Abstract
The identification of genes that modify pathological ocular phenotypes in mouse models may improve our understanding of disease mechanisms and lead to new treatment strategies. Here, we identify modifier loci affecting photoreceptor cell loss in homozygous Mfrprd6 mice, which exhibit a slowly progressive photoreceptor degeneration. A cohort of 63 F2 homozygous Mfrprd6 mice from a (B6.C3Ga-Mfrprd6/J × CAST/EiJ) F1 intercross exhibited a variable number of cell bodies in the retinal outer nuclear layer at 20 weeks of age. Mice were genotyped with a panel of single nucleotide polymorphism markers, and genotypes were correlated with phenotype by quantitative trait locus (QTL) analysis to map modifier loci. A genome-wide scan revealed a statistically significant, protective candidate locus on CAST/EiJ Chromosome 1 and suggestive modifier loci on Chromosomes 6 and 11. Multiple regression analysis of a three-QTL model indicated that the modifier loci on Chromosomes 1 and 6 together account for 26% of the observed phenotypic variation, while the modifier locus on Chromosome 11 explains only an additional 4%. Our findings indicate that the severity of the Mfrprd6 retinal degenerative phenotype in mice depends on the strain genetic background and that a significant modifier locus on CAST/EiJ Chromosome 1 protects against Mfrprd6-associated photoreceptor loss.
Keywords: eye disease, retinal degeneration, MFRP, QTL analysis, modifier genes
1. Introduction
Genetic approaches hold great promise toward revealing the molecular basis of degenerative diseases such as retinitis pigmentosa (RP). RP consists of a heterogeneous group of retinal dystrophies characterized by the progressive death of rod and cone photoreceptor cells (Ayuso and Millan, 2010; Ferrari et al., 2011). The clinical phenotype includes night blindness with deterioration of peripheral visual field and, in many cases, eventual loss of central vision. The causal association of 57 genes with RP (June 2013, Retinal Information Network, http://www.sph.uth.tmc.edu/Retnet/) highlights the success of genetic approaches in the 23 years since a mutation in the first gene associated with autosomal dominant RP, RHO, was documented (Dryja et al., 1990). Despite this success, the molecular processes that link RP-causing mutations and pathological progression remain obscure in most cases.
Part of the difficulty of elucidating the molecular mechanisms underlying the pathologies observed in RP is the considerable variation in the onset, severity and clinical presentation of affected individuals. A major source of variation is genetic heterogeneity. Mutations in any one of the 57 disease-causing genes can yield the clinical RP-like phenotype, each with distinct disease characteristics. Allelic variation, affecting different domains within a gene, is also thought to contribute to differences in phenotypic outcome. However, phenotypic variation is observed even among family members with identical mutations, suggesting an effect of environment or interaction of genetic modifiers with the disease gene (Ayuso et al., 1996; Bandah-Rozenfeld et al., 2010; Berson et al., 2001, 1991a,b; Chang et al., 2007; Jacobson et al., 2000; Langmann et al., 2010; Passerini et al., 2007; Paunescu et al., 2007; To et al., 2002). Modifier genes are able to influence the phenotypic expression of genes in both monogenic and multigenic traits, and may interact directly with the mutant gene or influence the pathological pathways that are induced by the disease allele(s) (Genin et al., 2008; Haider et al., 2002). Therefore, identifying modifier genes for RP may help to explain the variation in disease presentation and therapeutic response, improve our understanding of functional pathways that underlie the retinal degenerative phenotype, and reveal important targets for clinical intervention.
Identifying modifier genes in rare human diseases such as RP is challenging due to small population sizes, environmental variability, and heterogeneous genetic backgrounds. For example, the population size of patients carrying the most common autosomal dominant RP (adRP) allele in North America, the P23H mutation of rhodopsin, is estimated to be ~900 individuals (2013 estimate of North American population × RP incidence × fraction of RP attributed to adRP × fraction of adRP linked to RHO × fraction of RHO RP ascribed to P23H = 470,000,000 × 0.00033 × 0.2 × 0.27 × 0.11).[https://www.cia.gov/library/publications/the-world-factbook/] (Daiger et al., 2007; Sullivan et al., 2006). Population sizes are expected to be smaller for less common alleles and for autosomal recessive RP genes. Even if all individuals with the P23H RHO RP mutation were assessed, variation in phenotype due to non-genetic factors, including age, diet, light exposure history, and differences in clinical assessment would confound efforts to establish gene associations. These difficulties are compounded by the substantial genetic variation in human populations. Although modifier genes have recently been revealed in analyses of very large adRP families with similar genetic backgrounds (Venturini et al., 2012) and in X-linked RP (Fahim et al., 2011), success with identifying modifiers of autosomal recessive RP has been limited.
The identification of modifier genes in mouse models, which allows for the control of environmental, genetic and experimental variation, is an attractive complementary approach to human studies (Hamilton and Yu, 2012). Modifier genes can be discovered in relatively small cohorts (typically 50–300 animals) with well-characterized genetic backgrounds that can be readily generated by crossing inbred strains. A growing number of genes that modify ocular disease phenotypes have been successfully identified in mice, such as Mtap1a, which modifies retinal degeneration associated with Tub and Tulp1 mutations (Ikeda et al., 2002; Maddox et al., 2012); Rpe65, which modifies RP conferred by mutant Rho alleles (Samardzija et al., 2006); and Tyr, which modifies retinoschisis caused by Rs1 mutations (Johnson et al., 2008) [for additional examples, see (Hamilton and Yu, 2012)].
The study presented here identifies candidate genetic loci that modify the phenotype of homozygous Mfrprd6 mice, which exhibit a progressive retinal degeneration as observed in RP (Chang et al., 2002; Hawes et al., 2000; Kameya et al., 2002). A similar phenotype has been described in homozygous Mfrprdx mice (Fogerty and Besharse, 2011). In humans, MFRP mutations are associated with RP in an ocular syndrome that also includes posterior microphthalmos, foveoschisis and optic nerve head drusen (Ayala-Ramírez et al., 2006; Crespí et al., 2008; Mukhopadhyay et al., 2010; Neri et al., 2012; Zenteno et al., 2009). A distinct MFRP-associated syndrome characterized by nanophthalmos without RP has also been described (Sundin et al., 2008, 2005). A prominent feature of homozygous Mfrprd6 and homozygous Mfrprdx mice is the appearance of small discrete spots throughout the fundus, which are likely to correspond to subretinal innate immune cells (Fogerty and Besharse, 2011; Hawes et al., 2000). Similar spots have been documented in individuals with MFRP-associated disease (Neri et al., 2012; Zenteno et al., 2009), suggesting that innate immune cells may contribute to disease pathology. Mfrp encodes a transmembrane protein of un-known function that has been proposed to modulate or regulate Wnt/Frizzled signaling (Kameya et al., 2002; Katoh, 2001). In this study, we show that the severity of the Mfrprd6 retinal degenerative phenotype varies with genetic background. We identify modifier loci that account for this variation and apply bioinformatics to narrow the list of candidate genes that may explain the observed effects.
2. Methods
2.1. Experimental animals
Mice provided with acidified water and JL Rat and Mouse/Auto 4F (5K54) diet (LabDiet, St. Louis, MO) were housed in cages exposed to a 12 h light–dark cycle in The Jackson Laboratory Research Animal Facility. All mice were treated in accordance with the Animal Care and Use Committee at The Jackson Laboratory and in compliance with the Association for Research in Vision and Ophthalmology (ARVO) statement for ethical care and use of animals.
2.2. Mouse production and genotyping
Mutant F2 progeny from an intercross of F1 hybrids of homozygous B6.C3Ga-Mfrprd6/J and CAST/EiJ mice were genotyped by PCR to identify homozygous Mfrprd6 mice, which possess a 4 bp deletion in the splice donor sequence of Mfrp intron 4 (Kameya et al., 2002). The PCR primers rd6delF (5′-CACTACCACCCCAGCAAGGAC-3′) and rd6delR (5′-CTTCTCCAGAGAGTGCCCTTG-3′) flanking the mutation were used to generate 91 and 87 bp products from the Mfrp wild-type and rd6 alleles, respectively, using the following cycling conditions: 97 °C, 3 min; [95 °C, 15 s; 55 °C, 30 s; 72 °C, 30 s] × 50; 72 °C, 3 min; 11 °C hold. The resulting PCR products were resolved by gel electrophoresis in a mixture of 3% Metaphor® and 1% SeaKem® LE agarose (Lonza Rockland, Rockland, ME). Mice were additionally genotyped by allele-specific PCR as described (Chang et al., 2013) to test for Crb1rd8, a common mutation found in laboratory mice (Chang et al., 2013; Mehalow et al., 2003) that is known to influence ocular phenotypes (Mattapallil et al., 2012). For QTL analysis, mice were genotyped by KBio-sciences (Beverly, MA) with a panel of single nucleotide polymorphism (SNP) markers spaced ~20 cM apart on all chromosomes. Additional PCR analyses with the MIT markers D1Mit33 and D1Mit353 were used to refine the candidate modifier locus on Chromosome 1. Finally, to strengthen bioinformatics analysis, B6.C3Ga-Mfrprd6/J mice were screened with a 150-marker SNP panel developed at The Jackson Laboratory that distinguishes C57BL/6J and C57BL/6N.
2.3. Mouse phenotyping
Eyes from 20-week-old F2 Mfrprd6 homozygotes were assessed histologically as previously described (Maddox et al., 2012), with the exception that microscopy was performed with a 40× objective on a Leica DMLB microscope (Leica Microsystems, Buffalo, NY) and images were captured with a DMC-1 digital microscope camera (Polaroid, Minnetonka, MN). Retinal sections in which the optic nerve was at its widest were selected for imaging. Retinal degeneration was quantified by manually counting the number of nuclei in a 450-pixel wide region of the outer nuclear layer (ONL) positioned roughly parallel to the image frame at ~170–230 µm from the edge of the optic nerve.
2.4. QTL analysis
2.4.1. Genome-wide one-dimensional scan
Phenotype and genotype data were imported into R/qtl, version 1.23–16 (Broman et al., 2003), which enables genome-wide one-and two-dimensional (pair-wise) scans, multiple locus modeling and inclusion of covariates. The distribution of ONL nuclei counts was not statistically different from normality (Shapiro–Wilk goodness-of-fit, p = 0.367). Therefore the data were not transformed, as they met the modeling assumption criteria. One thousand permutations were performed to determine the thresholds for QTL detection in genome-wide scans (Doerge and Churchill, 1996). Four thresholds, 1%, 5%, 10% and 63%, were calculated from the permutation results. QTL with a log of the odds ratio (lod) score above the 5% threshold were considered significant, while those above the 63% threshold were considered suggestive (Lander and Kruglyak, 1995).
2.4.2. Genome-wide pair-wise scan
Pair-wise scans were performed using 2-cM spacing. All possible pairs of QTL locations on each chromosome were tested for association with the number of cell bodies remaining within a fixed region of the ONL as described above. As permutations are extremely resource intensive, genome-wide two-dimensional scan significance thresholds were based on previously recommended P < 0.05 thresholds for a mouse intercross (Broman and Sen, 2009).
2.4.3. Multiple QTL modeling
QTL along with possible QTL × QTL interactions derived from a single QTL scan and pair-wise scans were fit into multiple regression models in the presence of significant covariates, if any. By doing so, variations of the phenotype in the models were estimated. The p values for terms in the multiple regression models were calculated. Terms were dropped sequentially until all of the terms in the model were significant at the 1% level for both the main QTL effects and their interaction effects.
3. Results
3.1. Variation in Mfrprd6 retinal degeneration
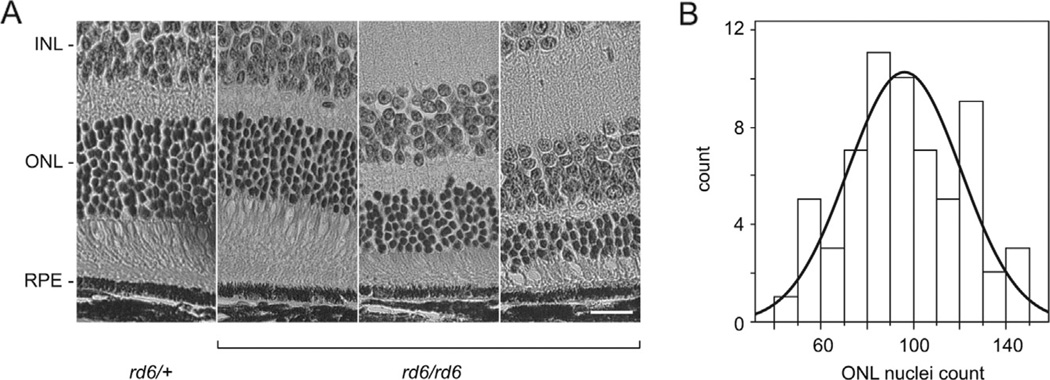
To test whether the Mfrprd6 phenotype varies with strain genetic background, we examined homozygous Mfrprd6 F2 mice from a (B6.C3Ga-Mfrprd6/J × CAST/EiJ) F1 intercross. At 20 weeks of age, the ONL of these mice was thinner than that of heterozygous control mice and varied in thickness (Fig. 1A). The number of nuclei in a 450-pixel wide region (67 µm) of the ONL at a constant distance centered ~200 µm from the optic nerve was counted manually in each of 63 specimens. The frequency distribution of the ONL nuclear count in these samples (Fig. 1B) is consistent with a normal distribution (see Methods). The use of ONL count as a quantitative phenotype does not correct for section obliqueness, which can affect the number of nuclei counted within a fixed area of the histological section. Experimental variation in ONL thickness due to section obliqueness is unlikely to exceed 15–20%, as eyes are oriented similarly prior to embedding, and variation of this magnitude is expected to have a minimal effect on QTL analysis.
Fig. 1.
Phenotypic analysis of retinas at 20 weeks of age. (A) ONL thickness of F2 homozygous Mfrprd6 mice varies in a segregating genetic background obtained from a (B6.C3Ga- Mfrprd6/J × CAST/EiJ) F1 intercross. Heterozygous mice (rd6/+, left panel) show normal retinal layers while homozygous Mfrprd6 progeny (rd6/rd6) showed differing degrees of ONL thinning. The Mfrp genotype is indicated. INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium. Bar: 20 µm. (B) Frequency distribution of ONL nuclear counts from 63 F2 homozygous Mfrprd6 mice. The data fit a normal distribution (solid curve).
3.2. QTL analysis
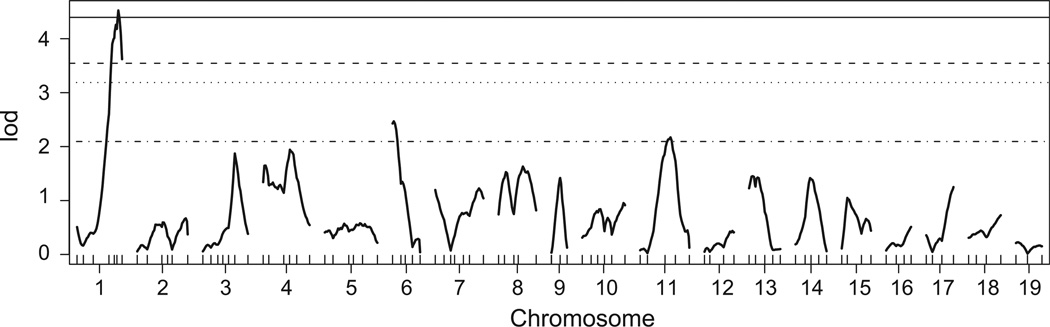
The genotypes of 100 genome markers in 63 F2 mice were determined by whole genome SNP analysis. Although genotyping quality was generally excellent, four of the markers showed a lod score of >3.0 with the checkAlleles() function of R/qtl and two resulted in a larger predicted recombination estimate than expected. These markers were excluded from further analysis. Three of the excluded markers were located on Chromosome 9 near the Mfrp gene, consistent with SNP variation due to the congenic region of B6.C3Ga-Mfrprd6/J. Two of the excluded markers were located on Chromosome 1, where preliminary studies had indicated a candidate modifier locus. Therefore, additional genotyping was performed with the MIT markers D1Mit33 and D1Mit353 and included in the final QTL analysis. A genome-wide one-dimensional scan (Fig. 2) of autosomal markers indicated a significant modifier locus on Chromosome 1 (exceeding the p=0.01 threshold calculated for a genome-wide scan; Fig. 2, solid line) and suggestivemodifiers on Chromosomes 6 and 11 (exceeding the p=0.63 threshold; Fig. 2, dashed-dotted line). Further testing failed to reveal an influence of sex on the phenotype, but left open the possibility that analysis of a larger cohort might reveal an influence of gender. There was no evidence for interaction among modifier loci, leading to a simple three-QTL model.
Fig. 2.
QTL analysis. Genome-wide one-dimensional scan analysis of 63 F2 Mfrprd6 homozygous mice at SNP intervals of ~20 cM yielded a significant modifier locus on Chromosome 1 and suggestive loci on Chromosomes 6 and 11. Sex chromosomes were not included in the analysis as there was no indication of gender effects on the disease phenotype. Genome-wide significance thresholds are indicated at p = 0.63 (dotted-dashed line), 0.10 (dotted line), 0.05 (dashed line) and 0.01 (solid line). lod: log of the odds ratio.
To test the relative importance of the modifier loci, the three-QTL model was examined by multiple regression analysis, in which the effect of eliminating terms one at a time on model quality was explored. As shown in Table 1, the candidate loci on Chromosomes 1 and 6 contribute significantly to the observed phenotypic variation and are required for the model, whereas that on Chromosome 11 may be omitted. Together, the three candidate loci are estimated to account for ~30% of the phenotypic variation, while the loci on Chromosomes 1 and 6 account for a combined 26%.
Table 1.
Multiple regression analysis of the QTL model.
| QTL | dfa | Type III SSb | lodc | %Vard | Fe | p-value (χ2)f | p-value (F)g |
|---|---|---|---|---|---|---|---|
| 1@70.1 cM | 2 | 5742 | 3.31 | 15.44 | 7.66 | 0 | 0.0012** |
| 6@14.1 cM | 2 | 3760 | 2.25 | 10.11 | 5.01 | 0.006 | 0.0099** |
| 11@52.9 cM | 2 | 1739 | 1.09 | 4.67 | 2.32 | 0.082 | 0.1079 |
Values reflect a comparison of the full three-QTL model to the sub-model with the indicated QTL term dropped.
Degree of freedom.
Type III sum of squares.
log of the odds ratio.
Percentage of variance explained.
F statistics.
p value for χ2 distribution.
p value for F distribution with significance threshold <0.01,**; <0.05,*.
3.3. Effect plot and confidence interval of the Chromosome 1 modifier locus
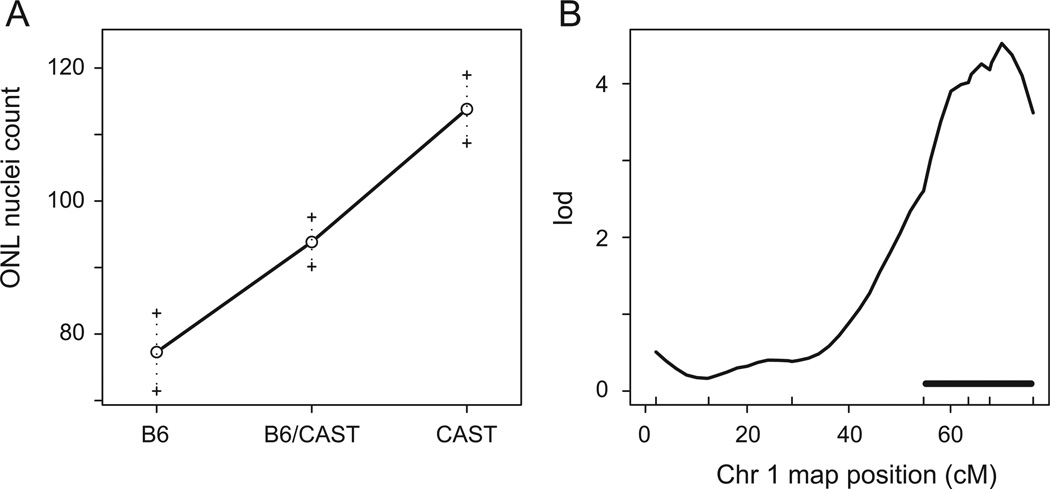
The effect plot (Fig. 3A) of D1Mit33, the marker closest to the Chromosome 1 modifier locus, indicates that the CAST/EiJ sequence protects against the loss of photoreceptors in homozygous Mfrprd6 mice. The plot linearity shows that the Chromosome 1 modifier effect is additive, arguing against dominance of the CAST/EiJ or C57BL/6J allele. The peak marker on Chromosome 1 likely to contain modifier genes at the 95% Bayes credible interval is shown in Fig. 3B. This interval maps to 54.7–76.3 centimorgans of Chromosome 1, which relates to a genomic distance of ~44 Mbp (Chromosome 1124 Mb–168 Mb, GRCm38 build).We designate this locus as retinal degeneration 6 modifier 1 (Rd6m1).
Fig. 3.
Effect plot and confidence interval for the significant modifier locus on Chromosome 1. (A) Effect plot of homozygous (B6, C57BL/6J; CAST, CAST/EiJ) and heterozygous (B6/ CAST) phenotypes indicated an additive, protective effect of the modifier locus on Chromosome 1. (B) A confidence interval of ~22 cM was found for the Chromosome 1 modifier locus. lod: log of the odds ratio; ONL: outer nuclear layer.
3.4. Identification of candidate modifier genes within the Rd6m1 locus
Bioinformatics analysis of the C57BL/6J and CAST/EiJ inbred strains was used to provide useful leads for modifier genes within the Rd6m1 locus. To justify an analysis based on C57BL/6J data, we first determined whether B6.C3Ga-Mfrprd6/J is congenic with this strain. Although B6.C3Ga-Mfrprd6/J was derived by backcrossing to C57BL/6J, the discovery of the Crb1rd8 mutation during introgression (Mehalow et al., 2003) suggests that at least one generation was conducted with a strain related to C57BL/6N, the apparent source of the Crb1rd8 allele (Mattapallil et al., 2012). We therefore examined B6.C3Ga-Mfrprd6/J with a collection of 150 SNP markers that differ in C57BL/6N relative to C57BL/6J and are distributed throughout the genome. Of 145 markers that yielded products, 143 were homozygous for the C57BL/6J allele. A single marker on Chromosome 6 was heterozygous for the C57BL/6N allele and a single marker close to the Mfrp locus was homozygous for the C57BL/6N allele. These results indicate that B6.C3Ga-Mfrprd6/J is at least 95% congenic with C57BL/6J, supporting bioinformatics analysis based on C57BL/6J data.
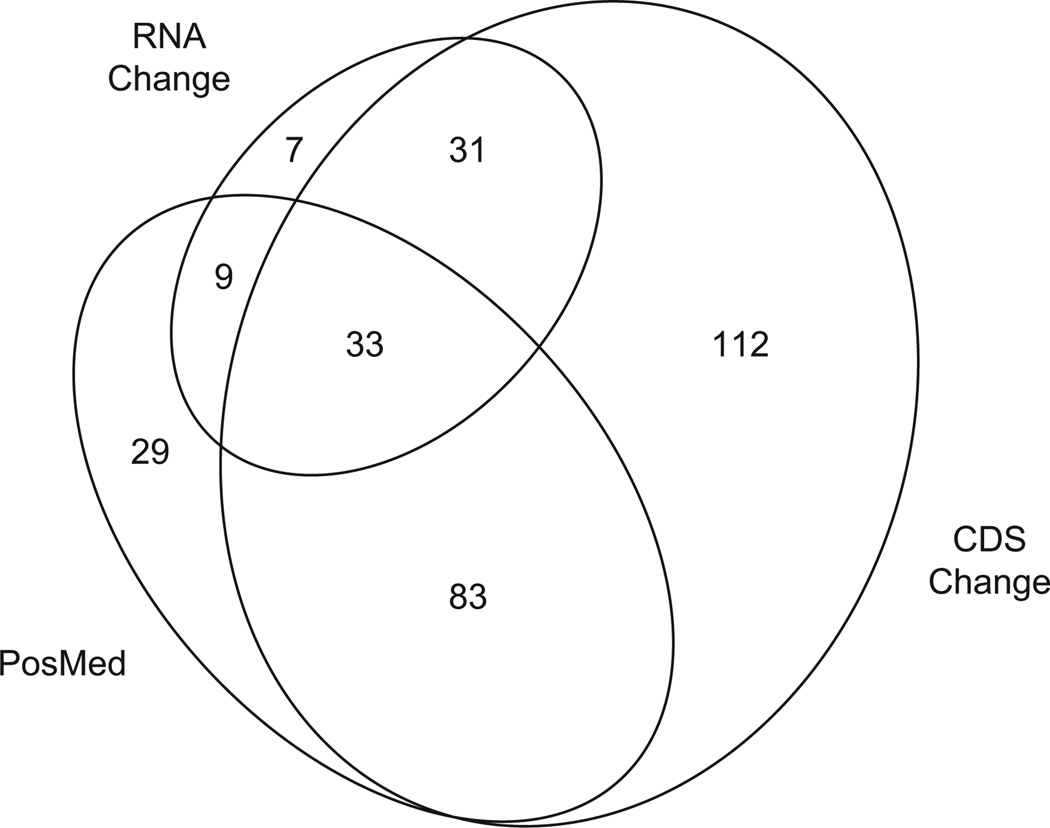
The Mouse Genome Informatics (MGI) Gene Query tool (GRCm38 build of the C57BL/6J mouse reference genome) indicated that 385 genes are located within the Rd6m1 modifier locus, including 343 protein-coding and 42 noncoding RNA genes (Supplemental Table 1). Comparison of the C57BL/6J (GRCm38 build) and CAST/EiJ genome sequences using the Wellcome Sanger Mouse SNP query tool (http://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1303) yielded 259 genes in the candidate locus with single nucleotide polymorphisms or insertion/deletions that are predicted to result in one or more protein coding sequence changes (CDS Change, Supplemental Table 1; Supplemental Data File 1). Fourteen genes containing a variation in a splice donor or acceptor sequence (Cd55, Tnnt2, RP23-302C16.3, Crb1, Cfh, Gm15486, Myoc, Ysk4, Elk4, Plekha6, Gm10530, Serpinc1, Cenpl, Rxrg), six genes predicted to contain a frameshift (Eif2d, Gm10188, Gm17678, Dennd1b, Tor1aip1, Blzf1), and two genes predicted to gain a premature stop codon (Gm4204, Gm10530) are of particular interest. These variations may be the most likely to alter the structure and function of the encoded polypeptides. An accession-based query of the 385 genes within the Rd6m1 modifier interval in PosMed (http://biosparql.org/PosMed/) using a keyword containing terms for retina, retinal pigment epithelium, Wnt/Frizzled signaling, or immune processes yielded 154 unique genes that may be relevant to Mfrp expression or function in the eye (PosMed, Supplemental Table 1). Of these, 116 genes overlapped with CDS Change genes (Fig. 4). To filter this list further, differentially expressed genes within the modifier locus were found by reanalyzing published microarray data comparing C57BL/6J and CAST/EiJ retinas (Jelcick et al., 2011). This subset contained 80 genes (RNA Change, Supplemental Table 1), 33 of which were common to both the CDS Change and PosMed gene subsets (Fig. 4).
Fig. 4.
Area-proportional diagram showing the intersection of gene subsets derived by filtering 385 annotated genes within the Rd6m1 modifier locus. The subsets include genes with nucleotide changes in CAST/EiJ compared to C57BL/6J that are likely to affect protein coding (CDS Change, obtained with the Sanger Mouse Genomes Project query tool); genes with a semantic link to the postulated function or tissue expression of Mfrp (PosMed); and retinal genes that are differentially expressed (Q value of false discovery rate <0.05) in C57BL/6J compared to CAST/EiJ (RNA Change, obtained from reevaluation of microarray data (Jelcick et al., 2011)). The number of genes in each area of the diagram is indicated. Proportioned ellipses were generated with eulerAPE v2.0.3 (http://www.eulerdiagrams.org/eulerAPE/).
4. Discussion
Our examination of an intercross of F1 hybrids between inbred B6.C3Ga-Mfrprd6/J and CAST/EiJ mice revealed that strain genetic background affects the retinal degenerative phenotype associated with homozygosity for the retinal degeneration 6 mutation, Mfrprd6. QTL analysis of the F2 intercross mice indicated the presence of a significant modifier locus on CAST/EiJ Chromosome 1, Rd6m1, which protects against the retinal degeneration phenotype of homozygous Mfrprd6 mice on the C57BL/6J genetic background in a primarily additive fashion. Suggestive modifier loci on Chromosomes 6 and 11 were also observed in the one-dimensional genome-wide scan. However, based on multiple regression analysis, only Rd6m1 and the modifier locus on Chromosome 6 were essential for the simplest QTL model.
The observation that genetic background affects the phenotype of Mfrprd6 mutant mice under conditions where environmental and experimental variation is minimized strongly supports the influence of one or more Mfrp modifier genes. Mfrp thus joins a growing list of ocular disease genes in mice that vary in phenotypic severity depending on genetic background, such as Crb1, Tub, Tulp1, Nr2e3, Rs1, Rp1, and Rd3 (Danciger et al., 2008; Haider et al., 2008; Ikeda et al., 2002; Johnson et al., 2008; Liu et al., 2009; Maddox et al., 2012; Mehalow et al., 2003). The existence of at least two ocular syndromes associated with human MFRP mutations (Mukhopadhyay et al., 2010; Neri et al., 2012; Sundin et al., 2005) may reflect an influence of genetic background. In support of this hypothesis, phenotypic variation has been noted among family members with the same MFRP mutation (Mukhopadhyay et al., 2010). However, a more recent report claimed that the phenotype of a common MFRP deletion variant was similar among individuals in the same family as well as between families, arguing against the hypothesis (Neri et al., 2012). In these studies, interpreting the role of genetic background in MFRP-associated disease is problematic due to the small population size. Our finding that the Mfrprd6 phenotype in mice depends on genetic background argues for future studies of larger patient cohorts to test if similar effects modify disease phenotypes in humans with MFRP mutations.
The discovery of Rd6m1 prompted us to explore bioinformatics approaches to find candidate modifier genes. From the 385 annotated genes within the modifier interval, 33 genes were identified that possess coding sequence changes in CAST/EiJ compared to C57BL/6J, have semantic association with pathways or tissue and cell types relevant to postulated Mfrp function, and are differentially expressed in the CAST/EiJ and C57BL/6J retinas. Although a broader subset of genes focused on differential expression changes or coding sequence changes might also be productive, these 33 genes may serve as a good starting point for future studies. For example, one of these genes, Cfh, is a mouse ortholog of CFH, which encodes complement factor H and is a risk determinant for age-related macular degeneration (AMD) (Hageman et al., 2005; Klein et al., 2005). Mutations in CFH that both increase and decrease AMD risk have been documented (Hageman et al., 2005; Klein et al., 2005; Tortajada et al., 2009). The CAST/EiJ gene includes 14 missense polymorphisms and a change in a putative splice acceptor site (Supplemental Data File 1), which may alter the CFH protein in away that protects against photoreceptor loss. Thus, the Cfh gene in CAST/EiJ mice may be among the most promising candidates within the critical region for modifying the Mfrprd6 phenotype.
Future work will refine the Rd6m1 locus and the modifier regions on Chromosomes 6 by generating congenics for use in additional mapping studies. Eventually, as the region is narrowed, combining mouse models of the candidate genes that fall within the critical modifier region with Mfrprd6 mice in the same genetic background may shed light on the genetic interactions. Identification of modifier(s), especially those that provide protection for photoreceptor loss, is arguably important as they may provide blueprints for therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Drs. Ron Korstanje and Ken Johnson for careful review of the manuscript and Jeanie Hansen for expert technical assistance. This study was supported by NIH grant EY011996 (to P. M. N.).
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.exer.2013.10.020.
References
- Ayala-Ramírez R, Graue-Wiechers F, Robredo V, Amato-Almanza M, Horta-Diez I, Zenteno JC. A new autosomal recessive syndrome consisting of posterior microphthalmos, retinitis pigmentosa, foveoschisis, and optic disc drusen is caused by aMFRP gene mutation. Mol. Vis. 2006;12:1483–1489. [PubMed] [Google Scholar]
- Ayuso C, Millan JM. Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med. 2010;2:34. doi: 10.1186/gm155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso C, Trujillo MJ, Robledo M, Ramos C, Benitez J, Martin-Oses F, del Rio T, Garcia-Sandoval B. Novel rhodopsin mutation in an autosomal dominant retinitis pigmentosa family: phenotypic variation in both heterozygote and homozygote Val137Met mutant patients. Hum. Genet. 1996;98:51–54. doi: 10.1007/s004390050158. [DOI] [PubMed] [Google Scholar]
- Bandah-Rozenfeld D, Mizrahi-Meissonnier L, Farhy C, Obolensky A, Chowers I, Pe’er J, Merin S, Ben-Yosef T, Ashery-Padan R, Banin E, Sharon D. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:382–391. doi: 10.1016/j.ajhg.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson EL, Grimsby JL, Adams SM, McGee TL, Sweklo E, Pierce EA, Sandberg MA, Dryja TP. Clinical features and mutations in patients with dominant retinitis pigmentosa-1 (RP1) Investig. Ophthalmol. Vis. Sci. 2001;42:2217–2224. [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Dryja TP. Ocular findings in patients with autosomal dominant retinitis pigmentosa and a rhodopsin gene defect (Pro-23-His) Arch. Ophthalmol. 1991a;109:92–101. doi: 10.1001/archopht.1991.01080010094039. [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Dryja TP. Ocular findings in patients with autosomal dominant retinitis pigmentosa and rhodopsin, proline-347-leucine. Am. J. Ophthalmol. 1991b;111:614–623. doi: 10.1016/s0002-9394(14)73708-0. [DOI] [PubMed] [Google Scholar]
- Broman KW, Sen S. New York: Springer; 2009. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chang B, Hawes NL, Hurd RE, Davisson MT, Nusinowitz S, Heckenlively JR. Retinal degeneration mutants in the mouse. Vis. Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- Chang B, Hurd R, Wang J, Nishina P. Survey of common eye diseases in laboratory mouse strains. Investig. Ophthalmol. Vis. Sci. 2013;54:4974–4981. doi: 10.1167/iovs.13-12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Ding Q, Tang Z, Liu P, Jiang F, Ke T, Ren X, Wang Z, Liu J, Wang QK, Liu M. A novel de novo frameshift mutation of RPGR ORF15 is associated with X-linked retinitis pigmentosa in a Chinese family. Mol. Vis. 2007;13:1548–1554. [PubMed] [Google Scholar]
- Crespí J, Buil JA, Bassaganyas F, Vela-Segarra JI, Díaz-Cascajosa J, Ayala-Ramírez R, Zenteno JC. A novel mutation confirms MFRP as the gene causing the syndrome of nanophthalmos-renititis pigmentosa-foveoschisisoptic disk drusen. Am. J. Ophthalmol. 2008;146:323–328. doi: 10.1016/j.ajo.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 2007;125:151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danciger M, Ogando D, Yang H, Matthes MT, Yu N, Ahern K, Yasumura D, Williams RW, Lavail MM. Genetic modifiers of retinal degeneration in the rd3 mouse. Investig. Ophthalmol. Vis. Sci. 2008;49:2863–2869. doi: 10.1167/iovs.08-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, Berson EL. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. doi: 10.1038/343364a0. [DOI] [PubMed] [Google Scholar]
- Fahim AT, Bowne SJ, Sullivan LS, Webb KD, Williams JT, Wheaton DK, Birch DG, Daiger SP. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One. 2011;6:e23021. doi: 10.1371/journal.pone.0023021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Di Iorio E, Barbaro V, Ponzin D, Sorrentino FS, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr.Genomics. 2011;12:238–249. doi: 10.2174/138920211795860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogerty J, Besharse JC. 174delG mutation in mouse MFRP causes photo-receptor degeneration and RPE atrophy. Investig. Ophthalmol. Vis. Sci. 2011;52:7256–7266. doi: 10.1167/iovs.11-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum. Genet. 2008;124:357–368. doi: 10.1007/s00439-008-0560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider NB, Ikeda A, Naggert JK, Nishina PM. Genetic modifiers of vision and hearing. Hum. Mol. Genet. 2002;11:1195–1206. doi: 10.1093/hmg/11.10.1195. [DOI] [PubMed] [Google Scholar]
- Haider NB, Zhang W, Hurd R, Ikeda A, Nystuen AM, Naggert JK, Nishina PM. Mapping of genetic modifiers of Nr2e3rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm. Genome. 2008;19:145–154. doi: 10.1007/s00335-008-9092-2. [DOI] [PubMed] [Google Scholar]
- Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes NL, Chang B, Hageman GS, Nusinowitz S, Nishina PM, Schneider BS, Smith RS, Roderick TH, Davisson MT, Heckenlively JR. Retinal degeneration 6 (rd6): a new mouse model for human retinitis punctata albescens. Investig. Ophthalmol. Vis. Sci. 2000;41:3149–3157. [PubMed] [Google Scholar]
- Ikeda A, Naggert JK, Nishina PM. Genetic modification of retinal degeneration in Tubby mice. Exp. Eye Res. 2002;74:455–461. doi: 10.1006/exer.2001.1139. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Iannaccone A, Weleber RG, Fishman GA, Maguire AM, Affatigato LM, Bennett J, Pierce EA, Danciger M, Farber DB, Stone EM. Disease expression of RP1 mutations causing autosomal dominant retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2000;41:1898–1908. [PubMed] [Google Scholar]
- Jelcick AS, Yuan Y, Leehy BD, Cox LC, Silveira AC, Qiu F, Schenk S, Sachs AJ, Morrison MA, Nystuen AM, DeAngelis MM, Haider NB. Genetic variations strongly influence phenotypic outcome in the mouse retina. PLoS One. 2011;6:e21858. doi: 10.1371/journal.pone.0021858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Aoyama N, Friedell NH, Ikeda S, Ikeda A. Genetic modification of the schisis phenotype in a mouse model of X-linked retinoschisis. Genetics. 2008;178:1785–1794. doi: 10.1534/genetics.107.084905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameya S, Hawes NL, Chang B, Heckenlively JR, Naggert JK, Nishina PM. Mfrp a gene encoding a frizzled related protein, is mutated in the mouse retinal degeneration 6. Hum. Mol. Genet. 2002;11:1879–1886. doi: 10.1093/hmg/11.16.1879. [DOI] [PubMed] [Google Scholar]
- Katoh M. Molecular cloning and characterization of MFRP a novel gene encoding a membrane-type Frizzled-related protein. Biochem. Biophys. Res. Commun. 2001;282:116–123. doi: 10.1006/bbrc.2001.4551. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Langmann T, Di Gioia SA, Rau I, Stohr H, Maksimovic NS, Corbo JC, Renner AB, Zrenner E, Kumaramanickavel G, Karlstetter M, Arsenijevic Y, Weber BH, Gal A, Rivolta C. Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:376–381. doi: 10.1016/j.ajhg.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Saveliev A, Pierce EA. The severity of retinal degeneration in Rp1h gene-targeted mice is dependent on genetic background. Investig. Ophthalmol. Vis. Sci. 2009;50:1566–1574. doi: 10.1167/iovs.08-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox DM, Ikeda S, Ikeda A, Zhang W, Krebs MP, Nishina PM, Naggert JK. An allele of microtubule-associated protein 1A (Mtap1a) reduces photoreceptor degeneration inTulp1 andTub Mutant Mice. Investig. Ophthalmol. Vis. Sci. 2012;53:1663–1669. doi: 10.1167/iovs.11-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Investig. Ophthalmol. Vis. Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum. Mol. Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Sergouniotis PI, Mackay DS, Day AC, Wright G, Devery S, Leroy BP, Robson AG, Holder GE, Li Z, Webster AR. A detailed phenotypic assessment of individuals affected by MFRP-related oculopathy. Mol. Vis. 2010;16:540–548. [PMC free article] [PubMed] [Google Scholar]
- Neri A, Leaci R, Zenteno JC, Casubolo C, Delfini E, Macaluso C. Membrane frizzled-related protein gene-related ophthalmological syndrome: 30-month follow-up of a sporadic case and review of genotype-phenotype correlation in the literature. Mol. Vis. 2012;18:2623–2632. [PMC free article] [PubMed] [Google Scholar]
- Passerini I, Sodi A, Giambene B, Menchini U, Torricelli F. Phenotypic intrafamilial variability associated with S212G mutation in the RDS/peripherin gene. Eur. J. Ophthalmol. 2007;17:1000–1003. doi: 10.1177/112067210701700624. [DOI] [PubMed] [Google Scholar]
- Paunescu K, Preising MN, Janke B, Wissinger B, Lorenz B. Genotype-phenotype correlation in a German family with a novel complex CRX mutation extending the open reading frame. Ophthalmology. 2007;114:1348–1357. e1341. doi: 10.1016/j.ophtha.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Samardzija M, Wenzel A, Naash M, Reme CE, Grimm C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur. J. Neurosci. 2006;23:1028–1034. doi: 10.1111/j.1460-9568.2006.04639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, Gire AI, Seaman R, Duzkale H, Spellicy CJ, Zhu J, Shankar SP, Daiger SP. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Investig. Ophthalmol. Vis. Sci. 2006;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin OH, Dharmaraj S, Bhutto IA, Hasegawa T, McLeod DS, Merges CA, Silval ED, Maumenee IH, Lutty GA. Developmental basis of nanophthalmos: MFRP Is required for both prenatal ocular growth and postnatal emmetropization. Ophthalmic Genet. 2008;29:1–9. doi: 10.1080/13816810701651241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin OH, Leppert GS, Silva ED, Yang J-M, Dharmaraj S, Maumenee IH, Santos LC, Parsa CF, Traboulsi EI, Broman KW, Dibernardo C, Sunness JS, Toy J, Weinberg EM. Extreme hyperopia is the result of null mutations in MFRP which encodes a Frizzled-related protein. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9553–9558. doi: 10.1073/pnas.0501451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K, Adamian M, Dryja TP, Berson EL. Histopathologic study of variation in severity of retinitis pigmentosa due to the dominant rhodopsin mutation Pro23His. Am. J. Ophthalmol. 2002;134:290–293. doi: 10.1016/s0002-9394(02)01545-3. [DOI] [PubMed] [Google Scholar]
- Tortajada A, Montes T, Martinez-Barricarte R, Morgan BP, Harris CL, de Cordoba SR. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum. Mol. Genet. 2009;18:3452–3461. doi: 10.1093/hmg/ddp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G, Rose AM, Shah AZ, Bhattacharya SS, Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012;8:e1003040. doi: 10.1371/journal.pgen.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenteno JC, Buentello-Volante B, Quiroz-González MA, Quiroz-Reyes MA. Compound heterozygosity for a novel and a recurrent MFRP gene mutation in a family with the nanophthalmos-retinitis pigmentosa complex. Mol. Vis. 2009;15:1794–1798. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.