Abstract
DNA self-assembly has produced diverse synthetic three-dimensional polyhedra. These structures typically have a molecular weight no greater than 5 megadaltons. We report a simple, general strategy for one-step self-assembly of wireframe DNA polyhedra that are more massive than most previous structures. A stiff three-arm-junction DNA origami tile motif with precisely controlled angles and arm lengths was used for hierarchical assembly of polyhedra. We experimentally constructed a tetrahedron (20 megadaltons), a triangular prism (30 megadaltons), a cube (40 megadaltons), a pentagonal prism (50 megadaltons), and a hexagonal prism (60 megadaltons) with edge widths of 100 nanometers. The structures were visualized by means of transmission electron microscopy and three-dimensional DNA-PAINT super-resolution fluorescent microscopy of single molecules in solution.
DNA nanotechnology has produced a wide range of shape-controlled nanostructures (1–10). Hollow polyhedra (1, 5, 11–26) are particularly interesting because they resemble natural structures such as viral capsids and promise applications for scaffolding and encapsulating functional materials. Previous work has constructed diverse polyhedra, such as tetrahedra (13, 16, 20, 24), cubes (1, 19, 23), bipyramids (15), truncated octahedra (11), octahedra (12), dodecahedra (16, 18), icosahedra (17, 21), nanoprisms (14, 22, 25, 26), and buckyballs (16), with ≤80-nm sizes and ≤5-MD molecular weights (such as Fig. 1A, structures 1 to 8). Assembly strategies include step-wise synthesis (1, 11, 21, 22), folding of a long scaffold (12, 19, 20, 24, 25), cooperative assembly of individual strands (13–15, 18, 26), and hierarchical assembly of branched DNA tiles (16, 17, 23).
Fig. 1. DNA-origami polyhedra.
(A) Polyhedra self-assembled from DNA tripods with tunable inter-arm angles, and comparison of their sizes and molecular weights with selected previous polyhedra (structures 1 to 9) (details are provided in fig. S1). (B) Design diagram of a tripod. Cylinders represent DNA double helices. Details of the arm connection at the vertex are provided in fig. S2. (C) Cylinder model illustrating the connection between two tripod monomers. (D and E) Connection schemes for assembling (E) the tetrahedron and (D) other polyhedra (represented here by the cube design).
A promising route to scaling up polyhedra is the hierarchical assembly of larger monomers. Previous work using small three-arm-junction (80 kD) (16, 23) and five-arm-junction (130 kD) (17) tiles has produced several ≤5-MD polyhedra (such as Fig. 1A, structures 5 to 7). Additionally, a 15-MD icosahedron (Fig. 1A, structure 9) (5) was assembled from three double-triangle–shaped origami monomers. Perhaps because of the lack of precise geometric control of the flexible double-triangle monomers, this icosahedron has low yield (5), and this method has not been generalized to construct more complex polyhedra.
We developed a more general strategy for hierarchical self-assembly of polyhedra from megadalton monomers using a DNA “tripod,” a 5-MD three-arm-junction origami tile [60 times more massive than previous three-arm tiles (16, 23)]. The tripod motif features inter-arm angles controlled by supporting struts and strengthened by vertex helices. Self-assembly of tripods into wireframe polyhedra is further facilitated by a dynamic connector design. We constructed a tetrahedron (~20 MD), a triangular prism (~30 MD), a cube (~40 MD), a pentagonal prism (~50 MD), and a hexagonal prism (~60 MD) (Fig. 1A and fig. S1) (27). With 100-nm edges, these polyhedra have a size comparable with those of bacterial microcompartments such as carboxysomes.
To characterize the three-dimensional (3D) single-molecule morphology of these polyhedra, we used a DNA-based super-resolution fluorescence imaging method (resolution below the diffraction limit) called DNA-PAINT (a variation of point accumulation for imaging in nanoscale topography) (28–30). Unlike traditional transmission electron microscopy (TEM), which images the samples in a vacuum under dried and stained conditions, 3D DNA-PAINT introduces minimal distortion to the structures in a “native” hydrated imaging environment.
Design
In one-pot annealing, the scaffold and staple strands first assemble into a tripod origami monomer, and then the tripods (without intermediate purification) assemble into the polyhedron (Fig. 1A). Diverse polyhedra can be constructed by using tripods with different designed inter-arm angles. The tripod has three equal-length (~50 nm) stiff arms connected at the vertex (connection details are available in fig. S2) with controlled inter-arm angles (Fig. 1B). To ensure stiffness, each arm contains 16 parallel double-helices packed on a honeycomb lattice (5) with twofold rotational symmetry. A supporting “strut” consisting of two double-helices controls the angle between the two arms. We name a tripod according to its three inter-arm angles (for example, the tetrahedron and the cube are respectively assembled from 60°-60°-60° and 90°-90°-90° tripods). To avoid potential unwanted aggregation resulting from blunt-end stacking of DNA helices (4), up to six short DNA double-helices (denoted “vertex helices”) are included at the vertex so as to partially conceal its blunt duplex ends (Fig. 1B). The number of helices and their lengths vary for different polyhedra (fig. S2). Additionally, the vertex helices are expected to help maintain inter-arm angles by increasing rigidity of the vertices. Two connection strategies are used to assemble tripods into polyhedra. To facilitate exposition, the three arms are denoted as X-arm, Y-arm, and Z-arm (Fig. 1B). Connecting X-arm to X-arm (Fig. 1C) and Y-arm to Z-arm produces polyhedra (such as a cube) (Fig. 1D) other than the tetrahedron, which is assembled by connecting X to X, Y to Y, and Z to Z (Fig. 1E).
Results
Tripod Conformation Control with Struts
First, we verified that the inter-arm angle was controlled by the length of the supporting strut. Gel electrophoresis of 60°-60°-60° and 90°-90°-90° tripods revealed a dominant band for each tripod (Fig. 2A), confirming their correct formation. Consistent with its more compact designed conformation, the 60°-60°-60° tripod migrated slightly faster than did the 90°-90°-90° one. The two tripod bands each were purified, were imaged with TEM, and showed designed tripod-like morphologies (Fig. 2B). The measured inter-arm angles were slightly smaller than designed [53 ± 5° (SD, n = 60 tripods) for 60°-60°-60° tripods; 87 ± 4° (SD, n = 60 tripods) for 90°-90°-90° tripods], possibly reflecting a small degree of strut bending.
Fig. 2. Self-assembly of DNA tripods and polyhedra.
(A) Gel electrophoresis and (B) TEM images of the 60°-60°-60° (lane 1 in the gel) and 90°-90°-90° (lane 2) tripods. Gel lane 3 is a 1-kb ladder. Gel electrophoresis involves 1.5% native agarose gel and ice water bath. (C and D) Two schemes of connector designs and corresponding gel electrophoresis results. For each scheme, the strand model depicts the connection between two pairs of DNA duplexes. The number above a gel lane denotes the number of connected helices between two adjacent arms. Lane L is a 1-kb ladder, and lane S is a scaffold. Arrowheads indicate the bands corresponding to assembled cubes. (C) Scheme i: long (30 nt) connector (red) including a 2-nt sticky end. The complete 30-nt connector is only shown on the left, with a 28-nt segment anchored on the left helices and a 2-nt exposed sticky end available for hybridization with the 90°-90°-90° right neighbor (dashed circle depicts hybridization site). (D) Scheme ii: short (11 nt) connector including a 2-nt sticky end. (E) Assembly yields of the cubes, calculated as intensity ratio between a cube band and the corresponding scaffold band. (F) Agarose gel electrophoresis of the polyhedra. Lane 1 is the 90°-90°-90° monomer. Lanes 2 to 6 are polyhedra. Lane 7 is the assembly reaction containing tripods without struts. Lane 8 is the assembly reaction containing 90°-90°-90° tripods without vertex helices. Lane 9 is a 1-kb ladder. Gel bands corresponding to desired products are marked with arrowheads. Gel electrophoresis involves 0.8% native agarose gel and ice water bath.
Connector Designs
The strands connecting the tripods are called “connectors.” Connector designs affected the polyhedra assembly yields. Two designs were tested for the cube. In scheme i, each 30-base connector spanned two adjacent tripods, with a 28-base segment anchored on one tripod and another 2-base (sticky end) anchored on the other (Fig. 2C and fig. S3). Gel electrophoresis (shown in Fig. 2C and quantified in Fig. 2E) revealed that the assembly yield was affected by the number of connected helices (n): a product band was only observed for 4 ≤ n ≤ 12; for n < 4, the dominant bands were monomers, likely reflecting overly weak inter-monomer connections; for n > 12, aggregations dominated.
In scheme i, the connectors were stably anchored (forming 28 base pairs) on tripods before inter-monomer connection occurred. In scheme ii, the connector was shortened from 30 to 11 bases so that it should only be anchored to two adjacent tripods by 9- and 2-base segments in the assembled cube (Fig. 2D) and only dynamically binds to a monomeric tripod. Compared with the stably attached connector design, the dynamic connector design could potentially help correct inter-monomer mismatches that occurred during the assembly because such mismatches would be less likely frozen in a kinetic trap. Indeed, scheme ii showed substantially increased assembly yield (Fig. 2E). It was thus used for subsequent polyhedra designs, except for the tetrahedron, in which scheme i produced sufficient yield for this relatively simple structure. The assembly yields were estimated from the gel (Fig. 2F). The 90°-90°-90° monomer sample (Fig. 2F, lane 1) showed a strong monomer band and a putative dimer band (not studied with TEM, ~27% intensity as compared with the monomer). We define the assembly yield of a polyhedron as the ratio between its product band intensity and the combined intensity of the 90°-90°-90° monomer and dimer bands (lane 1) and obtained yields of 45, 24, 20, 4.2, and 0.11% for the tetrahedron, the triangular prism, the cube, the pentagonal prism, and the hexagonal prism, respectively (Fig. 2F).
Polyhedra Assembly
The lengths and the attachment points of the struts varied for each polyhedron (Table 1). The tetrahedron, the triangular prism, the cube, the pentagonal prism, and the hexagonal prism should be assembled from monomers with designed 60°-60°-60°, 90°-90°-60°, 90°-90°-90°, 90°-90°-108°, and 90°-90°-120° angles, respectively (Fig. 1A). The first three monomers indeed produced tetrahedra, triangular prisms, and cubes [verified with gel electrophoresis (Fig. 2F) and TEM imaging (Fig. 3, A to C)], suggesting accurate control for angles within 90°. However, the pentagonal prism was assembled from monomers with designed angles of 90°-90°-120° (instead of 90°-90°-108°), and the hexagonal prism from 90°-90°-140° (instead of 90°-90°-120°). Thus, the assembly of these two polyhedra requires monomers with designed Y-Z angles greater than those of the design criteria. This requirement likely reflects slight bending of the relevant struts, which could be compensated by using longer struts.
Table 1. Strut designs of the polyhedra.
| i | ii | iii | iv | v | vi | |
|---|---|---|---|---|---|---|
| Tetrahedron | 28 | 28 | 28 | 29 | 29 | 29 |
| Triangular prism | 18 | 26 | 26 | 18 | 18 | 18 |
| Cube | 30 | 30 | 30 | 21 | 21 | 21 |
| Pentagonal prism | 32 | 26 | 26 | 19 | 18 | 18 |
| Hexagonal prism | 37 | 28 | 28 | 20 | 20 | 20 |
All units are nanometers. Entries are the designed length of the strut connecting the (i) Y-arm and Z-arm, (ii) X-arm and Z-arm, or (iii) X-arm and Y-arm or the designed distance from the vertex to the strut attachment point on the (iv) X-, (v) Y-, or (vi) Z-arm.
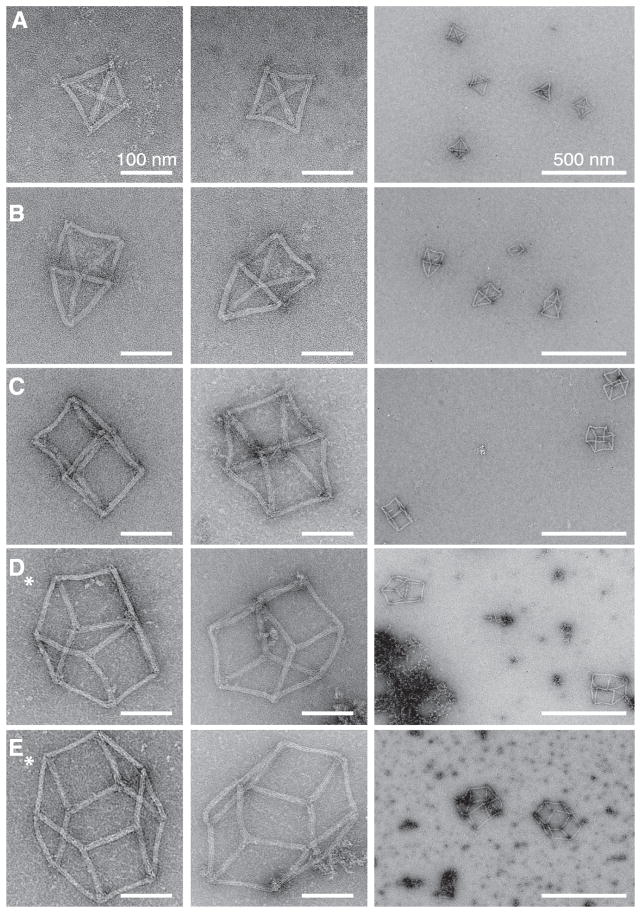
Fig. 3. TEM images of polyhedra.
The zoomed-in (columns 1 and 2) and zoomed-out (column 3) images are shown for (A) the tetrahedron, (B) the triangular prism, (C) the cube, (D) the pentagonal prism, and (E) the hexagonal prism. Images of the tetrahedron, the triangular prism, and the cube were acquired from purified samples. Images of the pentagonal prism and the hexagonal prism were collected from crude samples (denoted with an asterisk). Scale bars are 100 nm in the zoomed-in TEM images and 500 nm in the zoomed-out images. Aggregates are clearly visible for unpurified samples [such as in (D), right].
Effects of Struts and Vertex Helices
We next verified that both the struts and the vertex helices were required for the tripods to assemble into the designed polyhedron. Three samples were prepared for cube assembly by using tripods that contain (i) both the struts and the vertex helices (Fig. 2F, lane 4), (ii) the vertex helices but not the struts (lane 7), and (iii) the struts but not the vertex helices (lane 8); the latter samples were subjected to gel electrophoresis after annealing. The first sample showed a sharp strong band corresponding to the cube (verified with TEM) (Fig. 3C). The second failed to produce any clear product band. The third produced substantial aggregates and a clear but weak band with mobility comparable with that of the triangular prism. This band may correspond to a hexamer, but its molecular morphology was not investigated. On the basis of the above experiments, we included both the struts and the vertex helices in the tripods for subsequent polyhedra assembly.
TEM Characterization
Product bands were purified and imaged under TEM. For the tetrahedron, the triangular prism, and the cube, most structures appeared as intact polyhedra; a small fraction of broken structures (< 20%) were likely ruptured during the purification and imaging (Fig. 3, A to C). In contrast, few intact structures were observed for the purified pentagonal and hexagonal prisms. Thus, unpurified samples for these two were directly imaged, and the expected molecular morphologies were observed (Fig. 3, D and E). Additional images are provided in figs. S4 to S13. The struts are clearly visible in many images (a zoomed-in example is provided in fig. S14).
3D DNA-PAINT Super-Resolution Microscopy
Localization-based 3D super-resolution fluorescence microscopy (31–33) offers a minimally invasive way to obtain true single-molecule 3D images of DNA nanostructures in their “native” hydrated environment (distorted and broken tetrahedra, likely caused by the TEM imaging conditions, are shown in figs. S4 and S15). In stochastic reconstruction microscopy (34), most molecules are switched to a fluorescent dark (OFF) state, and only a few emit fluorescence (ON state). Each molecule is localized with nanometer precision by fitting its emission to a 2D Gaussian function. In DNA-PAINT, the “switching” between ON and OFF states is facilitated through the repetitive, transient binding of fluorescently labeled oligonucleotides (“imager” strands) to complementary “docking” strands (24, 28, 29, 35).
We extended DNA-PAINT to 3D imaging (29) using optical astigmatism (31, 36), in which a cylindrical lens used in the imaging path “converts” the spherical point spread function (PSF) of a molecule to an elliptical PSF when imaged out of focus. The degree and orientation of the elliptical PSF depends on the displacement and direction of the point source from the current focal imaging plane and is used to determine its z position (31, 36). We applied 3D DNA-PAINT to obtain subdiffraction-resolution single-molecule images of the polyhedra. To ensure all the vertices of a polyhedron will be imaged, each vertex is modified with multiple (~18) 9-nucleotide (nt) docking strands (Fig. 4A1) in a symmetric arrangement (fig. S2). For surface immobilization, a subset of strands along the polyhedron edges were modified with 21-nt extensions, which were hybridized to biotinylated complementary strands attached to a streptavidin covered glass slide [(27), DNA sequences].
Fig. 4. 3D DNA-PAINT super-resolution fluorescence imaging of polyhedra.
(A1) Staple strands at the vertices of each polyhedron were extended with single-stranded docking sequences for 3D DNA-PAINT super-resolution imaging. (A1 to E1) Schematics of polyhedra, with DNA-PAINT sites highlighted in green. (A2 to E2) 3D DNA-PAINT super-resolution reconstruction of typical polyhedra shown in the same perspective as depicted in A1 to E1. (A3 to E3) 2D x-y projection. (A4 to E4) 2D x-z projection. (A5 to E5) Height measurements of the polyhedra obtained from the cross-sectional histograms in the x-z projections. (F) A larger 2D super-resolution x-y projection view of tetrahedra and drift markers (bright individual dots). The diffraction-limited image is superimposed on the super-resolution image in the upper half. (G) Tilted 3D view of a larger-field-of-view image of the tetrahedron. Drift markers appear as bright individual dots. Scale bars, 200 nm. Color indicates height in the z direction.
Using 3D DNA-PAINT microscopy, all five polyhedra showed designed 3D patterns of vertices (Fig. 4, columns 1 to 4) with expected heights (Fig. 4, A5 to E5), suggesting that the solution shape of the structures is maintained during surface immobilization and imaging. We quantified the tetrahedra formation and imaging yields (Fig. 4, F and G, and fig. S16). Out of 285 structures, 253 (89%) contained four spots in the expected tetrahedral geometry. Height measurement yielded 82 ± 15 nm, which is consistent with the designed value (82 nm). Single DNA-PAINT binding events were localized with an accuracy of 5.4 nm in x-y and 9.8 nm in z [(27) describes how localization accuracy was determined]. This z localization accuracy almost completely accounts for the 15-nm spread in the height measurement distribution. The calculated localization precisions translate to an obtainable resolution of ~13 nm in x and y, and ~24 nm in z. Movies S1 and S2 are 3D DNA-PAINT videos. Design is provided in figs. S17 to S22, and tables S1 to S7 have sequence details.
Discussion
Previous work demonstrated diverse DNA polyhedra self-assembled from small three-arm-junction tiles (~80 kD) (16, 23), which consist of three double-helix arms connected by flexible single-stranded hinges. However, straightforward implementation of megadalton three-arm origami tiles by use of similar flexible inter-arm hinges (tripods with no struts or vertex helices) failed to produce well-formed polyhedra (Fig. 2F, lane 7). An origami tripod contains 50 times more distinct strands than do previous three-arm-junction tiles (formed from three distinct strands) and is 60 times more massive in molecular weight. Apart from the challenges associated with the more error-prone construction of the more complex monomers from individual strands, successful hierarchical assembly of such large monomers into polyhedra also needs to overcome much slower reaction kinetics, caused by the larger size and lower concentration of the tripod monomers. The stiff DNA tripods, with rationally designed inter-arm angles controlled by supporting struts and vertex helices, lead to successful construction of diverse polyhedra, suggesting that conformation control of branched megadalton monomers can facilitate their successful assembly into higher-order structures.
The design principles of DNA tripods may be extended to stiff megadalton n-arm (n ≥ 4) branched motifs with controlled inter-arm angles. Self-assembly with such n-arm motifs could be used to construct more sophisticated polyhedra and potentially extended 2D and 3D lattices with ≤100-nm tunable cavities. Such structures could potentially be used to template guest molecules for diverse applications—for example, spatially arranging multiple enzymes into efficient reaction cascades (37) or nanoparticles to achieve useful photonic properties (38, 39). Furthermore, the DNA polyhedra constructed here, with a size comparable with those of bacterial microcompartments, may potentially be used as skeletons for making compartments with precisely controlled dimensions and shapes by wrapping lipid membranes around their outer surfaces (40). Such membrane-enclosed microcompartments could potentially serve as bioreactors for synthesis of useful products or as delivery vehicles for therapeutic cargo (25).
For 3D characterization of DNA nanostructures, super-resolution fluorescence microscopy (such as 3D DNA-PAINT) provides complementary capabilities to present electron microscopy [such as cryogenic electron microscopy (cryo-EM) (12, 16, 17, 23)]. Whereas cryo-EM offers higher spatial resolution imaging of unlabeled structures, DNA-PAINT is less technically involved to implement, obtains true single-molecule images of individual structures (rather than relying on class averaging), and preserves the multicolor capability of fluorescence microscopy (29). Additionally, DNA-PAINT in principle allows for observation of dynamic structural changes of nanostructures in their “native” hydrated environment, which is currently suitable for slow changes on the minutes time scale (such as the locomotion of synthetic DNA walkers) and potentially for faster motions with further development.
Supplementary Material
Acknowledgments
We thank S. Woo and W. Shih for discussions. This work is supported by an Office of Naval Research (ONR) Young Investigator Program Award (N000141110914), ONR grants (N000141010827 and N000141310593), an Army Research Office grant (W911NF1210238), a National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD007292), a NIH Transformative Research Award (1R01EB018659), a NIH grant (5R21HD072481), a National Science Foundation (NSF) Faculty Early Career Development Award (CCF1054898), a NSF Expedition in Computing Award (CCF1317291), NSF grants (CCF1162459, CMMI1333215), a gift from JSR corporation, and a Wyss Institute for Biologically Inspired Engineering Faculty Startup Fund to P.Y. R.I. acknowledges support from JSR corporation. R.J. acknowledges support from the Alexander von Humboldt Foundation through a Feodor-Lynen Fellowship. R.I., Y.K., and R.J. contributed equally.
Footnotes
R.I. and Y.K. designed the system, conducted the experiments, analyzed the data, and wrote the paper. R.J. designed and performed the super-resolution study, analyzed the data, and wrote the paper. T.S. and J.B.W. performed the super-resolution experiments and analyzed the data. P.Y. conceived, designed, and supervised the study; interpreted the data; and wrote the paper. All authors commented on and approved the manuscript.
References and Notes
- 1.Chen JH, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 2.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 3.Rothemund PW, Papadakis N, Winfree E. PLOS Biol. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothemund PW. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 5.Douglas SM, et al. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng J, et al. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei B, Dai M, Yin P. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke Y, Ong LL, Shih WM, Yin P. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han D, et al. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 10.Linko V, Dietz H. Curr Opin Biotechnol. 2013;24:555–561. doi: 10.1016/j.copbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Seeman NC. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 12.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 13.Goodman RP, et al. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 14.Aldaye FA, Sleiman HF. J Am Chem Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 15.Erben CM, Goodman RP, Turberfield AJ. J Am Chem Soc. 2007;129:6992–6993. doi: 10.1021/ja071493b. [DOI] [PubMed] [Google Scholar]
- 16.He Y, et al. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, et al. Proc Natl Acad Sci USA. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann J, Cebulla MP, Mönninghoff S, von Kiedrowski G. Angew Chem Int Ed Engl. 2008;47:3626–3630. doi: 10.1002/anie.200702682. [DOI] [PubMed] [Google Scholar]
- 19.Andersen ES, et al. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 20.Ke Y, et al. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 21.Bhatia D, et al. Angew Chem Int Ed Engl. 2009;48:4134–4137. doi: 10.1002/anie.200806000. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, et al. Nat Chem. 2009;1:390–396. doi: 10.1038/nchem.290. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, et al. J Am Chem Soc. 2009;131:1413–1415. doi: 10.1021/ja809666h. [DOI] [PubMed] [Google Scholar]
- 24.Smith DM, et al. J Nucleic Acids. 2011;2011:360954. doi: 10.4061/2011/360954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 26.Nie Z, et al. Chem Commun (Camb) 2013;49:2807–2809. doi: 10.1039/c3cc39177a. [DOI] [PubMed] [Google Scholar]
- 27.Materials and methods are available as supplementary materials on Science Online.
- 28.Jungmann R, et al. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 29.Jungmann R, et al. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharonov A, Hochstrasser RM. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B, Wang W, Bates M, Zhuang X. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shtengel G, et al. Proc Natl Acad Sci USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmied JJ, et al. Nano Lett. 2013;13:781–785. doi: 10.1021/nl304492y. [DOI] [PubMed] [Google Scholar]
- 34.Hell SW. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 35.Lin C, et al. Nat Chem. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao HP, Verkman AS. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu J, Liu M, Liu Y, Woodbury NW, Yan H. J Am Chem Soc. 2012;134:5516–5519. doi: 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acuna GP, et al. Science. 2012;338:506–510. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- 39.Kuzyk A, et al. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 40.Langecker M, et al. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.