Abstract
Purpose
To identify differentially expressed proteins in the pterygium compared to healthy conjunctiva using a proteomic analysis.
Methods
Pterygial and healthy conjunctival tissues were obtained from 24 patients undergoing pterygium excision. Total proteins of the pterygia and healthy conjunctiva were analyzed with one-dimensional electrophoresis, and protein bands of interest were excised and subjected to liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-MS/MS) using Thermo’s Finnigan ProteomeX workstation LTQ linear ion trap MS/MS. Using bioinformatics, differentially expressed proteins were classified, and three proteins closely involved in the response to oxidative stress were selected for further validation. Differential expression of these proteins was confirmed with western blot and immunohistochemistry.
Results
A web-based gene ontology program, DAVID, was used to classify 230 proteins that were differentially expressed in pterygial tissues. Among these genes, we chose three proteins, aldehyde dehydrogenase, dimeric NADP-preferring (ALDH3A1), protein disulfide-isomerase A3 (PDIA3), and peroxiredoxin-2 (PRDX2), that were significantly upregulated in pterygium and further increased in recurrent pterygium. Immunohistochemistry and western blot analysis confirmed that these three proteins were mainly detected in the basal epithelial layer, and their expression was significantly increased in the pterygium compared to normal conjunctiva.
Conclusions
This study reported increased expression of ALDH3A1, PDIA3, and PRDX2 in pterygia using a proteomic approach. These proteins are presumed to have a protective role against oxidative stress-induced apoptosis. This result is consistent with the hypothesis that oxidative stress is a significant factor in the pathogenesis of pterygia.
Introduction
Pterygium refers to a wing-shaped fibrovascular tissue thought to be formed by the overgrowth of transformed conjunctival tissue into the cornea. The lesion is characterized by centripetal growth of a leading edge of altered limbal epithelial cells, followed by squamous cell metaplasia with goblet cell hyperplasia. Activated stromal fibroblasts, neovascularization, and extracellular matrix remodeling are also present [1-4].
Ultraviolet (UV) radiation damage, chronic irritation, and/or inflammation are hypothesized to play roles in the pathogenesis of pterygium. Patients with increased ultraviolet (UV) exposure, such as those living close to the equator or those with sunlight-related conditions, have an increased risk of pterygia formation [3-5]. Previous studies focused primarily on the clinical characteristics and surgical management of the disease, and therefore, its pathogenesis remains incompletely understood. However, considerable progress in this area has been achieved, and recent evidence implicates antiapoptotic mechanisms, oxidative stress, immunological mechanisms, various cytokines, and extracellular matrix (ECM) modulation as possible causative factors [3,6-10]. Regardless of the inciting agent, inflammatory mediators appear to be associated with pterygia. Interleukin (IL-6), IL-8, and tumor necrosis factor-α are increased in pterygia and induced after UV-B exposure in cultured pterygia epithelial cells [11]. Cyclooxygenase-2 is overexpressed in pterygia, contributing to a further increase in the inflammatory cascades [12]. Immunological mechanisms also likely contribute to the development of pterygia. An increase in mast cells, lymphocytes, plasma cells, dendritic cells, and CD4+ and CD8+ T cells in pterygia has also been documented [13]. The contribution of inflammatory mediators, immunological mechanisms, angiogenic stimulation, and ECM modulation are likely secondary in nature after actual pterygia formation [6].
In this study, we adopted a proteomics technique for identifying the proteins that were differentially expressed in pterygium compared to normal conjunctiva. Proteomics has expanded the opportunities to identify disease-specific proteins involved in many diseases, including ophthalmic diseases such as dry eye [14,15], cataract [16,17], keratoconus [18], myopia [19], and pterygia [20].
As in previous studies, hundreds of protein molecules were newly identified as candidate biomarkers of pterygia. To narrow our interests, we focused on proteins that show increased expression in primary pterygium and are further increased in recurrent pterygium. Among those proteins, we further validated increased expression of three proteins known to have roles related to oxidative stress, aldehyde dehydrogenase, dimeric NADP-preferring (ALDH3A1), protein disulfide-isomerase A3 (PDIA3), and peroxiredoxin-2 (PRDX2), using western blot analysis and immunohistochemistry.
Methods
Subjects and tissue preparation
Between May 2012 and Apr 2013, 24 subjects, 10 males and 14 females, with mean age 49±5.2 (38-62) years old who underwent pterygium excision in Hando general hospital (Ansan, Korea) were included in the study. Samples of healthy conjunctiva were obtained from the same patients who underwent pterygium excision. Pterygium tissues and remnants from the autologous upper bulbar conjunctiva grafts were collected immediately at the time of surgery and frozen in liquid nitrogen. This study protocol was approved by the institutional review board of our hospital and it conformed to the guidelines provided in the ARVO statement on human subjects in ophthalmic and vision research. Written informed consent to participate in this research was obtained from each patient.
One-dimensional gel electrophoresis and LC-ESI-MS/MS analysis
Pterygia tissues were homogenized using Tissue-Ruptor (Qiagen, Valencia, CA) with PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma-Aldrich, CA). Proteins were quantified with bicinchoninic acid (BCA) assay (Pierce, Cramlington, UK). Fifty micrograms of proteins were separated with gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 4–20% polyacrylamide gels, Novex gel, Invitrogen, Carlsbad, CA). After Coomassie staining, the protein bands of interest were excised and subjected to in-gel trypsin digestion as previously described [21]. Briefly, after destaining in 75 mM ammonium bicarbonate/40% ethanol (1:1), the gels were subjected to reducing (5 nM dithiothreitol [DTT] in 25 mM ammonium bicarbonate) and alkylating (55 mM iodoacetamide) conditions, each at room temperature for 30 min. After dehydration with acetonitrile, 20 μg/mL sequencing-grade trypsin (Roche Applied Science) containing 25 mM ammonium bicarbonate (pH 8.5) was added, and the slice was incubated at 37 °C overnight. The in-gel tryptic peptides were eluted with 0.1% formic acid. Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-MS/MS) analysis was performed using Thermo’s Finnigan ProteomeX workstation LTQ linear IT MS (Thermo Electron, San Jose, CA) equipped with a nanospray ionization (NSI) source (San Jose, CA).
Data-dependent acquisition mode (m/z 300–1800) was enabled, and each survey MS scan was followed by five MS/MS scans with the 30 s dynamic exclusion option on. The spray voltage was 1.9 kV and the temperature of the ion transfer tube was set at 1957C. The normalized collision energy was set at 35% [22]. MS/MS data were searched based on the IPI human protein database (version 3.29) using the SEQUEST algorithm (Thermo Electron). Scaffold ver. 01_07_00 was used to validate MS/MS-based peptides and protein identification.
Gene ontology analysis
Gene ontology analysis was performed using Integrated Discovery (DAVID) Functional Annotation Tool. Through DAVID, proteins with gene symbols were grouped according to biologic process.
Western blot analysis
Protein extracts were obtained from pterygia and normal conjunctiva tissue and were lysed by sonication in PBS containing 1 mM PMSF. Proteins were resolved with 12.5% SDS–PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Milan, Italy). The membrane was blocked for 1 h in a 5% skim milk solution and then incubated with mouse monoclonal antibodies against ALDH3A1 (1:2,000 dilution, AbFrontier, Seoul, Korea), PDIA3 (1:5,000 dilution, AbFrontier), and PRDX2 (1:2,000 dilution, AbFrontier) for 2 h at room temperature. The unbound primary antibodies were removed with one 15-min and two 5-min washes in PBS containing 0.1% (v/v) Nonidet P40. Then the membrane was incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (IgG) secondary antibody (Ab Frontier) for 2 h at room temperature. The target protein was detected with enhanced chemiluminescence (ECL) solution (Amersham Biosciences, Cleveland, OH) using X-ray film.
Immunohistochemistry
The tissue was fixed with 4% (v/v) buffered paraformaldehyde for 1 day and then washed in tap water for 1 day. The tissue was dehydrated with a series of graded ethanol and embedded in paraffin. Sections were cut to a thickness of 4 µm and deparaffinized. Then, the sections were hydrated using a series of decreasing concentrations of ethanol. The sections were incubated in blocking buffer for 1 h at room temperature to block non-specific binding. The primary antibodies used were against ALDH3A1 (1:200 dilution, AbFrontier), PDIA3 (1:400 dilution, AbFrontier), and PRDX2 (1:200 dilution, AbFrontier). The sections were incubated in primary antibody solution for 1 h at room temperature. All samples were incubated for 30 min with universal biotinylated secondary antibodies at room temperature. Samples were then washed twice, and final incubation was performed for 30 min using streptavidin-peroxidase. Signals were developed using 3,3-diaminobenzidine for 5 min and counterstained with Mayer’s hematoxylin (Dako, Glostrup, Denmark). Negative controls were prepared by leaving out the primary antibody.
Results
Protein Identification with LC-ESI-MS/MS analysis
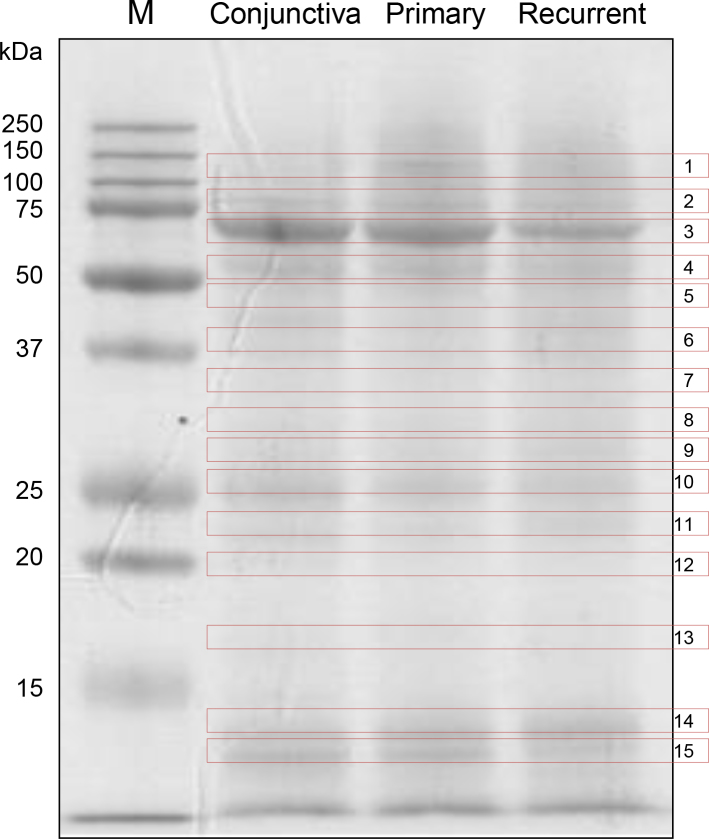
Pooled crude extracts of eight individuals from each group (normal conjunctiva, pterygium, and recurrent pterygia) were run on an SDS–PAGE gel, and the gel was stained with Coomassie brilliant blue G-250 (Figure 1). The stained SDS–PAGE gel was cut into 15 pieces according to molecular weight and subjected to in-gel digestion. Digested peptides were then fractionated and identified with LC-MS/MS. All the experiments were replicated twice. In the first experiment, 346 proteins were differentially expressed in the three groups. Among these, 249 protein expression patterns were replicated in the second experiment.
Figure 1.
Identification of differentially expressed proteins in pterygium compared to healthy conjunctiva. Crude extracts of healthy conjunctiva as well as primary and recurrent pterygial tissues of eight subjects from each group were pooled; 50 μg of protein from each pooled sample was separated with SDS–PAGE and stained with Coomassie brilliant blue. Fifteen bands of each sample were excised and subjected to in-gel digestion. Digested peptides were then fractionated and identified with LC-MS/MS analysis. Molecular weight markers are shown in the left side of the figure.
First, we analyzed the pterygia-specific proteins by comparing normal conjunctiva with primary and recurrent pterygium. The expression of 153 proteins was downregulated (Appendix 1), and 77 proteins were upregulated in pterygia (Appendix 2).
Ontology analysis of pterygium-specific proteins
Using a web-based gene ontology program, DAVID, 230 proteins that were up- or downregulated in pterygia tissues were classified. Molecular functions were attributed to 230 proteins using GOTERM_MF_FAT from the DAVID database. The related gene ontology categories were merged into a cluster using the DAVID functional clustering module. The major relevant categories were as follows: structural molecule activity, nucleotide binding, cytoskeletal protein binding, peptidase activity, guanyl ribonucleotide binding, structural constituent of eye, and oxidoreductase activity (Table 1). Interestingly, a group of proteins closely related to the structural molecule and peptidase activity were either up- or downregulated in pterygium. These data suggest that tissue rearrangement is strongly involved in pterygia formation and growth.
Table 1. DAVID analysis results for significantly up- or down- regulated proteins in pterygium tissues.
| Term | Count | % | P-Value | Benjamini |
|---|---|---|---|---|
| Upregulated proteins | ||||
| structural molecule activity |
32 |
21.9 |
7.60E-14 |
2.20E-11 |
| nucleotide binding |
32 |
21.9 |
3.10E-02 |
4.00E-01 |
| cytoskeletal protein binding |
17 |
11.6 |
3.90E-05 |
1.90E-03 |
| peptidase activity |
11 |
7.5 |
6.00E-02 |
5.30E-01 |
| guanyl ribonucleotide binding |
10 |
6.8 |
1.40E-02 |
2.40E-01 |
| structural constituent of eye lens |
6 |
4.1 |
5.00E-07 |
3.70E-05 |
| glycosaminoglycan binding |
6 |
4.1 |
1.30E-02 |
2.50E-01 |
| phosphotransferase activity |
3 |
2.1 |
4.20E-02 |
4.50E-01 |
| Downregulated proteins | ||||
| structural molecule activity |
12 |
17.4 |
1.60E-04 |
1.70E-02 |
| peptidase activity |
7 |
10.1 |
4.30E-02 |
4.70E-01 |
| identical protein binding |
7 |
10.1 |
7.90E-02 |
6.50E-01 |
| carbohydrate binding |
6 |
8.7 |
2.50E-02 |
3.70E-01 |
| protein binding, bridging |
5 |
7.2 |
9.80E-04 |
6.80E-02 |
| glycosaminoglycan binding |
5 |
7.2 |
4.20E-03 |
1.40E-01 |
| polysaccharide binding |
5 |
7.2 |
5.90E-03 |
1.70E-01 |
| oxidoreductase activity, acting on peroxide as acceptor |
3 |
4.3 |
9.80E-03 |
2.10E-01 |
| antioxidant activity |
3 |
4.3 |
2.00E-02 |
3.30E-01 |
| integrin binding |
3 |
4.3 |
3.10E-02 |
3.90E-01 |
| growth factor binding | 3 | 4.3 | 8.70E-02 | 6.60E-01 |
The Categories “GO-Molecular Function” were selected for testing. Term, specific terms within DAVID-categories; p value, p values for enrichment calculated by the EASE method used in DAVID; Benjamini, p values corrected for multiple testing by the Benjamini Hochberg method.
Selection of proteins for validation
The number of different proteins in the pterygium tissues was still too high to be validated. To narrow the possible candidate proteins, we further analyzed the proteomics data. Instead of comparing two groups (normal conjunctiva and pterygia), we compared proteome from three groups: normal conjunctiva, primary pterygium, and recurrent pterygium. Several fundamental profile patterns could be identified from these analyses, and we focused on increasing or decreasing patterns: upregulated in primary pterygium and further increased in recurrent pterygium or downregulated in primary pterygium and further decreased in recurrent pterygium. We thought that if the change in expression remained or increased in recurrent pterygium, the molecule should be considered to have a critical role in pterygia formation or growth. In this context, 40 proteins were differentially expressed in primary and recurrent pterygia. Among them, we chose three proteins, ALDH3A1, PDIA3, and PRDX2, for further validation. As shown in Table 2, the expression of all three proteins was significantly increased in pterygium and further increased in recurrent pterygium. Interestingly, all three proteins have some roles related to oxidative stress. PDIA3 and ALDH3A1 were classified into cell redox homeostasis (GO:0045454) and oxidation-reduction process (GO:0055114) groups, respectively. PRDX2 was also associated with cellular response to oxidative stress (GO:0034599).
Table 2. Proteins Identified by LC-MS/MS Analysis in Normal conjunctiva, primary and recurrent pterygium tissue.
| Identified Proteins | Accession Number | Peptide Hits |
||
|---|---|---|---|---|
| Normal Conjunctiva | Primary Pterygium | Recurrent Pterygium | ||
| ALDH3A1 Aldehyde dehydrogenase, dimeric NADP-preferring |
IPI00296183 |
5 |
11 |
17 |
| ATP5B ATP synthase subunit beta, mitochondrial |
IPI00303476 |
0 |
2 |
3 |
| COL1A1 Collagen alpha-1(I) chain |
IPI00297646 |
3 |
7 |
9 |
| COL1A2 Collagen alpha-2(I) chain |
IPI00304962 |
0 |
20 |
23 |
| EIF3H Eukaryotic translation initiation factor 3, isoform CRA_b |
IPI00647650 |
0 |
3 |
9 |
| FBLN5 Fibulin-5 |
IPI00294615 |
6 |
12 |
68 |
| FH Isoform Mitochondrial of Fumarate hydratase, mitochondrial |
IPI00296053 |
26 |
53 |
254 |
| IGHV4–31; Putative uncharacterized protein |
IPI00384938 |
0 |
8 |
36 |
| IGKC Ig kappa chain C region |
IPI00940069 |
0 |
10 |
14 |
| MANF mesencephalic astrocyte-derived neurotrophic factor |
IPI00328748 |
0 |
8 |
54 |
| OBSCN Isoform 1 of Obscurin |
IPI00288940 |
69 |
99 |
141 |
| POSTN Periostin, osteoblast specific factor |
IPI00410241 |
0 |
0 |
13 |
| PRDX2 Peroxiredoxin-2 |
IPI00027350 |
5 |
12 |
13 |
| RPSAP15;RPSA 33 kDa protein |
IPI00413108 |
0 |
9 |
11 |
| SLC4A1solutecarrierfamily4 |
IPI00791534 |
0 |
6 |
7 |
| TACSTD2 Tumor-associated calcium signal transducer 2 |
IPI00297910 |
5 |
12 |
34 |
| TGM2 Isoform 1 of Protein-glutamine gamma-glutamyltransferase 2 |
IPI00294578 |
11 |
21 |
39 |
| TYMP Thymidine phosphorylase |
IPI00292858 |
7 |
12 |
26 |
| PDIA3 Protein disulfide-isomerase A3 |
IPI00025252 |
0 |
9 |
10 |
| APEX1 DNA-(apurinic or apyrimidinic site) lyase |
IPI00215911 |
7 |
2 |
0 |
| ATP5J2 Putative uncharacterized protein ATP5J2 |
IPI00219291 |
12 |
9 |
3 |
| CALML3 Calmodulin-like protein 3 |
IPI00216984 |
13 |
8 |
3 |
| CLC Eosinophil lysophospholipase |
IPI00216071 |
6 |
3 |
0 |
| CRYBB2 Beta-crystallin B2 |
IPI00218748 |
42 |
29 |
9 |
| GOT1 Aspartate aminotransferase, cytoplasmic |
IPI00219029 |
7 |
5 |
0 |
| HIST1H2AC Histone H2A type 1-C |
IPI00216456 |
36 |
21 |
4 |
| HPRT1 Hypoxanthine-guanine phosphoribosyltransferase |
IPI00218493 |
6 |
4 |
2 |
| IGJ Immunoglobulin J chain |
IPI00178926 |
37 |
3 |
0 |
| LMNB1 Lamin-B1 |
IPI00217975 |
12 |
8 |
0 |
| NCL NCL protein |
IPI00183526 |
8 |
2 |
0 |
| PLS3 Plastin-3 |
IPI00216694 |
10 |
6 |
3 |
| PSMA2 Proteasome subunit alpha type-2 |
IPI00219622 |
150 |
117 |
58 |
| RPL22 60S ribosomal protein L22 |
IPI00219153 |
4 |
3 |
0 |
| RPL30 60S ribosomal protein L30 |
IPI00219156 |
4 |
3 |
0 |
| STOM Erythrocyte band 7 integral membrane protein |
IPI00219682 |
5 |
4 |
0 |
| TAGLN Transgelin |
IPI00216138 |
14 |
8 |
3 |
| TTN Isoform 7 of Titin | IPI00179357 | 18 | 3 | 0 |
Western blot analysis and immunohistochemistry
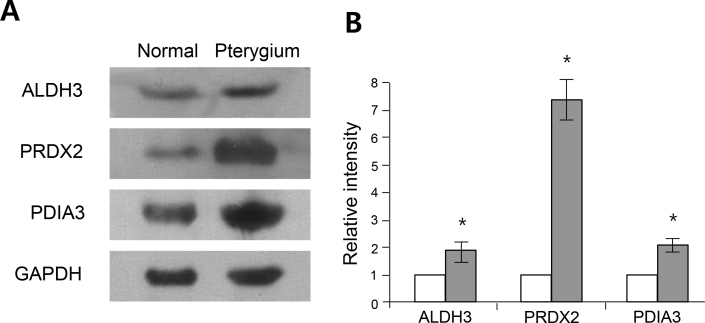
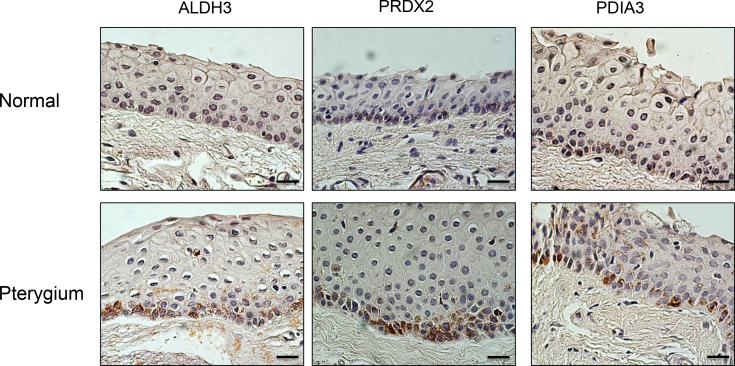
To validate the proteomic analysis data, we performed a western blot analysis of homogenates from normal conjunctiva and pterygium (100 μg of protein/lane). Our data showed that expression of the ALDH3A1, PDIA3, and PRDX2 proteins was significantly increased in the pterygia tissue compared to normal conjunctiva. This finding was consistent with the proteome analysis data (Figure 2). Using immunohistochemistry, upregulation of ALDH3A1, PDIA3, and PRDX2 expression in pterygia was also detected. As shown in Figure 3, all three proteins were mainly detected in the epithelial layer, and their intensities were increased in pterygium tissues compared to normal conjunctiva. Interestingly, these proteins were preferentially localized to the basal layer of the pterygial epithelium. These results confirmed that these three proteins were expressed at lower levels in normal conjunctiva and were present at considerably higher levels in the pterygium tissue.
Figure 2.
Western blot analysis of ALDH3A1, PDIA3, and PRDX2. A: Representative images of western blot analysis, examining the expression of ALDH3A1, PDIA3, and PRDX2 in pterygium and healthy conjunctiva tissue. GAPDH protein was used as an internal control. B: Statistically significant differences were observed between the pterygium and the healthy conjunctiva (p<0.05, n=6 in each group).
Figure 3.
Immunolocalization of ALDH3A1, PDIA3, and PRDX2 in pterygium. Immunohistochemistry assay showed the presence of ALDH3A1, PDIA3, and PRDX2 preferentially in the cytoplasm of the basal epithelium of the pterygium. Meanwhile, there was only faintly positive immunostaining of these proteins in the healthy conjunctiva.
Discussion
The mechanism of pterygia formation is not yet fully understood, and its origin remains controversial. Despite many publications showing the influence of UV radiation in the pathogenesis, the role of oxidative stress and the antioxidant system is still unclear. The presence of oxidative stress in pterygium has been proved by finding an increase in 8-hydroxydeoxyguanosine (8-OHdG), malonedialdehyde (MDA), and decreased activity of superoxide dismutase, catalase, and glutathione peroxidase [7,23].
In this study, a proteomic approach was used to identify putative differences in pterygium compared to healthy conjunctiva. The expression of 153 proteins was downregulated (Appendix 1), and 77 proteins were upregulated in pterygia (Appendix 2). Our results revealed that proteins associated with an increase in extracellular matrix, structural, and mitotic proteins and those involved in tissue invasion were significantly affected. A remarkable change was also found in the group of proteins associated with oxidative stresses and protection against them. Among them, we confirmed increased expression of ALDH3A1, PDIA3, and PRDX2 in pterygial epithelium.
ALDH3A1 is an abundant corneal crystalline in mammals, and it has been known to contribute to corneal defense by playing a multifunctional role in the protection of the cornea, and perhaps the entire eye, against UV-induced oxidative stress [24,25]. ALDH3A1 is a member of the ALDH superfamily of proteins that catalyze the NAD(P)-dependent oxidation of a wide range of endogenous and exogenous aldehydes. The central roles and mechanisms by which ALDH3A1 protects against UV-induced damage are thought to include the following: 1) metabolism of the toxic aldehydes, 2) direct absorption of UV light, 3) antioxidant function either directly through the scavenging of free radicals or indirectly through the production of NADPH, 4) maintaining corneal refractive and transparent properties as a corneal crystalline, 5) chaperon-like activity, and 6) cell cycle regulation [25-28]. DNA is one of the major targets of UV-induced oxidative stress, and modified DNA, if left unchecked, may result in genetic mutation or cell death. Cell cycle control by ALDH3A1 resonates with the antiapoptotic and cell growth regulating roles of the lens alpha-crystallins [29,30]. Despite the upregulated expression of ALDH3A1 in pterygial tissue, we noted decreased expression of other crystalline proteins (alpha-crystallin A chain, alpha-crystallin B chain, beta-crystallin A3, beta-crystallin B1, beta-crystallin B2, and beta-crystallin S) in pterygia. This result suggests specific roles of ALDH3A1 in pterygia compared to other crystallins that have structural roles in conjunctiva.
PDIA3, another biomarker in our study, is a member of the endoplasmatic reticulum (ER) stress signaling pathway also known as unfolded protein response (UPR). The expression level of PDIA3 increases in response to cellular stress due to its function as a chaperone [31,32]. Recently published data connected PDIA3 to the apoptotic process and demonstrated PDIA3 had an antiapoptotic effect in the melanoma cell line A375 after ER stress was induced [33]. PDIA3 possibly plays a role in the malignant transformation of prostate and cervical cancer [34,35]. The expression of this gene is induced during neoplastic transformation possibly leading to redox-dependent modulation of cancer-relevant regulatory factors [36,37]. These previous findings suggest that the observed increase of PDIA3 in pterygium is most likely due to elevated cellular stress, which may be induced by oxidative damage. In addition to a role in the ER stress pathway, PDIA3 has recently gained attention due to its function as a component of the peptide-loading complex of the major histocompatibility complex (MHC) class I pathway [38]. In Pdia3-deficient mice, this complex is impaired and negatively influences presentation of antigenic peptides [39]. This may help tumors escape from immune surveillance with cytotoxic T cells. It has been proposed that pterygium and neoplasia have common features, raising the possibility that pterygium is a neoplastic-like growth disorder. Whether PDIA3 plays a similar role in pterygium as in tumor formation is still unclear.
We also reported increased expression of PRDX2 in pterygium. This result confirmed the recent findings of Bautista-de Lucio et al. [20]; they also conducted a proteomic approach to identify disease-specific protein expression and identified upregulated expression of PRDX2 in the pterygium. PRDX2 is a cytoplasmic enzyme that reduces intracellular reactive oxygen species (ROS) levels. It has been described that PRDX2 overexpression protects significantly from peroxide-induced apoptosis and necrosis, while downregulation of this enzyme promotes injurious effects of oxidative stress in cardiomyocytes [40]. A recent report found that pterygia express high levels of cellular proliferation proteins and low levels of apoptosis markers, suggesting that there is a disruption in the equilibrium among proliferation and apoptosis favoring cell proliferation [41]. The antiapoptotic role of this enzyme has been demonstrated in red blood cells and pancreatic cells as well [42,43]. Many isoforms of the peroxiredoxin family have been implicated in the survival of several types of ocular cells [44-47]. These data support the proposal that PRDX2 could be involved in inhibiting apoptosis in the pterygium.
Another important aspect of this study is the immunohistochemical localization of ALDH3A1, PDIA3, and PRDX2 in pterygia. Interestingly, all three proteins are preferentially localized in the pterygium basal epithelium. Previous studies have reported that the expression of mutant p53 and other important apoptosis-related proteins, bax and bcl-2, was noted predominantly in the basal epithelial layer of pterygia [10,41]. Immunohistochemical studies demonstrated that apoptotic cells are found mainly confined to basal epithelial cells in pterygia [10,41], leading the chance that ALDH3A1, PDIA3, and PRDX2 are involved in regulating apoptosis and cell proliferation in pterygia. The antiapoptotic role of these three proteins has been demonstrated in various tissues [24,28,33,40,42,43]. Further studies are required to establish the functional roles of these proteins in pterygia formation.
Limited investigation regarding differences in the pathogenesis of primary and recurrent pterygia has been performed. We focused on proteins that show graded patterns of overexpression in primary and recurrent pterygia compared to normal conjunctiva. We presumed that these proteins might have more critical roles in pterygia formation and recurrence. We also focused on the difference in proteomic data between primary and recurrent pterygium, because the mechanisms that determine the genesis and the recurrence of a pterygia may not be the same. Interestingly, our result showed that periostin is a pathognomic marker for recurrent pterygium, and it was not expressed in primary pterygium or in normal conjunctiva. This finding confirms Kuo et al.’s transcriptome study [48,49]. It seemed reasonable to assume that recurrent pterygia possess molecular changes related to their recurrence.
In this study, we reported increased expression of ALDH3A1, PDIA3, and PRDX2 in pterygia using a proteomic approach. All three proteins are presumed to have significant roles related to oxidative stress. Our results are consistent with the hypothesis that oxidative stress is a significant factor in the pathogenesis of pterygia. We hypothesized that UV exposure and resultant oxidative stress leads to increased ALDH3A1, PDIA3, and PRDX2 levels, which provide resistance to UV-induced apoptosis and a hyperproliferative state.
Acknowledgments
This work was supported by the research fund of Hanyang University (HY-2009–419). Boram Lee was partly supported by NRF-2012M3A9D1054451.
Appendix 1.
Down-Regulated Proteins in pterygia, Identified by LC-MS/MS. To access the data, click or select the words “Appendix 1.”
Appendix 2.
Up-Regulated Proteins in pterygia, Identified by LC-MS/MS. To access the data, click or select the words “Appendix 2.”
References
- 1.Solomon AS. Immunologic basis for the pathogenesis of pterygium. Am J Ophthalmol. 1985;99:216–7. doi: 10.1016/0002-9394(85)90243-0. [DOI] [PubMed] [Google Scholar]
- 2.Hill JC, Maske R. Pathogenesis of pterygium. Eye (Lond) 1989;3:218–26. doi: 10.1038/eye.1989.31. [DOI] [PubMed] [Google Scholar]
- 3.Chui J, Di Girolamo N, Wakefield D, Coroneo MT. The pathogenesis of pterygium: current concepts and their therapeutic implications. Ocul Surf. 2008;6:24–43. doi: 10.1016/s1542-0124(12)70103-9. [DOI] [PubMed] [Google Scholar]
- 4.Todani A, Melki SA. Pterygium: current concepts in pathogenesis and treatment. Int Ophthalmol Clin. 2009;49:21–30. doi: 10.1097/IIO.0b013e3181924f62. [DOI] [PubMed] [Google Scholar]
- 5.Saw SM, Tan D. Pterygium: prevalence, demography and risk factors. Ophthalmic Epidemiol. 1999;6:219–28. doi: 10.1076/opep.6.3.219.1504. [DOI] [PubMed] [Google Scholar]
- 6.Bradley JC, Yang W, Bradley RH, Reid TW, Schwab IR. The science of pterygia. Br J Ophthalmol. 2010;94:815–20. doi: 10.1136/bjo.2008.151852. [DOI] [PubMed] [Google Scholar]
- 7.Balci M, Sahin S, Mutlu FM, Yagci R, Karanci P, Yildiz M. Investigation of oxidative stress in pterygium tissue. Mol Vis. 2011;17:443–7. [PMC free article] [PubMed] [Google Scholar]
- 8.Perra MT, Maxia C, Corbu A, Minerba L, Demurtas P, Colombari R, Murtas D, Bravo S, Piras F, Sirigu P. Oxidative stress in pterygium: relationship between p53 and 8-hydroxydeoxyguanosine. Mol Vis. 2006;12:1136–42. [PubMed] [Google Scholar]
- 9.Seet LF, Tong L, Su R, Wong TT. Involvement of SPARC and MMP-3 in the pathogenesis of human pterygium. Invest Ophthalmol Vis Sci. 2012;53:587–95. doi: 10.1167/iovs.11-7941. [DOI] [PubMed] [Google Scholar]
- 10.Tan DT, Tang WY, Liu YP, Goh HS, Smith DR. Apoptosis and apoptosis related gene expression in normal conjunctiva and pterygium. Br J Ophthalmol. 2000;84:212–6. doi: 10.1136/bjo.84.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Girolamo N, Kumar RK, Coroneo MT, Wakefield D. UVB-mediated induction of interleukin-6 and −8 in pterygia and cultured human pterygium epithelial cells. Invest Ophthalmol Vis Sci. 2002;43:3430–7. [PubMed] [Google Scholar]
- 12.Chiang CC, Cheng YW, Lin CL, Lee H, Tsai FJ, Tseng SH, Tsai YY. Cyclooxygenase 2 expression in pterygium. Mol Vis. 2007;13:635–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Beden U, Irkec M, Orhan D, Orhan M. The roles of T-lymphocyte subpopulations (CD4 and CD8), intercellular adhesion molecule-1 (ICAM-1), HLA-DR receptor, and mast cells in etiopathogenesis of pterygium. Ocul Immunol Inflamm. 2003;11:115–22. doi: 10.1076/ocii.11.2.115.15913. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen CQ, Peck AB. Unraveling the pathophysiology of Sjogren syndrome-associated dry eye disease. Ocul Surf. 2009;7:11–27. doi: 10.1016/s1542-0124(12)70289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soria J, Durán JA, Etxebarria J, Merayo J, González N, Reigada R, García I, Acera A, Suárez T. Tear proteome and protein network analyses reveal a novel pentamarker panel for tear film characterization in dry eye and meibomian gland dysfunction. J Proteomics. 2013;78:94–112. doi: 10.1016/j.jprot.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Hains PG, Truscott RJ. Proteome analysis of human foetal, aged and advanced nuclear cataract lenses. Proteomics Clin Appl. 2008;2:1611–9. doi: 10.1002/prca.200800085. [DOI] [PubMed] [Google Scholar]
- 17.Stastna M, Behrens A, McDonnell PJ, Van Eyk JE. Analysis of protein composition of rabbit aqueous humor following two different cataract surgery incision procedures using 2-DE and LC-MS/MS. Proteome Sci. 2011;9:8. doi: 10.1186/1477-5956-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011;92:282–98. doi: 10.1016/j.exer.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Frost MR, Norton TT. Alterations in protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Invest Ophthalmol Vis Sci. 2012;53:322–36. doi: 10.1167/iovs.11-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bautista-de Lucio VM, López-Espinosa NL, Robles-Contreras A, Pérez-Cano HJ, Mejía-López H, Mendoza G, Jiménez-Martínez MC, Garfias Y. Overexpression of peroxiredoxin 2 in pterygium. A proteomic approach. Exp Eye Res. 2013;110:70–5. doi: 10.1016/j.exer.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Sung HJ, Ahn JM, Yoon YH, Rhim TY, Park CS, Park JY, Lee SY, Kim JW, Cho JY. Identification and validation of SAA as a potential lung cancer biomarker and its involvement in metastatic pathogenesis of lung cancer. J Proteome Res. 2011;10:1383–95. doi: 10.1021/pr101154j. [DOI] [PubMed] [Google Scholar]
- 22.Heo SH, Lee SJ, Ryoo HM, Park JY, Cho JY. Identification of putative serum glycoprotein biomarkers for human lung adenocarcinoma by multilectin affinity chromatography and LC-MS/MS. Proteomics. 2007;7:4292–302. doi: 10.1002/pmic.200700433. [DOI] [PubMed] [Google Scholar]
- 23.Tsai YY, Cheng YW, Lee H, Tsai FJ, Tseng SH, Lin CL, Chang KC. Oxidative DNA damage in pterygium. Mol Vis. 2005;11:71–5. [PubMed] [Google Scholar]
- 24.Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–89. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 25.Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–92. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- 26.Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: a corneal crystallin with diverse functions. Exp Eye Res. 2007;84:3–12. doi: 10.1016/j.exer.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Estey T, Chen Y, Carpenter JF, Vasiliou V. Structural and functional modifications of corneal crystallin ALDH3A1 by UVB light. PLoS ONE. 2010;5:e15218. doi: 10.1371/journal.pone.0015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black W, Chen Y, Matsumoto A, Thompson DC, Lassen N, Pappa A, Vasiliou V. Molecular mechanisms of ALDH3A1-mediated cellular protection against 4-hydroxy-2-nonenal. Free Radic Biol Med. 2012;52:1937–44. doi: 10.1016/j.freeradbiomed.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamradt MC, Chen F, Sam S, Cryns VL. The small heat shock protein alpha B-crystallin negatively regulates apoptosis during myogenic differentiation by inhibiting caspase-3 activation. J Biol Chem. 2002;277:38731–6. doi: 10.1074/jbc.M201770200. [DOI] [PubMed] [Google Scholar]
- 30.Andley UP, Song Z, Wawrousek EF, Brady JP, Bassnett S, Fleming TP. Lens epithelial cells derived from alphaB-crystallin knockout mice demonstrate hyperproliferation and genomic instability. FASEB. 2001;15:221–9. doi: 10.1096/fj.00-0296com. [DOI] [PubMed] [Google Scholar]
- 31.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–51. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, Redfern CP, Piacentini M. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. Br J Cancer. 2007;96:1062–71. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aureli C, Gaucci E, Arcangeli V, Grillo C, Eufemi M, Chichiarelli S. ERp57/PDIA3 binds specific DNA fragments in a melanoma cell line. Gene. 2013;524:390–5. doi: 10.1016/j.gene.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Pressinotti NC, Klocker H, Schäfer G, Luu VD, Ruschhaupt M, Kuner R, Steiner E, Poustka A, Bartsch G, Sültmann H. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer. Mol Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Marco F, Bucaj E, Foppoli C, Fiorini A, Blarzino C, Filipi K, Giorgi A, Schininà ME, Di Domenico F, Coccia R, Butterfield DA, Perluigi M. Oxidative stress in HPV-driven viral carcinogenesis: redox proteomics analysis of HPV-16 dysplastic and neoplastic tissues. PLoS ONE. 2012;7:e34366. doi: 10.1371/journal.pone.0034366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cicchillitti L, Di Michele M, Urbani A, Ferlini C, Donat MB, Scambia G, Rotilio D. Comparative proteomic analysis of paclitaxel sensitive A2780 epithelial ovarian cancer cell line and its resistant counterpart A2780TC1 by 2D-DIGE: the role of ERp57. J Proteome Res. 2009;8:1902–12. doi: 10.1021/pr800856b. [DOI] [PubMed] [Google Scholar]
- 37.Coe H, Michalak M. ERp57, a multifunctional endoplasmic reticulum resident oxidoreductase. Int J Biochem Cell Biol. 2010;42:796–9. doi: 10.1016/j.biocel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Garbi N, Hammerling G, Tanaka S. Interaction of ERp57 and tapasin in the generation of MHC class I-peptide complexes. Curr Opin Immunol. 2007;19:99–105. doi: 10.1016/j.coi.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Garbi N, Tanaka S, Momburg F, Hammerling GJ. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol. 2006;7:93–102. doi: 10.1038/ni1288. [DOI] [PubMed] [Google Scholar]
- 40.Zhao W, Fan GC, Zhang ZG, Bandyopadhyay A, Zhou X, Kranias EG. Protection of peroxiredoxin II on oxidative stress-induced cardiomyocyte death and apoptosis. Basic Res Cardiol. 2009;104:377–89. doi: 10.1007/s00395-008-0764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang K, Jiang Z, Ding BQ, Cheng P, Huang DK, Tao LM. Expression of cell proliferation and apoptosis biomarkers in pterygia and normal conjunctiva. Mol Vis. 2011;17:1687–93. [PMC free article] [PubMed] [Google Scholar]
- 42.Nagababu E, Mohanty JG, Friedman JS, Rifkind JM. Role of peroxiredoxin-2 in protecting RBCs from hydrogen peroxide-induced oxidative stress. Free Radic Res. 2013;47:164–71. doi: 10.3109/10715762.2012.756138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao F, Wang Q. The protective effect of peroxiredoxin II on oxidative stress induced apoptosis in pancreatic beta-cells. Cell and Bioscience. 2012;2:22. doi: 10.1186/2045-3701-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fatma N, Kubo E, Sen M, Agarwal N, Thoreson WB, Camras CB, Singh DP. Peroxiredoxin 6 delivery attenuates TNF-alpha-and glutamate-induced retinal ganglion cell death by limiting ROS levels and maintaining Ca2+ homeostasis. Brain Res. 2008;1233:63–78. doi: 10.1016/j.brainres.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fatma N, Kubo E, Toris CB, Stamer WD, Camras CB, Singh DP. PRDX6 attenuates oxidative stress- and TGFbeta-induced abnormalities of human trabecular meshwork cells. Free Radic Res. 2009;43:783–95. doi: 10.1080/10715760903062887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tulsawani R, Kelly LS, Fatma N, Chhunchha B, Kubo E, Kumar A, Singh DP. Neuroprotective effect of peroxiredoxin 6 against hypoxia-induced retinal ganglion cell damage. BMC Neurosci. 2010;11:125. doi: 10.1186/1471-2202-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chhunchha B, Fatma N, Bhargavan B, Kubo E, Kumar A, Singh DP. Specificity protein, Sp1-mediated increased expression of Prdx6 as a curcumin-induced antioxidant defense in lens epithelial cells against oxidative stress. Cell Death Dis. 2011;2:e234. doi: 10.1038/cddis.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo CH Miyazaki D, Nawata N, Tominaga T, Yamasaki A, Sasaki Y, Inoue Y. Prognosis-determinant candidate genes identified by whole genome scanning in eyes with pterygia. Invest Ophthalmol Vis Sci. 2007;48:3566–75. doi: 10.1167/iovs.06-1149. [DOI] [PubMed] [Google Scholar]
- 49.Kuo CH, Miyazaki D, Yakura K, Araki-Sasaki K, Inoue Y. Role of periostin and interleukin-4 in recurrence of pterygia. Invest Ophthalmol Vis Sci. 2010;51:139–43. doi: 10.1167/iovs.09-4022. [DOI] [PubMed] [Google Scholar]