SUMMARY
Vertebrate centrioles lose their geometric scaffold, the cartwheel, during mitosis, concurrently with gaining the ability to recruit the pericentriolar material (PCM) and thereby function as the centrosome. Cartwheel removal has recently been implicated in centriole duplication, but whether “cartwheel-less” centrioles are intrinsically stable, or must be maintained through other modifications remains unclear. Here, we identify a newborn centriole-enriched protein, KIAA1731/CEP295, specifically mediating centriole-to-centrosome conversion but dispensable for cartwheel removal. In the absence of CEP295, centrioles form in S/G2 phase, and lose their associated cartwheel in mitosis, but cannot be converted to centrosomes, uncoupling the two events. Strikingly, centrioles devoid of both the PCM and cartwheel progressively lose centriolar components, whereas centrioles associating with either the cartwheel or PCM alone can exist stably. Thus, cartwheel removal can have grave repercussions to centriole stability, and centriole-to-centrosome conversion mediated by CEP295 must occur in parallel to maintain cartwheel-less centrioles for duplication.
INTRODUCTION
Centrioles in cycling cells are carefully maintained. They duplicate exactly once in S phase, and segregate equally through association with spindle poles during cell division. Centrioles can duplicate and associate with spindle poles only when they have been converted to centrosomes (Wang et al., 2011). The conversion of centrioles to centrosomes is a critical step of centriole modification in which newborn, unmodified centrioles acquire the competence to recruit the pericentriolar material (PCM) and thereby function as the microtubule-organizing center (MTOC) or centrosome. Modification starts in early mitosis depending on Plk1 activity, and completes at late mitosis, giving rise to one old/previously converted and one newly converted centriole that are both MTOC-competent to start the new cell cycle. The converted centriole duplicates and then carries the newly formed, unconverted daughter centriole to which it is engaged through the segregation process, ensuring centriole homeostasis. While Plk1 plays an essential role in centriole-to-centrosome conversion, it is also required for many other cellular processes during mitosis (Petronczki et al., 2008), including centriole disengagement (Wang et al., 2011). No centriolar factors specifically involved in centriole-to-centrosome conversion have been identified.
Centrioles are stable structures. The integrity of a centriole is in part contributed by a geometric scaffold known as the cartwheel, which defines the 9-fold symmetry of a centriole (Anderson and Brenner, 1971). The cartwheel is located at the proximal lumen of a centriole, coincident with centriolar proteins SAS-6 (Nakazawa et al., 2007) and STIL/SAS-5 (Stevens et al., 2010). In particular, SAS-6 has been shown to form the primary backbone of the cartwheel (Kitagawa et al., 2011; van Breugel et al., 2011). Surprisingly, while the cartwheel is essential for centriole assembly, it is removed from newborn centrioles at the end of the cell cycle (Vorobjev and Chentsov, 1980; Vorobjev and Chentsov Yu, 1982), when both SAS-6 and STIL are disassociated from centrioles during early mitosis (Arquint and Nigg, 2014), and degraded subsequently (Arquint and Nigg, 2014; Arquint et al., 2012; Strnad et al., 2007; Tang et al., 2011), generating “cartwheel-less” centrioles that appear to be stable.
The function of cartwheel removal is not clear. Recent studies showed that the empty proximal lumen of the cartwheel-less centriole can function as a template for SAS-6 assembly prior to daughter centriole formation, revealing a potential role of cartwheel elimination in centriole duplication (Fong et al., 2014). While the stability of centrioles can be maintained in the absence of the cartwheel, the underlying mechanism is unclear. Interestingly, cartwheel removal occurs during mitosis, when newborn centrioles are converted to centrosomes. When the conversion is blocked by Plk1 inhibition, cartwheel removal also fails to occur (Wang et al., 2011), revealing an intimate relationship between these two events.
RESULTS
Identification of CEP295 as a daughter centriole-enriched proximal end protein
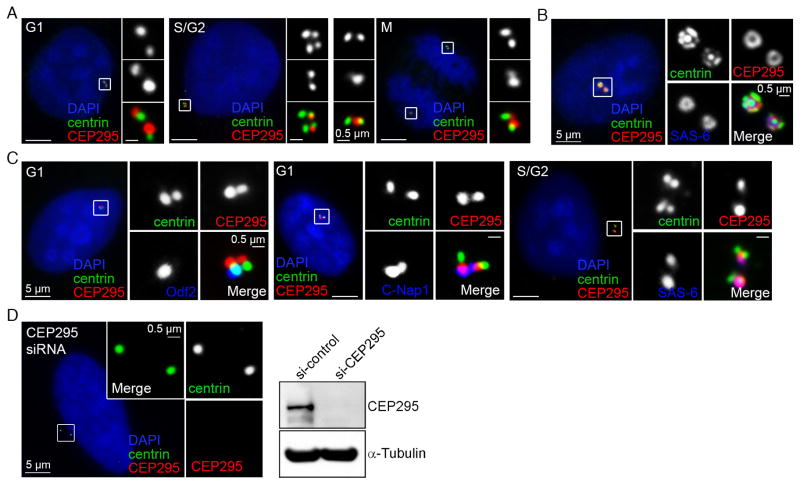
To screen for centriolar factors specifically involved in centriole-to-centrosome conversion, we focused on proteins that were enriched in newborn (unconverted) centrioles, using the “proteomic map” of the centrosome generated recently (Tanos et al., 2013; Wang et al., 2013). The study of one protein KIAA1731, hereafter named CEP295, was presented. KIAA1731/CEP295 has been previously characterized as an essential factor for centriole assembly or stability (Knorz et al., 2010), but its exact role in centriole biogenesis and maintenance remains unclear. Our immunofluorescence studies showed that CEP295 associates with both of the (converted) centrioles during G1 (Figure 1A), but becomes more enriched at the newly formed daughter (or unconverted) centrioles during S, G2 and early M phases (Figure 1A & C), a pattern distinct from other known daughter centriole proteins. Consistently, CEP295 was found to label the extra daughter centrioles induced by overexpression of Plk4 (Figure 1B, centriole rosettes). Detailed examination further revealed that CEP295 co-localizes with the proximal end markers C-Nap1 or SAS-6 (Figure 1C) but not the distal end proteins centrin or ODF2 (Figure 1C). Note that the signal recognized by CEP295 antibodies is specific, as it disappeared in cells depleted of CEP295 by RNAi (Figure 1D) or by CRISPR-mediated gene targeting (Figure 2B). Together, our data demonstrate that CEP295 localizes to all centrioles at the proximal end, with a strong preference at newly formed daughter centrioles.
Figure 1. CEP295 is a daughter-enriched proximal end protein.
(A) U2OS cells at different cell cycle stages were stained with indicated antibodies. DAPI (blue) marks nuclei.
(B) CEP295 localizes to the extra daughter centrioles of the rosette centrosome, inducibly formed by Plk4 overexpression in RPE1 cells.
(C) G1, or S/G2 centrioles were visualized with indicated antibodies, including the proximal end marker C-Nap1 or SAS-6, and distal end marker centrin or Odf2.
(D) CEP295 signals were lost in CEP295-RNAi cells, revealed by both immunostaining and western blot.
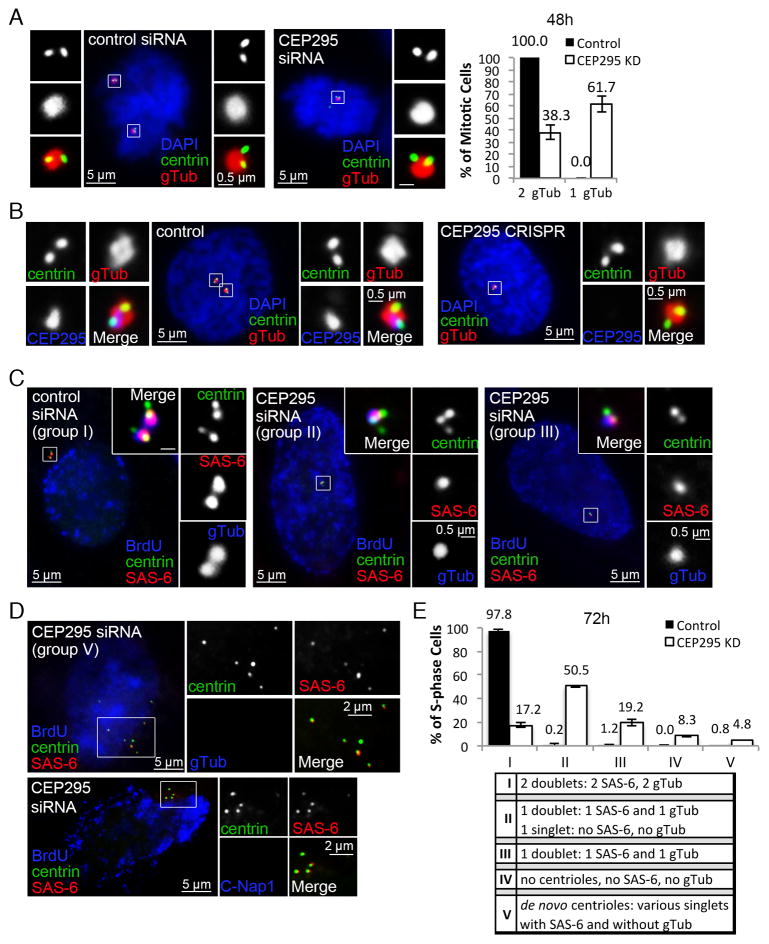
Figure 2. Loss of CEP295 does not block centriole assembly but leads to centrosome reduction.
(A) U2OS cells transfected with control or CEP295 siRNA for 48hr were stained with indicated antibodies. γ-tubulin (gTub) marks active centrosomes. Only mitotic centrosomes were examined (boxed and magnified), and quantified (n>50, N=3, right). Error bars represent standard deviation.
(B) RPE1 cells in which CEP295 gene was mutated by CRISPR/Cas9 gene targeting were analyzed and stained with indicated antibodies. Control cells were nucleofected with Cas9 and empty gRNA vectors. Early prophase cells are shown. ~15% of cells were effectively targeted by CRISPR (not shown).
(C) Centriole duplication was analyzed in control or CEP295-RNAi cells in S phase 72hr after transfection, with indicated antibodies. BrdU was added 30 min before fixation. Unlike control cells carrying two centriole doublets (group I), majority of CEP295-RNAi cells contained only one centriole doublet, with or without an additional centrin singlet devoid of SAS-6 (group II & III).
(D) De novo centrioles were found in a small percentage of CEP295-depleted cells lacking centrosomes (group V), labeled with centrin and SAS-6, but not with γ-tubulin or C-Nap1.
(E) Quantification of centriole duplication (n>160, N=3). Four groups of duplication defects were seen in CEP295-depleted cells as indicated (II–V). Error bars represent standard deviation.
See also Figure S1.
Loss of CEP295 has no effect on initial centriole assembly but leads to centrosome reduction
Consistent with the previous report (Knorz et al., 2010), knockdown of CEP295 by siRNA quickly resulted in centrosome number reduction (Figure 2A). The same defect was also seen by CRISPR/Cas9-mediated gene targeting (Figure 2B) (Mali et al., 2013). Specifically, only one γ-tubulin focus was seen in most M-phase cells depleted of CEP295 (Figure 2A and 2B), suggesting a potential defect in centriole duplication. Surprisingly, however, a pair of centrioles (or a centrin doublet), instead of a singlet, was always observed within the remaining γ-tubulin focus (Figure 2A and 2B), indicating that while the number of centrosomes (γ-tubulin foci) reduces, loss of CEP295 does not seem to abolish centriole assembly. To more carefully examine centriole duplication, S-phase cells treated with control or CEP295 siRNA for 72 hours were examined for centrin, γ-tubulin, and the cartwheel protein SAS-6. Control S-phase cells uniformly carried 2 centriole doublets (or 2 duplicated centriole pairs) each of which was marked by γ-tubulin and SAS-6 (Figure 2C and 2E; group I). In contrast, most of CEP295-depleted cells either had 1 doublet and 1 singlet (group II), or 1 doublet alone (group III), with SAS-6 and γ-tubulin present only on the doublet (Figure 2C and 2E). Other centriolar proteins such as CPAP, CP110 and Centrobin also properly localize to these centriole doublets formed in the absence of CEP295 (Figure S1A–C). Moreover, electron microscopy analyses also reveal the duplication of centrioles in the absence of CEP295 (Figure S1D). Interestingly, in a small percentage of CEP295-depleted cells, no γ-tubulin foci could be detected (Figure 2E; group IV & V); instead, de novo centrioles appeared to form in some of these acentrosomal cells (Figure 2D and 2E; group V), as indicated by the presence of variable number of centrin singlets marked with SAS-6 (Figure 2D), but lacking C-Nap1 or PCM-associated γ-tubulin (Figure 2D). (Wang et al., 2011). Note that these de novo centrioles are distinct from the centriole singlet described above in the group II of Figure 2C & E, which was devoid of SAS-6 (Figure 2C). Together, these suggest that CEP295 is not critically required for initial centriole assembly per se. Instead, the reduction of the centrosome number throughout the cell cycle suggests a potential role of CEP295 in centriole-to-centrosome conversion.
CEP295 is required for centriole-to-centrosome conversion, but dispensable for cartwheel removal or centriole disengagement
To test if CEP295 depletion blocks centriole-to-centrosome conversion that normally occurs at the end of mitosis, G1 centrioles exiting from mitosis were examined for γ-tubulin or C-Nap1, two markers of MTOC-competent centrioles (Wang et al., 2011). Control G1 cells displayed two γ-tubulin or C-Nap1 foci, one at each centriole, indicating that both centrioles are MTOC competent (Figure 3A). In CEP295-depleted cells, however, only the mother centriole that had previously acquired the MTOC competency, as marked by Odf2, carried the γ-tubulin or C-Nap1 signal (Figure 3A), whereas the majority of daughter centrioles born in the previous S phase remained devoid of C-Nap1 or γ-tubulin (Figure 3A). These results indicate that CEP295 is essential for the conversion of newborn centrioles to centrosomes, but is dispensable for centrioles that have already converted, similar to the function of Plk1 (Wang et al., 2011). Interestingly, however, while centriole disengagement and cartwheel removal at mitosis both depend on Plk1 (Wang et al., 2011), they occur normally in the absence of CEP295, as revealed by the extensive separation of the mother and daughter centrioles, and the loss of the SAS-6 signal from the daughter centriole (Figure 3A). Together, we demonstrate that the conversion of centrioles to centrosomes is specifically mediated by CEP295, and can be uncoupled from cartwheel removal or centriole disengagement.
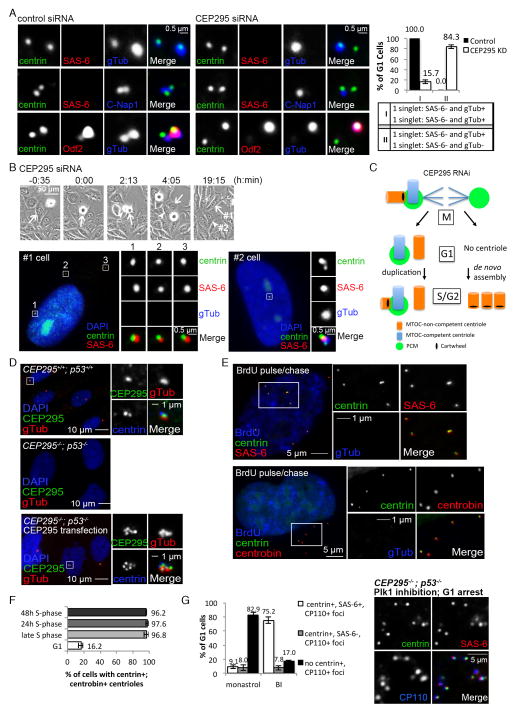
Figure 3. Loss of CEP295 blocks centriole-to-centrosome conversion, but has no effect on centriole disengagement and cartwheel removal.
(A) Control and CEP295 siRNA-treated cells were transiently labeled with BrdU before fixation. G1 cells, BrdU negative (not shown) and carrying two centrin singlets, were examined with indicated antibodies. Quantifications (n>100, N=3) are shown (right). Error bars represent standard deviation.
(B) Cells grown on a gridded coverslip were treated with CEP295 siRNA and filmed for 72 hours before fixation. A cell carrying de novo centrioles was identified (arrow; #1 cell), and its sister cell from the previous cell division was traced through the timelapse movie (arrowhead; #2 cell), and examined with indicated antibodies.
(C) Schematic summary of the phenotypes seen in CEP295-depleted cells.
(D) Unsynchronized wild-type or CEP295−/−; p53−/− RPE1 cells, and CEP295−/−; p53− cells transiently expressing full-length CEP295 were stained with indicated antibodies.
(E) Unsynchronized CEP295−/−; p53−/− cells pulsed with BrdU for 30 min and chased for 4 hours were stained with indicated antibodies.
(F) CEP295−/−; p53−/− cells in late S phase as described in (E), or arrested in S or G1 phase for 24–48 hours as indicated were examined with centrin and centrobin antibodies. Quantifications are shown (n>100, N=3).
(G) CEP295−/−; p53−/− cells were allowed to enter mitosis in the presence of the Plk1 inhibitor (BI-2536), or Eg5 inhibitor (monastrol) as a control, and release to and arrest at G1 by Cdk inhibition with Roscovitine for 16 hours before fixation and staining with indicated antibodies. Cells displaying donut-shaped, multilobed, or multiple small nuclei (Tsou et al., 2009) were identified for analyses. In Plk1-inhibited cells arrested in G1, centrioles were stably present, and retained the cartwheel and other markers, as shown in the panel on the right. Quantifications are shown (n>50, N=4).
We next explored the fate of CEP295-depleted cells entering mitosis with one centrosome (Figure 2A and 2B). If these cells manage to divide, daughter cells inheriting no centrosome/centriole would arise, which in turn may trigger de novo centriole assembly as described above (Figure 2D), and previously (Khodjakov et al., 2002). A correlated timelapse/immunofluorescence microscopy assay was performed to examine if de novo centrioles were indeed formed in this sequence of events. Cells grown on a gridded coverslip were depleted of CEP295 by RNAi and filmed for 72 hours before fixation. Cells carrying de novo centrioles were identified by immunofluorescence microscopy, and their sister cells were traced and located through the timelapse movie. In all 9 cells containing de novo centrioles whose sisters could be located, every sister was found to carry a duplicated centriole pair, i.e. a centrin doublet marked by SAS-6 (Figure 3B), with or without the extra centriole singlet (see below in Figure 4). This result supports the scenario (Figure 3C) that after cell division, one daughter cell inherits the only active centrosome in which the associated MTOC-competent centriole duplicates in the following S phase, whereas the sister cell that receives no centrosome undergoes de novo centriole assembly (Figure 3C).
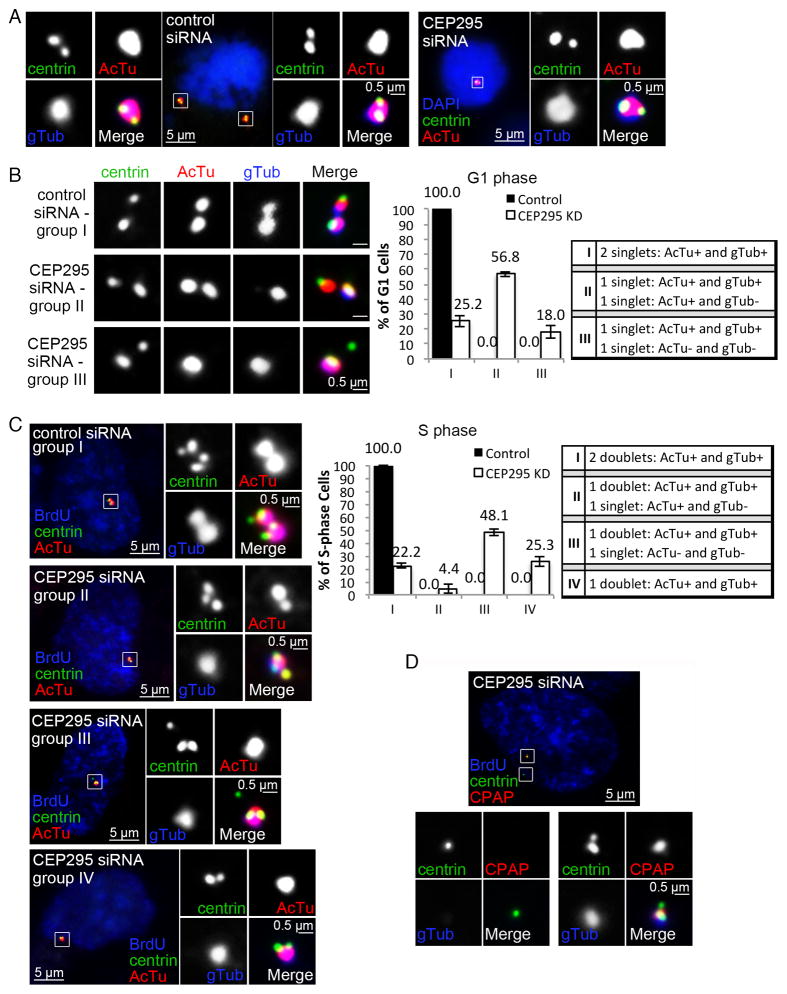
Figure 4. Centriole-to-centrosome conversion stabilizes centrioles devoid of the cartwheel.
(A) Mother centrioles and their engaged daughter centrioles in cells treated with control (left) or CEP295 siRNA (right) for 72 hours were examined at early mitosis with antibodies against centrin, γ-tubulin (gTub), and acetylated α-tubulin (AcTu).
(B) Cells treated with control or CEP295 siRNA for 72 hours were labeled with BrdU before fixation. G1 cells, BrdU negative (not shown) and inheriting a pair of disengaged centrioles, were examined with indicated antibodies. Quantifications are shown (right) (n>100, N=3). Error bars represent standard deviation. Note that in CEP295 siRNA-treated cells, majority of daughter centrioles failed to convert to centrosomes (group II & III; gTub-), some of which also lost the AcTu staining (group III) (gTub- & AcTu-).
(C) Cells were treated as described in (B). BrdU-positive cells inheriting active centrosomes (gTub foci) were examined with indicated antibodies. Cells inheriting no centrosomes were excluded. Quantifications are shown (right) (n>100, N=3). Note that in the majority of CEP295 RNAi cells, the inherited DCP/daughter centrioles either became non-detectable (group IV), or had lost the AcTu staining (group III) (gTub-; AcTu-).
(D) DCP centrioles in S-phase cells were examined for the localization of CPAP with indicated antibodies.
See also Figure S3.
To confirm that de novo centrioles form efficiently in the absence of CEP295, we generated a stable RPE1 cell line in which both CEP295 and p53 genes were mutated by CRISPR-mediated gene targeting (CEP295−/−; p53−/−), as cells devoid of centrosomes are not viable in the presence of p53 (Bazzi and Anderson, 2014). As expected, no centrosomes could be detected in proliferating CEP295−/−; p53−/− cells (Figure 3D; Figure S2), but strikingly, for cells that had entered S phase for at least 4 hours, nearly all (>90%, n>100) carried some numbers of de novo centrioles marked by centrin, SAS-6 and centrobin (Figure 3E), confirming that de novo centriole assembly initiates efficiently, but none are converted to centrosomes in the absence of CEP295. Note that the centrosome assembly defect in CEP295−/−; p53−/− cells can be rescued by putting back wild-type CEP295 (Figure 3D), confirming the specificity. Together, we conclude that while CEP295 is not required for initial centriole assembly, it enables centriole-dependent centriole duplication by converting centrioles to centrosomes.
Centriole-to-centrosome conversion stabilizes cartwheel-less centrioles
Vertebrate centrioles in cycling cells are associated with either the cartwheel or PCM, suggesting that neither plays a role in centriole stability, or alternatively, both elements function redundantly to stabilize centrioles. To differentiate between these two possibilities, centrioles devoid of both the cartwheel and PCM (or “DCP” centrioles for simplicity), which form at the end of mitosis in CEP295-depleted cells, were examined for their stability. We first examined if de novo centrioles formed in CEP295−/−; p53−/− cells are stable after mitosis (or cartwheel removal). Strikingly, while de novo centrioles were consistently present in nearly all late S-phase or prolonged S phase-arrested cells (Figure 3E, 3F, & Movie S1), they could not be detected in most post-mitotic cells arrested in G0/G1 (Figure 3F), revealing an intriguing phenotype that de novo centrioles are repeatedly formed (in S phase) and lost (after mitosis) every cell cycle in CEP295−/−; p53−/− cells. To test if cartwheel removal underlies the loss of these de novo centrioles, CEP295−/−; p53−/− cells were allowed to pass through mitosis in the absence of Plk1 activity, a treatment known to retain the cartwheel in centrioles (Tsou et al., 2009; Wang et al., 2011). Strikingly, in the presence of the cartwheel, de novo centrioles became stable in most of post-mitotic CEP295−/−; p53−/− cells arrested in G1 (Figure 3G).
To ensure that loss of stability is not just for de novo centrioles, we tracked the fate of the engaged, newly duplicated daughter centriole in CEP295-RNAi cells. In early mitosis, before being converted to DCP centrioles, newborn daughter centrioles devoid of CEP295 were positively stained for acetylated α-tubulin (100%, n=139) (Figure 4A, right), a marker for stabilized microtubules at the core of centrioles. Intriguingly, after mitosis, unlike converted mother centrioles that continued to be stable, DCP centrioles formed at the end of mitosis progressively lost centriolar components in the following interphase (Figure 4B and 4C). Specifically, in G1 phase, unlike control centrioles, which were all converted and associated with acetylated α-tubulin (Figure 4B; gTub+ & AcTu+ singlets in group I), DCP centrioles in 18% of RNAi-treated cells had lost the acetylated α-tubulin staining (Figure 4B; gTub− & AcTu− singlets in group III). More strikingly, by S phase, in contrast to converted centrioles seen both in control and CEP295-RNAi cells, which all stably duplicated (Figure 4C; doublets in group I–IV), DCP centriole singlets that remain positively stained for both centrin & acetylated α-tubulin were no longer seen in >90% of RNAi-affected cells (Figure 4C; group III & IV). Similarly, these DCP centrioles also lost the centriolar protein CPAP (Figure 4D) known to associate with centriolar microtubules (Hung et al., 2004). These results together indicate that while converted centrioles can exist stably in the absence of both the cartwheel and CEP295, unconverted centrioles formed in the absence of CEP295 become unstable after cartwheel removal.
Cartwheel is essential for the maintenance of newborn centrioles
To further test if the cartwheel is also essential for the stability of newborn centrioles formed in the presence of CEP295, we examined the effect of cartwheel removal in wild-type, S-phase cells in which centriole duplication had already occurred prior to SAS-6 depletion (Figure S3). In brief, cells transfected with control or SAS-6 siRNA were pulsed with BrdU at 32 hours after the transfection (32h post RNAi), followed immediately by S-phase arrest (Figure S3A). These BrdU labeled, S-phase arrested cells were then fixed at 36h, 48h, 60h, or 72h post RNAi, and examined for centrin, SAS-6, and C-Nap1. We found that at the 36h, 48h, and 60h after SAS-6 RNAi, majority of BrdU positive cells (~70%) still contained two duplicated centriole pairs (doublets), each of which is associated with SAS-6 (Figure S3B; group I, see quantifications). However, at the 72h time point, the same population of cells that still carried normal centriole pairs dropped abruptly, with a significant increase in the number of cells in which the SAS-6 signal (or cartwheel) was lost. More importantly, most of these cells losing the SAS-6 signal carried only two centriole singlets each of which was marked with C-Nap1 (group II; see Figure S3B quantifications), indicating that previously existed newborn centrioles are either detached, or become unstable upon SAS-6/cartwheel removal. Note that we also observed what appears to be an intermediate phenotype in a small percentage of cells (group IV), where newborn centrioles were seen nearby the mother centriole despite losing the SAS-6 signal (Figure S3B; group IV). To determine if the “lost” newborn centrioles were present somewhere in the cell, we examined centriole distal end markers centrin, CEP162, and CP110, which are known to associate with all centrioles in S phase (Figure S3C) (Tsang et al., 2008; Wang et al., 2013). Intriguingly, in contrast to mother centrioles that could be reliably detected, no detached newborn centrioles marked with centrin and CEP162 or CP110 signals could be found in the majority of SAS-6 depleted cells, as revealed by the maximum projection of a Z stack images (Figure S3C). Thus, the SAS-6 based cartwheel is essential for the proper maintenance of newborn centrioles, even when they are born with CEP295. These results together support the idea that vertebrate centrioles associate with either the cartwheel or PCM to maintain their stable composition, and that cartwheel removal can be detrimental if “cartwheel-less” centrioles are not converted to centrosomes.
DISCUSSION
Centriole to centrosome conversion at late mitosis, which promotes centriole to acquire the PCM, is essential for cycling cells to maintain centriole homeostasis, as both centriole duplication and segregation fully depend on it (Wang et al., 2011). Here we identify CEP295 as a newborn centriole-enriched factor mediating this process, and surprisingly discover that centriole-to-centrosome conversion also has an essential role in stabilizing cartwheel-less centrioles. In particular, we found that (i) when centrioles are devoid of both the PCM and cartwheel, they become unstable even in the presence of CEP295, and (ii) when centrioles are associated with either the PCM or cartwheel, they are perfectly stable even in the absence of CEP295. These results together suggest that cartwheel removal can be harmful to centrioles, and that centriole-to-centrosome conversion mediated by CEP295 is required to neutralize such negative impacts.
Our findings, however, raise a puzzling question as to why centrioles risk their stability to discard the cartwheel in the first place. The cartwheel is a geometric scaffold upon which all centrioles are assembled, but vertebrate centrioles naturally lose it before they support duplication in the next S phase. Both centriole disengagement and centriole-to-centrosome conversion occurring at the late mitosis are known to enable centriole duplication, raising a possibility that cartwheel removal may function in the same process. Our previous studies showed that unconverted (or unmodified) centrioles, which carry the cartwheel, cannot support duplication even when they are disengaged (Wang et al., 2011). This is consistent with the idea that perhaps cartwheel removal and centriole-to-centrosome conversion work together to enable newborn centrioles for duplication, whereas centriole disengagement merely “licenses” the re-duplication of mother centrioles that are otherwise competent to duplicate. Interestingly, recent studies showed that the empty lumen of the cartwheel-less centriole can function as a geometric template to shape SAS-6 assembly prior to centriole duplication (Fong et al., 2014), a process that may underlie the preservation of centriole number and structure in cycling cells. We thus speculate that loss of the cartwheel is specifically programmed in cycling cells to control centriole-dependent centriole duplication, while its repercussion, i.e. centriole destabilization, is shielded by centriole-to-centrosome conversion. More experiments are required to test these ideas.
EXPERIMENTAL PROCEDURES
CRISPR and p53−/−; CEP295−/− cell lines
RNA-guided targeting of CEP295 in human RPE1 cells was achieved through coexpression of the Cas9 protein with guide RNAs (gRNAs) as described by the Church group (Mali et al., 2013). RPE1 cells nucleofected with Cas9 plasmid and gRNA were examined for the loss of CEP295 at 5, 6, or 7 days after nucleofection. We consistently see loss of the CEP295 signal in ~15% of transfected cells. CEP295−/− cells can divide but eventually die/arrest depending on p53 (Bazzi and Anderson, 2014). To obtain stable cell lines lacking CEP295, the p53 gene (TP53) was mutated by the same CRISPR method prior to targeting of CEP295, generating p53−/−; CEP295−/− cells. CEP295−/−; p53−/− cells proliferate actively (Figure S2), although their mitosis are slightly lengthened due to lack of centrosomes.
Details of experimental procedures are provided in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank K. Rhee (Seoul National University, Korea) for antibodies; A. Hall and C. Haynes at MSKCC for comments on the manuscript. This work was supported by the National Institutes of Health grants GM088253 to M.-F. B. Tsou.
Footnotes
AUTHOR CONTRIBUTIONS
D.I. and M-F.B.T. designed experiments. D.I. performed most of the experiments. W-J.W. and K.U. performed EM analyses. W-J.W. helps to perform CRISPR/Cas9 gene targeting, and time-lapse fluorescence microscopy. D.I. and M-F.B.T. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson RG, Brenner RM. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. The Journal of cell biology. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arquint C, Nigg EA. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Current biology : CB. 2014;24:351–360. doi: 10.1016/j.cub.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. Journal of cell science. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1491–1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CS, Kim M, Yang TT, Liao JC, Tsou MF. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Developmental cell. 2014 doi: 10.1016/j.devcel.2014.05.008. http://dx.doi.org/10.1016/j.devcel.2014.1005.1008. [DOI] [PMC free article] [PubMed]
- Hung LY, Chen HL, Chang CW, Li BR, Tang TK. Identification of a novel microtubule-destabilizing motif in CPAP that binds to tubulin heterodimers and inhibits microtubule assembly. Molecular biology of the cell. 2004;15:2697–2706. doi: 10.1091/mbc.E04-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. The Journal of cell biology. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorz VJ, Spalluto C, Lessard M, Purvis TL, Adigun FF, Collin GB, Hanley NA, Wilson DI, Hearn T. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif-containing proteins C10orf90 and KIAA1731. Molecular biology of the cell. 2010;21:3617–3629. doi: 10.1091/mbc.E10-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Current biology : CB. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Developmental cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Developmental cell. 2010;19:913–919. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Developmental cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. The EMBO journal. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes & development. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Developmental cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Developmental cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. The ultrastructure of centriole in mammalian tissue culture cells. Cell Biol Int Rep. 1980;4:1037–1044. doi: 10.1016/0309-1651(80)90177-0. [DOI] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. The Journal of cell biology. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. The Journal of cell biology. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Tay HG, Soni R, Perumal GS, Goll MG, Macaluso FP, Asara JM, Amack JD, Bryan Tsou MF. CEP162 is an axoneme-recognition protein promoting ciliary transition zone assembly at the cilia base. Nature cell biology. 2013;15:591–601. doi: 10.1038/ncb2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.