Abstract
RNA polymerases from phage-infected bacteria and mammalian cells have been shown to bypass single-strand breaks (SSBs) with a single nucleotide gap in the template DNA strand during transcription elongation; however, the SSB bypass efficiency varies significantly depending upon the backbone end-chemistries at the break. Using a reconstituted T7 phage transcription system (T7 RNAP) and RNA polymerase II (RNAPII) in HeLa cell nuclear extracts, we observe a slight reduction in transcription arrest at SSBs with no gap as compared to those with a single nucleotide gap. We have shown that biotin and carbon-chain moieties linked to the 3′ side, and in select cases the 5′ side, of a SSB in the template strand strongly increase transcription arrest when compared to unmodified SSBs. We also find that a small carbon-chain moiety linked to the upstream side of a SSB aids transcriptional bypass of SSBs for both T7 RNAP and RNAP II. Analysis of transcription across SSBs flanked by bulky 3′ adducts reveals the ability of 3′ end-chemistries to arrest T7 RNAP in a size dependent manner. T7 RNAP is also completely arrested when 3′ adducts or 3′-phosphate groups are placed opposite 5′-phosphate groups at a SSB. We have also observed that a biotinylated thymine in the template strand (without a break) does not pose a strong block to transcription. Taken together these results emphasize the importance of the size of 3′, but usually not the 5′, end-chemistries in arresting transcription at SSBs, substantiating the notion that bulky 3′ lesions (e.g. topoisomerase cleavable complexes, 3′-phosphoglycolates and 3′-unsaturated aldehydes) pose very strong blocks to transcribing RNA polymerases. These findings have implications for the processing of DNA damage through SSB intermediates and the mechanism of SSB bypass by T7 RNAP and mammalian RNAPII.
Single strand breaks (SSBs) are disruptions in the DNA backbone that often include the loss of one nucleotide and damage to the 3′ and 5′ termini that surround the break. They are frequently caused by reactive oxygen species (ROS), which can disintegrate backbone sugars by direct attack or initiate SSB-inducing base excision repair (BER) by damaging DNA bases (most commonly guanine). Other types of DNA base incongruities that are processed into SSB intermediates during BER include uracil, which results from cytosine deamination, and alkylated base lesions such as 3-methyladenine. Aborted topoisomerase I (TOPI) activity also results in a SSB, leaving the covalently bound TOPI on the 3′-terminus at the break. SSBs along with abasic sites, which are processed through a SSB intermediate, represent the prevalent form of endogenous DNA damage in living cells.1-5
SSB repair begins with the processing of DNA end-groups at the break to a 5′-phosphate (5′-PO4) and a 3′-hydroxyl (3′-OH) group. There are several types of damaged DNA ends that require further processing. Direct oxidative damage of the DNA backbone by ROS results in 3′-phosphate (3′-PO4) and 3′-phosphoglycolate groups at a SSB. The removal of a damaged base during BER and the removal of covalently bound 3′-TOPI by the phosphodiesterase TDP1 also leaves a 3′-PO4 at a SSB. Other damaged SSB termini include 5′-hydroxyl (5′-OH) groups, which are products of TDP1 activity and direct oxidative backbone damage, and 3′-unsaturated aldehydes (4-hydroxy-2-pentenal), which can arise during BER.1-3, 6
SSBs in the template strand (TS) of a transcribed sequence are bypassed during transcription in vitro by phage, prokaryotic and eukaryotic RNA polymerases, at efficiencies that depend significantly on the 5′ and 3′ end-chemistries at the break. SSBs with 5′-OH∣3′-OH, 5′-PO4∣3′-OH, 5′-dRP∣3′-OH and 5′-OH∣3′-PO4 end-chemistries have been shown to cause less than 50% arrest of T7 RNA polymerase (T7 RNAP) in vitro.7 In contrast, 5′-PO4∣3′- PO4 SSBs pose a very strong block to transcribing polymerases, causing nearly complete arrest of T7 RNAP and E. coli RNA polymerase (E. coli RNAP) in vitro and 85% arrest of RNA polymerase II (RNAP II) in HeLa cell nuclear extracts.7-9 SSBs with 3′-aldehyde groups are also very strong blocks to T7 RNAP, RNAP II and E. coli RNAP.7-9
The extent of TS SSB bypass by RNA polymerases is also influenced by the size of the gap at the SSB. Most in vitro experiments that explore the effects of DNA end-chemistries on polymerase bypass have been carried out on substrates with SSBs at the site of a missing nucleotide.7-9 However, T7 RNAP has been shown to bypass SSBs in the TS with gaps up to 24 nt.10, 11 Transcription across gaps by T7 RNAP and E. coli RNAP in vitro results in shortened transcripts, lacking the code for the nucleotides that were missing at the SSB.7, 8, 10
In vivo, SSBs resulting from BER of 8-oxo-guanine as well as abasic sites, which arrest T7 RNAP and RNAP II transcription in vitro, enhance transcriptional arrest.12-14 SSBs with covalently bound 3′-TOPI lesions are blocks to transcription in vitro and in mammalian cells.15-18 When present in either the TS or the non-template strand (NTS), SSBs can also lead to an overall reduction in gene expression.19, 20
Since SSBs do not appear to be preferentially repaired in transcribed DNA21, 22, their impact on transcriptional miscoding and mutation is likely significant. In contrast to the in vitro data, the results for transcription in vivo across SSBs with one nucleotide gaps, abasic sites, uracil or 8-oxo-guanine in the TS show transcriptional miscoding opposite the lesion.20, 23, 24 This process, known as transcriptional mutagenesis (TM), has been documented during the transcription of damaged plasmids in E. coli and mammalian cells.20, 23, 24 In particular, there is evidence to suggest that SSBs in the TS with unprocessed 3′-PO4 termini, despite being strong blocks to transcription in vitro, are more likely to be bypassed and result in TM than other types of SSB.20
We set out to learn the characteristics of SSBs that influence transcriptional arrest and bypass at SSBs in the TS. To accomplish this, we have tested the effects of various bulky chemical DNA end adducts at SSBs on transcription by T7 RNAP and RNAP II. Furthermore, since this had not been studied systematically in vitro, we sought to compare the effect of a single nucleotide gap to that of a simple strand break with no gap in contributing to transcription arrest at SSBs. Our results shed light on some of the factors that contribute to the arrest of T7 RNAP and RNAP II at SSBs. In particular, we have shown that bulky adducts at 3′ damaged termini are influential in causing T7 RNAP and RNAP II transcription arrest at SSBs. These results raise speculation about whether transcription-coupled DNA repair might occasionally occur at these sites.
Materials and Methods
DNA Substrates
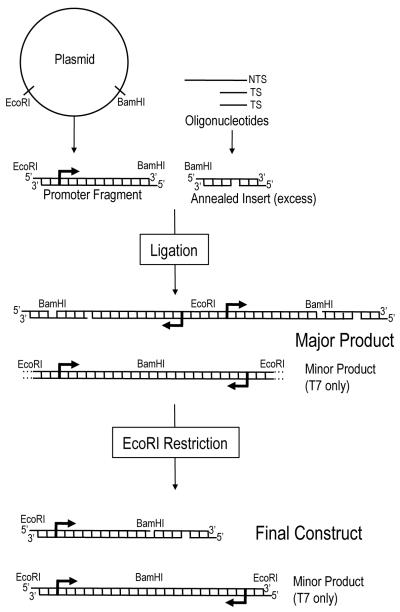
DNA transcription substrates were prepared using the protocol described previously (Figure 1).25 Substrates consisted of two parts – a promoter fragment isolated from plasmid DNA and an “insert” containing a SSB in the TS. Two different promoter fragments, containing either the T7 RNAP promoter or the RNAP II cytomegalovirus (CMV) promoter were prepared via EcoRI/BamHI restriction of the pWT or pWT-C plasmid or EcoRI/BseRI restriction of the pCMVbeta plasmid (Clontech) respectively.25, 26 Promoter-containing DNA fragments were then purified via agarose gel purification without exposure to UV or ethidium bromide, as described previously.25
Figure 1.
Protocol for making transcription substrates, which consisted of two parts: a promoter-containing fragment, which was obtained via restriction from plasmid DNA and an insert, which consisted of three oligonucleotides that were annealed such that a site-specific SSB with various flanking termini was present in the transcribed strand. Inserts were ligated to promoter fragments via BamHI sticky ends for T7 RNAP substrates and BseRI sticky ends for RNAPII substrates. Ligation was followed by EcoRI digestion to obtain the final substrates containing one promoter fragment (designated by the arrow) and one insert. For T7 RNAP substrates, NTS oligonucleotides were not phosphorylated, to prevent insert self-ligation, but this left a nick in the NTS of final constructs. Furthermore, a minor fraction of T7 RNAP substrates, consisting of promoter fragments self-ligated through BamHI sites still appeared but did not interfere with transcription experiments. NTS oligonucleotides were phosphorylated for RNAP II substrates because BseRI sticky ends were asymmetrical, thus they did not contain a nick in the NTS. This also prevented the formation of any minor products during RNAP II substrate fabrication. Sticky ends on the downstream side of substrates were not used in this study.
Annealed oligonucleotide inserts (Figure S1A) consisted of three PAGE-purified oligonucleotides from Integrated DNA Technologies (IDT): a long NTS oligo, a short complementary TS oligo upstream of the SSB and a short complementary TS oligo downstream of the SSB. Some NTS oligos contained a thymine opposite the SSB site, such that SSBs in these substrates encompassed a single nucleotide gap (black dot, Figure S1A). Other NTS oligos did not include this thymine, permitting the construction of substrates containing a SSB without a gap. The TS custom oligos were made with different 5′ (for the upstream oligo) and 3′ (for the downstream oligo) end-chemistries so that SSBs with various termini could be generated. Oligos were annealed by incubating the NTS oligo (2 μM) with an excess of both TS oligos (4 μM) in a solution containing 10 mM Tris-HCl and 10 mM MgCl2 at 65°C for 20 min, followed by 1 hour at room temperature.
Annealed inserts were ligated to T7 promoter fragments in reactions containing T7 promoter fragment (14 nM), annealed insert (565 nM), 1X T4 DNA ligase buffer (NEB) and T4 phage DNA ligase (100 U/μL) in a total volume of 20 μL overnight at 16 °C. The excess of insert was necessary during the T7 ligation reaction in order to reduce the probability of promoter fragments ligating to themselves via BamHI sticky ends (Figure 1, minor products). Annealed inserts were ligated to CMV promoter fragments in reactions containing CMV promoter fragment (30 nM), annealed insert (100 nM), 1X T4 DNA ligase buffer (NEB) and T4 phage DNA ligase (100 U/μL) in a total volume of 20 μL overnight at 16°C. Inserts were not present in significant excess for CMV reactions because BseRI sticky ends were asymmetrical. Thus there was no concern that CMV promoters or inserts would ligate to themselves. Following overnight incubation, both CMV and T7 reaction mixtures were heated at 65°C for 30 minutes to inactivate ligase, and then digested with EcoRI to obtain constructs containing one promoter fragment and one insert (Figure 1, final construct). In the case of T7 substrates NTS oligos were not 5′ phosphorylated (which prevents self-ligation of the insert via symmetrical sticky ends) and final constructs contained a nick at the site of ligation in the NTS. This NTS nick was shown to have no significant effect on transcription.25 For substrates containing 5′- PO4∣3′-OH nicks, the TS oligo downstream of the SSB was added to the construct mixture after T4 DNA ligase had been heat-inactivated. This ensured that the SSB of interest would not be ligated during construction. Final constructs were analyzed by gel electrophoresis on 1.5% agarose gels and visualized by EtBr staining and exposure to UV light (Figure S2).
The double-strandedness of inserts downstream of SSB sites was confirmed by SalI restriction. This quality check ensured that the majority of SSB-induced blockage during in vitro transcription was not due to incomplete annealing. Restriction assays were carried out on radiolabeled, annealed oligonucleotide inserts (not ligated to promoter fragments) except in the case of 5′-PO4∣3′-OH substrates, where final constructs were used. Radiolabeling reactions were performed in a total volume of 12.5μL containing 208 nM annealed inserts, 1X T4 kinase forward reaction buffer (Invitrogen), 12μCi [©-32P] ATP and 5 U of T4 polynucleotide kinase (Invitrogen). The reaction was incubated for 1-min at 37 °C. Radiolabeled products were purified using the QIAquick nucleotide removal kit. The restriction assay was carried out in a total volume of 50 μL containing 0.83 pmol of radiolabeled annealed insert, 1X NEBuffer 3 (NEB) and 1X BSA (NEB) with or without 20 U of SalI. Reactions were incubated for 2hrs at 37°C. EDTA was added to a concentration of 20 mM and the restriction enzyme was inactivated by heating at 65°C for 20min. Solutions were diluted 1:20 in water and run on an 8% polyacrylamide gel containing 8M of urea for 2 hours at 2000V. Results were visualized using a BioRad personal molecular imager system. SalI restriction percentage was calculated as the percent of restricted NTS oligo (Figure S1B/C).
T7 RNAP Transcription
Reactions were performed in a total volume of 12μl, containing 2μl of 5X transcription buffer (Promega, 40mM Tris (pH 7.9), 6mM MgCl2, 2mM spermidine, 10mM NaCl), 4mM DTT, 16U of RNAsin (Promega), 0.17mM each of ATP, GTP and UTP, 0.017mM of CTP, 10μCi of [α-32P] CTP, 20U of T7 RNAP (Promega) and ≈1nM of corresponding DNA substrate. The reactions were carried out at 37°C for 30min. The reaction was stopped and transcription products were precipitated as described previously.26 Samples were run in a 5% polyacrylamide gel containing 8M of urea for 1.5 hours at 2000V. Results were visualized using a BioRad personal molecular imager system.
RNAPII Transcription in HeLa Cell Nuclear Extracts
Reactions were performed in two stages – initiation and elongation. Initiation reactions were performed in a total volume of 18μL containing 4nM CMV transcription substrate, 3μL of 1X HeLa nuclear extract transcription buffer (20 mM HEPES (pH 7.9 at 25°C), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol), 4.17mM MgCl2 and 8 units of HeLaScribe nuclear extract (Promega). Initiation reactions were incubated at 28°C for 30min. 7μL of elongation mixture was then added to the initiation reaction for a final elongation reaction volume of 25μL containing, in addition to the reagents present in the initiation reaction, 400 nM ATP, GTP and UTP, 40μCi of [α-32P] CTP, 40U of RNAsin (Promega) and 4% DMSO. Elongation reactions were carried out at 28°C for 30min. To stop the reactions EDTA was added to a final concentration of 6 mM and RNA transcripts were purified using the Qiagen RNeasy MinElute cleanup kit. Samples were run in a 5% polyacrylamide gel containing 8M of urea for 1.5 hours at 2000V. Results were visualized using a BioRad personal molecular imager system.
To test whether the enzymatic activity of proteins in the HeLa cell nuclear extract altered the composition of SSB end-chemistries, incubation was carried out under the same conditions as for the transcription experiments using radiolabeled annealed oligonucleotide inserts (without promoters) instead of CMV transcription substrates. Following the reaction, inserts were isolated using the Qiagen Nucleotide Removal kit and run in a 20% polyacylamide gel containg 8M of urea for 4.5 hours at 2000V. Alterations to SSB end-groups were visualized as a change of gel shift in 20% gel, or a loss of radioactive 5′-PO4 label (Figure S3). To further test the effects of the HeLa cell nuclear extract on SSB end-chemistries, RNAPII transcription initiation reactions were incubated for 1hr. at 28°C and compared side-by-side with reactions that underwent the standard 30min. initiation incubation. This allowed us to observe whether alterations to SSB end-chemistries caused by increased incubation time with enzymes in the nuclear extract altered SSB-induced transcription arrest. All changes in transcription arrest observed from the longer initiation time were less than 5% (data not shown).
Results
T7 Transcription Arrest at SSBs flanked by Hydroxyl and Phosphate groups
To test the effects of different SSB end-chemistries on elongating T7 RNAP, annealed DNA oligonucleotides containing a site-specific SSB were ligated to a DNA fragment containing a T7 promoter sequence (Figure 1). In the case of double stranded DNA substrates that did not contain a SSB the one, well-defined band corresponds to full-length transcription run off (324nt) (Figure 2, Lane 9). Truncated transcription products, which were interpreted as a consequence of transcription arrest, were observed when 5′-OH∣3′-OH, 5′-PO4∣3′-OH, 5′-OH∣3′-PO4 and 5′-PO4∣3′-PO4 SSBs were present in the TS with or without a single nucleotide gap (Figure 2, lanes 1-8). The lengths of these truncated transcripts correspond to the site of the SSB (284nt) in the TS. While all of these SSBs were blocks to transcription, the extent to which they arrested transcription varied (measured as the percentage of truncated transcript in relation to the overall transcription products, normalized by the number of radiolabeled cytosine residues expected in each transcript, see numbers under each lane). All SSBs were tested at least twice and the average percentage arrest is indicated in figure 2. 5′-PO4∣3′-PO4 SSBs were a complete block to T7 transcription (Figure 2, Lanes 7,8) and 5′-OH∣3′-OH SSBs arrested transcription 46% of the time when there was no gap (Figure 2, Lane 1). The 5′-PO4∣3′-OH SSBs arrested transcription ~15% less than the 5′-OH∣3′-PO4 SSBs (Figure 2, Lanes 3-6). There was also a noticeable difference between SSBs containing a 1nt gap and no gap, with no gap causing 6-10% less blockage depending on end-chemistry (Figure 2).
Figure 2.
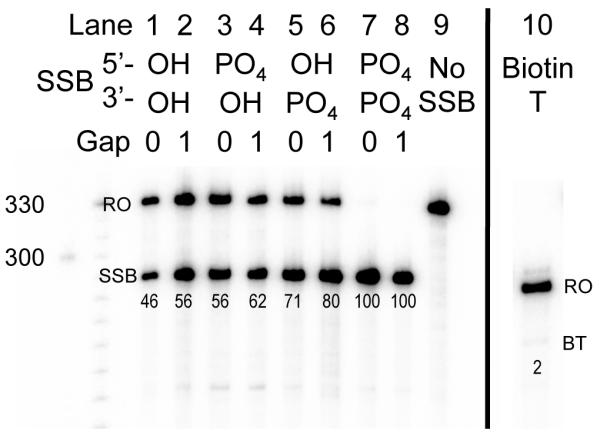
T7 RNAP transcription arrest at SSBs with naturally occurring end-chemistries. Runoff products (324nt) are indicated by RO. Truncated transcripts (SSB) correspond to transcription arrest at the site of the SSB in the TS of transcription substrates (284nt). Percentage of transcription arrest (indicated underneath each lane) was calculated as the radioactive volume of truncated transcripts divided by that volume plus the radioactive volume of runoff products. Each radioactive volume was normalized for the number of radioactively labeled cytosine residues expected in the corresponding transcript. Lane 10 contains transcription products using a transcription substrate that contained biotinylated thymine. This substrate had a runoff (RO) that was 40nt shorter than SSB-containing substrates. Biotinylated thymine (BT) caused negligible arrest.
Bulky Groups at the 3′ Side of a SSB Cause Greater Transcription Arrest Than Those at the 5′ Side
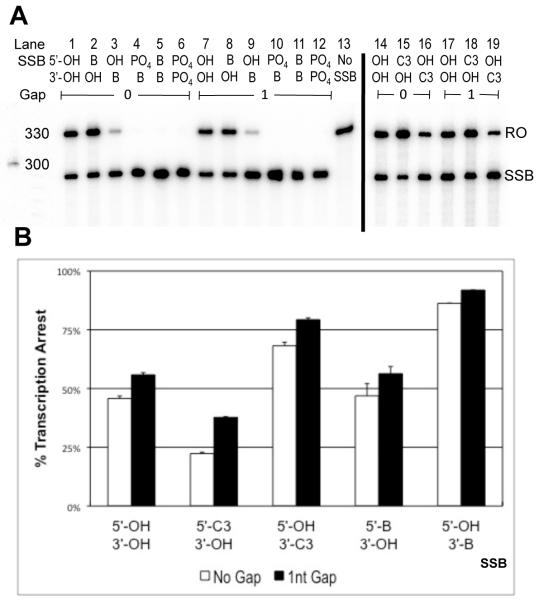
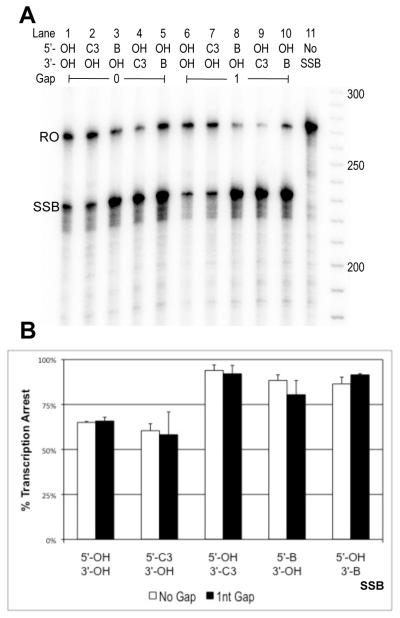
In light of the finding that a phosphate group on the 3′ side of a SSB (5′-OH∣3′-PO4) is a stronger block to T7 transcription than a phosphate group on the 5′ side (5′-PO4∣3′- OH), experiments were performed to determine whether the same trend is observed for other DNA end adducts for both T7 RNAP and RNAPII. Transcription substrates were designed using two different synthetic end-groups (available from IDT), such that mirror image SSBs existed in the TS. One synthetic end-group consisted of a biotin group that was attached to 5′ (5′-biotin, m.w.=393.4) or 3′ (3′-biotin, m.w.=437.4) phosphate DNA ends. The other was a 3-carbon-containing 1,3-propanediol end adduct that was also connected to either 5′ (5′-C3, m.w.=138.1) or 3′ (3′-C3, m.w=138.1) phosphate DNA ends (HO-(CH2)3-O-P(=O)(O−)-3′/5′). For T7 RNAP, 5′-C3∣3′-OH SSBs with no gap and a 1nt gap caused 22% and 38% transcription arrest, respectively, while 5′-OH∣3′-C3 SSBs with no gap and a 1nt gap caused 68% and 79% arrest, respectively (Figure 3, lanes 15, 16, 18, 19). Similarly, 5′-biotin∣3′-OH SSBs with no gap and a 1nt gap caused 47% and 56% arrest respectively as compared to 5′-OH∣3′-biotin SSBs with no gap and a 1nt gap, which caused 86% and 92% arrest respectively (Figure 3, lanes 2, 3, 8, 9). For RNAPII, the C3 synthetic end groups showed asymmetrical blockage patterns. 5′-C3∣3′-OH SSBs with no gap and a 1nt gap caused 60% and 58% arrest for RNAPII respectively (Figure 4, lanes 2, 4, 7, 9). In contrast, 5′-OH∣3′-C3 SSBs caused 94% and 92% arrest (Figure 4, lanes 3, 5, 8, 10). 5′-C3∣3′-OH SSBs caused ~20% less transcription arrest than 5′-OH∣3′-OH SSBs for T7 RNAP and ~5% less for RNAPII. There was also a noticeable increase in arrest caused by substrates with 1nt as compared to those with no gap (Figures 3B, 4B)
Figure 3.
3′, but not 5′, moieties increase T7 transcription arrest at SSBs. A) Radiolabeled T7 RNAP transcription products. The letters RO indicate runoff products (324nt). The letters SSB indicate truncated transcripts with length corresponding to arrest at the SSB in the TS of transcription substrates (284nt). B) Percent blockage was calculated as described before. Each SSB was tested at least twice and the bar graph depicts average arrest for the corresponding SSB end-chemistries. Error bars indicate standard error. SSBs containing 3′ bulky adducts show noticeably greater levels of arrest when compared to their mirror images.
Figure 4.
3′-C3, but not 5′-C3, moieties increase RNAP II transcription arrest at SSBs. A) Radiolabeled RNAP II transcription products. The letters RO indicate runoff products (270nt). The letters SSB indicate truncated transcripts with length corresponding to arrest at the SSB in the TS of transcription substrates (230nt). B) Percent blockage was calculated as described before. Each SSB was tested at least twice and the bar graph depicts average arrest for the corresponding SSB end-chemistries. Error bars indicate standard error. SSBs containing 3′-C3 show noticeably greater levels of arrest when compared to their mirror images, but 5′-OH∣3′-biotin and 5′-biotin∣3′-OH SSBs exhibit similar levels of transcription arrest.
Bulky 3′ Adducts Cause T7 RNAP Transcription Arrest in a Size Dependent Manner
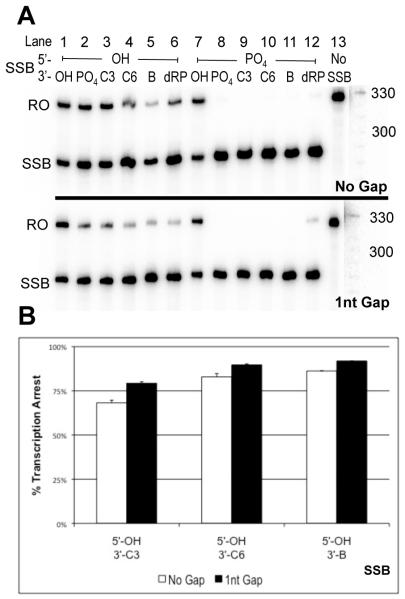
To test the influence of the size of end-termini on T7 RNAP transcription arrest at SSBs, transcription assays were carried out on substrates containing various 3′ synthetic groups opposite 5′-hydroxyl termini at a SSB. The modified 3′ termini consisted of the biotin and C3 groups described above, as well as a 6-carbon 1,6-hexanediol adduct (HO-(CH2)6-O-P(=O)(O−)-3′) (3′-C6) (m.w.=18-.1g/mol) and a 1′-2′-deoxyribose phosphate (3′-dRP) (m.w.=180.1g/mol). The level of SSB-induced transcription arrest observed when these 3′ groups were opposite 5′-OH at an SSB increases with increasing length of the chain 3′ groups (C3<C6<Biotin) (Figure 5B). Furthermore, these adducts cause 100% transcription arrest for T7 RNAP when they are located on the 3′ side of a SSB opposite a 5′-PO4 group in our system (Figure 5A, lanes 9-12). In the case of RNAPII, all synthetic 3′ groups caused very strong arrest, and their quantitative ranking is difficult to evaluate. A decrease in the level of arrest caused by 1nt gap SSBs versus SSBs with no gap was observed for these substrates as well (Figure 5B).
Figure 5.
Transcription arrest increases with increasing size of bulky 3′ groups at SSBs with 5′-OH groups. A) In this figure two gels containing T7 RNAP transcription products are stacked; substrates with no gap in the top panel and substrates with a 1nt gap in the bottom panel. Runoff products (324nt) appear as expected (RO). Truncated transcripts (SSB) have length corresponding to transcription arrest at the site of the SSB in the TS of transcription substrates (284nt). B) Percent blockage was calculated as described before. Each SSB was tested at least twice and the bar graph depicts average arrest for the corresponding SSB end-chemistries. Error bars indicate standard error. SSBs are ordered by increasing size of 3′-adducts from left to right.
An internal biotin modification, without a SSB, causes very minor blockage for T7 RNAP
Since biotin-containing end-groups caused such strong blockage at SSBs, we wanted to determine whether an internal biotin moiety (not at an SSB) in the TS would cause transcription arrest as well. To test this, we prepared T7 transcription substrates containing a biotinylated thymine (BT) in the TS. BT consisted of a thymine with the 6-carbon linker/biotin group described before attached to the C5 carbon of thymine. These substrates were 40 nt shorter than other T7 transcription substrates and did not contain a SSB in the TS. Like the linear control substrates, the one well-defined transcription product from the BT substrates corresponds to the transcription runoff (Figure 2, lane 10). There was ~2% arrest at the site of BT.
Discussion
We found that large adducts on the 5′ side of a SSB in the TS (5′-biotin∣3′-OH, 5′-C3∣3′-OH) cause considerably less transcription arrest than large 3′ adducts (5′-OH∣3′-biotin, 5′-OH∣3′-C3) for T7 RNAP and, to some extent, RNAP II. This result is in accord with the previous studies of Zhou and Doetsch, who reported that SSBs with bulky 3′-termini (5′- OH∣3′-aldehyde, 5′-PO4∣3′-aldehyde) are strong blocks to T7 RNAP, while SSBs with bulky 5′ groups (5′-dRP∣3′-OH) were less significant sources of transcription arrest.7 Thus it is clear that bulky 3′, but not 5′, termini at a SSB have a significant influence on SSB-induced transcription arrest.
This asymmetry in the effect of bulky adducts flanking a SSB on transcription arrest might occur because the RNAP must capture the 3′ (downstream) terminus at the break and reinsert it into the catalytic site to continue transcribing. In contrast, a bulky adduct on the 5′ (upstream) side of a break may cause little interference with transcription because the RNAP is already bound and translocating along the DNA backbone on the upstream side of a SSB. It is also possible that bulky 5′ adducts do not interfere with SSB bypass because they are extruded from the transcription complex during 3′ end-capture, especially if this capture occurs from the post-translocated state of the RNAP, which would leave the 5′-terminus of the SSB farther away from the catalytic center of the polymerase.27-29 This suggests that a bulky backbone adduct, in contrast to a bulky base,30 produces relatively little interference with transcription because it is extruded from the transcription complex. In accordance with that explanation, we found that thymine with a biotinylated linker at the C5 position (which is on the side of the base opposite Watson-Crick base pairing) within an intact template strand produces almost no transcription blockage for T7 RNAP (Figure 2, lane 10). It has also been shown that bulky aminofluorene adducts in the TS do not cause significant arrest of T7 RNAP.33 Both thymine biotinylated at the C5 carbon and aminofluorene contain bulky/flexible adducts that face away from the region of Watson-Crick base pairing. It is possible that this orientation of a bulky base adduct allows for T7 RNAP bypass.
In the case of a 5′-OH∣3′-biotin or 5′-OH∣3′-C3 SSB, 3′ end-capture could be inhibited because the biotin or carbon group is too large or unrecognizable to be efficiently captured and reinserted into the polymerase. In contrast, at a 5′-biotin∣3′-OH or 5′-C3∣3′-OH SSB, the smaller 3′-OH group is probably more easily recognized and captured by T7 RNAP, allowing for a greater extent of SSB bypass. The notion that size plays a role in 3′ terminus recognition and capture is supported by our finding that, at 5′-OH SSBs, T7 RNAP blockage increases with increased size of 3′ end groups (C3<C6<biotin) (Figure 5). Our proposed model for transcription arrest at SSBs supports the hypothesis of Doetsch and coworkers, from template strand “thread-in” experiments, which suggested that T7 RNAP has difficulty restarting transcription on the downstream side of a large gap when there is a 3′-PO4 terminus because of charge and/or size.10
In some instances, we found that bulky 5′ adducts enhance transcription arrest at SSBs. We observed that the substitution of a 5′-PO4 for a 5′-OH group at SSBs containing bulky 3′ groups exacerbates transcription arrest for T7 RNAP (in most cases increasing blockage to 100%) (Figure 5, lanes 8-12). Thus, it seems that, in the case of inhibited 3′ end-capture, the presence of a bulky 5′ adduct at a TS SSB exacerbates transcription arrest. Interestingly, 5′-C3∣3′-OH SSBs caused less transcription arrest than 5′-OH∣3′-OH SSBs for both T7 RNAP and RNAP II, suggesting that the 1,3-propanediol adduct aids SSB bypass when linked to the 5′ side of a break. It is possible that the 5′-C3 adduct, which is less bulky than 5′-biotin, somehow stabilized the transcription complex, permitting more efficient bypass or tracking across the SSB. This sort of interaction may not be possible on the downstream side of the break if the polymerase must actively engage the downstream terminus to continue translocation. We also found that, in contrast to the T7 RNAP results, RNAPII was arrested to nearly the same extent at 5′-biotin∣3′-OH and 5′-OH∣3′-biotin SSBs. These results, however, might be affected by enzymatic reactions with biotin in HeLa extracts (Figure S3).
In addition to our findings regarding end-chemistry effects on transcription arrest at SSBs, we detected a difference between the level of transcription arrest caused by SSBs with no gap and SSBs with a 1nt gap. SSBs with no gap occur most frequently as DNA repair intermediates, either preceding the final ligation step of excision repair pathways or following TDP1 cleavage after abortive TOPI activity. Such SSBs contain 5′-PO4∣3′-OH and 5′-OH∣3′-PO4 end groups respectively. In our study, 5′- PO4∣3′-OH and 5′-OH∣3′-PO4 SSBs with no gap blocked T7 transcription 56% and 71% of the time respectively, suggesting such repair intermediates may pose a block to transcription in vivo. All of the gapless SSBs tested (both naturally occurring and unnatural) caused a slight decrease in transcription arrest when compared to 1nt gap SSBs with the same end-chemistries. This consistent finding suggests that the 1nt gap present at most naturally occurring SSBs increases the likelihood of transcription arrest at those sites.
At the physiological level, our findings with T7 RNAP have implications for the mechanism of arrest and bypass at SSBs by RNAPs, especially human mitochondrial RNA polymerase, which is similar in structure and mechanism to T7 RNAP.34 This relevance is highlighted by the fact that mitochondrial DNA is prone to SSBs due to the elevated levels of ROS observed in mitochondria.35 Furthermore, the observed patterns of enhanced arrest at SSBs with bulky 3′ adducts also arise with RNAP II. We conclude that SSB bypass by T7 RNAP and RNAP II is significantly hindered at SSBs with large 3′ end-chemistries. This blockage may play a part in alterations in gene expression and transcriptional mutagenesis due to SSBs. The RNAP arrest might also trigger a gratuitous form of transcription-coupled DNA repair that could be deleterious.
Supplementary Material
Acknowledgement
We thank Viviana Salinas-Rios for providing experimental materials and we have appreciated helpful discussions with all members of the Hanawalt lab group.
Funding Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number R01CA077712 (to P.CH.), and a UAR Major Grant (to A.J.N.) at Stanford University.
Abbreviations
- SSB
single-strand break
- 5′0PO4
5′ phosphate DNA end-chemistry
- 3′-PO4
3′ phosphate DNA end-chemistry
- 5′-OH
5′ hydroxyl DNA end-chemistry
- 3′-OH
3′ hydroxyl DNA end-chemistry
- 5′-dRP
5′ deoxyribose sugar DNA end-chemistry
- 3′-dRP
3′ deoxyribose sugar DNA end-chemistry
- 3′-C3
3′ 1,3-propanediol DNA end-chemistry
- 3′-C6
3′ 1,6-hexanediol DNA end-chemistry
- TS
transcribed strand
- NTS
non-transcribed strand
- BER
base excision repair
- TOPI
topoisomerase I
- RNAP
RNA polymerase
Footnotes
Supporting Information Available Supporting information contains figures relating to the construction and confirmation of the structure of transcription substrates. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 2.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demple B, DeMott MS. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene. 2002;21:8926–8934. doi: 10.1038/sj.onc.1206178. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Doetsch PW. Transcription bypass or blockage at single-strand breaks on the DNA template strand: effect of different 3′ and 5′ flanking groups on the T7 RNA polymerase elongation complex. Biochemistry. 1994;33:14926–14934. doi: 10.1021/bi00253a032. [DOI] [PubMed] [Google Scholar]
- 8.Zhou W, Doetsch PW. Effects of abasic sites and DNA single-strand breaks on prokaryotic RNA polymerases. Proc Natl Acad Sci U S A. 1993;90:6601–6605. doi: 10.1073/pnas.90.14.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathe SD, Shen GP, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J Biol Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Reines D, Doetsch PW. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995;82:577–585. doi: 10.1016/0092-8674(95)90030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Doetsch PW. Template strand gap bypass is a general property of prokaryotic RNA polymerases: implications for elongation mechanisms. Biochemistry. 1996;35:14999–15008. doi: 10.1021/bi961455x. [DOI] [PubMed] [Google Scholar]
- 12.Khobta A, et al. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair (Amst) 2009;8:309–317. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Kitsera N, et al. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem Res Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 15.Desai SD, et al. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bendixen C, et al. Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry. 1990;29:5613–5619. doi: 10.1021/bi00475a028. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996;17:31–35. doi: 10.1093/carcin/17.1.31. [DOI] [PubMed] [Google Scholar]
- 19.Khobta A, et al. Mouse CSB protein is important for gene expression in the presence of a single-strand break in the non-transcribed DNA strand. DNA Repair (Amst) 2010;9:985–993. doi: 10.1016/j.dnarep.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Clauson CL, et al. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:3657–3662. doi: 10.1073/pnas.0913191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ljungman M. Repair of radiation-induced DNA strand breaks does not occur preferentially in transcriptionally active DNA. Radiat Res. 1999;152:444–449. [PubMed] [Google Scholar]
- 22.May A, Bohr VA. Gene-specific repair of gamma-ray-induced DNA strand breaks in colon cancer cells: no coupling to transcription and no removal from the mitochondrial genome. Biochem Biophys Res Commun. 2000;269:433–437. doi: 10.1006/bbrc.2000.2264. [DOI] [PubMed] [Google Scholar]
- 23.Bregeon D, et al. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 24.Saxowsky TT, et al. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salinas-Rios V, Belotserkovskii BP, Hanawalt PC. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic Acids Res. 2011;39:7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belotserkovskii BP, et al. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 27.Steitz TA. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr Opin Struct Biol. 2004;14:4–9. doi: 10.1016/j.sbi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, Sousa R. T7 RNA polymerase elongation complex structure and movement. J Mol Biol. 2000;303:347–358. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 29.Durniak KJ, Bailey S, Steitz TA. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulrich S, Kool ET. Importance of steric effects on the efficiency and fidelity of transcription by T7 RNA polymerase. Biochemistry. 2011;50:10343–10349. doi: 10.1021/bi2011465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi YB, Gamper H, Hearst JE. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J Biol Chem. 1988;263:527–534. [PubMed] [Google Scholar]
- 32.Choi DJ, et al. Site-specific benzo[a]pyrene diol epoxide-DNA adducts inhibit transcription elongation by bacteriophage T7 RNA polymerase. Biochemistry. 1994;33:780–787. doi: 10.1021/bi00169a020. [DOI] [PubMed] [Google Scholar]
- 33.Chen YH, Bogenhagen DF. Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J Biol Chem. 1993;268:5849–5855. [PubMed] [Google Scholar]
- 34.Ringel R, et al. Structure of human mitochondrial RNA polymerase. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 35.Sykora P, Wilson DM, 3rd, Bohr VA. Repair of persistent strand breaks in the mitochondrial genome. Mech Ageing Dev. 2011 doi: 10.1016/j.mad.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.