhoA, and GR in co-immunoprecipitation assays (Fig. 3). RhoA was successfully coprecipitated with the wild type FLAG-Brx but not with the GEF domain-defective FLAG-BrxY769F (Fig. 3A, top gel, lanes 4 and 3, respectively). The RhoA band did not appear when FLAG-Brx was not transfected or control serum was used for the immunoprecipitation (Fig. 3A, top gel, lanes 1 and 2, respectively). Transfected FLAG-Brx and endogenous RhoA were similarly expressed throughout the experiment (Fig. 3A, middle and bottom gels, respectively). These results indicate that RhoA interacts with Brx in a GEF domain-dependent fashion. Furthermore, both wild type FLAG-Brx and FLAG-BrxY769F were associated with GR in a dexamethasone-dependent fashion (Fig. 3B, top gel, lanes 4 and 2, respectively). RhoA was coprecipitated with wild type FLAG-Brx but not with FLAG-BrxY769F (Fig. 3B, second gel, lanes 3–4 and 1–2, respectively). RhoA was also coprecipitated with GR in the presence of wild type FLAG-Brx, but not the FLAG-BrxY769F mutant, in a dexamethasone-dependent fashion (Fig. 3B, third gel, lanes 4 and 2, respectively). Endogenous RhoA and GR and expressed FLAG-Brx were detected similarly throughout the experiment (Fig. 3B, bottom three gels). Taken together, these results indicate that Brx forms a complex with RhoA via its GEF domain in vivo, and GR interacts with this protein complex in a ligand-dependent fashion, leading to formation of a ternary complex consisting of Brx, RhoA, and GR. Brx-independent interaction of RhoA and GR was not observed in the co-immunoprecipitation experiments.
FIGURE 3. Brx is associated with RhoA and GR in vivo.
A, Brx interacts with RhoA through the GEF domain. HeLa cells were transfected with FLAG-Brx wild type-or Y769F mutant-expressing plasmid, and a co-immunoprecipitation assay was performed with anti-FLAG (M2) or control antibodies. Samples were run on 12% (RhoA) and 6% (Brx) SDS-PAGE gels, and blotted RhoA and FLAG-Brx were visualized with anti-RhoA or-FLAG (M2) antibodies. Transfected plasmids and antibodies used for precipitation (IP) are indicated in the top of the figure, whereas antibodies used for visualization of bands (Blot) are shown in the right side. RhoA coprecipitated with transfected FLAG-Brx is shown in the top gel, whereas amounts of wild type Brx and mutant BrxY769F expressed from their plasmids are shown in the second gel. Endogenously expressed RhoA is shown in the bottom gel. Ab, antibody; Cont. S, control serum; IP, immunoprecipitation. B, Brx forms a ternary complex with GR and RhoA in a dexamethasone- and Brx GEF domain- dependent fashion. HeLa cells were transfected with FLAG-Brx wild type- or Y769F mutant-expressing plasmid, and a co-immunoprecipitation assay was performed with anti-FLAG (M2) or -GR antibody. Samples were run on 12% (RhoA), 8% (GR), and 6% (FLAG-Brx) SDS-PAGE gels, and blotted RhoA, GR, and Brx were visualized with their specific antibodies. Transfected plasmids and dexamethasone treatment are indicated at the top of the figure, whereas antibodies used for immunoprecipitation (IP) or visualization of bands (Blot) are shown in the right side. RhoA and GR, coprecipitated with transfected wild type FLAG-Brx or mutant BrxY769F, or endogenous GR are shown in the top three gels, whereas amounts of endogenously expressed RhoA, GR, and wild type FLAG-Brx or mutant BrxY769F expressed from their plasmids are shown in the bottom three gels.
GR Attracts Brx and RhoA to GREs of the MMTV Promoter in Vivo
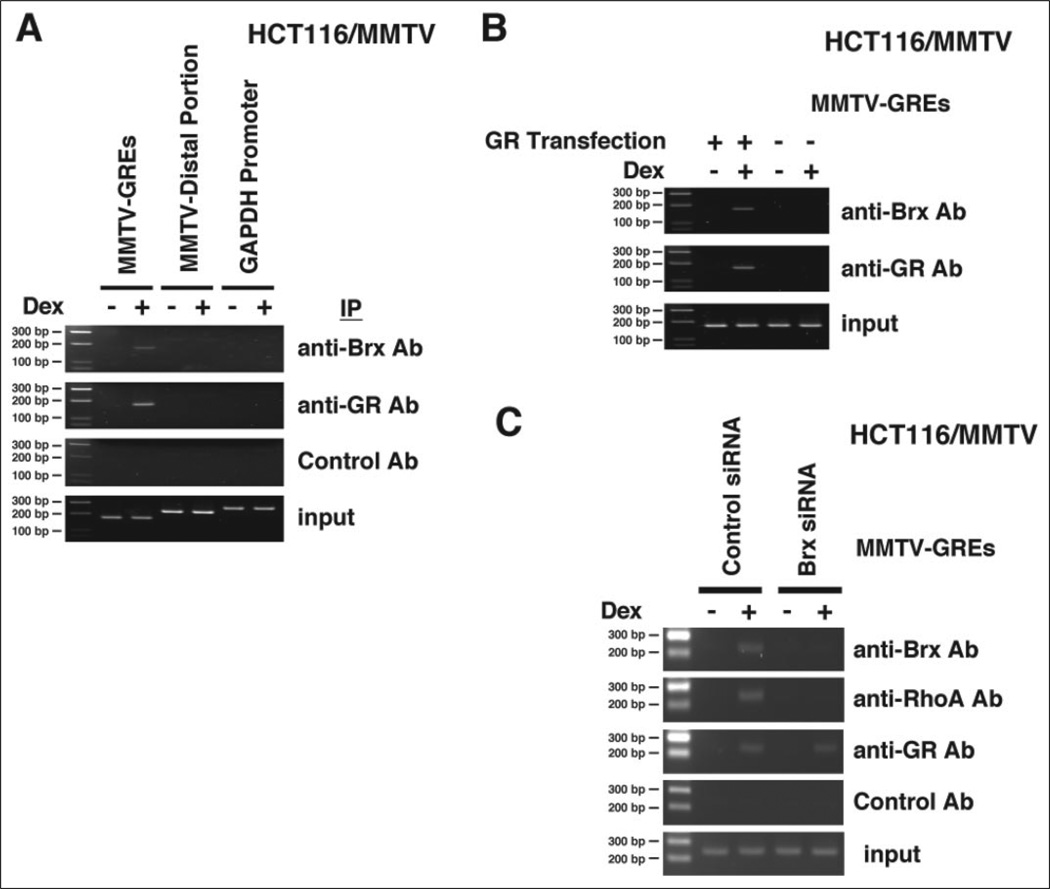
These results prompted us to examine whether GR also attracts Brx and RhoA to its target sequence GREs located in the genomic DNA using a ChIP assay in HCT116/MMTV cells, which contain the full-length MMTV promoter integrated in their chromatin. Because these cells do not express functional GR, we transfected them with a GR-expressing plasmid. In these experiments endogenous Brx was attracted to the proximal region of the MMTV promoter that contained two GREs in a dexamethasone-dependent fashion (Fig. 4A). In contrast, Brx was not attracted to the distal region of the MMTV promoter or a fragment of the GAPDH promoter, neither of which contain GREs. Brx was precipitated with GREs of the MMTV promoter when the cells were transfected with GR-expressing plasmids, whereas Brx attraction disappeared when the control plasmid was used (Fig. 4B), indicating that accumulation of Brx to the promoter is dependent upon the presence of GREs and ligand-activated GR. Similarly, endogenous Brx and RhoA were attracted to GREs of the MMTV promoter, whereas transfection of the Brx siRNA completely abolished this attraction (Fig. 4C). Together with the results obtained in the co-immunoprecipitation experiments, these findings indicate that Brx is attracted to GREs of the chromatin-integrated MMTV promoter via physical interaction with the GR. Attraction of RhoA to GREs was dependent on the presence of Brx, possibly through interaction with the Brx GEF domain. Thus, Brx and RhoA may regulate GR-induced transcriptional activity by directly influencing the GR-attracted transcriptional complex formed on the GRE-driven promoters.
FIGURE 4. GR attracts Brx and RhoA to GREs of the chromatin-integrated MMTV promoter.
A, Brx is attracted to the region of the MMTV promoter, which contains two GREs, in a dexamethasone (Dex)-dependent fashion. Brx is not associated with the GRE-devoid distal portion of the MMTV promoter or with the glucocorticoid-unresponsive and GRE-devoid GAPDH promoter. HCT116/MMTV cells were transfected with the GR-expressing plasmid, and the ChIP assay was performed with anti-Brx, -GR, or control antibody. The portion of the MMTV promoter that contains two GREs (MMTV-GREs), the distal portion of the MMTV promoter that does not contain GREs (MMTV-Distal Portion), and the glucocorticoid-unresponsive GAPDH promoter (from −777 to −516) (GAPDH Promoter) were amplified by PCR with their specific primer pairs. Ab, antibody; IP, immunoprecipitation. B, attraction of Brx to GREs of the chromatin-integrated MMTV promoter is dependent on the presence of ligand-activated GR in HCT116/MMTV cells. HCT116/MMTV cells were transfected with GR-expressing or control plasmid, and the ChIP assay was performed with anti-Brx or -GR antibodies. The portion of the MMTV promoter that contains 2 GREs was amplified by PCR with a specific primer pair. C, knock-out of endogenous Brx by its siRNA abolished attraction of Brx and RhoA to GREs of the chromatin-integrated MMTV promoter in HCT116/MMTV cells. HCT116/MMTV cells were transfected with control or Brx siRNA together with the GR-expressing plasmid, and the ChIP assay was performed with anti-Brx, -RhoA, or -GR antibodies. The portion of the MMTV promoter that contains two GREs was amplified by PCR with a specific primer pair.
Brx/RhoA Mediates LPA-induced Potentiation of GR Transactivation
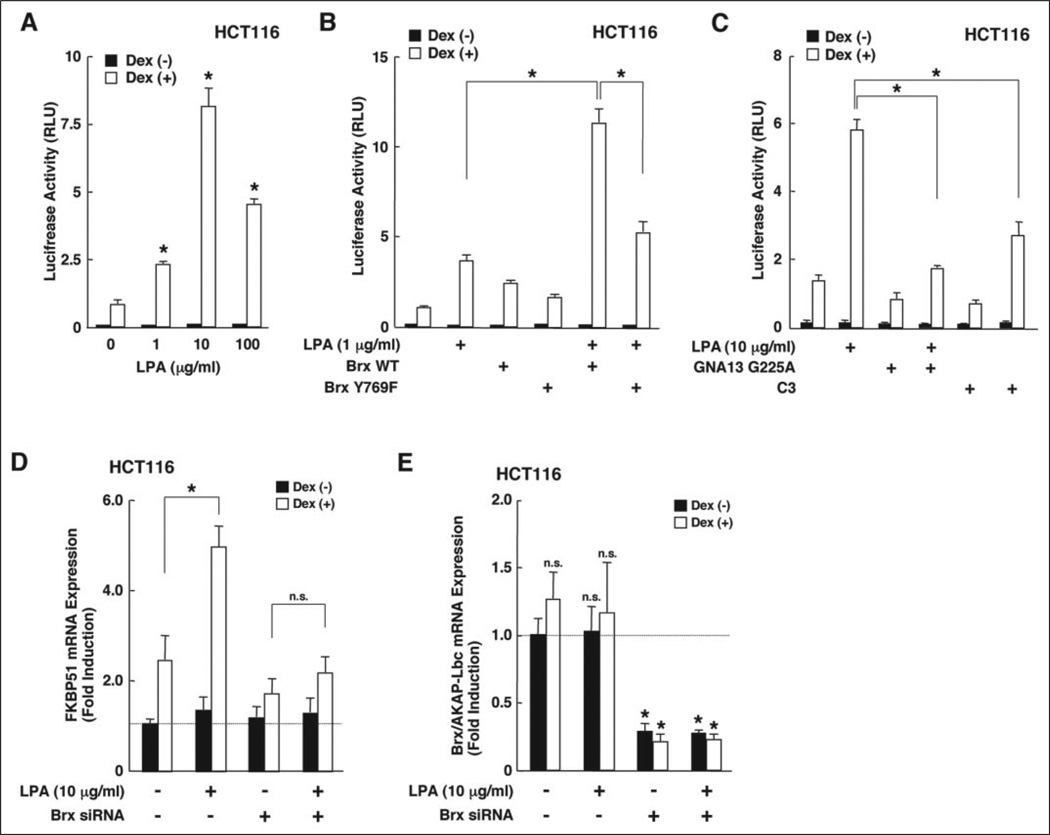
The observation that Brx enhanced glucocorticoid activity prompted us to examine the effect of LPA on GR-induced transcriptional activity for two reasons. First, because LPA activates RhoA through a specific cell surface, 7-transmembrane receptor domain coupled with Gα12 and/or Gα13 stimulation (27, 37–39) and AKAP-Brx mediates LPA-induced signal to the formation of stress fibers operated by RhoA (27). Second, because LPA is produced from activated platelets that are frequently observed in patients with “dysmetabolic” syndrome, this syndrome is associated with increased sensitivity to glucocorticoids in adipose tissues (8, 40). We first tested the effects of increasing concentrations of LPA on GR-induced transactivation (Fig. 5). In HCT116 cells, LPA potentiated GR-induced transactivation of the MMTV promoter in a dexamethasone-dependent fashion (Fig. 5A). The concentration of 100 µg/ml LPA produced a morphologically toxic effect on the cells (data not shown), explaining reduction of the LPA-induced stimulation at this concentration of this compound. Expression of wild type Brx synergistically enhanced LPA-induced potentiation of GR transactivation, whereas expression of BrxY769F failed to cooperate with LPA (Fig. 5B). Co-expression of a transdominant negative form of Gα13 (GNA13 G225A) or the botulinum toxin C3 almost completely abolished LPA-induced potentiation of GR transactivation, indicating that activation of the Gα13/RhoA pathway by LPA is likely to have mediated the observed effect of LPA on GR transactivation in this cell line (Fig. 5C). Taken together these results indicate that LPA potentiates GR-induced transcriptional activity, and Brx acts to enhance LPA function by activating RhoA, possibly through its GEF activity.
FIGURE 5. LPA potentiates GR-induced transcriptional activity by activating the Gα13/RhoA pathway, and Brx acts as an enhancer of LPA-induced potentiation.
A, LPA potentiates GR-induced transactivation of the MMTV promoter in a dose-dependent fashion in HCT116 cells. HCT116 cells were transfected with pMMTV-Luc and pSV40-β-Gal, and the cells were treated with the indicated concentrations of LPA. Bars represent the mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone (Dex). *, p < 0.01, compared with the base line. RLU, relative luminescence units. B, wild type Brx, but not its Y769F mutant, potentiates LPA-induced enhancement of GR transactivation. HCT116 cells were transfected with FLAG-Brx wild type- or Y769F mutant-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal, and the cells were treated with 1 µg/ml LPA. Bars represent the mean ± S.E. values of luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01, compared with the base line. C, LPA potentiates GR transactivation by activating Gα13 and RhoA. HCT116 cells were transfected with Gα13G225A- or C3-expressing plasmid together with pMMTV-Luc and pSV40-β-Gal, and the cells were treated with 10 µg/ml LPA. Bars represent the mean ± S.E. values of the luciferase activity normalized for β-galactosidase activity in the absence or presence of 10−6m dexamethasone. *, p < 0.01, compared with the base line. C3, botulinum toxin C3; GNA13, G-protein α13. D and E, endogenous Brx/AKAP-Brx is necessary for LPA-mediated GR-induced expression of the glucocorticoid-responsive FKBP51 mRNA. HCT116 cells were transfected with control (lamin A/C) or Brx siRNA, and the cells were treated with 10 µg/ml LPA. Total RNA was purified from the cells, and the amounts of FKBP51, Brx/AKAP-Brx, or RPLP0 mRNAs were determined by real-time PCR. Relative expression levels of FKBP51 and Brx/AKAP-Brx mRNA shown as -fold induction to the base line are shown in D and E, respectively. *, p < 0.01; n.s., statistically non-significant, compared with the base line.
To further explore a physiologic role of Brx on LPA-induced potentiation of GR activity, we used siRNA experiments to examine Brx/AKAP-Brx ablation on LPA-induced mRNA expression of the endogenous glucocorticoid-responsive gene FKBP51 in HCT116 cells (Fig. 5, D and E). FKBP51 is an immunophilin that binds the FK506 immunosuppressant, and its expression is positively regulated by glucocorticoids (35, 41–43). We treated/transfected HCT116 cells with 10 µg/ml LPA and/or Brx siRNA together with the GR-expressing plasmid, and measured the mRNA levels of FKBP51, Brx/AKAP-Brx, and control RPLP0 with real-time PCR using specific primer pairs. More than 60% of HCT116 cells were transfected by using x-tremeGENE siRNA transfection reagent (data not shown). LPA treatment enhanced dexamethasone-induced FKBP51 mRNA expression by 2-fold, whereas this effect disappeared in the cells transfected with Brx siRNA (Fig. 5D). Transfection of Brx siRNA also reduced the Brx/AKAP-Brx mRNA expression by about 70% (Fig. 5E). These results indicate that LPA enhances the transcriptional activity of the GR on the endogenous glucocorticoid-responsive FKBP51 gene and that Brx/AKAP-Brx is necessary for mediating the effect of LPA to the GR-induced transactivation.
DISCUSSION
We demonstrated that reduction of the endogenous Brx, a Rho family small G-protein GEF, by genetic manipulation in mice attenuated the effectiveness of dexamethasone in splenic lymphocytes ex vivo, indicating that Brx physiologically acts as an enhancer of glucocorticoid actions in these cells. Consistent with this result, Brx enhanced the transcriptional activity of GR on the glucocorticoid-responsive MMTV promoter in HeLa and CV-1 cells.
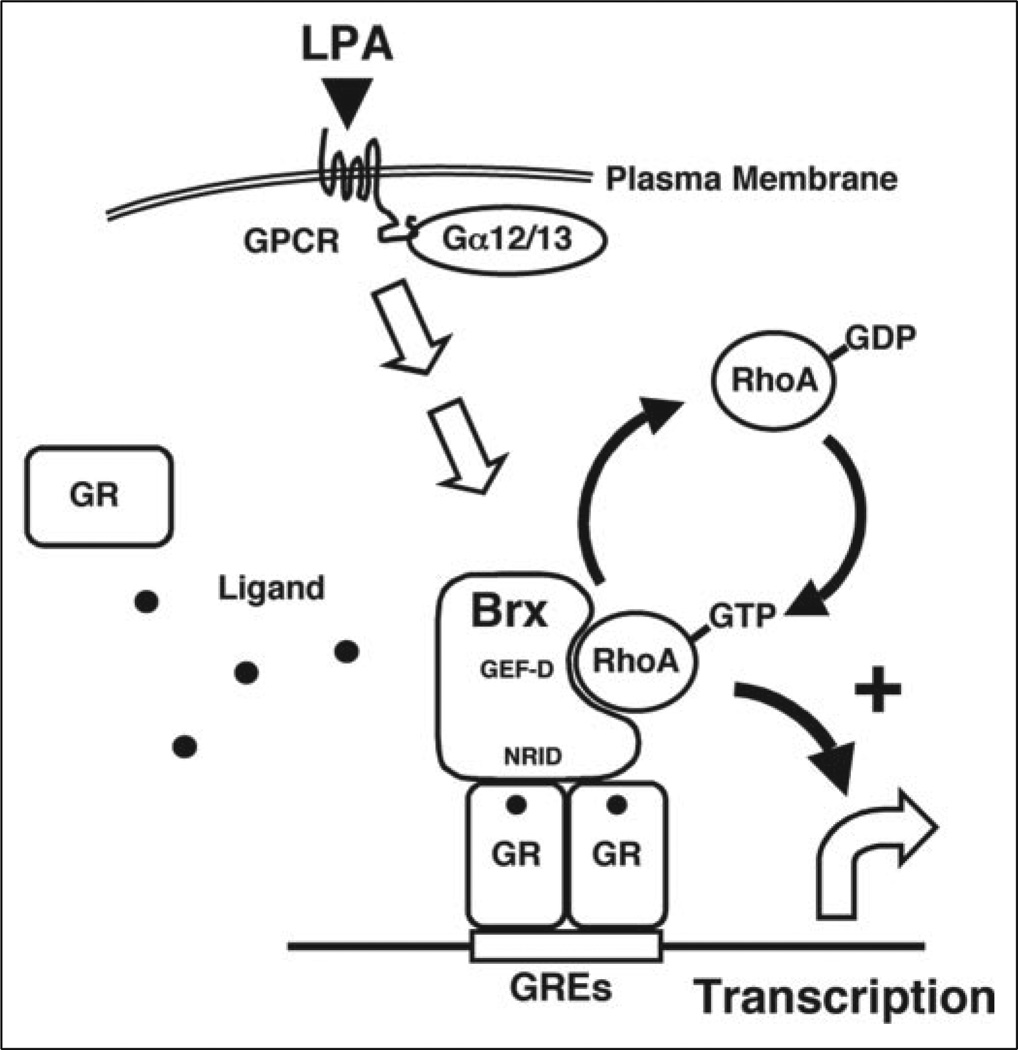
The mechanism required GEF enzymatic activity, since BrxY769F and Brx-(1042–1429), which are devoid of GEF activity, showed reduced enhancement of GR transactivation compared with that of wild type Brx. In agreement with this, constitutively active forms of RhoA and Cdc42 strongly enhanced GR transactivation; the enhancement appeared maximal. Furthermore, Brx promoted complex formation with RhoA and GR in vivo. The C-terminal NRID was required for bridging Rho family small G-proteins to GR. It appears that Brx acts as a bridging factor between RhoA and GR, facilitating the interaction between the GR and activated Rho family small G-proteins (Fig. 6). Although constitutively active forms of RhoA and Cdc42 strongly enhanced GR transcriptional activity in the overexpression experiment, attraction and local activation of RhoA governed by Brx was necessary for small G-proteins to enhance GR transactivation in physiologic situations. Thus, Brx may provide a subcellular microenvironment in which GR and possibly attracted transcriptional components can efficiently interface with the activated small G-proteins.
FIGURE 6. Proposed model of Brx-induced enhancement of GR transcriptional activity.
Brx mediates LPA-induced potentiation of GR transcriptional activity by interacting with RhoA and ligand-bound GR at GRE-containing glucocorticoid-responsive promoters. Brx converts RhoA from an inactive, GDP-bound form to an active, GTP-associated type and communicates the latter to the GR inside the transcription initiation complex. GEF-D, GEF domain; Gα12/13, G-protein α 12 and/or 13; GPCR, G-protein-coupled receptor.
In a previous publication we demonstrated that Brx enhanced the ERα-mediated transcriptional activity by cooperating with Cdc42 (25). In this manuscript we showed that Brx potentiated GR transactivation mainly through RhoA but not via Cdc42. It is possible that differential specificity of RhoA- or Cdc42-associated Brx complexes to these two steroid hormone receptors may account for this difference. Because Cdc42, but not RhoA, activates the p38 mitogen-activated protein kinase (44), this kinase may be also functional in the case of ERβ but perhaps not with GR.
Our results obtained in the ChIP assay indicate that Brx and RhoA directly enhance GR transcriptional activity in the nucleus. We speculate that Rho family small G-proteins, such as Rac1 and Cdc42, translocate from the cytoplasm into the nucleus through use of their putative nuclear localization signal in their C-terminal portion (45, 46). Brx, on the other hand, may also shuttle between the cytoplasm and the nucleus, possibly through its physical association with 14-3-3, a cellular regulator protein that interacts with several partner molecules, including the GR, modulating their activity by changing their subcellular localization (32, 47, 48). Because GTP-bound RhoA functions as a molecular switch, it is possible that Brx influences the transcriptional activity of components of the GR-generated transcriptosome via attraction of GTP-bound RhoA. Recently, 14-3-3 was reported to physically interact with histones and to regulate its activity on transcription and chromosome condensation (49). Thus, it is also possible that Brx additionally regulates GR-induced transcriptional activity on chromatin through its association with 14-3-3. Interestingly, 14-3-3 is also an interactant of the GR, with effects on the transcriptional activity of the GR (32, 50),
We also demonstrated that LPA potentiated GR-induced transactivation of the MMTV promoter by stimulating the Gα13/RhoA pathway. Indeed, this lipid compound enhanced dexamethasone-induced mRNA expression of the endogenous glucocorticoid-responsive FKBP51 gene, and depletion of Brx/AKAP-Brx by treatment with Brx siRNA attenuated LPA-induced enhancement of FKBP51 mRNA expression in HCT116 cells. These results suggest that Brx/AKAP-Brx directs LPA-induced biological activities toward GR in tissues that have abundant endogenous expression of these molecules (Fig. 6). It is known that LPA is produced from activated platelets, which are frequently observed in pathologic conditions associated with increased sensitivity to glucocorticoids in local tissues, such as visceral-type obesity, hypertension, hyperlipidemia, and insulin resistance/overt diabetes mellitus type 2 (8, 22). Thus, it is possible that LPA is one of the factors that up-regulates tissue glucocorticoid sensitivity in these pathologic problems. Because LPA promotes atherosclerosis and hypertension, increased glucocorticoid action by LPA might further contribute to the development/worsening of these pathologic states caused by this lipid. It is also possible that up-regulation of glucocorticoid action by LPA in local tissues contributes to some of the known biological activities of LPA, such as those on cellular morphology, proliferation, migration, and survival.
Acknowledgments
We thank Drs. R. M. Evans, J. S. Gutkind, and G. L. Hager for providing plasmids and K. Zachman for superb technical assistance. We remain appreciative of the assistance of A. Grinberg, K. McDaniel, K. McLean, K. Pagilai, and M. Venere, all of whom contributed to development of the brx knockout mice.
Footnotes
This study was funded in part by NICHD, National Institutes of Health, Bethesda, MD.
The abbreviations used are: GR, glucocorticoid receptor; WT, wild type; GEF, guanine nucleotide exchange factor; NRID, nuclear receptor-interacting domain; PH, pleck-strin homology; LPA, lysophosphatidic acid; GRE, glucocorticoid response element; MMTV, murine mammary tumor virus; ChIP, chromatin immunoprecipitation; siRNA, small interfering RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
J. Wadell, C. Mayers, M. Venere, K. McLean, K. Sarber, P. Driggers, K. Pagliai, M. Gorivodsky, H. Westphal, and J. Segars, manuscript in preparation.
REFERENCES
- 1.Chrousos GP. N. Engl. J. Med. 1995;332:1351–1362. doi: 10.1056/NEJM199505183322008. [DOI] [PubMed] [Google Scholar]
- 2.Miller WL, Chrousos GP. In: Endocrinology and Metabolism. 4th Ed. Felig P, Frohman LA, editors. New York: McGraw-Hill Book Co; 2001. pp. 387–524. [Google Scholar]
- 3.Chrousos GP. In: Endocrinology and Metabolism. 4th Ed. Felig P, Frohman LA, editors. New York: McGraw-Hill Book Co; 2001. pp. 609–632. [Google Scholar]
- 4.Enmark E, Gustafsson JÅ. Trends Pharmacol. Sci. 2001;22:611–615. doi: 10.1016/s0165-6147(00)01859-9. [DOI] [PubMed] [Google Scholar]
- 5.Kino T, De Martino MU, Charmandari E, Mirani M, Chrousos GP. J. Steroid Biochem. Mol. Biol. 2003;85:457–467. doi: 10.1016/s0960-0760(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blum-berg B, Kastner P, Mark M, Chambon P, Evans RM. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kino T, Chrousos GP. J. Allergy Clin. Immunol. 2002;109:609–613. doi: 10.1067/mai.2002.123708. [DOI] [PubMed] [Google Scholar]
- 8.Chrousos GP. Am. J. Med. 2004;117:204–207. doi: 10.1016/j.amjmed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Schaaf MJ, Cidlowski JA. J. Steroid Biochem. Mol. Biol. 2002;83:37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 10.Ihle JN, Kerr IM. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 11.Krupnick JG, Benovic JL. Annu. Rev. Pharmacol. Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 12.Mosesson Y, Yarden Y. Semin. Cancer Biol. 2004;14:262–270. doi: 10.1016/j.semcancer.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Takai Y, Sasaki T, Matozaki T. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 14.Kaibuchi K, Kuroda S, Amano M. Annu. Rev. Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 15.Aznar S, Lacal JC. Cancer Lett. 2001;165:1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- 16.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Liu M, Kozasa T, Rothstein JD, Sternweis PC, Neubig RR. J. Biol. Chem. 2004;279:28831–28834. doi: 10.1074/jbc.C400105200. [DOI] [PubMed] [Google Scholar]
- 18.Moolenaar WH. Ann. N. Y. Acad. Sci. 2000;905:1–10. doi: 10.1111/j.1749-6632.2000.tb06532.x. [DOI] [PubMed] [Google Scholar]
- 19.Fukata M, Kaibuchi K. Nat. Rev. Mol. Cell Biol. 2001;2:887–897. doi: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 20.Mills GB, Moolenaar WH. Nat. Rev. Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 21.Graler MH, Goetzl EJ. Biochim. Biophys. Acta. 2002;1582:168–174. doi: 10.1016/s1388-1981(02)00152-x. [DOI] [PubMed] [Google Scholar]
- 22.Siess W. Biochim. Biophys. Acta. 2002;1582:204–215. doi: 10.1016/s1388-1981(02)00173-7. [DOI] [PubMed] [Google Scholar]
- 23.Cherfils J, Chardin P. Trends Biochem. Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt A, Hall A. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 25.Rubino D, Driggers P, Arbit D, Kemp L, Miller B, Coso O, Pagliai K, Gray K, Gutkind S, Segars J. Oncogene. 1998;16:2513–2526. doi: 10.1038/sj.onc.1201783. [DOI] [PubMed] [Google Scholar]
- 26.Driggers PH, Segars JH, Rubino DM. J. Biol. Chem. 2001;276:46792–476797. doi: 10.1074/jbc.M106927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diviani D, Soderling J, Scott JD. J. Biol. Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 28.Boguski MS, McCormick F. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel K, Mayers C, Venere M, Driggers P, Westphal H, Gorivodsky M, Segars J. J. Soc. Gynecol. Investig. 2004;11(suppl):199. (Abstr. 375) [Google Scholar]
- 30.De Martino MU, Bhattachryya N, Alesci S, Ichijo T, Chrousos GP, Kino T. Mol. Endocrinol. 2004;18:820–833. doi: 10.1210/me.2003-0341. [DOI] [PubMed] [Google Scholar]
- 31.Kino T, Gragerov A, Kopp JB, Stauber RH, Pavlakis GN, Chrousos GP. J. Exp. Med. 1999;189:51–62. doi: 10.1084/jem.189.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kino T, Souvatzoglou E, De Martino MU, Tsopanomihalou M, Wan Y, Chrousos GP. J. Biol. Chem. 2003;278:25651–25656. doi: 10.1074/jbc.M302818200. [DOI] [PubMed] [Google Scholar]
- 33.Ichijo T, Voutetakis A, Cotrim AP, Bhattachryya N, Fujii M, Chrousos GP, Kino T. J. Biol. Chem. 2005;280:42067–42077. doi: 10.1074/jbc.M509338200. [DOI] [PubMed] [Google Scholar]
- 34.Kino T, Tiulpakov A, Ichijo T, Chheng L, Kozasa T, Chrousos GP. J. Cell Biol. 2005;169:885–896. doi: 10.1083/jcb.200409150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. J. Clin. Endocrinol. Metab. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 36.Karl M, Lamberts SW, Koper JW, Katz DA, Huizenga NE, Kino T, Haddad BR, Hughes MR, Chrousos GP. Proc. Assoc. Am. Physicians. 1996;108:296–307. [PubMed] [Google Scholar]
- 37.Ridley AJ, Hall A. EMBO J. 1994;13:2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jalink K, van Corven EJ, Hengeveld T, Morii N, Narumiya S, Moolenaar WH. J. Cell Biol. 1994;126:801–810. doi: 10.1083/jcb.126.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kranenburg O, Poland M, van Horck FP, Drechsel D, Hall A, Moolenaar WH. Mol. Biol. Cell. 1999;10:1851–1857. doi: 10.1091/mbc.10.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colwell JA, Nesto RW. Diabetes Care. 2003;26:2181–2188. doi: 10.2337/diacare.26.7.2181. [DOI] [PubMed] [Google Scholar]
- 41.Baughman G, Wiederrecht GJ, Campbell NF, Martin MM, Bourgeois S. Mol. Cell. Biol. 1995;15:4395–4402. doi: 10.1128/mcb.15.8.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vermeer H, Hendriks-Stegeman BI, van Suylekom D, Rijkers GT, van Buul-Offers SC, Jansen M. Mol. Cell. Endocrinol. 2004;218:49–55. doi: 10.1016/j.mce.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Baughman G, Wiederrecht GJ, Chang F, Martin MM, Bourgeois S. Biochem. Biophys. Res. Commun. 1997;232:437–443. doi: 10.1006/bbrc.1997.6307. [DOI] [PubMed] [Google Scholar]
- 44.Minden A, Lin A, Claret FX, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 45.Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL. J. Biol. Chem. 2004;279:44197–44210. doi: 10.1074/jbc.M404977200. [DOI] [PubMed] [Google Scholar]
- 46.Williams CL. Cell. Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 47.Diviani D, Abuin L, Cotecchia S, Pansier L. EMBO J. 2004;23:2811–2820. doi: 10.1038/sj.emboj.7600287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu H, Subramanian RR, Masters SC. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 49.Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC. Mol. Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 50.Wakui H, Wright AP, Gustafsson J, Zilliacus J. J. Biol. Chem. 1997;272:8153–8156. doi: 10.1074/jbc.272.13.8153. [DOI] [PubMed] [Google Scholar]