Abstract
Study Objectives:
To examine association between periodic leg movements (PLM) and 13 single nucleotide polymorphisms (SNPs) in 6 loci known to increase risk of restless legs syndrome (RLS).
Setting:
Stanford Center for Sleep Sciences and Medicine and Clinical Research Unit of University of Wisconsin Institute for Clinical and Translational Research.
Patients:
Adult participants (n = 1,090, mean age = 59.7 years) from the Wisconsin Sleep Cohort (2,394 observations, 2000-2012).
Design and Interventions:
A previously validated automatic detector was used to measure PLMI. Thirteen SNPs within BTBD9, TOX3/BC034767, MEIS1 (2 unlinked loci), MAP2K5/SKOR1, and PTPRD were tested. Analyses were performed using a linear model and by PLM category using a 15 PLM/h cutoff. Statistical significance for loci was Bonferroni corrected for 6 loci (P < 8.3 × 10-3). RLS symptoms were categorized into four groups: likely, possible, no symptoms, and unknown based on a mailed survey response.
Measurements and Results:
Prevalence of PLMI ≥ 15 was 33%. Subjects with PLMs were older, more likely to be male, and had more frequent RLS symptoms, a shorter total sleep time, and higher wake after sleep onset. Strong associations were found at all loci except one. Highest associations for PLMI > 15/h were obtained using a multivariate model including age, sex, sleep disturbances, and the best SNPs for each loci, yielding the following odds ratios (OR) and P values: BTBD9 rs3923809(A) OR = 1.65, P = 1.5×10-8; TOX3/BC034767 rs3104788(T) OR = 1.35, P = 9.0 × 10-5; MEIS1 rs12469063(G) OR = 1.38, P = 2.0 × 10-4; MAP2K5/SKOR1 rs6494696(G) OR = 1.24, P = 1.3×10-2; and PTPRD(A) rs1975197 OR = 1.31, P = 6.3×10-3. Linear regression models also revealed significant PLM effects for BTBD9, TOX3/BC034767, and MEIS1. Co-varying for RLS symptoms only modestly reduced the genetic associations.
Conclusions:
Single nucleotide polymorphisms demonstrated to increase risk of RLS are strongly linked to increased PLM as well, although some loci may have more effects on one versus the other phenotype.
Citation:
Moore H, Winkelmann J, Lin L, Finn L, Peppard P, Mignot E. Periodic leg movements during sleep are associated with polymorphisms in BTBD9, TOX3/BC034767, MEIS1, MAP2K5/SKOR1, and PTPRD. SLEEP 2014;37(9):1535-1542.
Keywords: periodic leg movements, restless legs syndrome, polysomnography, Wisconsin Sleep Cohort, genetics
INTRODUCTION
Periodic limb movements (PLMs) are episodic, involuntary muscle contractions that occur during sleep, most notably NREM sleep. Restless legs syndrome (RLS) is often associated with PLMs, with four out of five patients diagnosed with RLS exhibiting PLMs in one study (≥ 5/h)1 and 85% to 95% in others.2,3 PLM can occur during sleep (PLMS) or wake (PLMW), and much research has been conducted to examine and understand the physiology of PLM in RLS (e.g., circadian effect, significance of PLMW, higher periodicity index).1,4–7 However, PLMs can also occur without RLS symptoms, though little is known about the cause of PLMs or their impact on daytime sleepiness or insomnia symptoms in the absence of RLS. A study review of cardiac risk for RLS and PLMS found associations between PLMS and congestive heart failure.8 Additionally, patients with RLS were at higher risk for heart disease and hypertension.8 While this association is controversial, another recent prospective study found increased mortality in men with RLS.9 Further, PLMs have been associated with increased risk of atrial fibrillation10 and coronary heart disease.11 PLM are known to be associated with several other disorders and pathologies such as depression, cardiovascular disease, REM behavior disorder, narcolepsy, Parkinson disease, and multiple system atrophy.12–15 Patients with REM behavior disorder and narcolepsy have more PLMs occurring in REM.16,17
Recent investigations have revealed strong genetic components for RLS and PLMI. Most notably, single nucleotide polymorphisms (SNPs) were identified as RLS susceptibility markers in a genome-wide association study (GWAS) described in 2011 (with related work shown earlier19–22), identifying nine genome wide significant loci with best effects at rs2300478 in MEIS1; rs3923809 in BTBD9; rs1975197 in PTPRD; rs12593813 in MAP2K5/SKOR1; rs6747972 in intergenic region on chromo-some 2 (presumably another distant regulator of MEIS1); and rs3104767 in TOX3/BC034767.18 Association of the BTBD9 and MEIS1 genes with RLS and end-stage renal disease was reported in a German study (200 RLS cases, 443 controls) with replication in a Greek sample (141 RLS cases, 393 controls).23 Another team, using an Icelandic discovery sample with replications in Icelandic and U.S. samples found association of BTBD9 in PLM in sleep (PLMS)22 apart from RLS diagnosis as adopted from a 1995 criteria.24 PLMS were measured using a small tri-axial accelerometer worn on the ankle that could not distinguish sleep from wake. PLMS were classified whenever the ambulatory device detected five or more PLM during an hour while participants were recumbent during their major rest period.22 These studies did not use PSG measures of PLM or account for additional effects on PLMS, such as use of medication that can aggravate or inhibit factors contributing to RLS.
In this study, we report on the genetic association of PLM with and without the presence of RLS symptoms from DNA samples and automatically obtained PLM metrics from the Wisconsin Sleep Cohort, an epidemiological sample totaling 1,086 subjects and 2,394 nocturnal polysomnography (PSG) based sleep studies conducted between 2000 and 2012.
METHODS
Cohort Used in the Analysis
A total of 2,394 nocturnal PSG (NPSG) studies from 1,086 Wisconsin Sleep Cohort (WSC) participants were used in this study. Electroencephalography (EEG), electrooculography (EOG), and chin electromyography (EMG) were used to score sleep stages for each 30-s epoch using standard Rechtschaffen and Kales criteria.25
The WSC is a longitudinal study of sleep habits and disorders in the general population that was established in 1988 from a sample of employees of 4 state agencies in south central Wisconsin, USA, aged 30-60 years.26 In 2000, WSC began digitally storing PSGs, thus the study dates for this paper are from 2000 and 2012. Participants in this time line without ≥ 2 h scored sleep were excluded. NPSGs were exported as EDF files and paired with scoring files of stages and events (e.g., apneas and hypopneas). PLM and SDB were characterized using a 16-channel PSG recording system (16-channel Grass-Telefactor Heritage digital sleep system Model 15). Arterial oxyhemoglobin saturation was measured by pulse oximetry using a 3-s averaging rate. Oral and nasal airflow were measured using thermocouples (ProTech). Nasal air pressure was measured with a pressure transducer (Validyne, Northridge). Thoracic cage and abdominal respiratory motion were measured with inductance plethysmography (Respitrace, Ambulatory Monitoring). These signals were used to identify SDB events. Apnea was defined as a cessation of airflow lasting ≥ 10 sec. Hypopnea was defined as a decrease in airflow accompanied by ≥ 4% reduction in oxyhemoglobin saturation and is close to the 2007 AASM recommended (Medicare) criteria for scoring hypopneas.27
A single-leg EMG channel, combined from the left and right anterior tibialis (LAT/RAT) EMGs, was used to determine significant leg movements according to the 2007 AASM Manual for Scoring Sleep (AASM Manual) with slight modifications. The AASM Manual defines a significant leg movement (LM) as a period of 0.5-10 s where LAT/RAT EMG activity exceeds 8 μV above baseline and then falls below 2 μV from baseline for 0.5 sec or longer.28 PLM are further defined as the consecutive sequence of ≥ 4 LMs whose inter-movement intervals are between 5 and 90 sec. This definition of PLM is consistent with the clinical guidelines set for by the World Association of Sleep Medicine in 2006 and the American Sleep Disorders Association in 1993.29,30
The AASM Manual does not allow LMs occurring 0.5 s before, during, or 0.5 s after a respiratory event to be included in PLM.28 Our PLM classifier modifies these rules to only exclude LM detected at the edges of respiratory events. Any LM occurring 5 s prior to the onset of a respiratory event until 0.5 sec after its onset are excluded from PLM, as are any LM occurring 0.5 s prior to offset of a respiratory event through 5.0 s after offset. This adjustment is based on empirical analysis of time-locked respiratory events and leg EMG activity in patients stratified by an apnea-hypopnea index (AHI) of 15. The analysis defined AHI as the average number of apneas and hypopneas per hour of objectively measured sleep.31
Data pertaining to the intake of drugs known to modulate RLS symptoms were also collected, and variable created if the subject took any of these drugs. RLS symptoms aggravators included antidepressants (e.g., selective serotonin reuptake inhibitors, tricyclics), antipsychotics, and antihistamines. RLS inhibitors include benzodiazepines, opiates, antiseizure medication, and medication for Parkinson disease.
RLS Symptoms
WSC participants were stratified according to RLS symptoms based on questionnaire responses from a previous study of the cohort in 2003.32 Patients were asked to provide the frequency with which they felt (a) repeated urge to move legs, (b) strange and uncomfortable feelings in the legs, (c) periods of several leg jumps or jerks. The choice of frequency included never, less than once a month, monthly, weekly, and nightly. Two further questions were asked to describe responses other than “never”: (d) Do these feelings just mentioned get better when you get up and start walking? (e) Do these feelings just mentioned disrupt your sleep? The questionnaire did not completely address all RLS diagnostic criteria put forth by the National Institutes of Health (NIH)31; notably it did not ask for the symptoms to be worse at night. As the circadian component of RLS is essential to the diagnosis, the term “RLS symptoms” is used rather than RLS.
RLS symptoms were split into 4 categories (Table S1, supplemental material). Category A (n = 186, observations = 360), likely RLS symptoms, was defined as response to (a) as weekly or more often, (d) yes, and (e) yes. The term RLS(A) is used to refer to this category. Category B (n = 190, observations = 384), possible RLS symptoms, was defined as response (a) monthly or more frequent, and (d) yes. Category B did not include members already in Category A. The term RLS(B) is used to refer to this category. Category C (n = 523, observations = 997), no RLS symptoms, was defined by response (a) as less than monthly and response (b) as either missing or less than monthly. The term RLS(C) is used to refer to this category. Remaining subjects (n = 171, observations = 321) were excluded from RLS symptom stratification (i.e., missing responses or responses that did not fit into categories A, B, or C).
RLS symptom categories were used as a single ordinal variable. As few differences were found between categories A and B, notably in terms of PLMI variable and association (Table S1), RLS A and B categories were most often combined in all analysis, although the effect of category A alone was also systematically studied. The term RLS(AB) is used to refer to the combination of these two categories.
PLM Phenotype
PSGs were processed for PLM using our described Stanford PLM automatic detector (S-PLMAD)31 that uses manually scored staging and respiratory event files in determining PLMI. The PLM detector was implemented in the SEV, a MATLAB toolbox for automating pattern recognition and biomarker classification algorithms in NPSG sleep cohorts.31
We define PLMI as the average number of PLMs occurring per hour of sleep and intermittent wake (i.e. wake after sleep onset [WASO]). PLMI is calculated automatically using a previously designed detection algorithm that was validated against manually scored data from 2 cohorts,31 and which is briefly described here. First, the algorithm removes cardiac interference from the EMG channel using an adaptive filter that reduces false positive detections. Next, a two-pass noise floor calculation is performed and variable amplitude thresholds generated to account for changes in the baseline noise. Candidate LMs are identified where the filtered signal (i.e., adaptively cleaned and root-mean-square filtered) meets the amplitude and duration which fall in line with AASM Manual criteria for significant LM as described in the original manuscript.31 LMs whose area under the curve is too small or that fall within the respiratory exclusion window of respiratory events described above are removed from candidacy. Remaining LMs are deemed significant and scored for PLM using AASM Manual criteria and inclusion of WASO.
A PLMI cutoff of 15 was used to distinguish between presence (PLMI ≥ 15) and absence (PLMI < 15) of the PLM phenotype (i.e., PLM+ or PLM−). Tables S1 and S2 (supplemental material) show the high association of PLMI with the presence of RLS symptoms, confirming the validity of our detector and the association of these phenotypes. The cutoff of 15 was used because OR were similar (∼2.0) across PLMI indices cutoffs above 10, 15, and 30, and because population frequency was approximately 33%, a reasonable portion of the population. Further, a PLMI cutoff of 15 is frequently used clinically in older individuals. A cutoff of ≥ 5 would have meant including more than 50% of the population as affected. Results with this cutoff did not change substantially the results, but led to lower ORs.
Table 1 describes the WSC stratified by PLMI ≥ 15 and shows several additional PLM metrics that were automatically obtained using this algorithm in order to determine a suitable PLM phenotype. The periodicity index is the number of inter-PLM-movement intervals > 10 s divided by the total number of inter-movement intervals (i.e. the total LM count less one). It is based on the observation that RLS patients exhibit longer inter-movement intervals than non-RLS patients.1 The PLM night ratio quantifies the circadian or diurnal effect observed in PLM26,27 and is presented here as the PLM count from the first half of the PSG sleep study divided by the PLM count measured in the second half. The PLM ratio uses sleep onset to define a study's start, and total sleep time plus WASO to determine the study midpoint. PLM attrition is the per hour change in PLM as determined by linear regression.
Table 1.
Characteristics of the Wisconsin Sleep Cohort observations stratified by the presence of PLM.
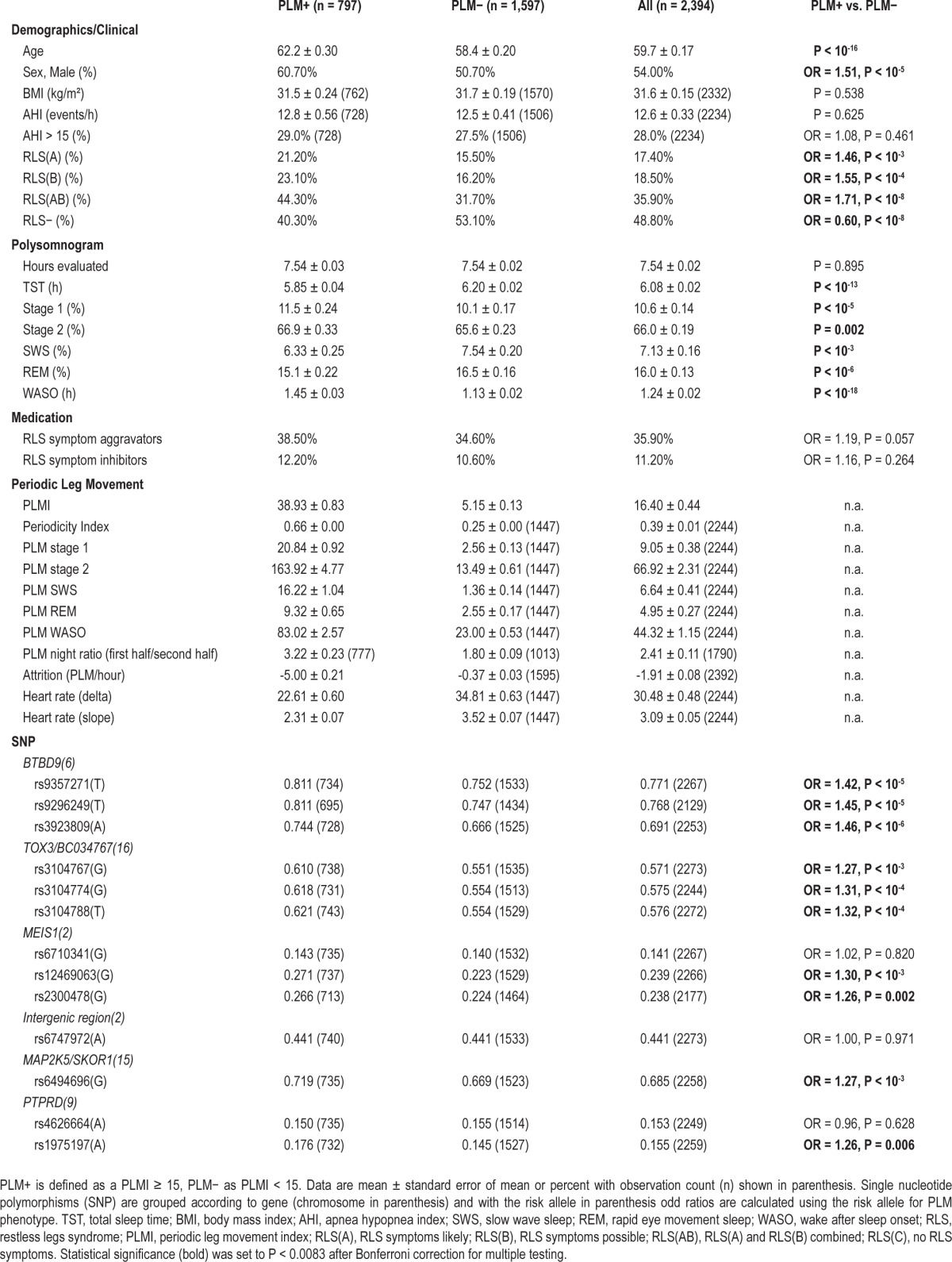
Genotypes
Thirteen SNPs from 6 genes are examined. SNPs from BTBD9 on chromosome 6 include rs935271, rs9296249, and rs392809. TOX3/BC034767 (chromosome 16) SNPs include rs3104767, rs3104774, and rs3104788. Two genes from chromosome 2 are examined: MEIS1 (rs6710341, rs12469063, rs2300478), and “no gene” (rs6747972, a SNP located a megabase away from MEIS1 but involved in its regulation). Also examined are chromosome 15 gene MAP2K5/SKOR1 (rs649469) and chromosome 9 gene PTPRD (rs4626664 and rs1975197). All 13 SNPs have previously been shown to increase susceptibility for RLS,18 while BTBD9 SNP rs392809 has been shown to increase susceptibility for PLM apart from RLS.22 Genotyping was performed on the MassARRAY system using MALDI-TOF mass spectrometry with the iPLEX Gold chemistry (Sequenom Inc, San Diego, CA, USA).
Analysis
PLM observations (more versus less than 15 PLM/h [PLM+ versus PLM−]) were first compared using t-test and χ2 for basic demographic variables and other variables that could influence the phenotype (Table 1). These basic comparisons were used for descriptive purposes and to identify variables to potentially control for in subsequent models. Table 1 also compares allele frequencies by group and allelic OR derived for each SNP.
Following these basic comparisons, allelic ORs of each SNP were calculated using logistic regressions on repeated measure observations with allele dose of the RLS risk allele as a linear variable, and controlling for covariates identified in Table 1. This was done in 2 adjusted models predicting PLM, with and without additional adjustment of RLS symptoms (Table 2). The models are solved with generalized estimating equations (GEEs) using a Markov correlation assumption for repeated measures as implemented in GEEQBOX, a MATLAB toolbox.33 To account for multiple observations, we used the Markov structure, a generalization of the first-order autoregressive that assumes the correlation between repeated measures is based on the timing between those measures (which can be unequal in the case of the WSC). Confidence intervals and P-values are determined using a robust estimate of the covariance matrix (i.e., sandwich method). Of notes, these results were similar using a model-based estimate, further confirming our choice of this correlation structure.
Table 2.
Associations of various SNPs with PLMs (PLMI ≥ 15 versus PLMI < 15)
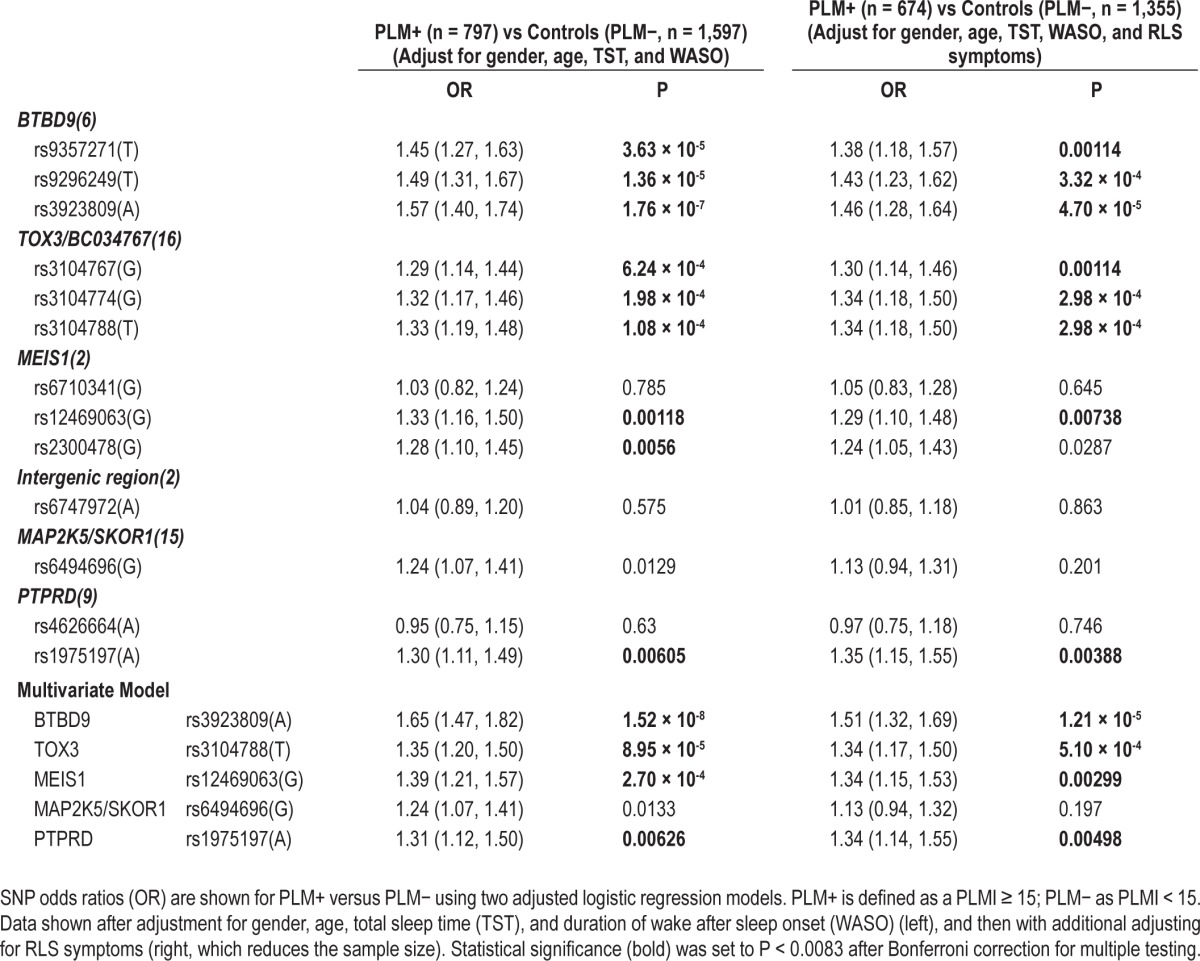
Finally, a linear trend test of each SNP on PLMI in repeated observations was done by linear regression and selected covariates, including RLS symptoms (ordinal categories, or considering likely RLS or likely and possible RLS as positive for RLS symptoms).
RESULTS
Prevalence and Associations of PLM in the Wisconsin Sleep Cohort
Prevalence of PLMI ≥ 15/h was 33% (Table 1). As expected, subjects with PLM were significantly older (about 4 years as a mean). They were also more frequently male (OR = 1.5) and significantly reported RLS symptoms—OR = 1.46 to 1.71, P < 10-8 for RLS(AB) versus RLS(C)—more frequently. Finally, we found that these subjects had a shorter total sleep time (TST) and higher wake after sleep onset (WASO) (P < 10-13 and 10-18, respectively), possibly reflecting disturbed sleep.
Unadjusted SNP Associations with PLM
PLM+ versus PLM− revealed association for almost all SNPs (Table 1): rs9357271(T), rs9296249(T), rs3923809(A) for BTBD9 (OR = 1.42-1.46, strongest for rs3923809); rs3104767(G), rs3104774(G), rs3104788(T) for TOX3/BC034767 (OR = 1.27-1.32, strongest for rs3104788); rs12469063(G), and rs2300478(G) for MEIS1 (OR = 1.25-1.30, strongest for rs12469063 but more significant for rs2300478); rs6494696(G) for MAP2K5/SKOR1 (OR = 1.27) and rs1975197(A) for PTPRD (OR = 1.26). The SNP in the intergenic region of Chromosome 2 known to regulate MEIS1 was not significantly associated. The top association and allelic directions revealed here with rs3923809(A) in BTBD9; rs3104788(T) in TOX3/BC034767; rs2300478(G) in MEIS1; and rs1975197(A) in PTPRD are all in the same direction as those associated with these loci in RLS.18 Regarding MAP2K5/SKOR1, the highest reported SNP in the Winkelmann study,18 rs12593813(G) was not tested, but we found a similarly high association with rs6494696(G) a SNP with almost complete linkage disequilibrium (LD) with it across ethnic groups (r2 = 0.91).
SNP Associations with PLM Adjusted for Age, Sex, and Sleep Disturbances
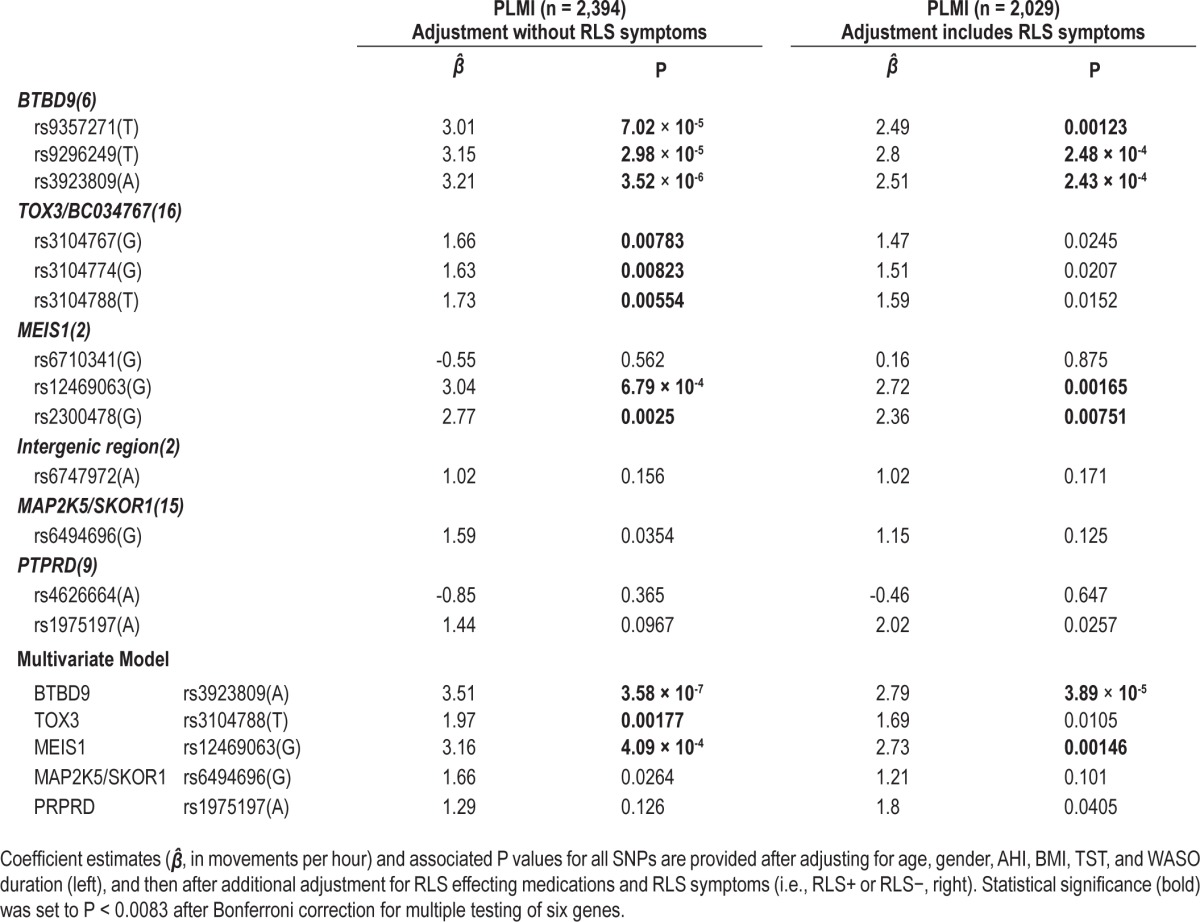
Categorical PLM associations with the various SNPs had similar effect sizes and P values to unadjusted models (Table 2). Association was most remarkable at rs39238809(A) when adjusted for age, sex, TST, and WASO (Table 2). In multivariate analysis where all significant SNPs (one per locus) except rs6747972(A) (no gene, a region presumably regulating MEIS1 but never significant in any of our models) were added in addition to age, sex, TST, and WASO, significance was improved in most cases, although rs6747972(A) remained nonsignificant (Table 2). Finally, the effects of each SNP (as a linear dose variable) on PLM index were tested using a linear regression models with adjustment of covariates with very similar effects (Table 3).
Table 3.
Associations of various SNPs with PLMs using PLMI as a linear outcome
SNP Associations with RLS Symptoms Adjusted for Covariates
SNP associations with RLS were next tested after adjustment of covariates detected in Table S1, with and without further adjustment with PLM (Table S2). The presence of RLS symptoms in these models was tested using multiple definitions for RLS, including RLS(A) and RLS(AB) versus RLS(C). Surprisingly, none of the SNPs were found to be associated with RLS symptoms in our cohort (Table S2A and S2B).
SNP Associations with PLM after Adjustment for the Presence of RLS Symptoms
Significant SNP associations with PLM were next tested after adjustment for RLS symptoms (Tables 2 and 3). The presence of RLS in these models was tested using multiple definitions for RLS, including RLS(A) and RLS(AB), and using RLS symptom categories as an ordinal variable, as presented in Tables 2 and 3. In all cases, the presence of RLS in the model only modestly reduced strong SNP associations with PLMs.
The sample was next stratified by PLM and RLS status (Table S3, supplemental material). Subjects reporting no RLS symptoms and with a low PLM index ≤ 15/h (PLM−) constituted the reference group. Subjects with possible RLS symptoms and PLM index > 15/h (PLM+), subjects with possible RLS symptoms and PLM−, and subjects with PLM+ and possible RLS were compared to this group, with adjustment of covariates. The same risk alleles previously reported for RLS and found to be associated with PLM in Tables 1–3 were used here. Similar models were also run with RLS(A) versus RLS(C), with similar results, although power was decreased due to smaller sample size.
As expected, subjects with both RLS and PLM showed stronger association with the BTBD9 locus, and similar associations with TOX3/BC034767 (Table S3). As these two loci together with MEIS1 are the most strongly associated in this study (see Tables 2–4), the result was expected. In subjects with PLM but no RLS symptoms, associations with BTBD9 and TOX3/BC034767 were present but with a reduced effect. Surprisingly however, MEIS1 SNPs were more associated in subjects with PLM without RLS symptoms, suggesting this locus may regulate PLM more strongly than RLS symptoms. Another unusual observation concerned the TOX3/BCO34767 locus, where PLM-associated SNP alleles were weakly protective in subjects with RLS and no PLM, an unusual combination that many researchers may consider a misdiagnosis.
Table 4.
Comparative effects of loci on PLM (this study) and RLS (from Winkelmann et al., 2011)
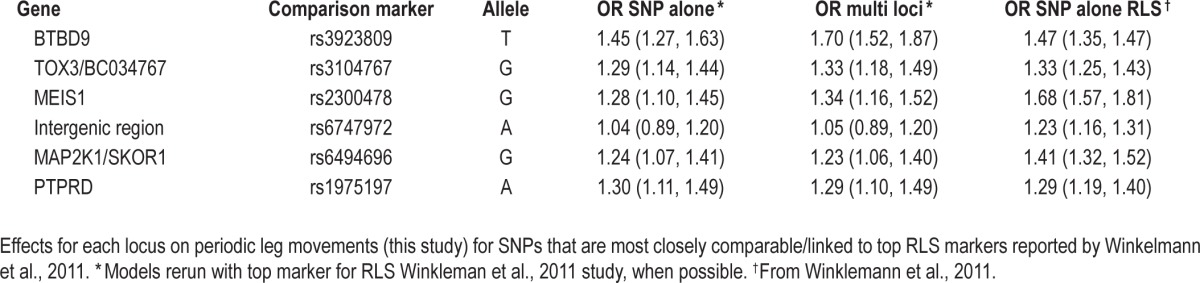
Comparison of the Effects of These Loci on PLMs (This Study) versus RLS (Prior Studies)
Table 4 contrasts the effects of the SNPs most associated with RLS in recent studies18,21 with corresponding data obtained for PLM as typed in this study. For better comparison, models obtained in prior tables for PLM most associated SNPs were recalculated using the previously reported top RLS markers rather than our top PLM markers, if applicable. As these SNPs are identical or in high LD (r2 > 0.9) with each other however, OR reported for each locus are essentially identical to those reported in prior tables and all these can be considered as proxies of each other.
OR reported for RLS are derived from Winkelmann et al. (2007)21 for rs3923809 in BTBD9 and from Winkelmann et al. (2011)18 for other loci, namely rs3104767 for TOX3/BC034767, rs2300478 for MEIS1, rs6747972 for the intergenic region suggested to regulate MEIS1, rs6494696 for MAP2K1/SKOR1, and rs1975197 for PTPRD. The rationale for using rs3923809 data from the 2007 study21 rather than the 2011 bigger sample study18 is that it is known rs3923809 is the most associated BTBD9 marker,21 and only rs9357271, a marker partially associated with rs3923809 (r2 = 0.429) was typed in the larger 2011 sample.18
As can be seen effects of rs9357271 are similar for PLM and RLS, but the effect of our top SNP (rs3923809) is stronger for PLM. Ranking the effects of the same SNPs on RLS and PLM, the effects of the various loci on RLS were: MEIS1 > BTBD9 ≥ MAP2K1/SCOR1 > TOX3/ BC034767 ≥ PTPRD and intergenic region, while ranking for PLM was BTBD9 ≥ TOX3/BC034767 and MEIS1 > PTPRD and MAP2K1/SCOR1.
DISCUSSION
In this study, we found that all loci known to be associated with RLS in prior studies were also associated with PLM except for rs6747972, an SNP located in an intergenic region distal of MEIS1 that has been suggested to modulate this gene. The stronger association with PLM was found with BTBD9 [rs3923809(A) OR = 1.57 in the single SNP model and 1.65 in the multi SNP model]. A similar association with PLM (PLMS > 5/h) was reported in Stefansson et al.22 for rs3923809(A), a marker also associated with RLS. In addition to this observation, we also found that MEIS1 [rs12469063(G) OR = 1.33 and 1.39], TOX3/BC034767 [rs3104788(T) OR = 1.33 and 1.35], PTPRD [rs1975197(A) OR = 1.30 and 1.31] and MAP2K1/SKOR1 [rs6494696(G) OR = 1.24] loci were also associated with PLM. Among the markers tested for these loci, the most associated markers were similar to those reported for RLS, except for MEIS1 (Table 4). At this locus, rs12469063 was slightly more associated with PLM than rs2300478. As the two markers are in high linkage disequilibrium (r2 = 0.93), however, this is likely chance variation. These results strongly suggest that RLS and PLM are part of the same disease continuum, with many more patients having PLMs and no significant RLS.
In contrast to these results, none of the 13 SNPs were found to be associated with “RLS symptoms” in the WSC, whether defined narrowly or more broadly using our questionnaire (Tables S1–S3), a result we believe is due to the poor ascertainment of RLS in this cohort. Indeed, RLS was poorly defined in this study, as we did not ask whether symptoms were worse in the evening, an important criterion of all classifications. The very high percentage of subjects reporting RLS symptoms (21% for likely and 41% for possible) and the lack of difference across gender of prevalence also argue in favor of a poorly defined phenotype, even in this older cohort expected to have higher prevalence of RLS. This problem notwithstanding, we believe that subjects reporting no symptoms are likely to be genuinely RLS free, and thus worth investigation. Further, RLS symptoms in this study were strongly associated with PLM, suggesting some validity (Table S1).
Interestingly, the ranking of these loci in term of effects on PLMs versus previously published effects on RLS somewhat differed, even when the same markers or proxies were used (Table 4), although this comparison suffers from the fact populations are different. This issue notwithstanding, MEIS1 and MAP2K1/SKOR1, for example, may have stronger effects on RLS than other loci, whereas BTBD9, TOX3/BC034767, and PTRD effects on RLS and PLM were similar. In our own data, after stratification of subjects by PLM and RLS symptom categories, we found that in most cases effects were stronger in subjects with PLM plus RLS symptoms, although some effect remained in subjects with PLM without RLS reports (Table S3). An outlier, however, was MEIS1, which surprisingly and opposite to previously published papers, had stronger effects of PLM without RLS. Overall, these results suggest that some loci may have stronger or equivalent effects on sensory versus motor component of the PLM-RLS disease spectrum.
In conclusion, we found that PLMs are strongly associated with loci known to be associated with RLS. Further fine mapping studies and genetic studies in subjects with PLM alone, or of sensory versus motor complaints in RLS patients, may reveal differential effects of these various loci.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Moore's work was supported by the Veteran Affair's Post 9/11 GI Bill. There was also support through National Institutes of Health grants NS23724, R01HL62252, and 1UL1RR025011. Dr. Winkel-mann has filed a patent application in relation to Nature Genetics (see reference 21), and has been a consultant and speaker on scientific meetings for UCB and Vifor. The authors have indicated no financial conflicts of interest. The work was performed at the Center for Sleep Sciences and Medicine, Stanford University School of Medicine, Palo Alto, CA.
ACKNOWLEDGMENTS
The authors thank all participants of the Wisconsin Sleep Cohort, the team who scored the data over the years, and three staff members in particular without whose care and effort in handling the PSG data and answering innumerous questions this project would not have been possible: Oscar Carrillo, Robin Stubbs, and Amanda Rasmuson.
SUPPLEMENTAL MATERIAL
Description of WSC participants stratified by RLS symptom category
SNP odds ratios (OR) for RLS(A) versus RLS(C) and for PLM+ versus PLM− with two model adjustments each.
SNP odds ratios (OR) are shown for RLS(AB) versus RLS(C) using two adjusted models each.
Effects of SNPs on PLM stratified by RLS status
REFERENCES
- 1.Ferri R, Zucconi M, Manconi M, Plazzi G, Bruni O, Ferini-Strambi L. New approaches to the study of periodic leg movements during sleep in restless legs syndrome. Sleep. 2006;29:759–69. [PubMed] [Google Scholar]
- 2.Rye DB, Trotti LM. Restless legs syndrome and periodic leg movements of sleep. Neurol Clin. 2012;30:1137–66. doi: 10.1016/j.ncl.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Kryger MH, Roth T, Dement WC. Principles and practice of sleep medicine. 5th ed. Philadelphia, PA: Saunders/Elsevier; 2011. [Google Scholar]
- 4.Hening WA, Walters AS, Wagner M, et al. Circadian rhythm of motor restlessness and sensory symptoms in the idiopathic restless legs syndrome. Sleep. 1999;22:901–12. doi: 10.1093/sleep/22.7.901. [DOI] [PubMed] [Google Scholar]
- 5.Trenkwalder C, Hening WA, Walters AS, Campbell SS, Rahman K, Chokroverty S. Circadian rhythm of periodic limb movements and sensory symptoms of restless legs syndrome. Mov Disord. 1999;14:102–10. doi: 10.1002/1531-8257(199901)14:1<102::aid-mds1017>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 6.Boehm G, Wetter TC, Trenkwalder C. Periodic leg movements in RLS patients as compared to controls: Are there differences beyond the PLM index? Sleep Med. 2009;10:566–71. doi: 10.1016/j.sleep.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Earley CJ, Allen RP, Hening W. Restless legs syndrome and periodic leg movements in sleep. Handb Clin Neurol. 2011;99:913–48. doi: 10.1016/B978-0-444-52007-4.00015-1. [DOI] [PubMed] [Google Scholar]
- 8.Walters AS, Rye DB. Review of the relationship of restless legs syndrome and periodic limb movements in sleep to hypertension, heart disease, and stroke. Sleep. 2009;32:589–97. doi: 10.1093/sleep/32.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Wang W, Winkelman JW, Malhotra A, Ma J, Gao X. Prospective study of restless legs syndrome and mortality among men. Neurology. 2013;81:52–9. doi: 10.1212/WNL.0b013e318297eee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirza M, Shen WK, Sofi A, et al. Frequent periodic leg movement during sleep is an unrecognized risk factor for progression of atrial fibrillation. PLoS One. 2013;8:e78359. doi: 10.1371/journal.pone.0078359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo BB, Blackwell T, Ancoli-Israel S, Stone KL, Stefanick ML, Redline S. Association of incident cardiovascular disease with periodic limb movements during sleep in older men: outcomes of sleep disorders in older men (MrOS) study. Circulation. 2011;124:1223–31. doi: 10.1161/CIRCULATIONAHA.111.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boivin DB, Montplaisir J, Poirier G. The effects of L-dopa on periodic leg movements and sleep organization in narcolepsy. Clin Neuropharmacol. 1989;12:339–45. doi: 10.1097/00002826-198908000-00012. [DOI] [PubMed] [Google Scholar]
- 13.Lapierre O, Montplaisir J. Polysomnographic features of REM sleep behavior disorder: development of a scoring method. Neurology. 1992;42:1371–4. doi: 10.1212/wnl.42.7.1371. [DOI] [PubMed] [Google Scholar]
- 14.Ancoli-Israel S, Kripke DF, Mason W, Kaplan OJ. Sleep apnea and periodic movements in an aging sample. J Gerontol. 1985;40:419–25. doi: 10.1093/geronj/40.4.419. [DOI] [PubMed] [Google Scholar]
- 15.Wetter TC, Collado-Seidel V, Pollmacher T, Yassouridis A, Trenkwalder C. Sleep and periodic leg movement patterns in drug-free patients with Parkinson's disease and multiple system atrophy. Sleep. 2000;23:361–7. [PubMed] [Google Scholar]
- 16.Godbout R, Montplaisir J, Poirier G, Bedard M. Distinctive electrographic manifestations of periodic leg movements during sleep in narcoleptic vs insomniac patients. Sleep Res. 1988;17:182. [Google Scholar]
- 17.Fantini ML, Michaud M, Gosselin N, Lavigne G, Montplaisir J. Periodic leg movements in REM sleep behavior disorder and related autonomic and EEG activation. Neurology. 2002;59:1889–94. doi: 10.1212/01.wnl.0000038348.94399.f6. [DOI] [PubMed] [Google Scholar]
- 18.Winkelmann J, Czamara D, Schormair B, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winkelmann J, Muller-Myhsok B. Genetics of restless legs syndrome: a burning urge to move. Neurology. 2008;70:664–5. doi: 10.1212/01.wnl.0000302178.53759.92. [DOI] [PubMed] [Google Scholar]
- 20.Winkelmann J, Lichtner P, Schormair B, et al. Variants in the neuronal nitric oxide synthase (nNOS, NOS1) gene are associated with restless legs syndrome. Mov Disord. 2008;23:350–8. doi: 10.1002/mds.21647. [DOI] [PubMed] [Google Scholar]
- 21.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 22.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–47. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 23.Schormair B, Plag J, Kaffe M, et al. MEIS1 and BTBD9: genetic association with restless leg syndrome in end stage renal disease. J Med Genet. 2011;48:462–6. doi: 10.1136/jmg.2010.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walters AS. Toward a better definition of the restless legs syndrome. The International Restless Legs Syndrome Study Group. Mov Disord. 1995;10:634–42. doi: 10.1002/mds.870100517. [DOI] [PubMed] [Google Scholar]
- 25.Rechtschaffen A, Kales A. Los Angeles: Brain Information Service/Brain Research Institute; 1968. A manual of standardized terminology, techniques and scoring system of sleep stages in human subjects. [Google Scholar]
- 26.Young T, Palta M, Dempsey J, Skatrud J, Weber S, Badr S. The occurrence of sleep-disordered breathing among middle-aged adults. N Engl J Med. 1993;328:1230–5. doi: 10.1056/NEJM199304293281704. [DOI] [PubMed] [Google Scholar]
- 27.Ruehland WR, Rochford PD, O'Donoghue FJ, Pierce RJ, Singh P, Thornton AT. The new AASM criteria for scoring hypopneas: impact on the apnea hypopnea index. Sleep. 2009;32:150–7. doi: 10.1093/sleep/32.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iber C, Ancoli-Israel S, Chesson A, Quan SF. Westchester, IL: American Academy of Sleep Medicine; 2007. The AASM manual for the scoring of sleep and associated events: rules, terminology and technical specifications. [Google Scholar]
- 29.Atlas Task Force. Recording and scoring leg movements. Sleep. 1993;16:748–59. [PubMed] [Google Scholar]
- 30.Zucconi M, Ferri R, Allen R, et al. The official World Association of Sleep Medicine (WASM) standards for recording and scoring periodic leg movements in sleep (PLMS) and wakefulness (PLMW) developed in collaboration with a task force from the International Restless Legs Syndrome Study Group (IRLSSG) Sleep Med. 2006;7:175–83. doi: 10.1016/j.sleep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Moore H., IV . Stanford: Stanford; 2013. Signal processing and pattern recognition for nocturnal polysomnography sleep studies [Dissertation] [Google Scholar]
- 32.Winkelman JW, Finn L, Young T. Prevalence and correlates of restless legs syndrome symptoms in the Wisconsin Sleep Cohort. Sleep Med. 2006;7:545–52. doi: 10.1016/j.sleep.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Ratcliffe SJ, Shults J. GEEQBOX: A MATLAB toolbox for generalized estimating equations and quasi-least squares. J Stat Softw. 2008;25:2–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of WSC participants stratified by RLS symptom category
SNP odds ratios (OR) for RLS(A) versus RLS(C) and for PLM+ versus PLM− with two model adjustments each.
SNP odds ratios (OR) are shown for RLS(AB) versus RLS(C) using two adjusted models each.
Effects of SNPs on PLM stratified by RLS status


