SUMMARY
Understanding the structure-function relationships at cellular, circuit, and organ-wide scale requires 3D anatomical and phenotypical maps, currently unavailable for many organs across species. At the root of this knowledge gap is the absence of a method that enables whole-organ imaging. Herein we present techniques for tissue clearing in which whole organs and bodies are rendered macromolecule-permeable and optically-transparent, thereby exposing their cellular structure with intact connectivity. We describe PACT, a protocol for passive tissue clearing and immunostaining of intact organs; RIMS, a refractive index matching media for imaging thick tissue; and PARS, a method for whole-body clearing and immunolabeling. We show that in rodents PACT, RIMS, and PARS are compatible with endogenous-fluorescence, immunohistochemistry, RNA single-molecule FISH, long-term storage, and microscopy with cellular and subcellular resolution. These methods are applicable for high-resolution, high-content mapping and phenotyping of normal and pathological elements within intact organs and bodies.
INTRODUCTION
Facile and physiologically informative optical access to intact tissues has long been a goal of biologists. As early as the 1800s, work by scientists such as Werner Spalteholz revealed the utility of rendering tissue optically transparent for anatomical and biomedical studies (Spalteholz, 1914). Although the Spalteholz technique and its variants incur damage to tissue integrity and morphology, they are still in use a century later (Steinke and Wolff, 2001), highlighting barriers to the adoption of more recent tissue-clearing methods and modern microscopy techniques. While separate tissue clearing protocols have strengths in an application-specific context, none is able to fully surmount the most common challenges: confirmed generalizability across organs other than the brain or embryo, difficulties in execution, and incompatibility with endogenous fluorescence and/or post-hoc immunohistochemistry (Table S1). Thus motivation to improve tissue clearing protocols is sustained around three main objectives: 1) efficient clearing of both central and peripheral organs; 2) preservation of cellular and subcellular structures of multiple organ types; and 3) compatibility with endogenous fluorescent protein expression and post-hoc detection of DNA, RNA, and proteins.
The payoffs of such a method are optical access throughout large volumes of tissues, enabling the study of cell-to-cell spatial relationships and long-range neural connectivity in the context of preserved tissue morphology (Chung and Deisseroth, 2013; Chung et al., 2013; Kim et al., 2013; Zhang et al., 2014). In conjunction with fluorescent tracers, tissue clearing facilitates the identification of interacting cellular structures, including diverging or converging nerves and vasculature at their target sites throughout the body. Fine-scale subcellular analysis of cleared specimens using standard protein and nucleic acid probes should also be achievable in the context of cleared tissues.
We have developed a methodology for whole-organism clearing, building upon previous techniques such as CLARITY, SCALE, SeeDB, ClearT, 3DISCO, CUBIC, dibenzyl ether (DBE), and BABB (Murray’s Clear) (Becker et al., 2012; Chung et al., 2013; Dodt et al., 2007; Erturk et al., 2012; Hama et al., 2011; Ke et al., 2013b; Kuwajima et al., 2013a; Susaki et al., 2014b). Each of these has made a clear contribution: hydrogel embedding to stabilize tissue structures (Chung et al., 2013), fluorescent protein-compatible clearing reagents (Susaki et al., 2014b), and imaging approaches for large or challenging tissue samples (Becker et al., 2013; Tseng et al., 2009). Although a comprehensive discussion of their respective strengths and weaknesses is beyond the scope of this text, a few critical points merit mention. First, in the original proof-of-principle for each of these techniques, the detailed methods and optimized protocols were only presented for clearing brain tissue, and occasionally for the spinal cord (Erturk et al., 2012; Zhang et al., 2014) or whole embryo (Dodt et al., 2007; Hama et al., 2011). 3DISCO represents, to date, the most complete elucidation of a clearing method in peripheral tissues. However, as is the case with many prior clearing protocols (Table S1), 3DISCO’s clearing reagents (tetrahydrofuran and DBE) substantially quench fluorescent signals in tissue samples (Erturk et al., 2012). CLARITY (Chung et al., 2013) and CUBIC (Susaki et al., 2014b) bypass the fluorescence quenching problem, but CLARITY in its original form used electrophoretic tissue clearing (ETC) to extract lipids from large samples, which can be challenging to implement and can cause variability in final tissue quality, including epitope and fine processes damage and tissue browning due to heating (forum.claritytechniques.org). This led to variations of CLARITY using passive lipid extraction (Zhang et al., 2014, with protocol described in detail in Tomer et al., 2014), along with thermal acceleration of clearing and improved imaging. CUBIC also achieves tissue transparency by passively clearing phospholipids and is compatible with hydrogel embedding. The main weakness of passive clearing methods is their slow speed, which makes them unsuited for clearing large tissue volumes or whole organisms.
We here propose a methodology to facilitate fast, whole-brain and whole-body clearing using the circulatory system or the cerebrospinal fluid route to directly deliver clarifying agents. A first step was to optimize the hydrogel embedding, clearing, and imaging reagents, which resulted in PACT, for PAssive CLARITY Technique, for quicker passive lipid extraction of 1–3mm thick tissues. To image PACT-cleared tissue we have developed a Refractive Index Matching Solution (RIMS) - a custom economic recipe, with outcome similar to FocusClear™ (Chung et al., 2013; Moy et al., 2013; Tseng et al., 2009). The PACT reagents were then delivered either intracranially or via the vasculature to achieve whole brain and body clearing and labeling. We term the latter PARS, for Perfusion-assisted Agent Release in Situ. All steps for PARS, including preservation, clearing, and labeling, are performed in situ prior to tissue extraction. We demonstrate below that PARS, together with RIMS, transform opaque, intact, whole-organisms into optically transparent, fluorescently labeled samples for visualization with conventional confocal microscopy and phenotypic analysis at the cellular, subcellular, and even single-molecule transcripts level.
RESULTS
Optimized Method for Passive Clearing and Immunostaining of Whole Organs in the Rodent
Similar to the CLARITY method (Chung et al., 2013; Tomer et al., 2014), we render thick tissue optically transparent for imaging in three main steps. First, tissue is cross-linked and hybridized to hydrogel monomers to stabilize biomacromolecules. Second, tissue lipids are extracted from the tissue-hydrogel matrix with ionic detergents. Third, cleared tissue is embedded in RIMS for imaging, or for long-term storage. Although whole-body clearing was our primary goal, we recognized that the processing of small or particularly fragile specimens and organs would best be accomplished by a mild, passive clearing protocol. We developed PACT for rendering rodent whole organs, their 1–3 mm thick sections, including brain, spinal cord, kidney, heart, lung, and intestine, or human tissue biopsies transparent. The clearing speed depends in part on the rate of lipid solvation by detergent micelles, and the rate of diffusion of detergent micelles in tissue (Hoffman, 2002). However, unless an applied force accelerates their diffusion through tissue, such as the electric field in CLARITY’s ETC (Chung et al., 2013; Tomer et al., 2014), lipid extraction by large micelles is slow. We tested different detergents at various concentrations for their ability to passively clear 3 mm coronal mouse brain blocks over a 3-day incubation. Sodium dodecyl sulfate (SDS) at all concentrations was superior for lipid solvation and removal from brain tissue relative to other detergents, and moreover, only the 8% SDS concentration achieved uniform clearing throughout the entire 3 mm block (Figure S1A).
We hypothesized that a decrease in the cross-link density of the tissue-hydrogel would facilitate both lipid extraction and macromolecule penetration into thick, highly myelinated or fibrous tissue during subsequent immunohistochemistry. To test this, 1mm brain sections were infused with varying combinations and concentrations of formaldehyde, acrylamide, and bis-acrylamide, degassed, and polymerized at 37 °C. The efficiency of tissue clearing (Figure 1A) and the depth of antibody penetration (Figure 1B) increased significantly when lower concentrations of formaldehyde and acrylamide were used, and when bis-acrylamide, an acrylamide cross-linking agent used in CLARITY (Tomer et al., 2014), was excluded from the cocktail of hydrogel monomers. Upon observing a qualitative increase in tissue transparency in the tissue-hydrogels prepared with lower acrylamide concentrations (Figure 1A), we assayed the different PACT tissue preparations for protein loss, tissue integrity, and changes in weight and volume during clearing to ensure that a minimal crosslinking scheme was sufficient to preserve tissue morphology and molecular information. The amount of protein that leached out of tissue into SDS clearing buffer was statistically indistinguishable between 4% PFA-fixed, uncleared tissue samples (A0P4) that were incubated in PBS as a control, and those cleared tissue-hydrogel matrices prepared with 4% acrylamide (A4P0) or with 4% acrylamide plus 4% PFA (A4P4) (Figure 1C). Notably, the amount of protein recorded in the 8% SDS clearing bath solutions for all hydrogel-embedded samples was less than the protein loss (0.57 ± 0.11 mg per mg gross weight) for the samples preserved only with 4% PFA and incubated in PBS-0.1% TritonX-100, a mild detergent-containing buffer. This implies that hydrogel monomers effectively crosslink and stabilize tissue protein, which is further supported by our finding that unpolymerized, PFA-fixed tissue incubated in 8% SDS showed poor protein retention (0.63 ± 0.02 mg protein loss per mg gross weight) (Figure 1C).
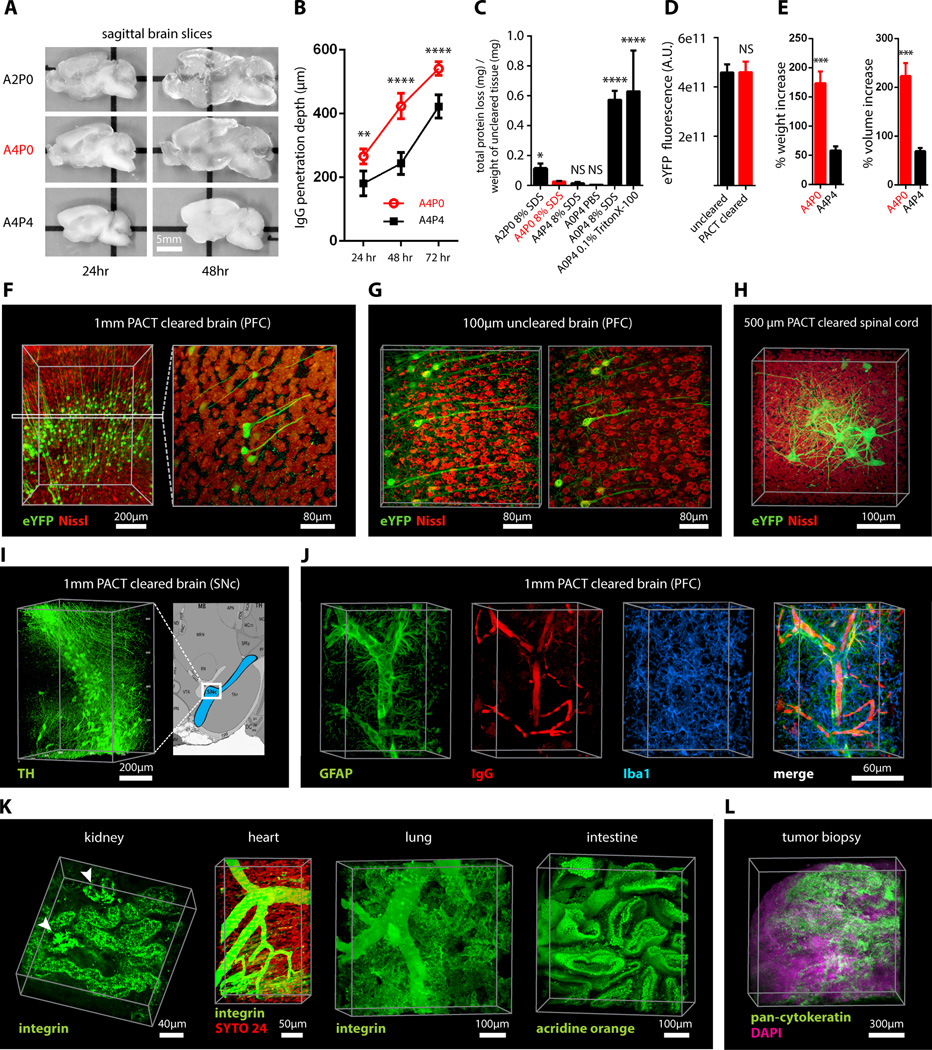
Figure 1. PACT clearing of A4P0 tissue-hydrogel hybrid achieves optimal transparency and immunohistochemistry compatibility across organs.
(A) Optical transparency comparison of 3 mm adult mouse sagittal blocks of A2P0, A4P0, and A4P4 tissue-hydrogel hybrid cleared for 24 h and 48 h. (B) Compared to A4P4, A4P0 tissue-hydrogel hybrid showed faster antibody penetration (n=6 fields of view per sample). (C) The percentage of protein loss from1 mm mouse brain slices (n=6 slices for each clearing condition); statistical significance is shown for each condition vs. A4P0 8% SDS (red). (D) The integrated eYFP fluorescence intensity in arbitrary units (A.U.) of uncleared and cleared 1 mm Thy1-eYFP mouse brain slices (n=6 slices). (E) Compared to A4P4, the A4P0 hydrogel-tissue hybrid showed higher tissue expansion and weight gain post clearing. (F–H) Thy1-eYFP mouse sections stained with Nissl: (F) 1 mm cleared brain slice, prefrontal cortex (PFC) area (left: z = 1 mm imaging stack depth); (G) 1 mm uncleared brain slice, PFC (left: z = 100 µm imaging stack depth); (H) 1 mm spinal cord slice (z = 500 µm). (I) Substantia nigra pars compacta (SNc) of 1 mm mouse brain slice stained with anti-tyrosine hydroxylase (TH) antibody (z = 1 mm). (J) PFC of 1 mm adult mouse brain slices stained with antibodies against GFAP, mouse-IgG, and Iba1 (z = 1 mm). (K) 1 mm section of mouse kidney (z = 150 µm; arrowheads show glomeruli), heart (z = 320 µm), lung (z = 550 µm) and intestine (z = 350 µm) stained with anti-integrin antibodies, SYTO24, and acridine orange. (L) PACT-cleared human tissue biopsy from basal cell carcinoma (BCC) was stained with anti-pan-cytokeratin (AE1/AE3) Alexa Fluor 488 primary antibody to label endothelial cells and DAPI (700 µm imaging stack depth). All graphs are shown in mean ± SEM Statistical significance: for paired samples: 2-tailed Student’s t test; for multiple comparisons: one-way ANOVA followed by Bonferroni posthoc (*p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.0001). All confocal imaging; for objectives see Supplemental Methods. Also see Figure S1–3 and Tables S1–2.
To corroborate these results on the preservation of molecular content in PACT tissue, the relative levels of native eYFP fluorescence were visualized and quantified in PACT brain samples from Thy1-eYFP transgenic mice. While a decrease in mean fluorescence intensity was observed under both hydrogel formulations (A4P0, A2P0), PACT samples showed comparable total intensity relative to uncleared tissue (Figure 1D) once the fluorescent measurements were normalized for tissue expansion (Figure S1C). Indeed, tissue-hydrogel matrices that were prepared using acrylamide alone (A4P0) exhibited tissue weight and volume changes of ~174% and ~223%, respectively (Figure 1E) over A4P4 counterparts. But, upon the transfer of tissue samples from clearing solution to mounting media, PACT samples shrank back to their original size within a few hours (Figure S3D). This tissue expansion-contraction has been documented in previous brain clearing protocols (Chung et al., 2013; Hama et al., 2011; Susaki et al., 2014b), wherein it was concluded that these size changes, though suboptimal, did not appear to negatively influence gross tissue morphology or cellular architecture. To visualize the effect of PFA on cross-link density in the tissue-hydrogel matrix, which is hypothesized to limit tissue expansion, PACT-cleared brain slices were imaged via scanning electron microscopy (SEM) (Figure S1B). We noted that A2P0 matrices had the largest pore sizes, followed by A4P0, while A4P4 had the smallest visualized pore sizes; pore size directly affects diffusion rate with faster macromolecular diffusion times in tissue-hydrogel matrices with larger pores. Tissue deformity (i.e. expansion and contraction) during PACT processing and mounting did not appear to affect the overall cellular organization or protein content of samples relative to conventional histological processing (Figure 1F–1L).Thus, we selected A4P0 for PACT given its balance between clearing speed, protein retention, and intermediate pore size tissue, which is condusive to macromolecule tissue penetration during histology.
PACT Reagents are Compatible with Histology and Endogenous Fluorochromes
To ensure that the signal intensity from genetically encoded fluorescent proteins was preserved throughout PACT processing, 1 mm-thick Thy1-eYFP tissue sections were A4P0-hybridized, PACT-cleared, and imaged using confocal microscopy. Despite PACT clearing, and importantly, the slow image acquisition time for thick samples the genetically expressed eYFP was readily detected throughout the samples (Figures 1F, 1H). Furthermore, the tissue-hydrogel matrix still permitted uniform Nissl staining of thick, cleared sections (Figure 1F, compared to uncleared 80 µm sections in Figure 1G). The overall tissue architecture remained constant between cleared and uncleared sections, as revealed by Nissl staining (red), which assuages concern that successive swelling and then shrinking of tissue caused permanent tissue deformity.
Not only were native proteins including those maintaining the structural integrity of tissue samples, retained by the tissue-hydrogel matrix during clearing (Figures 1C, 1F, 1H), but also the cleared tissue blocks were sufficiently macromolecule-permeable to permit labeling of peptidic and nucleic acid epitopes using a variety of common histological markers (e.g. antibodies, small-molecules, mRNA probes). For example, aside from Nissl, 1 mm PACT sections from the mouse brain and spinal cord were immunolabeled with antibodies against anti-tyrosine hydroxylase (TH) (Figure 1I); glial fibrillary acidic protein (GFAP), murine immunoglobulin G (IgG), and ionized calcium binding adaptor molecule 1 (Iba1) (Figure 1J). These targeted moieties represent antigens occupying a wide variety of cellular locations: membrane-localized and cytosolic, neuronal and non-neuronal antigens. PACT clearing decreased light scattering in tissue samples such that all labels were easily resolved across the entire 1 mm section during single-photon fluorescence imaging.
To confirm that PACT methodologies were effective on peripheral tissues as well, the kidney, heart, lung, and intestine of Thy1-eYFP mice were excised, cleared, and labeled with anti-integrin antibodies, acridine orange (AO), and/or SYTO24 (Figure 1K). As observed in the central organ samples (Figure 1F, 1H–J), small-molecule dyes and antibodies alike rapidly diffused through 1–3 mm thick A4P0-crosslinked and PACT-cleared sections of peripheral organs. While the time for complete immunolabeling of thick sections depends on several factors, including the tissue type, hydrogel pore size (Figure S1B), and the extent of lipid removal (Figures 1A, S1A), we achieved uniform antibody penetration throughout PACT samples with a 7–12 day incubation. However, for studies that only require labeling with small molecule fluorescent dyes one may obtain rapid staining of 1–3 mm PACT brain sections with a single overnight to 3-day incubation, respectively. Some peripheral tissues were stained even faster, wherein AO labeling of individual nuclei in unsectioned mouse intestinal tissue (~ 400um thick) was attained in under one hour (Figure 1K). We then determined if PACT can be applied to pathological samples. Human skin cancer biopsies (Figure S2A) were cleared and stained with pan-cytokeratin to visualize tumor cells (Figure 1L, Figure S2B–C). In sum, the entire PACT-cleared tissue block was accessible down to the subcellular level to molecular interrogation using standard immunohistochemical methods and conventional fluorescence microscopy.
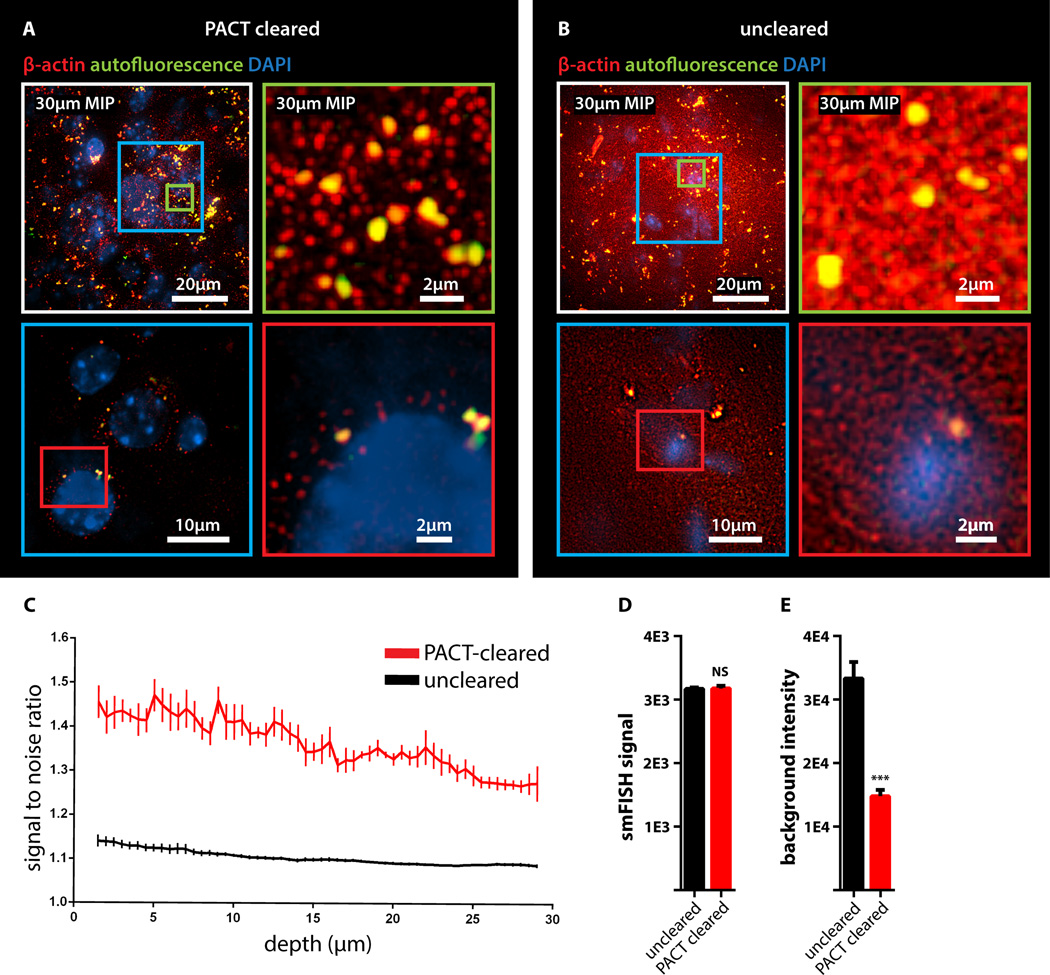
To determine if PACT is compatible with established procedures to visualize single mRNA transcripts, we subjected PACT-processed tissue to single-molecule fluorescent in situ hybridization, smFISH (Femino et al., 1998; Raj et al., 2008). The methodology of smFISH is capable of detecting single RNA molecules with high specificity in fixed cells and its high sensitivity allows for measurements of RNA abundance and subcellular localization. However, smFISH in tissue sections remains challenging due to low signal to noise ratio caused by tissue autofluorescence. Herein, β-actin transcripts in 100 µm-thick cleared mouse brain sections were labeled using 24 Alexa 594-labelled 20mer oligonucleotide probes towards β-actin mRNA. Tissue samples were slide-mounted in media containing 4',6-diamidino-2-phenylindole (DAPI) and imaged via single-photon microscopy. β-actin transcripts were indeed retained in the cytoplasm of neurons throughout PACT and smFISH processing, and single points of fluorescence could be distinguished despite the high copy number of β-actin in cells and the considerable thickness of imaged brain section (Buxbaum et al., 2014; Raj et al., 2008) (Figure 2A). PACT tissue exhibited significantly increased contrast of diffraction-limited spots throughout the tissue relative to uncleared tissue (Figure 2C). We found that smFISH intensity showed very little difference between PACT cleared and uncleared tissue, while background intensity was significantly reduced (Figure 2 A–B, 2D and 2E, S2D). These findings, taken together with the increase in smFISH signal to noise ratio seen in PACT cleared tissues, suggests that background autofluorescence in thick samples is the main factor obscuring smFISH signal in uncleared tissue.
Figure 2. Detection of individual mRNA transcripts in PACT tissue sections by smFISH.
100µm-thick mouse brain slices were hybridized with twenty-four 20mer oligonucleotide probes towards β-actin mRNA labeled with Alexafluor 594. (A) PACT-cleared smFISH brain slices. Upper panel shows 30 µm maximum intensity projection. An abundant number of diffraction limited spots corresponding to single beta-actin mRNAs (red) were readily detected up to 30 µm in depth under 589nm illumination. Note bright amorphous granules (yellow) are background lipofuscin vesicles that show up in both 589nm(red) and 532nm autofluorescence (green) channels, whereas smFISH signals are in the red channel only. (B) Compared to PACT cleared slices, smFISH in uncleared brain slices showed significantly decreased contrast. (Lower panels in A and B show single slices of 0.5 um at 12 um depth; the images were processed from raw data using the same contrast scale and Laplacian of Gaussian filtering; for raw data see Figure S2D) (C) Signal to noise ratio as a function of depth shows PACT-clearing tissue increases the signal to noise ratio of smFISH throughout the thickness of the sample as compared to uncleared tissue. (D) smFISH intensities show no appreciable differences between uncleared and PACT-cleared tissue. p = 0.8722; 2-tailed Student’s t test. (E) Comparison of background intensity between uncleared and PACT-cleared tissue illustrates the significant reduction of background fluorescence in PACT-cleared tissue. p = 0.0006; 2-tailed Student’s t test. All graphs are shown in mean ± s.d. For microscopy see Supplemental Methods.
Recipe for Refractive Index Matching Solution for Imaging and Long-Term Storage of Cleared Tissue
Effective imaging relies on sample immersion in a mounting media that reduces the refractive index (RI) variations within heterogeneous tissue and that alleviates the RI mismatch between tissue, mounting media and lens immersion media interfaces. In response to the prohibitive cost and limited availability of FocusClear, we formulated an affordable substitute: RIMS, for Refractive Index Matching Solution, with an RI appropriate for tissue imaging (RI = 1.46), biological safety, and biocompatibility for tissue preservation (see supplemental experimental procedures). To test RIMS, PACT-processed samples were mounted in 80% glycerol, FocusClear, or RIMS, and then imaged under identical conditions (Figure S3A). RIMS provided good optical clarity for fluorescence microscopy (Figure S3C) and caused minimal quenching of the eYFP signal over a 3-month period (Figure S3E). Since its performance appeared to be on par with or exceeds FocusClear (Figure S3A) and provided a >10-fold reduction in mounting costs, RIMS was employed for all subsequent PACT and PARS experiments. The exact RIMS formulation can be optimized in a case-specific manner to the RI of tissue samples (Figure S3B).
Whole-Body Clearing using the Vasculature in Adult Rodents
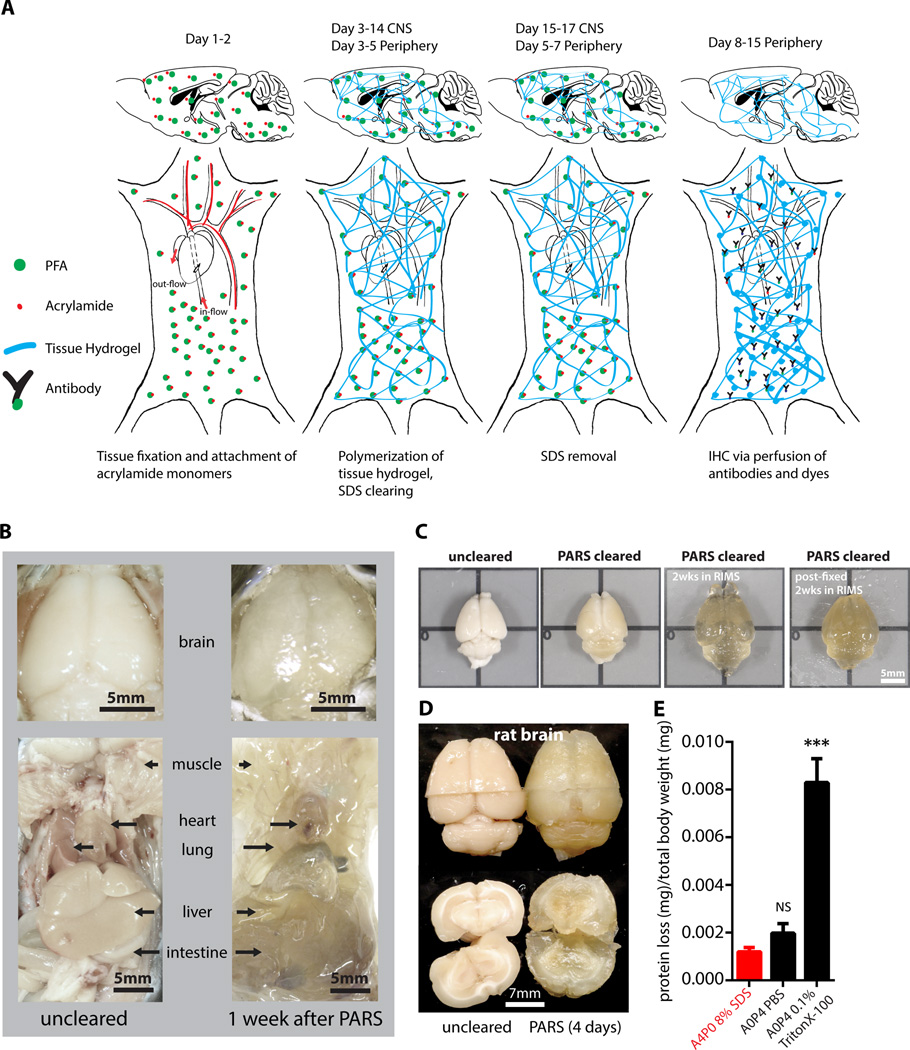
The PACT protocol uses a 4% acrylamide monomer solution to generate the final tissue hydrogel and results in a good combination of protein preservation, speed and ease of clearing, and optical clarity. However passive diffusion is slow, prohibitive for large volume or whole organism clearing. We also noted that the acrylamide hydrogels markedly swelled during the detergent clearing phase (Figure 1A, S1C). These two drawbacks, common to most clearing protocols, prompted us to develop an alternate methodology to speed up clearing and also to minimize tissue expansion during clearing. We decided to utilize the existing vasculature networks, as is done regularly in cardiac perfusion-fixation (Gage et al., 2012; Jonkers et al., 1984), to introduce agents directly to tissue by performing the entire fixation and clearing procedure in situ. We term this method Perfusion-Assisted Agent Release in Situ (or PARS). PARS utilizes the intact vasculature of the animal to infuse the hydrogel monomer and clearing solutions directly, which then diffuse throughout the tissues of interest.
To investigate both whether major blood vessels and whole-organism microvasculature was accessed by the perfusate (Leong and Ling, 1990; Li et al., 2012), AlexaFluor 647-conjugated antibodies against mouse immunoglobulin (Figure S4B, right) or Atto 647-conjugated nanobodies against GFAP (Figure S4B, left) were perfusion-recirculated through cardiac catheters for 24 hours. The mouse brain vasculature was extensively labeled, illustrating the accessibility of blood vessels to perfusate (Figure S4B). Perfusate was also observed to diffuse into surrounding tissue, as shown by the extravasculature GFAP labeling (Figure S4B, Supplementary Video 1).
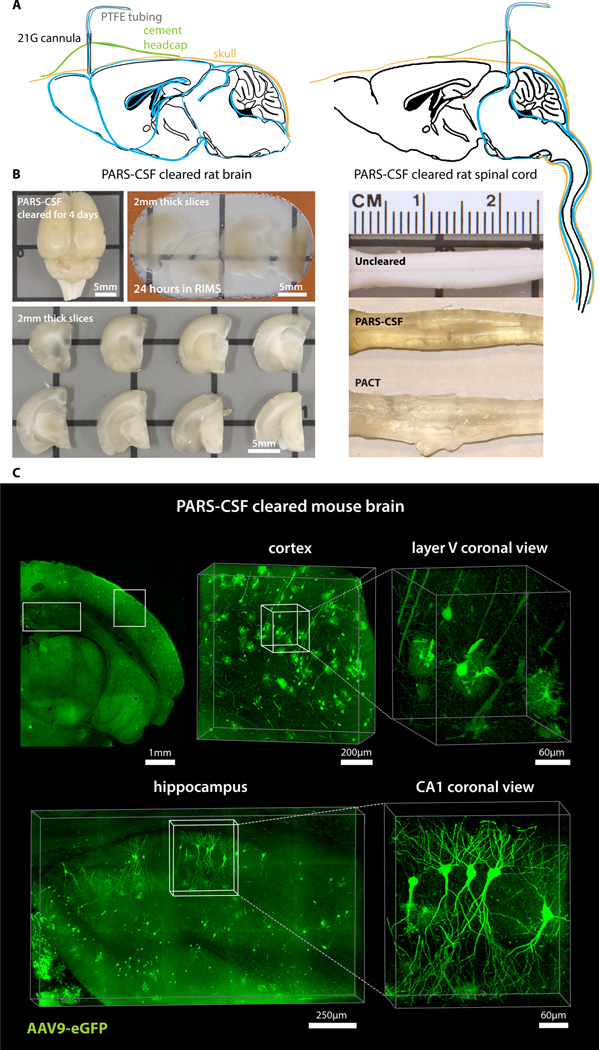
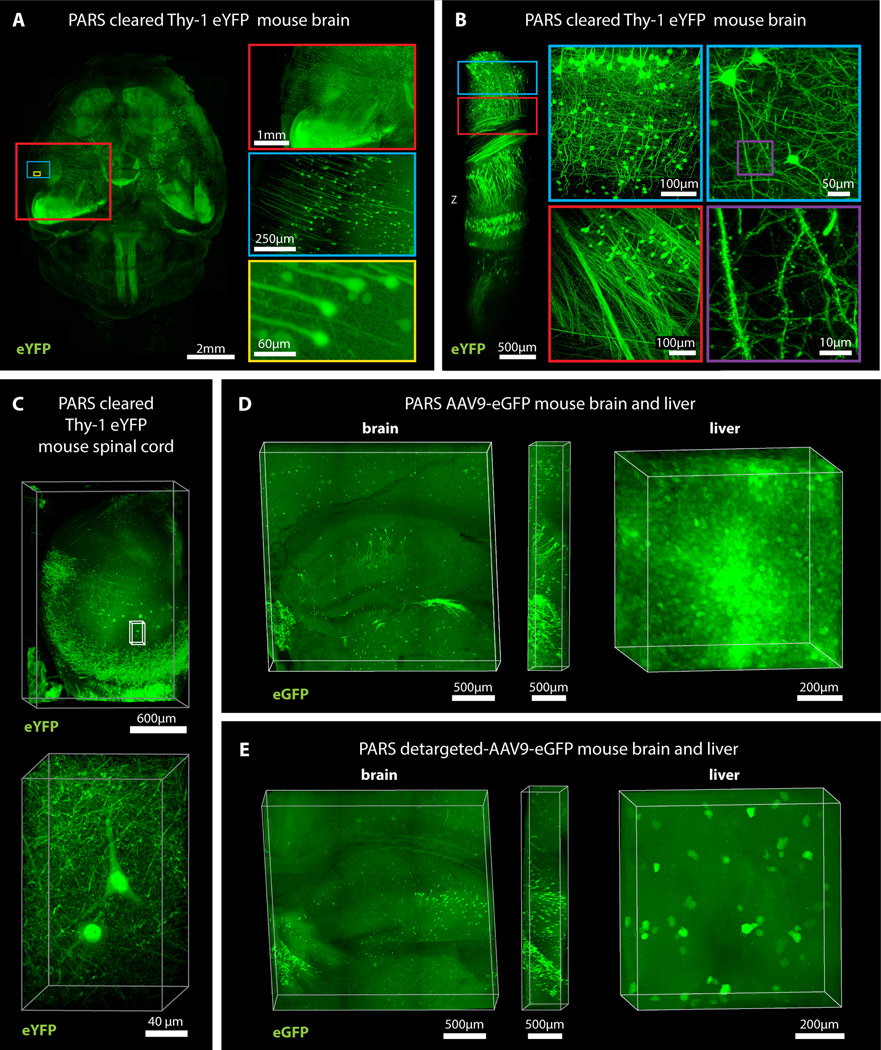
To recirculate PACT reagents into brain CSF or through whole-body vasculature for several days-to-weeks we developed a closed-loop perfusion system. Using this custom PARS chamber (Figure S4A), continuous intracranial perfusion of 8% SDS into CSF, via a method we termed PARS-CSF (Figure 3), attained whole-brain clearing in 4 days (Figure 3A–3B). Inserting the cannula more caudally into the cisterna magna (Figure 3A, right) granted clearing of the entire length of the rat spinal cord (Figure 3B). Next, AAV9-eGFP injected adult mice were prepared with a subdural cannula inserted directly above the olfactory bulb (Figure 3A, left), and after 4 days of recirculating 8% SDS at 37 °C, both unmyelinated and densely myelinated mouse brain regions near CSF circulation (most parts of the cortex, hypothalamus, regions near the ventricles and spinal cord) were transparentized). GFP-labeling of individual neurons, neuronal processes, and glial cells was clearly visible throughout the brain (Figure 3C).
Figure 3. PARS-CSF: a protocol for rapid whole-brain or spinal cord clearing and labeling via the cerebrospinal fluid route (CSF) using perfusion-assisted agent release in situ (PARS).
(A) CNS tissue may be rendered transparent optically transparent by the direct perfusion of all PARS reagents into the CSF via an intracranial brain shunt inserted either (left) below the dura in the region directly above the olfactory bulb, or into the cisterna magna (or placed directly above the dorsal inferior colliculus, right). The cannula, which is connected to the perfusion lines may be cemented into position with dental acrylic. (B) Whole-brain and the corresponding 2 mm thick slices (left) and whole-spinal cord (right) from PARS-CSF rats that were cleared at 37 °C for 4-days (brain) or for 2-weeks (spinal cord) are shown. The extent of whole-brain clearing is dependent on brain tissue proximity to the cannula: the frontal lobe was rendered optically transparent, whereas the mid-hind brain were only weakly cleared (see 2 mm slices on right side of panel). After 24-hour incubation in RIMS, PARS-CSF brain slices were sufficiently cleared for imaging without further sectioning. C) Images show native eGFP fluorescence in 500 µm PARS-CSF cleared coronal brain slices prepared from mice that, 6-months prior to clearing, received IV injections with AAV9:CAG-eGFP. Representative sections of cortex and hippocampus are presented at higher magnification in image boxes (right). In the layer V coronal view, an AAV9 transduced eGFP-expressing glial cell and eGFP-neuron adjacent to a blood vessel are clearly visible. In the hippocampus (bottom), the finer neuronal processes of eGFP-expressing CA1 neurons may be visualized with high resolution, which suggests that PARS-CSF may be completed without severe damage to cellular morphology. For microscopy see Supplemental Methods. Also see Figure S4.
Herein, we surmised that the same perfusion-clearing method of PARS-CSF could be extended to clearing whole-bodies in situ. Furthermore, the application of a pressure gradient on tissue during the lipid extraction and antibody diffusion, respectively, might hold the added benefit of accelerating the clearing and immunolabeling steps relative to PACT-based clearing of individual excised whole-organs. Clearing reagents were cycled through the whole-body vasculature (see timeline, Figure 4A), with complete clearing of all peripheral organs and of central nervous system accomplished within 1 week and 2 weeks, respectively, for mice and rats alike (Figure 4B–4D, S7). The minimal protein content of the PARS perfusate, and the higher protein content of perfusate from A0P4-infused mice (Figure 4E) suggested that the whole-organism hydrogel polymerization was both necessary and sufficient to stabilize gross organ structure and macromolecular content. To confirm that PARS was compatible with visualizing localized fluorescent protein expression in sparsely labeled cells in multiple organs, we delivered a GFP transgene by systemic administration adeno-associated virus (AAV). AAV9:CAG-eGFP (Figure 5D) or AAV9BD1:CAG-eGFP, a variant of AAV9 that transduces CNS neurons to a similar extent as AAV9, but exhibits reduced astrocyte and hepatocyte transduction (Figure 5E) were delivered via the vasculature in adult mice. In both the brain and the liver, native eGFP expression was readily detectable and the reduced transduction of liver hepatocytes by AAV9BD1 as compared with AAV9 was easily detected (Figure 5D versus 5E).
Figure 4. PARS achieves whole-body clearing.
(A) Schematic of PARS clearing and immunostaining. (B) A comparison of optical transparency of mouse brains and peripheral organs before and after PARS clearing. (C) Representative images of relative mouse brain size before (first box, from left) and after (second box) 2 weeks of PARS clearing shows that PARS circumvents hydrogel swelling and brain tissue expansion during the clearing process. Brain tissue expands gradually after immersion in RIMS (third box); this volume change may be mitigated via post-fixing PARS samples in 4% PFA overnight prior to RIMS mounting (fourth box). (D) Representative images of relative rat brain size before (right) and after (left) 4-days of PARS clearing, showing how PARS is a scalable method. Coronal slices of rat whole-brain samples show gross tissue morphology, highlighting that unmyelinated areas may be cleared within 4-days of PARS-based clearing. (E) Protein loss of PARS clearing compared to other clearing methods (n = 4 mice for each); graph shows mean ± s.e.m.; one way ANOVA followed by Bonferroni posthoc test was used to determine statistical significance in comparison to A4P0 8% SDS PARS clearing. * indicates p<0.05 and ** indicates p<0.01. Images for (B–D) were taken using bright field camera. Also see Figure S4, S7, S3E, and Supplemental Movie 1.
Figure 5. PARS enables whole-brain mapping of widespread and sparse genetically encoded fluorescent signals with subcellular resolution.
(A) Whole brain image (z = 6 mm), and (B) deep-brain imaging (z = 4 mm) of adult Thy1-eYFP mouse after PARS clearing for 10 days. The boxes on the right show high magnification images of indicated areas. (C) Spinal cord image of adult Thy1-eYFP mouse after PARS clearing for 2 weeks (z = 2 mm). Lower panel shows high magnification images of indicated region (z = 1.2 mm). (D) Images show native eGFP fluorescence in 1 mm coronal brain slices (left) and liver (right) prepared from the PARS cleared mice that received IV injections of AAV9:CAG-eGFP. Image columns to the right of each coronal brain image show the orthogonal views (z = 0.5 mm). (E) Native eGFP fluorescence in 1 mm coronal brain slices (left) and liver (right) prepared from PARS cleared mice injected with a liver detargeted variant, AAV9BD1:CAG-eGFP. Image columns to the right of each coronal brain image show the orthogonal views (z = 0.5 mm). For microscopy see Supplemental Methods. Also see Figure S5 and Supplemental Movie 2 (for 5B).
In comparison to PACT, we predicted that tissue volume changes during PARS processing would be reduced since musculoskeletal structures, such as the skull, the vertebral column, and muscle walls, would physically constrain tissue expansion. Indeed, PARS-based clearing of rodent brains was accomplished with limited hydrogel swelling and tissue expansion during clearing (Figures 4C–4D, S5A). Although PARS-processed brains do swell slightly following their extraction from the skull and placement in PBS or RIMS (Figure S5A), there was no evidence to suggest that gross changes in neuronal morphology occurred as a result of PARS processing and post-PARS expansion (Figure 5B). Nevertheless, we attempted to mitigate tissue swelling in RIMS through post-fixing PARS samples in 4% PFA overnight prior to RIMS mounting. To assess the extent to which overall tissue architecture was altered by volume changes, the intercellular distance and the average cell size within different brain regions (cortex, striatum, thalamus) of uncleared, PARS-cleared, and post-fixed PARS-cleared samples was measured (Figure S5B). It was predicted that individual regions may be differentially affected by PARS processing or RIMS incubation; for example, any sheer forces originating from perfusion-related intracranial pressure may exert a greater insult on less myelinated tissue or cause ventricle collapse. Post-fixing PARS samples significantly prevented the increased cell-sizes and intercellular distances that were detected throughout PARS samples. There were no significant differences in cell size or intercellular spacing between uncleared and post-fixed samples in all brain regions assayed (Figure S5B).
Whole-Organism PARS Enables Phenotyping and Imaging in an Organ-by-Organ Fashion
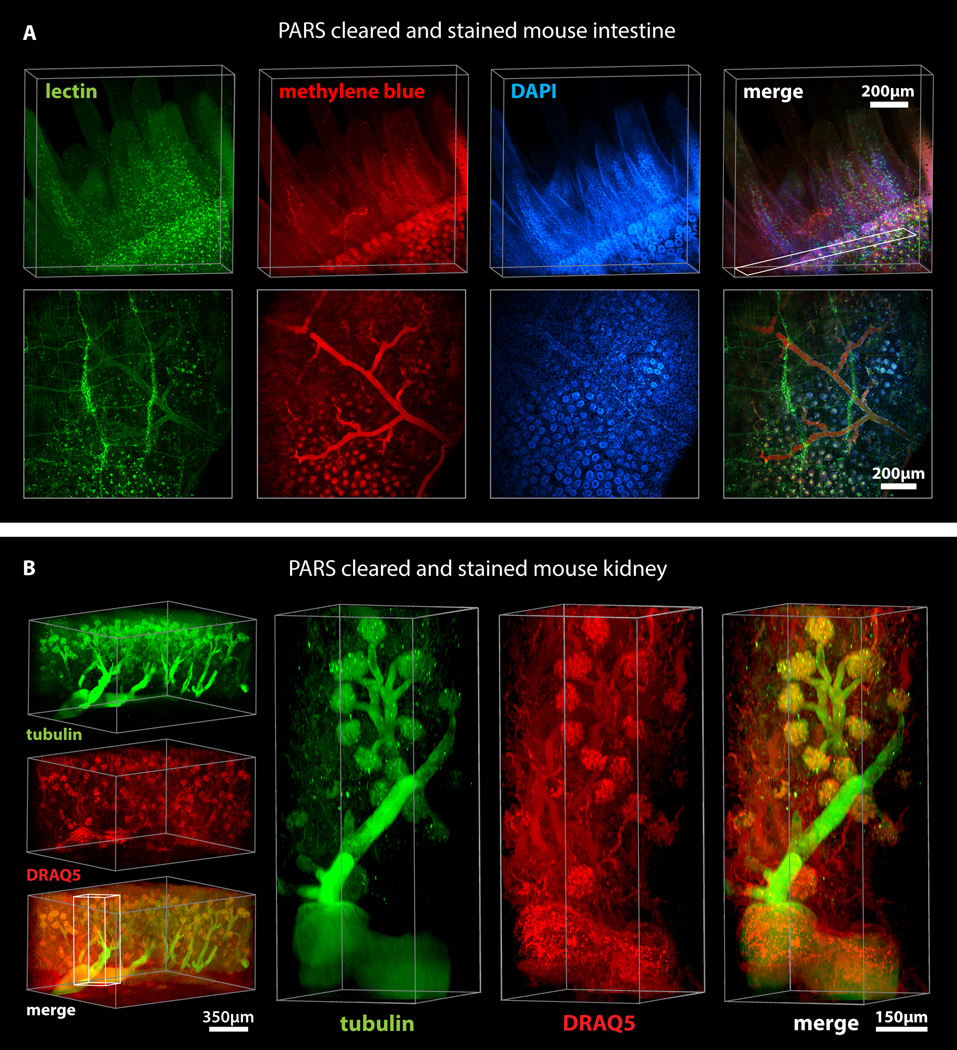
Following whole-body PARS processing and labeling, major organs were excised, thick-sectioned and imaged using confocal microscopy (Figures 5, 6, S6). The PARS-cleared whole-brain (Figures 5A–B) and spinal cord (Figure 5C) of Thy1-eYFP mice were imaged, and we concluded that PARS processing rendered entire organs optically transparent to the extent that visualizing deep-tissue structures with cellular resolution was possible. Through visualizing individual neurons and nephrons throughout the cleared whole-brain (Figures 5A–B; Supplementary Video 2) and kidney (Figure 6B; Supplementary Video 3) respectively, we may posit that this optical clarity was achieved while leaving fine cellular structures intact, in part due to the success of in situ tissue-hydrogel polymerization in stabilizing tissue architecture, preserving protein content and endogenous fluorescence, and maintaining the spatial relationships between subcellular and cellular tissue components (Figure 5A–B). For example, we could resolve individual fluorescently-labeled glomeruli of individual nephrons within 1 mm kidney sections, which establishes the ability of PARS to access peripheral organs through intact vasculature (Figure 6B, S6). Importantly, this includes the delivery of all immunohistochemical solutions as well, including blocking solutions, primary and fluorescently-labeled secondary antibody cocktails, or fluorescently-labeled small-molecules, and wash buffers. Immunolabeling using PARS was target-specific, uniformly distributed throughout peripheral organs, and exhibited low background, as illustrated by the tubulin and DRAQ5 labeling in PARS-processed mouse kidney sections (Figure 5B, Supplementary Video 2) and by the perfusion-based labeling of blood vessels in the liver, lung, pancreas with lectin, the filamentous actin probe phalloidin, and the nucleic acid stain DAPI (Figure S6).
Figure 6. PARS allows rapid and uniform clearing and immunolabeling of peripheral organs.
Clearing and immunohistochemical labeling was achieved in whole mice through PARS alone. (A) PARS-cleared mouse intestine was stained with lectin, methylene blue, and DAPI, and imaged through a depth of 500 µm. Lower panels shows maximum intensity projection of above rendering, z = 50 µm. (z = 500 µm). (B) A 1 mm thick kidney section was imaged (left) for anti-tubulin antibody and DRAQ5 labeling. Right panels show high magnification images of the indicated region and the structure of glomeruli, demonstrating that PARS enables antibody-based labeling throughout the kidney (z = 1.2 mm). For microscopy see Supplemental Methods. Also see Figure S6, S7, and Supplemental Movie 3 (for 6B).
DISCUSSION
Herein we introduce PARS, a method that renders intact whole-organisms transparent for imaging with single-cell resolution while preserving fluorescent and protein-based signals and tissue architecture. Our starting point, the CLARITY method (Chung et al., 2013) provided scientists with a brain-processing platform for elucidating the 3D cellular arrangement and connectome in toto. Numerous laboratories have previously reported on new clearing reagents in the decade before CLARITY, however many of these reagents were highly application- or tissue-specific (summarized in Table S1). In contrast, CLARITY introduced two broadly applicable techniques pertaining to tissue preservation (hydrogel embedding) and clearing efficiency (electrophoretic tissue clearing, ETC), both of which could be incorporated into the design, or redesign, of other clearing procedures. Traditionally, making tissue transparent was a process that demanded solvent incubations on the order of weeks-to-months, as reported in other clearing protocols (Hama et al., 2011). ETC, however, challenged the prevailing view that the rate of tissue clearing could only be accelerated through assaying large panels of organic solvents for their ability to solubilize tissue rapidly. Oftentimes, candidate solvents tested in these screens achieved rapid tissue clearing, but compromised tissue structure (Hama et al., 2011) or quenched native fluorescence (Becker et al., 2012; Erturk et al., 2012; Susaki et al., 2014b). Although the reagents introduced by CLARITY are gentler by comparison, the needed ETC step for fast clearing is complex to implement and causes tissue degradation from sample heating. Although these challenges can be bypassed by the use of passive CLARITY (Tomer et al., 2014) the slow rate of clearing make the technique impractical for scaling up or for whole-body mapping.
With the goal of rapidly clearing whole organisms while still using mild detergents and fluorescence non-quenching reagents throughout, we evolved PARS on the basic principles of CLARITY, but aimed to bypass the need for ETC while maintaining faster clearing than through passive diffusion. First we optimized the clearing agents for passive CLARITY by removing bisacrylamide and increasing the detergent concentration to 8% SDS (PACT reagents). The tissue clearing step was redesigned such that the electrophoretic force used by CLARITY to drive fast lipid extraction was replaced with a perfusion-based pressure gradient. Controlled flow of PACT reagents throughout intact tissue vasculature transforms most peripheral organs into optically transparent tissue within 2–3 days, while whole-mouse and whole-rat brains are rendered transparent within 1 – 2 weeks. Additionally, the self-contained nature of clearing in situ also reduced tissue expansion during the monomer infusion and lipid removal.
PARS opens up the possibility of whole-organ and whole organism mapping with high phenotypic content. With this in mind, quick, low resolution scanning of large tissue blocks can direct investigators to restricted areas worthy of slow, high phenotypic content analysis, including smFISH; a method that preserves fluorescent markers long-term is particularly valuable in this respect. Both PACT and PARS methodologies are scalable, cost-effective relative to the original CLARITY process and theoretically transferrable to other model organisms or human tissue. Indeed, while PARS was depicted using cardiac perfusion in rodents, the overall methodology may also be applicable to instances in which sufficiently large vessels are available for creating a perfusion route, such as whole-organ perfusion in larger, higher order mammals, including isolated human tissue. While PARS does achieve increased speed of clearing and reduced swelling without tissue damage (Table S1) the method’s unique strength lies in its scalability. Our data demonstrate, for example, that PARS can be employed to assess AAV-mediated transduction at the cellular level in multiple organs after systemic delivery. By eliminating the need to section individual tissues, the PARS approach could expedite efforts to screen numerous AAV serotypes and/or gene regulatory elements for optimal expression in the cell types of interest. In addition to improving screening throughput and speed, a PARS-based whole-body method could also counteract the risk of underestimating AAV transduction in target tissues due to undersampling errors. Similarly, PARS holds the potential to refine our understanding of peripheral nerves at their target whole-organs. Accurate maps of complex long-range fiber bundles, such as for the vagus nerve (George et al., 2000), could help inform improvements in existing therapies or spur the development of entirely novel therapeutic strategies, such as for bioelectronics medicines (Famm, 2013). PARS can also facilitate biomedical work in brain-to-body interconnections, in whole-body screening experiments for off- and on-target agents, and in whole-organ mapping for sparse elements such as tumor cells or stem cells. Lastly, the PARS method is compatible with cell-filling endoskeletal structures. By combining PARS with TEMPEST – a precursor to CLARITY (Deisseroth and Gradinaru, 2014) – the in vivo expression of long-lasting keratin filaments (that outlive the cells themselves while keeping a loyal blueprint of the morphology) within populations of interest can facilitate accurate post-mortem quantification and mapping of long-degenerated cells throughout the brain.
The methods we introduce here builds upon our prior work in CLARITY to expand tissue clearing and phenotyping to whole organisms by using the intrinsic circulatory system. Because the vascular network is not homogeneous, leading to non-uniform perfusive flow, organs of interest will clear at different rates. To achieve optimal clearing while retaining high tissue content, further validation for specific applications and technical improvements will be necessary. The blood-brain barrier may present a challenge to efficient perfusion-based transport of particularly large molecules such as antibodies (150 kDa) to the brain relative to the periphery. To improve perfusion efficiency, one solution likely will be to develop (or utilize when already available) smaller antibody scaffolds for immunolabeling; these include the fragment-antigen-binding format of immunoglobulins (Fab ~50 kDa), and nanobodies, single domain antibodies derived from camelid antibodies, whose smaller size (~12–15 kDa) promotes tissue permeability (Harmsen and De Haard, 2007).
Improved imaging platforms will also be needed to take full advantage of all the recent tissue clearing work, ours and others (Becker et al., 2012; Chung et al., 2013; Dodt et al., 2007; Ertürk et al., 2012; Ertürk and Bradke, 2013; Hama et al., 2011; Ke et al., 2013a; Kuwajima et al., 2013b; Susaki et al., 2014a). In order to obtain cellular and subcellular information in thick cleared tissue, it is necessary to utilize long-working distance objectives while still preserving high magnification and numerical aperture. Scanning speed is an additional barrier with cleared tissue blocks taking many days to be fully imaged – resonant scanners or light-sheet microscopy (Tomer et al., 2014) can accelerate the process while retaining high-resolution data.
Given increasing interest in the link between the brain and peripheral organs (Birmingham et al., 2014), it will be critical to have an unsegmented view of the whole-body, with structural connections between the brain and peripheral organs left intact. Through the development of PARS and enabling technologies (nanobodies, imaging platforms) it is becoming possible not only to facilitate neuroscientists’ overarching goal of creating a brain connectome, but also to facilitate elucidation of a brain-to-body-and-back connectome as well as the phenotyping of every other organ system in the body, healthy or diseased.
METHODS
PACT Clearing
4% paraformaldehyde (PFA)-fixed tissue sections were incubated at 4°C overnight in the hydrogel monomer solution A4P0 (4% acrylamide in PBS) supplemented with 0.25% photoinitiator 2,2'-Azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride (VA-044, Wako Chemicals USA, Inc.). A4P0-infused samples were degassed with nitrogen for 1–5 minutes and then incubated for 2–3 hours at 37 °C to initiate tissue-hydrogel hybridization. After removing excess hydrogel via brief PBS washes, tissue-hydrogel matrices were transferred into 50 mL conical tubes containing 8% SDS in 0.1M PBS (pH 7.5), and depending on tissue size, were incubated for 2–5 days at 37 °C with shaking. For immunostaining, 1–3 mm thick PACT-processed samples were washed in PBS with 4–5 buffer changes over the course of a day and then transferred to buffer containing small-molecule dyes or primary antibodies followed by fluorescently-conjugated secondary antibody (1:200–400, in PBS containing 2% normal donkey serum, 0.1% TritonX-100 and 0.01% sodium azide) for 3–7 days or with small-molecule dyes for 1–3 days. Antibody or small molecule dye solutions need to be replaced every day. Unbound antibody was removed via PBS washes, as before, and then samples were incubated with secondary antibodies (Fab fragment secondary antibodies are preferred, 1:200–400) for 2–5 days then washed for 1 day in PBS or phosphate buffer (PB) prior to incubation in imaging media (RIMS). All staining and mounting steps were conducted at room temperature with gentle shaking.
RIMS Imaging Media (RI 1.47)
40 g of Sigma D2158 (Histodenz) in 30ml of 0.02M PB with 0.1% tween-20 and 0.01% sodium azide, pH to 7.5 with NaOH – which results in a final concentration of 88% Histodenz w/v. Samples are incubated in RIMS until transparent (~ 1 day for PACT samples, up to 1 week for PARS cleared brains), followed by mounting in fresh RIMS.
smFISH
100 µm PACT sections were ethanol-permeabilized, labeled with 24 Alexa 594-labeled 20mer oligo probes towards B-actin (overnight incubation at 37 °C), washed and coverslipped with Slowfade Gold + DAPI according to published protocols (Buxbaum et al., 2014; Lyubimova et al., 2013). Laplacian of Gaussian filtering with a radius of 3 was applied to visualize transcripts in both cleared and uncleared samples.
PARS Protocol
Immediately following standard cardiac perfusion with 4% PFA (in PBS, pH 7.4), the fixed rodent was transferred onto a perfusion chamber (Figure S4A) which recirculated all subsequent PACT and immunolabeling reagents (as above) continuously (1 ml/min) through rodent vasculature via a peristaltic pump. Perfusion tubing connected the chamber to a feeding needle inserted through the left ventricle into the aorta and loosely sutured in place. The rodent was post-fixed with 4% PFA for 1 hour and then perfusion-washed with PBS for 1 hour. A4P0 monomer was cycled through vasculature overnight, followed by a 2 hour PBS perfusion wash. Before polymerization and without disconnecting perfusion lines, the perfusion chamber was placed into a ziplock bag (Figure S4A), and the bag containing the chamber with rodent was degassed for 2 minutes under nitrogen gas. Polymerization was initiated via perfusion-recirculation of 200mL of 0.25% VA-044 initiator in PBS at 37°C for 2–3 hours. The whole-body was cleared through a ≤ 2-week perfusion with 8% SDS in PBS, pH 7.5 at 37–42 °C followed by extensive PBS perfusion-washing over 2–3 days. Antibodies and small-molecule dyes (as above in PACT) were then delivered via a 3-day perfusion and 1-day wash.
For the PARS-CSF variation of brain or spinal cord clearing (Figure 3A–B), transcardially-fixed rodents were decapitated and a subdural cannula was inserted above the region of interest and cemented to the skull. All PARS reagents are delivered in the same order and timeframe as PARS at 1ml/min
AAV production and systemic delivery
Single stranded ssAAV-CAG-eGFP vectors packaged into AAV9 or the AAV9 variant capsid, AAV2/9BD1, was generated and purified as described (Lock et al., 2010). The AAV2/9BD1 capsid was modified from AAV2/9 (U. Penn) with, amongst others, an N498Y mutation to reduce liver transduction (Pulicherla et al., 2011). 1×1012 vector genomes (vg) of either virus was delivered intravenously into mice and tissue assessed 6 months later by PARS for native eGFP fluorescence.
Fluorescence Microscopy
Cleared tissue samples were mounted in RIMS at room temperature using spacers from 0.5 mm – 7 mm depending on sample thickness (iSpacer, SunJin Lab Co.; Silicone Isolator, Electron Microscopy Sciences, PA) and coverslipped. For Figure 3B, the samples were imaged by Leica Microsystems using a Leica TCS SP8 two-photon microscope with the Leica HC FLUOTAR L 25×/1.00 IMM CORR objective (working distance, w.d. 6.0 mm). Other images were taken using a Zeiss LSM 780 single-photon microscope with either the Fluar 5×/0.25 M27 dry objective (w.d. 12.5 mm), Plan-Apochromat 10×/0.45 M27 air objective (w.d 2.0 mm), LD SC Plan-Apochromat 20×/1.0 Corr M32 85mm scale-immersion objective (w.d. 5.6 mm), or LD LCI Plan-Apochromat 25×/0.8 Imm Corr DIC M27 multi-immersion objective (w.d 0.57 mm). Image reconstructions were performed using Imaris imaging software (Bitplane). After imaging, samples were stored in RIMS at room temperature.
For more detailed methods see Extended Supplemental Procedures and Table S2 for a detailed list of reagents/buffers.
Supplementary Material
HIGHLIGHTS.
PACT: passive tissue clearing and immunolabeling protocol for intact thick organs
RIMS: compatible storage and imaging media preserves fluorescent markers over months
Single-molecule FISH compatible 3D phenotyping enabled for thick tissue samples
PARS: whole-body clearing and phenotyping compatible with endogenous fluorescence
ACKNOWLEDGEMENTS
We thank the entire Gradinaru lab for helpful discussions. We also thank Drs. Dianne Newman and David Anderson for helpful discussions and suggestions. We thank Drs. Amir Arbabi and Andrei Faraon for assistance with SEM imaging. We thank Leica Microsystems for acquiring some of the images presented with a Leica TCS SP8 with Leica HC FLUOTAR L 25×/1.00 IMM CORR objective designed for CLARITY imaging.
BD and VG especially wish to acknowledge the contributions of Paul H. Patterson (- 2014), whose desire to apply tissue clearing to unresolved questions in neuroscience pushed us to further evolve these methods.
This work was funded by: the NIH / NINDS New Innovator (NIH IDP20D017782-01); NIH 1R01AG047664-01; Startup funds from the President and Provost of California Institute of Technology and the Biology and Biological Engineering Division of California Institute of Technology; the Beckman Institute of Caltech; the Pew Charitable Trust; Kimmel Foundation (to VG). And NIH R01HD075605 (to LC).
VG is also supported by: Human Frontiers in Science Program, the Mallinckrodt Foundation, the Gordon and Betty Moore Foundation, the Michael J. Fox Foundation, Caltech-GIST, NIH 1R01NS085910-01, NIH 1R21MH103824-01.
C-K.C., S.S., E.L., acknowledge support from the Caltech Biology Division Training grant (NIH/NRSA 5T32GM07616). R.P.K. acknowledges support from NIH/NIAMS (5T32AR058921). J.B.T. acknowledges the Colvin Postdoctoral Fellowship and CALTECH Division of BBE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
BY, JT, and VG conceived the project. BY, JT, RPK, BD, CKC, EL, SS, LC, and VG planned and executed experiments. BY, JT, VG made the figures and wrote the paper with input from all other authors. VG supervised all aspects of the work.
REFERENCES
- Becker K, Jahrling N, Saghafi S, Dodt HU. Ultramicroscopy: light-sheet-based microscopy for imaging centimeter-sized objects with micrometer resolution. Cold Spring Harbor protocols. 2013;2013:704–713. doi: 10.1101/pdb.top076539. [DOI] [PubMed] [Google Scholar]
- Becker K, Jahrling N, Saghafi S, Weiler R, Dodt HU. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum AR, Wu B, Singer RH. Single β-Actin mRNA Detection in Neurons Reveals a Mechanism for Regulating Its Translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nature methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth KA, Gradinaru V. Functional Targeted Brain Endoskeletonization. 2014 [Google Scholar]
- Dodt H-U, Leischner U, Schierloh A, Jahrling N, Mauch CP, Deininger K, Deussing JM, Eder M, Zieglgansberger W, Becker K. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nature methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt H-U. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nature protocols. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Ertürk A, Bradke F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO) Experimental neurology. 2013;242:57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Famm K. Drug discovery: a jump-start for electroceuticals. Nature. 2013;496:300. doi: 10.1038/496159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of Single RNA Transcripts in Situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Gage GJ, Kipke DR, Shain W. Whole animal perfusion fixation for rodents. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MS, Sackeim HA, Rush AJ, Marangell LB, Nahas Z, Husain MM, Lisanby S, Burt T, Goldman J, Ballenger JC. Vagus nerve stimulation: a new tool for brain research and therapy. Biological psychiatry. 2000;47:287–295. doi: 10.1016/s0006-3223(99)00308-x. [DOI] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature neuroscience. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Applied Microbiology and Biotechnology. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AS. Hydrogels for biomedical applications. Advanced Drug Delivery Reviews. 2002;54:3–12. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- Jonkers B, Sterk J, Wouterlood F. Transcardial perfusion fixation of the CNS by means of a compressed-air-driven device. Journal of neuroscience methods. 1984;12:141–149. doi: 10.1016/0165-0270(84)90013-x. [DOI] [PubMed] [Google Scholar]
- Ke M-T, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nature neuroscience. 2013a;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Kim SY, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends in Cognitive Sciences. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. Clear(T): a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013a;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SK, Ling EA. Labelling neurons with fluorescent dyes administered via intravenous, subcutaneous or intraperitoneal route. Journal of neuroscience methods. 1990;32:15–23. doi: 10.1016/0165-0270(90)90067-p. [DOI] [PubMed] [Google Scholar]
- Li T, Bourgeois JP, Celli S, Glacial F, Le Sourd AM, Mecheri S, Weksler B, Romero I, Couraud PO, Rougeon F, et al. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:3969–3979. doi: 10.1096/fj.11-201384. [DOI] [PubMed] [Google Scholar]
- Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z, Wilson JM. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, van Oudenaarden A. Single-molecule mRNA detection and counting in mammalian tissue. Nat Protocols. 2013;8:1743–1758. doi: 10.1038/nprot.2013.109. [DOI] [PubMed] [Google Scholar]
- Moy AJ, Wiersma MP, Choi B. Optical histology: a method to visualize microvasculature in thick tissue sections of mouse brain. PLoS One. 2013;8:e53753. doi: 10.1371/journal.pone.0053753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov S, Juneja V, Isaacson AB, Mohler WA, Campagnola PJ. Optical clearing for improved contrast in second harmonic generation imaging of skeletal muscle. Biophysical journal. 2006;90:328–339. doi: 10.1529/biophysj.105.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulicherla N, Shen S, Yadav S, Debbink K, Govindasamy L, Agbandje-McKenna M, Asokan A. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Meth. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalteholz W. Über das Durchsichtigmachen von menschlichen und tierischen Präparaten. Leipzig: S. Hierzel; 1914. [Google Scholar]
- Steinke H, Wolff W. A modified Spalteholz technique with preservation of the histology. Annals of anatomy = Anatomischer Anzeiger : official organ of the Anatomische Gesellschaft. 2001;183:91–95. doi: 10.1016/S0940-9602(01)80020-0. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S. Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell. 2014a doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis. Cell. 2014b doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature protocols. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SJ, Lee YH, Chen ZH, Lin HH, Lin CY, Tang SC. Integration of optical clearing and optical sectioning microscopy for three-dimensional imaging of natural biomaterial scaffolds in thin sections. Journal of biomedical optics. 2009;14:044004. doi: 10.1117/1.3158998. [DOI] [PubMed] [Google Scholar]
- Zhang MD, Tortoriello G, Hsueh B, Tomer R, Ye L, Mitsios N, Borgius L, Grant G, Kiehn O, Watanabe M, et al. Neuronal calcium-binding proteins 1/2 localize to dorsal root ganglia and excitatory spinal neurons and are regulated by nerve injury. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1149–E1158. doi: 10.1073/pnas.1402318111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.