SUMMARY
Basement membranes (BMs) are resilient polymer structures that surround organs in all animals. Tissues, however, undergo extensive morphological changes during development. It is not known whether the assembly of BM components plays an active morphogenetic role. To study in vivo the biogenesis and assembly of Collagen IV, the main constituent of BMs, we used a GFP-based RNAi method (iGFPi) designed to knock down any GFP-trapped protein in Drosophila. We found with this method that Collagen IV is synthesized by the fat body, secreted to the hemolymph (insect blood), and continuously incorporated into the BMs of the larva. We also show that incorporation of Collagen IV determines organ shape, first by mechanically constricting cells and second through recruitment of Perlecan, which counters constriction by Collagen IV. Our results uncover incorporation of Collagen IV and Perlecan into BMs as a major determinant of organ shape and animal form.
INTRODUCTION
Basement membranes (BMs) are layered polymers of extracellular matrix proteins that underlie epithelia in all animals and surround organs, including muscles, fat, endothelium, and nervous tissue (Timpl, 1989; Yurchenco and Schittny, 1990). Among BM components, Collagen IV is the most abundant, comprising 50% of the proteins of the BM (Kalluri, 2003). Collagen IV molecules consist of α chains bound in long helical trimers that assemble into a network through lateral and enddomain interactions (Yurchenco and Ruben, 1987). Stacked layers of this polymer provide the main structural feature of BMs.
The biogenesis of functional collagen is very complex. Deficient or aberrant production is crucially involved in many diseases, including syndromes caused by mutations in collagens and collagen-modifying enzymes (Myllyharju and Kivirikko, 2004). Among the first steps in collagen biogenesis, α chains undergo extensive posttranslational modification in the endoplasmic reticulum (ER). A number of chaperones and enzymes assist folding and trimerization, including lysyl- and prolyl-hydroxylases, which require vitamin C as a cofactor (Lamandé and Bateman, 1999). CopII coated vesicles, which mediate ER-to-Golgi transport of secreted proteins, are typically 60– 80 nm in diameter, whereas collagen trimers are 300 nm long. Consequently, studies on collagen secretion have fuelled a controversy as to whether alternative mechanisms exist for the secretion of large proteins (Fromme and Schekman, 2005). It is not clear either how collagen trimers avoid self-aggregation and aggregation with other BM components inside the producing cell or at the plasma membrane. Therefore, while collagens are the most abundant proteins in the human body (30% of its protein mass), many of the steps of their biogenesis are poorly understood.
Collagen IV is found in all animals, from sponges to humans, indicating a central role of BMs in the development of complex body plans (Hynes and Zhao, 2000). It is the ancestral type of collagen, from which the 27 remaining vertebrate types have evolved. Type IV Collagens are divided into two subfamilies, α1-like and α2-like, split already in Cnidaria (Aouacheria et al., 2006). Drosophila has two genes encoding α chains of Collagen IV, named viking (vkg) and Collagen at 25C (Cg25C) (Le Parco et al., 1986; Natzle et al., 1982; Rodriguez et al., 1996; Yasothornsrikul et al., 1997), belonging to the α2-like and α1-like subfamilies respectively. vkg and Cg25C are adjacently located head-to-head in the genome, an arrangement conserved in the three α1-like/α2-like pairs of Collagen IV genes in mammals. Drosophila Collagen IV genes are essential, as loss of function of either of them causes embryonic lethality (Borchiellini et al., 1996; Rodriguez et al., 1996). In addition, Cg25C and Vkg modulate TGF-β gradient formation in the blastoderm embryo before a BM exists (Wang et al., 2008). Early lethality, although informative of the importance of Collagen IV for normal development, has precluded further analysis of its function in later stages, when organs and systems increase in size and complexity.
Apart from Collagen IV, the three other major components of BMs are Laminin, Nidogen, and the heparan-sulfate proteoglycan Perlecan, all conserved in Drosophila as well (Hynes and Zhao, 2000). Multiple interactions between BM components have been mapped in vitro. However, little evidence exists in vivo to show which interactions are crucial for BM assembly, maintenance and regeneration. Furthermore, a study of mice homozygous for a targeted deletion of the Collagen IV α1/α2 pair reported embryonic lethality but no defect in deposition of other BM components (Pöschl et al., 2004). This led to the proposal that Collagen IV is a terminal BM component, dispensable for BM assembly.
In this study, we have used a GFP protein trap inserted into the Drosophila vkg gene, producing a functional GFP-tagged version of Vkg, to study Collagen IV biogenesis and function. We developed an approach (in vivo GFP interference, iGFPi) to specifically knock down Vkg-GFP. Using iGFPi, we found that Collagen IV is synthesized in the larva by the fat body and continuously incorporated into BMs, where it exerts a constricting force on tissues. We additionally found that Collagen IV is not a terminal BM component, but, on the contrary, essential for deposition of Perlecan into BMs. Surprisingly, we found that Perlecan incorporation counters, rather than reinforces, constriction by Collagen IV.
RESULTS
iGFPi Is an Effective Method to Knock Down the Expression of GFP-Trapped Proteins
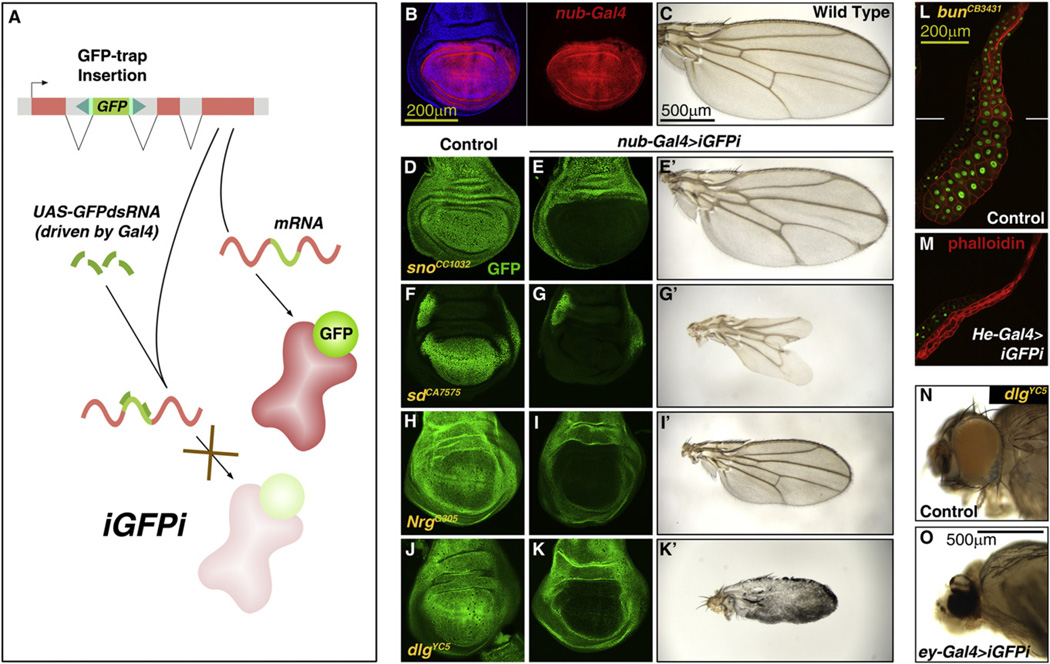
Protein trapping strategies have been developed in several model systems for large scale tagging of proteins and visualization of their subcellular localizations (Jarvik et al., 2002). In Drosophila, several insertional trapping screenings have been conducted by mobilizing transposons carrying the GFP-coding sequence flanked by a splice acceptor and donor (Buszczak et al., 2007; Clyne et al., 2003; Morin et al., 2001; Quiñones-Coello et al., 2007). As a result, more than 550 transgenic lines expressing GFP-tagged versions of proteins under control of their normal promoters are currently available. We found these trapping constructs are highly efficient (between 1.31 and 0.24% of transcripts skipped the artificial GFP exon in tested lines; see Figures S1A–S1C available online). We therefore reasoned that targeting through RNAi the GFP-encoding portion of the mRNA could be a versatile method to silence the expression of any GFP-trapped protein. To test the feasibility of this approach (in vivo GFP interference or iGFPi, Figure 1A), we expressed under control of the Gal4/UAS system an RNA inverted repeat targeting the GFP coding sequence (Brand and Perrimon, 1993; Roignant et al., 2003). iGFPi efficiently silenced expression of GFP-trapped proteins (examples in Figures 1B–1M and Figures S1D and S1E; 99% of the GFP-trapped product is knocked down, Figure S2E). This reduction was tissue specific, dependent on the expression pattern of the Gal4 line used to drive iGFPi. Furthermore, iGFPi was able to elicit loss-of-function phenotypes when the only allele of the gene present was GFP-trapped (i.e., flies homozygous or hemizygous for the GFP-trap insertion) (Figures 1B–1O and Figures S1F–S1I). These results show that iGFPi silences expression of GFP-trapped alleles specifically and that the method can be used for functional studies of GFP-trapped proteins.
Figure 1. iGFPi Is an Effective Method to Knock Down the Expression of GFP-Trapped Proteins.
(A) Strategy of in vivo GFP interference (iGFPi). Knockdown of GFP-trapped proteins is achieved through RNAi targeting the GFP-encoding sequence.
(B) Expression of nub-Gal4 in the blade region of a third instar (L3) wing disc, revealed by Gal4-dependent expression of RFP (UAS-myrRFP, red). Cell nuclei stained with DAPI (blue) on left.
(C) Wild-type adult wing.
(D and E) iGFPi driven by nub-Gal4 (nub>iGFPi) reduces expression of GFP-trapped Strawberry notch (Sno) (E, compared to control in D; GFP in green) and causes delta-shaped termination of veins at the margin (E’).
(F and G) nub>iGFPi reduces expression of GFP-trapped Scalloped (Sd) and causes loss of distal wing tissue (G’).
(H and I) nub>iGFPi reduces expression of GFP-trapped Neuroglian (Nrg) and causes reduction in wing size (I’).
(J and K) nub>iGFPi reduces expression of GFP-trapped Discs large (Dlg) and prevents correct differentiation (K’).
(L and M) iGFPi driven by He-Gal4 in the salivary gland reduces expression of GFP-trapped Bunched (Bun) and causes reduction in cell and organ size.
(N and O) iGFPi driven by ey-Gal4 in the eye of flies expressing GFP-trapped Discs large (Dlg) prevents correct differentiation.
See also Figure S1.
iGFPi Reveals a Postembryonic Requirement of Vkg
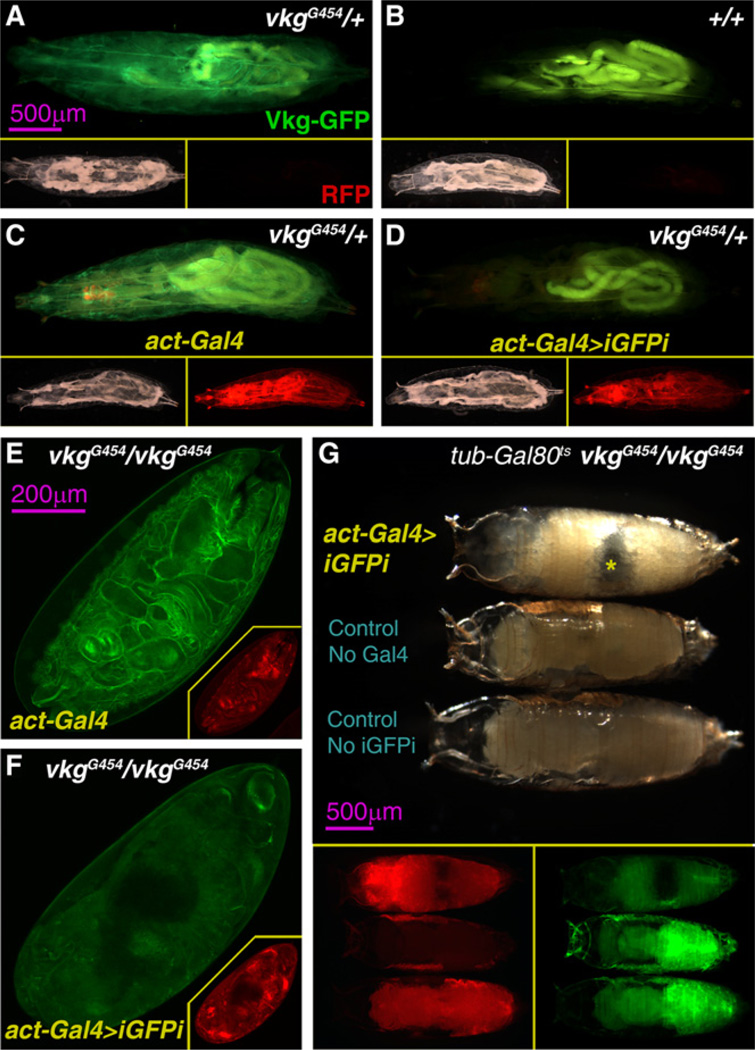
In order to study the biogenesis and function of Collagen IV, we made use of a Collagen IV GFP-trap line, vikingG454 (vkgG454) (Morin et al., 2001). Vkg-GFP localizes to BMs and has accordingly been used by us and others to monitor BM dynamics (Medioni and Noselli, 2005; Srivastava et al., 2007; Pastor-Pareja et al., 2008; Haigo and Bilder, 2011). iGFPi under control of the ubiquitous driver actin-Gal4 efficiently decreased Vkg-GFP expression in third instar (L3) heterozygous larvae (vkgG454/+; Figures 2A–2D and Figure S2E). iGFPi in vkgG454 homozygotes (vkgG454/vkgG454) caused in addition embryonic lethality (Figures 2E and 2F). This, together with the fact that vkgG454/vkgG454 flies are phenotypically wild-type, indicates that the Vkg-GFP fusion protein produced by the vkgG454 allele is functional.
Figure 2. iGFPi Reveals a Postembryonic Requirement of Vkg.
(A–D) Vkg-GFP expression (green) in a vkgG454 heterozygous (vkgG454/+) L3 larva (A), a wild-type larva (B), an actin-Gal4 vkgG454/+ larva (C) and an actin-Gal4>iGFPi vkgG454/+ larva (D). Lower right and left subpanels show, respectively, bright-light and red fluorescence pictures of the same larvae (UAS-myrRFP, driven by actin-Gal4).
(E and F) Vkg-GFP expression (green) in 24-hr-old vkgG454/vkgG454 embryos in the presence of actin-Gal4 (E) and actin-Gal4>iGFPi (F). Red fluorescence pictures in the bottom right corner subpanels (UAS-myrRFP, red, driven by actin-Gal4).
(G) vkgG454/vkgG454 pupae of the genotypes actin>iGFPi (upper specimen), UAS-GFP.dsRNA (middle specimen) andactin-Gal4 (lower), imaged 24 hr after puparium formation. actin>iGFPi animals arrest development in prepupa stage and die (notice air bubble in abdomen indicated by asterisk). Lower subpanels show green fluorescence (Vkg-GFP) and red fluorescence (UAS-myrRFP, driven by actin-Gal4) in the same pupae.
The previous result is consistent with embryonic lethality reported for vkg mutants (Rodriguez et al., 1996). It is not known, however, whether the function of Vkg, very abundant in BMs throughout development, is required after embryogenesis. To test a postembryonic requirement of Vkg, we made use of a ther-mosensitive form of the Gal4 repressor Gal80 (Gal80ts) to modulate Gal4-driven iGFPi. In presence of Gal80ts, vkgG454/vkgG454 animals developed into wild-type adults at 18°C (act-Gal4+-Gal80ts>iGFPi; permissive temperature). In contrast, shifting from 18°C to 29°C after completion of embryonic development resulted in pupal lethality (Figure 2G). These results show that Vkg expression is required for normal development after embryonic stages and confirm that the vkgG454 GFP trap can be used to study Collagen IV biogenesis in vivo.
Vkg Is Synthesized by the Fat Body and Continuously Incorporated into BMs during Larval Development
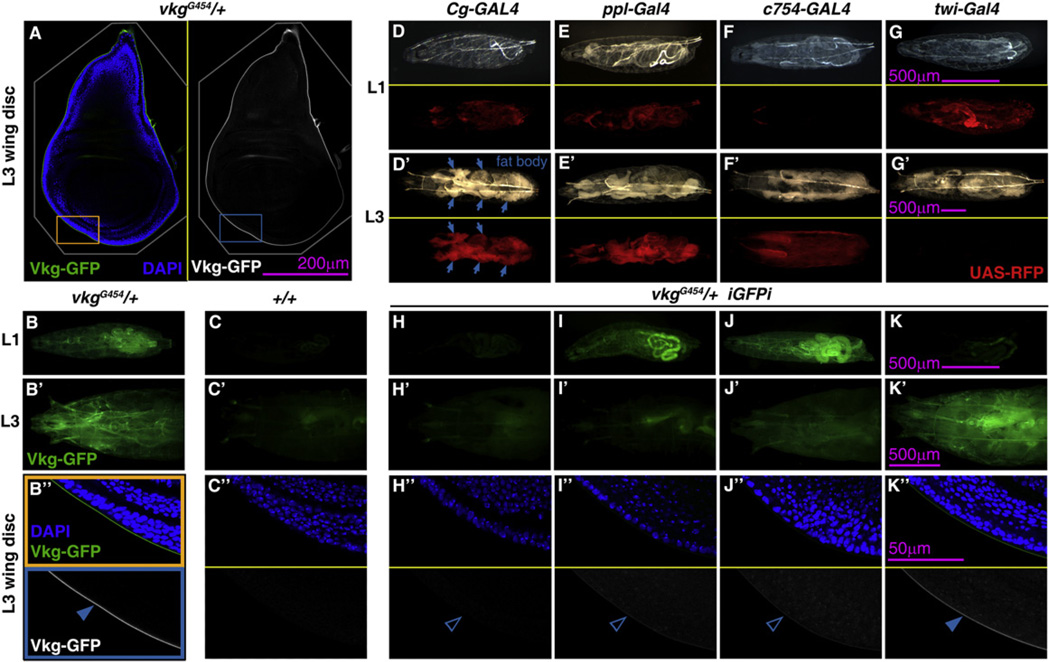
Once established a role for Viking in postembryonic development, we asked where the expression of Viking takes place during larval stages. In the embryo, even though Collagen IV is found in all tissues, mRNA in situ and enhancer traps suggest high expression in mesodermal derivatives (Le Parco et al., 1986; Rodriguez et al., 1996). In order to ascertain the source of Vkg protein, we made use of iGFPi in vkgG454/+ larvae and knocked down expression of GFP-trapped Viking using Gal4 drivers with restricted expression patterns. iGFPi under control of the wing disc drivers nub-Gal4 (data not shown) and Hh-Gal4 (Figures S2A–S2D) had no effect in the amount of Vkg-GFP present in the BM of the disc. In contrast, iGFPi driven by Cg-Gal4, expressed in hemocytes and fat body, drastically decreased Vkg-GFP expression in the whole animal, including imaginal discs (Figures 3A–3D and 3H; quantified in Figure S2E). Similar results were obtained when iGFPi was induced with ppl-Gal4, a fat body-specific driver (Figures 3E and 3I). iGFPi under control of hemocyte-specific drivers, however, did not reduce Vkg-GFP accumulation in BMs (Figure S2E) and neither did hemocyte ablation (data not shown). These results show that the fat body is the main source of the Vkg protein present in larval BMs.
Figure 3. Vkg Is Synthesized by the Fat Body and Continuously Incorporated into BMs during Larval Development.
(A) Confocal image of a wing imaginal disc of a vkgG454/+ L3 larva. Vkg-GFP in green (left) and white (right). Cell nuclei stained with DAPI (blue on left).
(B and C) Vkg-GFP expression in vkgG454/+ larvae (B) and control wild-type larvae (C). Images show L1 larvae (B and C), L3 larvae (anterior half, B’ and C’) and wing discs (B’’ and C’’; posterior ventral hinge, corresponding to orange and blue rectangles in A).
(D–G) Expression of Cg-Gal4 (D), ppl-Gal4 (E), c754-Gal4 (F), and twi-Gal4 (G) revealed with UAS-myrRFP in L1 (D, E, F, and G) and L3 larvae (D’, E’, F’, and G’). Upper subpanels show bright-light images and lower subpanels red fluorescence.
(H–K) Vkg-GFP expression in L1 (H–K), L3 (H’–K’), and L3 wing discs (H’’–K’’) from vkgG454/+ larvae where iGFPi is driven by Cg-Gal4 (H), ppl-Gal4 (I), c754-Gal4 (J), and twi-Gal4 (K).
See also Figure S2.
We also induced iGFPi under control of two mesodermal drivers with temporally opposite expression patterns: c754-Gal4, expressed in second and third instar larvae (L2 and L3) (Figure 3F) and twi-Gal4, expressed in the embryo and first instar (L1; Figure 3G). iGFPi under control of c754-Gal4 did not affect Vkg-GFP expression in L1; however, Vkg-GFP expression greatly decreased in L3 (Figure 3J). iGFPi under control of twi-Gal4, in contrast to c754-Gal4, reduced Vkg-GFP expression in L1, but Vkg-GFP visibly reappeared in L3 (Figure 3K). All these results together indicate that during larval development Vkg is produced by fat body cells and continuously incorporated into the BMs of the animal.
Multiple Requirements for Correct Incorporation of Collagen IV into BMs
The fat body is an organ formed by large polyploid cells (adipocytes), with known roles in lipid storage, metabolic regulation and immunity. To confirm that the fat body is the source of Viking and further investigate Collagen IV biogenesis, we decided to knock down in fat body cells the expression of several genes and examine the effects on Vkg-GFP localization.
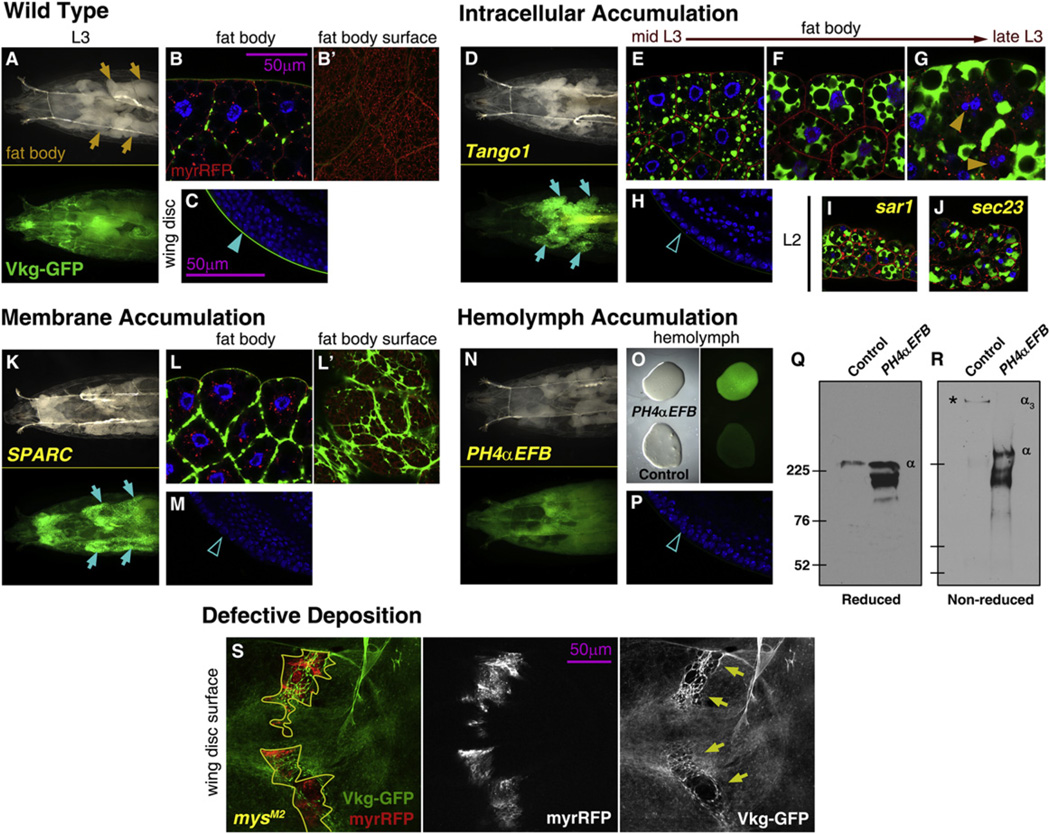
Secretion of collagen in human cells involves components of the CopII coatomer, required for ER-to-Golgi transport in the secretory pathway (Stephens and Pepperkok, 2002). It has been shown as well in human cells that Tango1, a CopII cargo adaptor, is required for secretion of Collagen VII (Saito et al., 2009). We knocked down expression of Tango1 and observed that this caused retention of Vkg-GFP in fat body cells (Figures 4A and 4D). Confocal imaging revealed that Vkg-GFP accumulated in growing intracellular aggregates (Figures 4B and 4E). The aggregates eventually coalesced and occupied most cytoplasm between the lipid droplets, affecting cell viability (Figures 4E –4G). These larvae lacked Vkg-GFP in the BM of other organs, such as the wing disc (Figures 4C and 4H; quantified in Figure S3C). We obtained the same results with ppl-Gal4 (data not shown). Additionally, knockdown of the CopII coat components Sar1 or Sec23 caused similar intracellular accumulation of Vkg-GFP (Figures 4I and 4J). These results show that fat body cells produce large amounts of Vkg protein and indicate that secretion of Collagen IV in Drosophila requires CopII coated vesicles and Tango1.
Figure 4. Multiple Requirements for Correct Collagen IV Incorporation into BMs.
(A) Anterior half of a vkgG454/+ L3 larva. Green fluorescence in lower subpanel.
(B) Confocal image of fat body cells of a vkgG454/+ L3 larva. Nuclei in blue (DAPI), Vkg-GFP in green and membranes in red (UAS-myrRFP). B’ shows a surface section.
(C) Confocal image of the wing disc (posterior ventral hinge) of a vkgG454/+ L3 larva. Nuclei in blue (DAPI), Vkg-GFP in green.
(D–H) Images of vkgG454/+ L3 larvae where expression in the fat body of the cargo adaptor Tango1 has been knocked down, showing the anterior half of the larva (D, compare to A), fat body cells (E–G, compare to B) and wing disc (H, compare to C).
(I and J) Confocal images of the fat body of vkgG454/+ L2 larvae where expression of the CopII components Sar1 (I) and Sec23 (J) has been knocked down.
(K–M) Images of vkgG454/+ larvae where SPARC expression in the fat body has been knocked down, showing the anterior half of the larva (K), fat body cells (L, surface in L’), and wing disc (M).
(N–P) Images of vkgG454/+ L3 larvae where PH4-αEFB expression in the fat body has been knocked down, showing the anterior half of the larva (N), bled hemolymph (O, bright-light image on left and green fluorescence on right) and wing disc (P).
(Q and R) Anti-GFP Western blots of hemolymph from control larvae and larvae where PH4αEFB expression in the fat body has been knocked down, both vkgG454/+ (as in O). SDS-PAGE gels were run under reducing conditions (Q) which separate Collagen IV monomers (α) or nonreducing conditions (R), which preserve the Collagen IV trimers (α3). Predicted molecular weight of a Vkg-GFP monomer is 221 kDa (Vkg 194 kDa + GFP 27 kDa).
(S) Clones of mysM2 (integrin βPS) mutant cells in vkgG454/+ wing discs. Clones, outlined in yellow, are labeled by myrRFP expression (red on left and white in middle). Arrows point to scars in the Vkg-GFP layer (green on left and white on right).
See also Figure S3.
We next investigated the effect of SPARC, the Drosophila homolog of BM40/SPARC/osteonectin, required for Collagen IV secretion by embryonic blood cells (Martinek et al., 2008). Knockdown of SPARC also caused retention of Vkg-GFP in the fat body (Figures 4K and 4L) and its absence in wing discs (Figure 4M). However, unlike loss of Tango1, sar1, or sec23, accumulation of Vkg-GFP occurred not intracellularly, but as thick fibers outside the plasma membranes of fat body cells (Figures 4B’ and 4L’;). These results show a requirement for SPARC in fat body cells for correct secretion of Collagen IV, suggesting that in its absence Collagen IV is not soluble.
A critical step in Collagen IV biogenesis is catalyzed by Prolyl-4-Hydroxylase (PH4). In the absence of this enzyme or its cofactor ascorbate (vitamin C), collagen chains cannot form functional trimers (Lamandé and Bateman, 1999). Among ten predicted Drosophila PH4 genes, we knocked down PH4αEFB, abundantly expressed in the fat body (Abrams and Andrew, 2002). In these larvae, we found most Vkg-GFP signal in the hemolymph that fills the body cavity (Figures 4N–4P). Western blotting of hemolymph in nonreducing conditions, known to preserve trimeric collagen, showed that wild-type Vkg migrated with a higher apparent weight than Vkg from PH4α-EFB-deficient larvae (Figures 4Q and 4R). These results show that in PH4α-EFB-deficient larvae Collagen IV is secreted into the hemolymph in monomeric form and cannot be incorporated into BMs.
Finally, we created in wing discs clones of cells mutant for myospheroid (mys), encoding the βPS subunit of the transmembrane integrin receptor. We found that loss of mys function caused scars in the BM underlying the mutant clones (Figure 4S and Figures S3A and S3B). This shows that integrins are involved in the capture by tissues of Collagen IV from the contacting hemolymph.
Altogether, our results, showing defective deposition of Vkg-GFP into BMs and accumulation at different locations, indicate that Collagen IV is synthesized by fat body cells in the larva, secreted as a trimer to the hemolymph and from there incorporated into BMs (Figure S3D).
Collagen IV Incorporation into BMs Maintains the Shape of Larval Organs
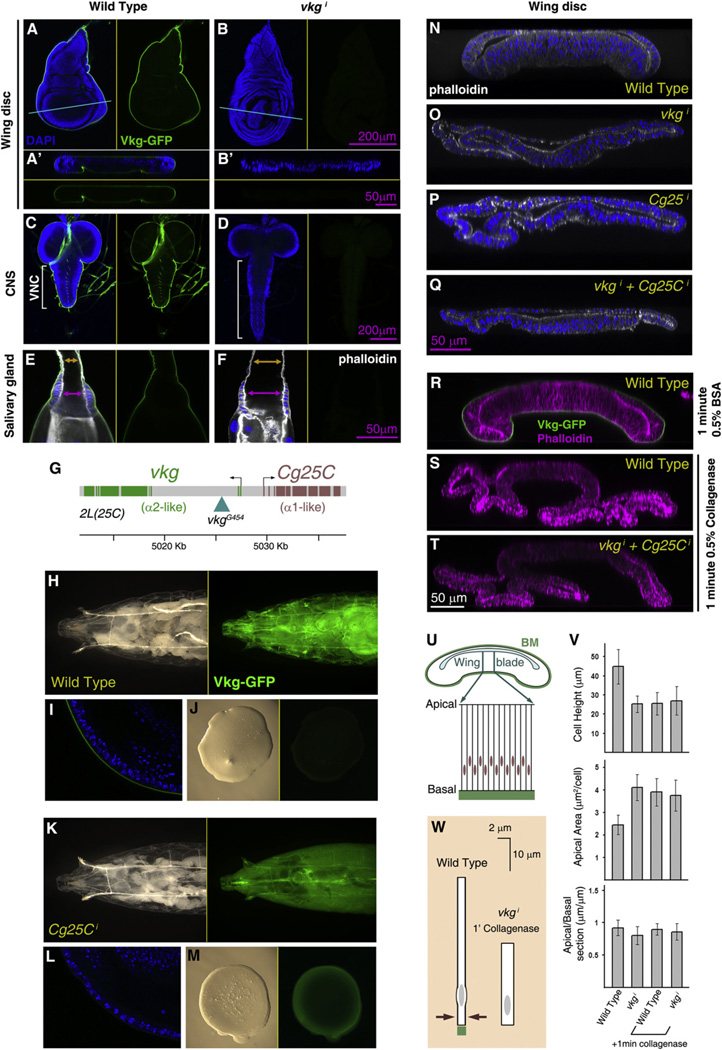
To address next the function of Collagen IV, we knocked down in the fat body the expression of vkg. We used Gal80ts again to limit knockdown to larval stages and thus avoid early lethality. At permissive temperature, CgGal4+Gal80ts>vkgi flies developed into normal adults, but pupal lethality resulted when larvae were transferred to restrictive temperature after completion of embryogenesis. Dissection of these larvae 96 hr after the temperature shift revealed highly aberrant shape in several organs. Imaginal discs, consisting of highly columnar epithelial cells, appeared flattened (Figures 5A and 5B), The ventral nerve cord (VNC) portion of the central nervous system was strikingly elongated (Figures 5C and 5D), reminiscent of embryonic defects in VNC condensation (Olofsson and Page, 2005). Additionally, the ducts and imaginal rings of the salivary glands were dilated, resulting in an expanded lumen (Figures 5E and 5F). Similar results were obtained when we knocked down in the same way expression of Cg25C, encoding a second Collagen IV chain (Figure 5G), and both vkg and Cg25C simultaneously (Figures 5N–5Q and Figures S4A–S4D).
Figure 5. Collagen IV Incorporation into BMs Maintains the Shape of Larval Organs.
(A and B) Confocal images of vkgG454/+ wing discs from control larvae (A) and larvae where expression of vkg in the fat body has been knocked down during larval stages (Cg-Gal4+Gal80ts>vkgi +96 hr at 29°C). A’ and B’ show transversal sections of the same discs, as indicated by the blue lines. Vkg-GFP in green in all subpanels. Nuclei in blue (DAPI) in left (A, B) and upper (A’, B’) subpanels.
(C and D) Confocal images of the central nervous system in vkgG454/+ larvae of control (C) and Cg-Gal4+Gal80ts>vkgi genotype (D, +96 hr). Nuclei in blue (DAPI) in left subpanels. Vkg-GFP in green in all subpanels.
(E and F) Confocal images of the salivary glands in vkgG454/+ larvae of control (C) and Cg-Gal4+Gal80ts>vkgi genotype (D, +96 hr). Nuclei in blue (DAPI) and actin in white (phalloidin) in left subpanels. Vkg-GFP in green in all subpanels. Arrows indicate the width of the lumen at the duct (orange) and imaginal ring (purple). (G) Head-to-head genomic arrangement of the two Drosophila Collagen IV genes.
(H-M) Images of vkgG454/+ control larvae (H-J) and vkgG454/+ larvae where expression of Cg25C has been knocked down (K-M; CgGal4+Gal80ts>Cg25C +96 hr). Images show the anterior half of the larva (H and K), a confocal section of the wing disc (I and L) and a drop of hemolymph (J and M). Vkg-GFP in green.
(N-Q) Transversal confocal sections across wing discs from a wild-type larva (N) and larvae where expression in the fat body of vkg (O), Cg25C (P), or both (Q) has been knocked down (Cg-Gal4+Gal80ts +96 hr). Nuclei (DAPI) in blue and F-actin (phalloidin) in white.
(R-T) Transversal confocal sections across a control wing disc (R) and wing discs treated with collagenase for 1 min (S and T). Discs were from wild-type (R and S) or Cg-Gal4+Gal80ts>vkgi +96 hr (T) larvae. F-actin (phalloidin) in magenta and Vkg-GFP in green.
(U) Schematic drawing of a section through the wing disc, showing the columnar epithelial organization of the wing blade.
(V) Cell shape changes caused in the wing blade by Collagen IV knockdown and collagenase treatment. Graphs represent height of the epithelium (measured in transversal sections), apical area (measured in planar sections), and apical-to-basal length ratios (length in section of a region measured apically divided by the basal length of the same region, both in transversal sections). Measurements from at least five specimens per genotype and treatment were averaged. Differences in cell height and apical area between wild-type and the other conditions are all significant (p < 0.05 in t tests). Error bars represent standard deviations.
(W) Schematic summary of cell shape changes caused by absence of Collagen IV and collagenase treatment. See also Figure S4.
Given the similar vkg and Cg25C loss-of-function phenotypes and the fact that Collagen IV molecules in mammals are heterotrimers (Hudson et al., 1993), we examined the effect of Cg25C knockdown on Vkg-GFP localization. Knockdown of Cg25C caused accumulation of Vkg-GFP in the hemolymph and prevented its deposition into BMs (Figures 5H –5M), similar to PH4-aEFB knockdown (Figures 4N–4R). This indicates that, in the absence of Cg25C,Vkg is secreted to the hemolymph in nonfunctional monomeric form, which supports the existence of all or most Drosophila Collagen IV as α1-like/α2-like heterotrimers despite suggestions to the contrary (Lunstrum et al., 1988).
The phenotypes obtained through inhibition of Collagen IV production suggested to us the possibility that a loss of tension provided by the BM could underlie the observed deformations. To explore this,we treated wing discs ex vivo with collagenase to degrade the BM (Figures S4E –S4H). We observed that 1 min of collagenase treatment was sufficient to elicit changes in cell and tissue shape that phenocopied loss of Collagen IV (Figures 5R and 5S). Furthermore, collagenase treatment of discs from Collagen IV-deficient larvae did not cause additional flattening (Figure 5T). These fast shape changes following collagenase treatment (apico-basal shortening and planar expansion; Figures 5U–5W) are consistent with the BM exerting a basal constricting force that contributes to the highly columnar shape of disc cells. We additionally confirmed tissue flattening after degradation of the BM by Matrix Metalloprotease 2(Mmp2) over-expression (Domínguez-Giménez et al., 2007), with cell shape changes reverting upon BM healing (Figures S4I–S4Q). Altogether, our experiments indicate a role of fat body-secreted Collagen IV in maintaining cell and organ shape by producing a basally constricting force.
Collagen IV Is Required for Incorporation into BMs of Perlecan, but Not Laminin or Nidogen
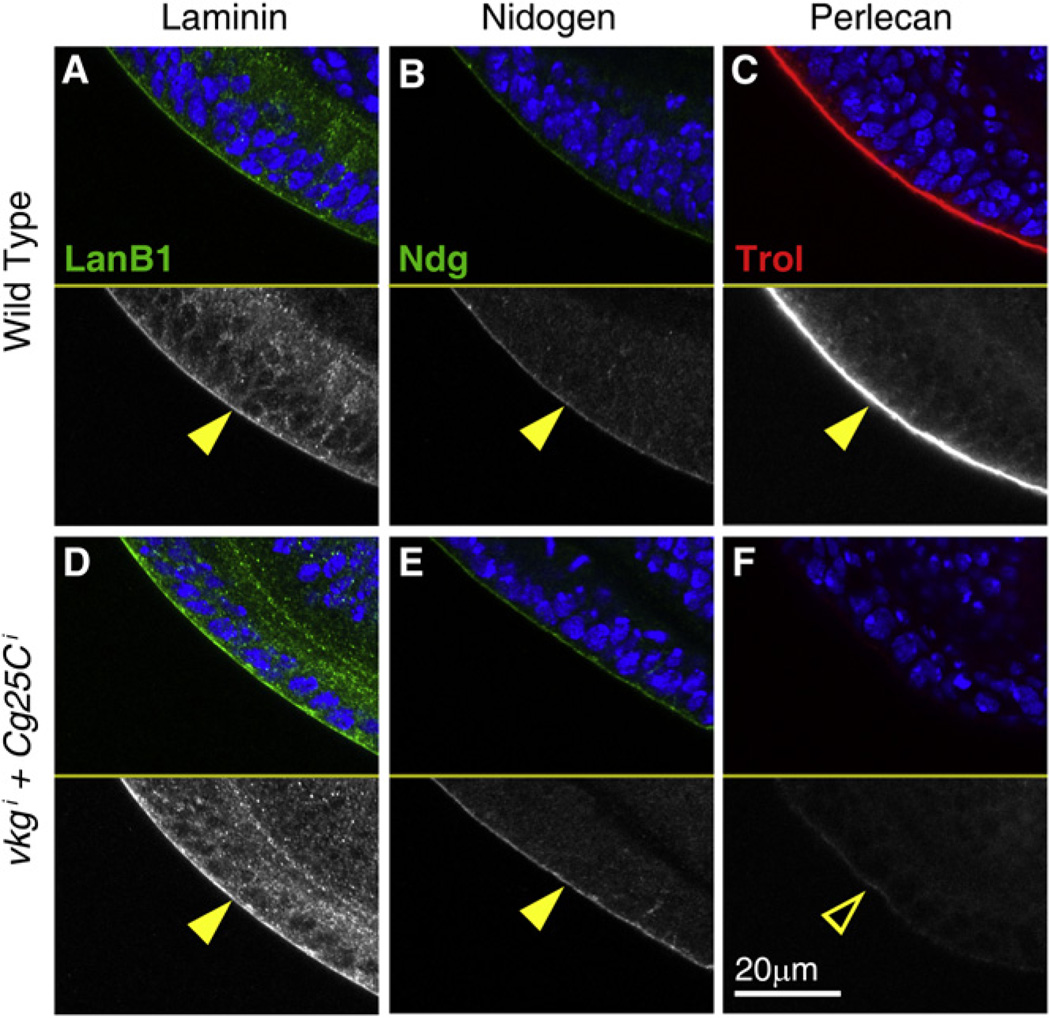
Apart from Collagen IV, the other three main BM constituents are Laminin, Nidogen, and the heparan sulfate proteoglycan (HSPG) Perlecan. Having established a role of Collagen IV in maintenance of cell and organ shape, we decided to investigate the effect of lack of Collagen IV in the deposition of other BM components. To this end, we examined with antibodies the expression of Laminin, Nidogen, and Perlecan in larvae where expression of both vkg and Cg25C had been knocked down (Cg-Gal4+ Gal80ts>vkgi+Cg25Ci). Presence of Nidogen and the Laminin B1 subunit was unaffected by the absence of Collagen IV (Figures 6A, 6B, 6D, and 6E). Basal localization of Myospheroid, the βPS subunit of the integrin receptor, was similarly unaffected (Figures S5J and S5K). In contrast, Perlecan, encoded by the gene trol, was absent from BMs in these animals (Figures 6C and 6F). These data show that Collagen IV deposition into BMs, while not required for localization of Nidogen or Laminin, is essential for Perlecan incorporation. Similar results were obtained in embryos homozygous for a deficiency deleting both vkg and Cg25C (Figures S5A –S5G), suggesting a requirement of Collagen IV for Perlecan incorporation also during initial BM assembly.
Figure 6. Perlecan Incorporation into BMs Requires Collagen IV.
Confocal images ofwing discs from wild-type L3larvae (A–C) and larvae where expression of vkg and Cg25C was knocked down in the fat body (D–F; Cg-Gal4+Gal80ts>vkgi+Cg25Ci+96 hr). Discs were stained with antibodies against Laminin B1 (A and D, green in upper subpanels and white in lower ones), Nidogen (B and E, green in upper subpanels, white in lower ones) and Perlecan (C and F, red in upper subpanels, white in lower ones). Cell nuclei stained with DAPI (blue) in upper subpanels. See also Figure S5.
Perlecan Incorporation into BMs Counters Constriction by Collagen IV
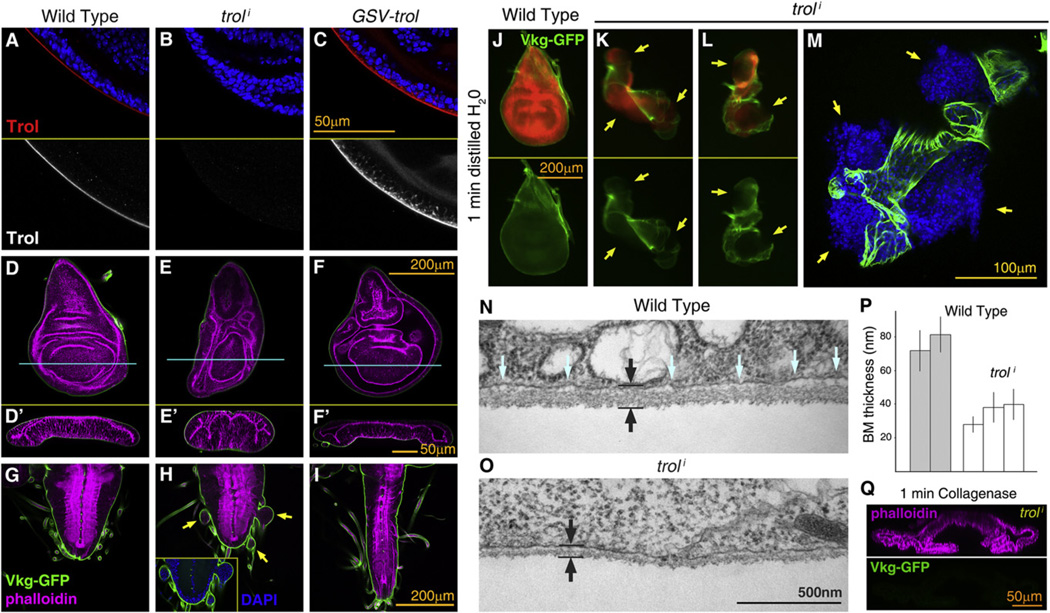
Having determined a requirement for Collagen IV in the incorporation of Perlecan to BMs, we finally decided to investigate the effects in larval tissues and BMs of both loss and excess of Perlecan. To reduce Perlecan expression, we knocked down trol expression through RNAi controled by actin-Gal4. This effectively abrogated the presence of Perlecan in BMs (anti-Trol staining; Figures 7A and 7B). To overexpress Perlecan, we used a unidirectional Gene Search Vector insertion (GSV2) upstream of trol (Figure 7C). Perlecan consists of a large core protein decorated by heparan-sulfate chains. Consistent with anti-Trol stainings, staining with the anti-heparan-sulfate antibody 3G10 disappeared from BMs when we knocked down trol and increased upon trol overexpression (Figures S6G –S6I).
Figure 7. Perlecan Counters Collagen IV BM Constriction.
(A–C) Confocal imagesof wing discs stained with anti-Perlecan (anti-Trol; redinupper subpanels, white inlower ones), dissected from a wild-type larva (A),alarva where expression of Perlecan was knocked down (B; act>troli) and a larva overexpressing Perlecan (C; act>GSV-trol).
(D–I) Confocal images of wing discs (D–F) and ventral nerve cords (G–I) dissected from wild-type (D and G), act>troli (E and H) and act>GSV-trol larvae (F and I). F-actin in magenta (phalloidin) and Vkg-GFP in green. Transversal sections of wing discs in (D’)–(F’) as indicated by blue lines. Arrows in (H) point to herniations of the VNC (inset shows cell nuclei stained with DAPI).
(J–L) Wing discs dissected from wild-type (J) and act>troli larvae (K and L) imaged in distilled water after 1 min of immersion. Yellow arrows point to breaks in the BM through which the underlying tissue expands. Cells express RFP (red in upper subpanels, driven by act-Gal4). Vkg-GFP in green.
(M) Confocal image of an act>troli wing disc after 1 min immersion in water. Vkg-GFP in green and cell nuclei (DAPI) in blue.
(N and O) Electron micrographs showing sections through the BM in wild-type (P) and act>troli (Q) wing discs. Black arrows indicate BM thickness. Blue arrows point to space between the BM and the cell membrane, reduced in the absence of Perlecan.
(P) BM thickness measured from electron micrographs of wild-type and act>troli wing discs. Thickness was measured infive discs from different larvae (two wild-type, three act>troli). Measurements from five micrographs were averaged per disc. Error bars represent standard deviations.
(Q) Transversal confocal section of an act>troli wing disc after 1 min collagenase treatment. Vkg-GFP in green. F-actin in magenta (phalloidin, upper subpanel). See also Figure S6.
We next examined larval organs in animals lacking or overex-pressing Perlecan, finding in both cases severe deformations. Upon trol knockdown imaginal discs appeared more compact than wild-type discs, with sections showing elongation of cells in the apico-basal axis (Figures 7D and 7E; quantified in Figure S6J). We found the same phenotypes with a second RNAi construct, and in trol8 (Park et al., 2001) and trolG0271 (Voigt et al., 2002) mutant larvae (Figures S6A–S6F). In addition, the central nervous system showed signs of hyperconstriction, with vesicles of tissue herniating from the VNC (Figures 7G and 7H). In contrast to these loss-of-function phenotypes, Perlecan overexpression caused flattening of wing discs (Figure 7F and Figure S6J) and elongation of the ventral nerve cord (Figure 7I), similar to loss of Collagen IV (Figure 5D).
The above data led us to hypothesize that presence of Perlecan in the BM opposed the effect of Collagen IV. Moreover, the compacted shape of discs lacking Perlecan suggested an increase in BM tension and a likely decrease in elasticity. To test this, we increased the pressure on BMs by immersing wing discs in distilled water, causing by osmosis an increase in disc volume through entrance of water into the tissue. Unlike wild-type BMs, the BMs of discs lacking Perlecan broke under pressure from the expanding tissue after just 1 min in distilled water (Figures 7J–7M; n = 6). Wild-type discs reached maximum turgor at around 5 min of water immersion with their BMs intact (n = 6). Further supporting increased tension in the absence of Perlecan, the BM of wing discs appeared thinner and in closer proximity to basal cell surfaces in electron micrographs (Figures 7N and 7O, quantified in Figure 7P). In addition, we found that 1 min of collagenase treatment flattened Perlecan-deficient wing discs all the way down to the same baseline as Collagen IV-deficient discs or collagenase-treated wild-type discs (Figure 7Q and Figure S6J), indicating that loss of Perlecan increases BM tension in a Collagen IV-dependent manner.
DISCUSSION
Functional Study of GFP-Trapped Proteins using iGFPi
GFP protein traps, producing GFP-tagged versions of proteins expressed from their endogenous loci, are excellent tools for the study of the expression patterns and subcellular localization of proteins. Here, we introduced in vivo GFP interference (iGFPi), a method to study GFP-trapped proteins in Drosophila. iGFPi involves specific RNAi knockdown of the GFP-trapped protein through expression of a double-stranded RNA targeting the GFP-encoding sequence. We showed that iGFPi is generally applicable to Drosophila GFP traps, producing loss-of-function phenotypes when flies were homozygous or hemizygous for wild-type GFP-trap insertions. Furthermore, iGFPi allowed us to investigate in vivo Collagen IV biogenesis and temporal requirements using a GFP protein-trap insertion into the Drosophila Collagen IV α2-encoding gene viking (vkg).
iGFPi significantly adds to the tools available in Drosophila to study gene function. Advantages of iGFPi are the easy assessment of knockdown effectiveness by examining GFP expression, and versatility, since a single RNAi construct can be used to target all GFP traps. Ongoing trapping projects, with improved techniques such as less biased transposons and recombineering-mediated tagging (Ejsmont et al., 2009; Venken et al., 2009), will increase the number of trap lines and make the iGFPi approach of broader application. Insertional protein trapping has proved successful in mammalian cells too (Jarvik et al., 2002). Therefore, iGFPi, or RNAi targeting any other tag, can be a powerful method providing a convenient avenue from protein expression/localization to functional studies in Drosophila and other organisms.
Collagen IV Biogenesis by the Fat Body
The fat body, formed by polyploid adipocytes, has essential roles in metabolic regulation (Arrese and Soulages, 2010). It is also an important effector of immune responses though secretion of antibacterial peptides to the hemolymph (Lemaitre and Hoffmann, 2007). To these functions, we add now the production of Collagen IV. Our experiments using iGFPi show that the fat body is the source of this protein in the larva. In contrast, there is no contribution from overlying cells to the Collagen IV content of a BM. Similar nonautonomy in the production of Collagen IV was observed in C. elegans (Graham et al., 1997). It has been shown that hemocytes, are the main source of Collagen IV earlier in the Drosophila embryo (Bunt et al., 2010; Martinek et al., 2008; Olofsson and Page, 2005). Since fat body and hemocytes are twin mesodermal lineages, it seems likely that the fat body takes over the task of Collagen IV production when the need of protein increases, as poliploidy makes fat body cells especially suited for synthesis of large amounts of protein. Secretion of Collagen IV by the fat body, in addition, raises the possibility that immune or metabolic inputs might regulate its synthesis.
We obtained further evidence that the larval fat body is the source of BM Collagen IV from experiments in which we silenced several genes and studied the effect on Vkg localization. Loss of SPARC caused retention of Vkg in the membranes of fat body cells in the form of thick fibers, reminiscent of fibrosis. Loss of CopII coatomer components, mediating vesicular traffic from the ER to the Golgi,or the Copll cargo adaptor Tango1 caused largeintra-cellular cumules of Vkg. Finally, loss of the prolyl-hydroxylase PH4αEFB prevented trimerization of Collagen IV, causing accumulation in the hemolymph of monomeric Vkg. These data, showing accumulation of Vkg at different locations when different steps of its biogenesis are prevented, indicate that Collagen IV is secreted by the fat body to the hemolymph as a soluble trimer and from there incorporated into BMs. Importantly, our results provide a genetic model to advance our understanding of collagen biogenesis and secretion. Due to their large size, fat body cells are ideal for subcellular imaging and localization studies, allowing detailed analysis of Collagen IV biogenesis in vivo.
Function of Collagen IV in Shaping Cells and Organs
The role of the cytoskeleton in shaping cells has received extensive attention. The effect of the BM on cell shape, in contrast, is less well dissected. Our results evidence a major role of Collagen IV in shaping Drosophila larval organs. Loss of Collagen IV caused deformations consistent with Collagen IV and the BM exerting a constricting physical force on the ensheathed tissues. It has been recently shown that Collagen IV constricts Drosophila eggs into an elliptical shape, becoming spheric instead in its absence (Haigo and Bilder, 2011). We showed here that in the wing imaginal disc, an epithelium consisting of highly columnar cells, knockdown of Collagen IV caused planar expansion and shortening in the apico-basal axis, resulting in flattening of the tissue. Similar changes in cell and organ shape were quickly elicited by collagenase treatment.
Our data also show that Collagen IV is required for Perlecan incorporation into the BM. Interactions among BM components have been mapped in vitro, but the relevance of these interactions in vivo remains to be established. It has been shown that Laminin, another BM component, is required for presence of Collagen IV in BMs of the embryo (Urbano et al., 2009). We showed here that the reverse is not true, with Laminin unaffected by absence of Collagen IV. We additionally found that Collagen IV was still present in BMs in the absence of Perlecan and that integrin expression in disc cells is required for Collagen IV to be correctly deposited. These results suggest that a strict hierarchy of assembly of BM components exists, as opposed to self-assembly guided by the simultaneous effect of multiple component interactions. Genetic data in model organisms like Drosophila will be essential to complement biochemical studies and help finally unravel the hierarchy of assembly of BM components during development and regeneration.
Lastly, when we addressed the function of Perlecan, we found that its presence counters BM constriction. Both loss- and gain-of-function experiments show that the effect of Perlecan is opposite to that of Collagen IV. Therefore, Collagen IV has a dual role in shaping organs: first, it constricts tissues, and, second, it mediates incorporation into the BM of Perlecan, which alleviates this constriction. Additional effects of BM components on morphogenesis are possible. Both Collagen IV and Perlecan affect signaling pathways (Kalluri, 2003; Wang et al., 2008) and, indeed, intercellular signaling has been implicated in cell shape changes in imaginal discs (Widmann and Dahmann, 2009). Our data, nonetheless, indicate a major role of the BM in shaping cells and organs through mechanical tension.
Further studies are needed to explain how Perlecan opposes Collagen IV and whether the core protein, the heparan sulfate chains or both mediate this effect. From our results, however, we can hypothesize that the ratio of the contents in Collagen IV and Perlecan will determine the tension exerted by a BM on a tissue. In this way, opposition between Collagen IV and Perlecan could be a major mechanism regulating cell and organ shape. It has been shown that Perlecan can regulate myosin localization in the follicle cells of the Drosophila ovary (Mirouse et al., 2009). It would be interesting to know to what extent cells passively change shape under tension from the BM or, on the contrary, reform their cytoskeleton in response. Also, by manipulating the amount of Collagen IV or Perlecan, it will be possible now to test recent models of wing disc growth that postulate a role of physical tension in controlling proliferation (Aegerter-Wilmsen et al., 2007; Hufnagel et al., 2007).
EXPERIMENTAL PROCEDURES
Drosophila Strains and Culture
Strains used and detailed genotypes of animals in each experiment are shown in the Supplemental Information. Cultures were maintained at 25°C on standard medium, except for experiments where tub-Gal80ts was used, in which cultures were transferred from restrictive (18°C) to permissive (29°C) temperature or vice versa at indicated times. Whenever staging of the larvae was required, parental flies were transferred to a fresh culture vial and left to lay eggs for 1 day; we considered the time of removing the flies from the vial 12 ± 12 hr AEL (hours after egg laying). Clones of mys mutant cells were induced using the MARCM system by heat-shocking 36 ± 12 hr AEL larvae at 37°C for 1.5 hr.
Stainings and Imaging
Samples were fixed, stained, and mounted in DAPI-Vectashield (Vector Labs) following standard procedures for imaginal discs and embryos. The following antibodies and dyes were used: rabbit anti-Ndg (1:2000; Wolfstetter et al., 2009), rabbit anti-Trol (1:2000; Friedrich et al., 2000), rabbit anti-LanB1 (1:500, Abcam), mouse Anti-heparan-sulfate 3G10 (1:100, Seikagaku), mouse anti-GFP (1:200, Roche), mouse anti-Dlg (1:1000), mouse anti-Mys (1:200), anti-rabbit and anti-mouse IgG conjugated to Alexa-488, Alexa-633, or Alexa 568 (1:200, Molecular Probes), phalloidin coupled to Texas red or Alexa-633 (1:100, Molecular Probes). No less than 20 discs were examined per experiment and genotype. For measurements of Vkg-GFP intensity, higher magnifi- cation images were taken of the posterior ventral hinge; fluorescence intensity and length of BM imaged was quantified with Image J software in five images per genotype. To image GFP and RFP in embryos (Figures 2E and 2F), these were dechorionated with bleach, mounted in PBS with coverslip spacers, and imaged in vivo without devitellinization. Confocal images were taken in a Zeiss LSM510 Meta confocal microscope. Adults were stored in a 1:1 mixture of ethanol and glycerol, and wings were mounted after letting ethanol evaporate on the slide. Images of larvae, pupae, adults, and water-immersed discs (Figures 7J –7L) were obtained with a Leica DFC300FX camera in a MZ FLIII stereomicroscope.
Western Blotting of Larval Hemolymph
Ten larvae of each genotype were bled in 20 µl of PBS containing 0.1 mg/ml of phenylthiourea to avoid melanization and 1 mg/ml of antiprotease cocktail. Four microliters of sample was loaded per genotype into a 10% SDS-PAGE gel (samples reduced with DTT) or a BioRad precast 3%–8% Tris-Acetate SDS-PAGE gel (nonreduced samples). High Range Rainbow (GE) was used as a molecular weight marker. Gels were blotted with a mouse anti-GFP antibody (Roche) and revealed with anti-mouse-HRP (Jackson Immunoresearch) and a Plus ECL kit (Perkin Elmer).
RT-PCR and Real-Time PCR
Total RNA from fat body, wing discs, or whole larvae was isolated using Trizol (Invitrogen). cDNA was synthesized from 2–5 µg of RNA with the SuperScriptIII First-Strand Synthesis System (Invitrogen). For RT-PCR analysis, 50 ng of cDNA per reaction was subjected to 30 amplification cycles. Real-time PCR analysis was performed in a Step One Real-Time PCR System (Applied Biosystems), using 50–200 ng of cDNA with SYBR green fast kit (Applied Biosystems) according to the manufacturer’s instructions. rp49 expression was used as an internal control for normalization. Three experiments for each genotype were averaged. Isoform-specific primers for real-time PCR were designed following recommendations in Brosseau et al. (2010).
Collagenase Treatment
Wing discs were dissected in PBS and immediately incubated in Collagenase Type I (Sigma-Aldrich, 0.5% in PBS) at 37°C for 1 min.
Cell Measurements in the Wing Blade
For quantification of cell shape changes, we used confocal images of discs stained with phalloidin. Height of the epithelium in the wing blade (length in the apico-basal axis) was measured in perpendicular sections (Z sections). Apical surface was measured in planar sections by counting the number of cells in a 10×10 µm square. Apical-to-basal length ratios were calculated from Z sections. Measurements from at least 5 discs were averaged per genotype and experiment.
Transmission Electron Microscopy
Ultrathin sections were prepared following standard procedures. Briefly, larvae were turned inside out in PBS, leaving wing discs attached to carcasses to facilitate later handling. Specimens were fixed using 2.5% gluteraldehyde/ 2% paraformaldehyde in 0.1M sodium cacodylate buffer, postfixed in 1% osmium tetroxide, and stained in 2% uranyl acetate. After embedding in epoxy resin, transversal ultrathin sections of wing discs were cut and imaged in a Tecnai Biotwin TEM microscope.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Cooley, A. Spradling, S. Datta, S. Baumgartner, D. Brower, the Bloomington Stock Center, the Developmental Studies Hybridoma Bank, the Kyoto National Institute of Genetics Fly Stock Center, the Vienna Drosophila RNAi Center, and the Drosophila Genetic Resource Center for providing fly strains and antibodies. We thank Hermann Steller for pointing out the effect of collagenase treatment on imaginal discs. We also thank M. Rojas, S. Landrette, and M. Graham for their assistance with western blots, real-time PCR, and electron microscopy. This work was supported by a grant from NIH/ NCI. T.X. is a Howard Hughes Medical Institute Investigator.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.devcel. 2011.06.026.
REFERENCES
- Abrams EW, Andrew DJ. Prolyl 4-hydroxylase alpha-related proteins in Drosophila melanogaster: tissue-specific embryonic expression of the 99F8–9 cluster. Mech Dev. 2002;112:165–171. doi: 10.1016/s0925-4773(01)00636-0. [DOI] [PubMed] [Google Scholar]
- Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech. Dev. 2007;124:318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Aouacheria A, Geourjon C, Aghajari N, Navratil V, Delé age G, Lethias C, Exposito JY. Insights into early extracellular matrix evolution: spongin short chain collagen-related proteins are homologous to basement membrane type IV collagens and form a novel family widely distributed in invertebrates. Mol. Biol. Evol. 2006;23:2288–2302. doi: 10.1093/molbev/msl100. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchiellini C, Coulon J, Le Parco Y. The function of type IV collagen during Drosophila muscle development. Mech. Dev. 1996;58:179–191. doi: 10.1016/s0925-4773(96)00574-6. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, Gervais-Bird J, Tremblay K, Perreault JP, Elela SA. High-throughput quantification of splicing isoforms. RNA. 2010;16:442–449. doi: 10.1261/rna.1877010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt S, Hooley C, Hu N, Scahill C, Weavers H, Skaer H. Hemocyte-secreted type IV collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev. Cell. 2010;19:296–306. doi: 10.1016/j.devcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Brotman JS, Sweeney ST, Davis G. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Giménez P, Brown NH, Martín-Bermudo MD. Integrin-ECM interactions regulate the changes in cell shape driving the morphogenesis of the Drosophila wing epithelium. J. Cell Sci. 2007;120:1061–1071. doi: 10.1242/jcs.03404. [DOI] [PubMed] [Google Scholar]
- Ejsmont RK, Sarov M, Winkler S, Lipinski KA, Tomancak P. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods. 2009;6:435–437. doi: 10.1038/nmeth.1334. [DOI] [PubMed] [Google Scholar]
- Friedrich MV, Schneider M, Timpl R, Baumgartner S. Perlecan domain V of Drosophila melanogaster. Sequence, recombinant analysis and tissue expression. Eur. J Biochem. 2000;267:3149–3159. doi: 10.1046/j.1432-1327.2000.01337.x. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Schekman R. COPII-coated vesicles: flexible enough for large cargo? Curr. Opin. Cell Biol. 2005;17:345–352. doi: 10.1016/j.ceb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 2011;331:1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BG, Reeders ST, Tryggvason K. Type IV collagen: structure, gene organization, and role in human diseases. Molecular basis of Goodpasture and Alport syndromes and diffuse leiomyomatosis. J. Biol. Chem. 1993;268:26033–26036. [PubMed] [Google Scholar]
- Hufnagel L, Teleman AA, Rouault H, Cohen SM, Shraiman BI. On the mechanism of wing size determination in fly development. Proc. Natl. Acad. Sci. USA. 2007;104:3835–3840. doi: 10.1073/pnas.0607134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J. Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Fisher GW, Shi C, Hennen L, Hauser C, Adler S, Berget PB. In vivo functional proteomics: mammalian genome annotation using CD-tagging. Biotechniques. 2002;33:852–854. doi: 10.2144/02334rr02. 856, 858–860 passim. [DOI] [PubMed] [Google Scholar]
- Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- Lamandé SR, Bateman JF. Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin. Cell Dev. Biol. 1999;10:455–464. doi: 10.1006/scdb.1999.0317. [DOI] [PubMed] [Google Scholar]
- Le Parco Y, Knibiehler B, Cecchini JP, Mirre C. Stage and tissue-specific expression of a collagen gene during Drosophila melanogaster development. Exp. Cell Res. 1986;163:405–412. doi: 10.1016/0014-4827(86)90071-6. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lunstrum GP, Bächinger HP, Fessler LI, Duncan KG, Nelson RE, Fessler JH. Drosophila basement membrane procollagen IV.I. Protein characterization and distribution. J. Biol. Chem. 1988;263:18318–18327. [PubMed] [Google Scholar]
- Martinek N, Shahab J, Saathoff M, Ringuette M. Haemocytederived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J. Cell Sci. 2008;121:1671–1680. doi: 10.1242/jcs.021931. [DOI] [PubMed] [Google Scholar]
- Medioni C, Noselli S. Dynamics of the basement membrane in invasive epithelial clusters in Drosophila. Development. 2005;132:3069–3077. doi: 10.1242/dev.01886. [DOI] [PubMed] [Google Scholar]
- Mirouse V, Christoforou CP, Fritsch C, St Johnston D, Ray RP. Dystroglycan and perlecan provide a basal cue required for epithelial polarity during energetic stress. Dev. Cell. 2009;16:83–92. doi: 10.1016/j.devcel.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Natzle JE, Monson JM, McCarthy BJ. Cytogenetic location and expression of collagen-like genes in Drosophila. Nature. 1982;296:368–371. doi: 10.1038/296368a0. [DOI] [PubMed] [Google Scholar]
- Olofsson B, Page DT. Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol. 2005;279:233–243. doi: 10.1016/j.ydbio.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Park Y, Fujioka M, Kobayashi M, Jaynes JB, Datta S. even skipped is required to produce a trans-acting signal for larval neuroblast proliferation that can be mimicked by ecdysone. Development. 2001;128:1899–1909. doi: 10.1242/dev.128.10.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis Model Mech. 2008;1:144–154. doi: 10.1242/dmm.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, Mayer U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development. 2004;131:1619–1628. doi: 10.1242/dev.01037. [DOI] [PubMed] [Google Scholar]
- Quiñones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, Hudson AM, Wang S, Castiblanco C, Buszczak M, Hoskins RA, Cooley L. Exploring strategies for protein trapping in Drosophila. Genetics. 2007;175:1089–1104. doi: 10.1534/genetics.106.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Zhou Z, Tang ML, Meller S, Chen J, Bellen H, Kimbrell DA. Identification of immune system and response genes, and novel mutations causing melanotic tumor formation in Drosophila melanogaster. Genetics. 1996;143:929–940. doi: 10.1093/genetics/143.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Carré C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc. Natl. Acad. Sci. USA. 2007;104:2721–2726. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DJ, Pepperkok R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 2002;115:1149–1160. doi: 10.1242/jcs.115.6.1149. [DOI] [PubMed] [Google Scholar]
- Timpl R. Structure and biological activity of basement membrane proteins. Eur. J. Biochem. 1989;180:487–502. doi: 10.1111/j.1432-1033.1989.tb14673.x. [DOI] [PubMed] [Google Scholar]
- Urbano JM, Torgler CN, Molnar C, Tepass U, López-Varea A, Brown NH, de Celis JF, Martín-Bermudo MD. Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development. 2009;136:4165–4176. doi: 10.1242/dev.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Pflanz R, Schäfer U, Jäckle H. Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev. Dyn. 2002;224:403–412. doi: 10.1002/dvdy.10120. [DOI] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- Widmann TJ, Dahmann C. Dpp signaling promotes the cuboidal-to-columnar shape transition of Drosophila wing disc epithelia by regulating Rho1. J. Cell Sci. 2009;122:1362–1373. doi: 10.1242/jcs.044271. [DOI] [PubMed] [Google Scholar]
- Wolfstetter G, Shirinian M, Stute C, Grabbe C, Hummel T, Baumgartner S, Palmer RH, Holz A. Fusion of circular and longitudinal muscles in Drosophila is independent of the endoderm but further visceral muscle differentiation requires a close contact between mesoderm and endoderm. Mech. Dev. 2009;126:721–736. doi: 10.1016/j.mod.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Yasothornsrikul S, Davis WJ, Cramer G, Kimbrell DA, Dearolf CR. viking: identification and characterization of a second type IV collagen in Drosophila. Gene. 1997;198:17–25. doi: 10.1016/s0378-1119(97)00274-6. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Ruben GC. Basement membrane structure in situ: evidence for lateral associations in the type IV collagen network. J. Cell Biol. 1987;105:2559–2568. doi: 10.1083/jcb.105.6.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.