SUMMARY
Long-distance communication between leaves and roots are key to properly regulate the uptake of trace metals from the soil. The molecular basis of this shoot-to-root signaling is currently unknown. In this manuscript, we describe the role of OPT3 in the shoot-to-root signaling of the iron status in Arabidopsis. We also show that reduced expression of OPT3 induces an over-accumulation of the toxic metal cadmium, but not other metals, in seeds.
Key words: phloem transport, seed loading, metal homeostasis, iron deficiency, ionomics.
Abstract
Plants and seeds are the main dietary sources of zinc, iron, manganese, and copper, but are also the main entry point for toxic elements such as cadmium into the food chain. We report here that an Arabidopsis oligopeptide transporter mutant, opt3-2, over-accumulates cadmium (Cd) in seeds and roots but, unexpectedly, under-accumulates Cd in leaves. The cadmium distribution in opt3-2 differs from iron, zinc, and manganese, suggesting a metal-specific mechanism for metal partitioning within the plant. The opt3-2 mutant constitutively up-regulates the Fe/Zn/Cd transporter IRT1 and FRO2 in roots, indicative of an iron-deficiency response. No genetic mutants that impair the shoot-to-root signaling of iron status in leaves have been identified. Interestingly, shoot-specific expression of OPT3 rescues the Cd sensitivity and complements the aberrant expression of IRT1 in opt3-2 roots, suggesting that OPT3 is required to relay the iron status from leaves to roots. OPT3 expression was found in the vasculature with preferential expression in the phloem at the plasma membrane. Using radioisotope experiments, we found that mobilization of Fe from leaves is severely affected in opt3-2, suggesting that Fe mobilization out of leaves is required for proper trace-metal homeostasis. When expressed in yeast, OPT3 does not localize to the plasma membrane, precluding the identification of the OPT3 substrate. Our in planta results show that OPT3 is important for leaf phloem-loading of iron and plays a key role regulating Fe, Zn, and Cd distribution within the plant. Furthermore, ferric chelate reductase activity analyses provide evidence that iron is not the sole signal transferred from leaves to roots in leaf iron status signaling.
INTRODUCTION
Heavy metals such as iron (Fe), zinc (Zn), copper (Cu), and manganese (Mn) are essential micronutrients for all organisms, acting as co-factors in a variety of biological processes. These heavy metals are extremely reactive and can become toxic at high concentrations; therefore, the intracellular concentration of these essential metals must be tightly regulated (Palmer and Guerinot, 2009; Mendoza-Cozatl et al., 2011). Other heavy metals such as cadmium (Cd), lead, mercury, and the metalloid arsenic (As) do not have biological functions in plants and are toxic even in trace amounts, disrupting several biochemical activities by displacing essential metals from their respective binding sites (Clemens et al., 1998; Mendoza-Cozatl et al., 2011). In humans, Cd exposure has been linked to cancer in the kidneys, lungs, and prostate, and severe Cd poisonings can result in neurological disorders and pulmonary and renal failure (Hendrick, 1996; Il’yasova and Schwartz, 2005; Nawrot et al., 2006). While occupational exposure and tobacco products are associated with a high risk of Cd poisoning, consumption of contaminated plant-based foods represents the major source of Cd exposure in the general public (Franz et al., 2008; Satarug et al., 2010). Many cases of widespread cadmium poisonings have been attributed to consumption of contaminated seeds in Thailand, China, Japan, and Australia (McLaughlin et al., 1997; Clemens et al., 2013). However, the molecular mechanisms and genes mediating the loading of both essential and non-essential heavy metals into seeds remain largely unknown.
Metal accumulation and distribution in plants consist of several mechanisms, including: (1) metal uptake into roots, (2) xylem-loading and transport to the shoot, and (3) phloem-mediated redistribution of metals from mature leaves to sink tissues, including younger leaves, roots, and seeds (reviewed in Mendoza-Cozatl et al., 2011; Palmer and Guerinot, 2009; Verbruggen et al., 2009). Cadmium enters the root through the Fe transporter IRT1, which shows broad substrate specificity towards divalent metals including Fe2+, Zn2+, Mn2+, and Cd2+ (Eide et al., 1996; Rogers et al., 2000). Once inside the cell, metals bind to different ligands, according to specific affinities, and these metal–ligand complexes can be stored in different cellular compartments or distributed to other tissues through the vasculature (Verbruggen et al., 2009; Mendoza-Cozatl et al., 2011).
Because of the broad substrate specificity of IRT1 for divalent metals, transcriptional regulation of the Fe-deficiency response, including up-regulation of IRT1, will also have an impact on the uptake of non-essential heavy metals such as Cd. In plants, the root iron-deficiency response is regulated by local signals within the root and also by systemic signals originating from leaves (Vert et al., 2003; Walker and Connolly, 2008; Hindt and Guerinot, 2012). Two major transcriptional networks have been identified to mediate the Fe-deficiency response at the root level in Arabidopsis: the FIT network and the POPEYE network (Walker and Connolly, 2008; Long et al., 2010; Hindt and Guerinot, 2012). The components of the systemic shoot-to-root Fe signaling on the other hand remain largely unknown. The identification of mutants showing a constitutive Fe-deficiency response even when Fe is supplied in sufficient amounts plus experiments where the constitutive root response is restored by foliar application of Fe suggest that mobile Fe (likely through the phloem) is required for proper shoot-to-root signaling (Vert et al., 2003; Garcia et al., 2013). However, the transporters, ligands, and the chemical speciation of the putative phloem-mobile molecule mediating the systemic Fe signaling have not yet been clearly identified.
Here, we report that opt3-2, an Arabidopsis mutant carrying an insertion in the 5’ UTR of the oligopeptide transporter gene OPT3 (Stacey et al., 2008), over-accumulates significant levels of Cd in seeds. We present evidence suggesting that this Cd over-accumulation may be the result of an enhanced transport of Cd through the plant, making opt3-2 a suitable background for studying long-distance transport of non-essential heavy metals. We further show that OPT3 is targeted to the plasma membrane and is preferentially expressed in the phloem. The Fe/Zn/Mn uptake transporter IRT1 and other iron-starvation-induced genes are constitutively up-regulated in opt3-2. Interestingly, shoot-specific expression of OPT3 restores metal homeostasis and IRT1 up-regulation in roots showing that OPT3 is the first identified molecular component of the network transferring information on the iron status from leaves to roots. Moreover, Fe mobilization between leaves is impaired in opt3-2, suggesting that OPT3 mediates the movement of Fe out of the leaves, and this transport is required for proper communication between leaves and roots and maintenance of the trace-metal homeostasis in Arabidopsis. Understanding phloem-mediated signaling, transport, and seed-loading mechanisms of both essential and non-essential heavy metals will help to develop strategies for excluding toxic metals from seeds and enhance the nutritional value of grains and plant-based products.
RESULTS
opt3-2 Over-Accumulates Cd in Seeds and Shows an Altered Cd Partitioning within Plant Tissues
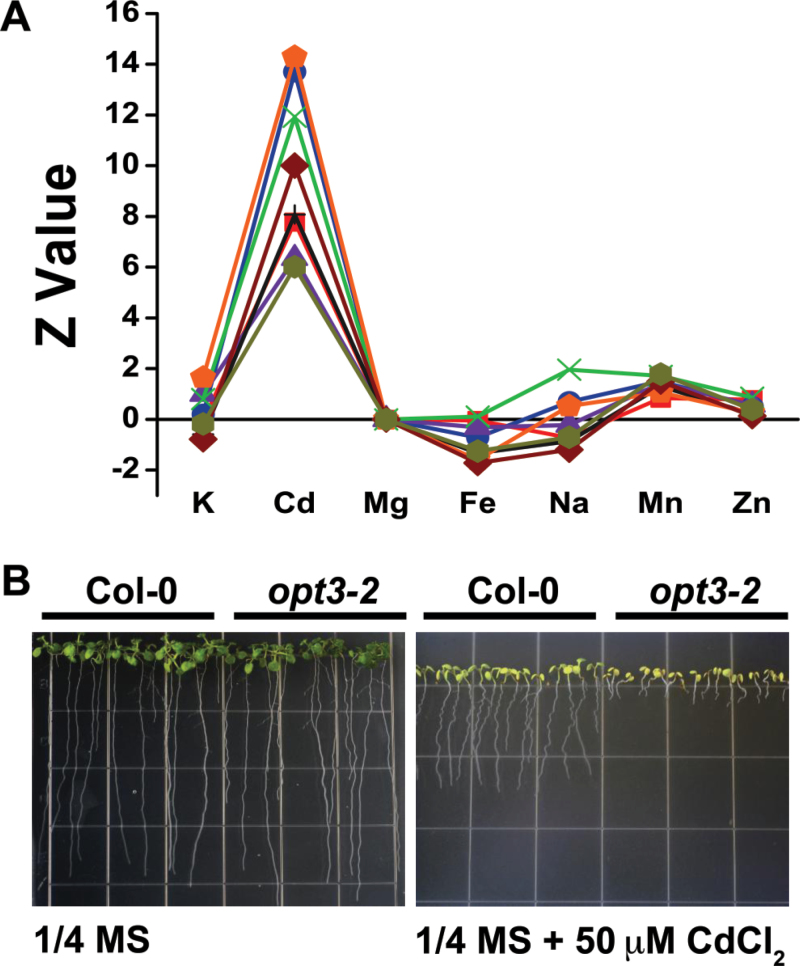
Members of the Arabidopsis oligopeptide transporter family (OPT) have been shown to mediate the transport of a broad spectrum of peptides (Koh et al., 2002; Pike et al., 2009). Glutathione (GSH) and phytochelatins are peptides that mediate tolerance and long-distance transport of heavy metals (Mendoza-Cozatl et al., 2008, 2011); therefore, we screened mutants in the Arabidopsis OPT family for differential accumulation of Cd in seeds. A mutant of the Arabidopsis OPT3 gene, opt3-2, showed the strongest over-accumulation of Cd in seeds (Figure 1A). To test whether this Cd over-accumulation had an effect on seedling growth, assays were performed on plates in the presence and absence of Cd. Figure 1B shows that opt3-2 is hypersensitive to Cd when grown on medium containing 50 μM CdCl2. To determine whether the increased Cd concentration in opt3-2 seeds was due to a systemic over-accumulation of Cd throughout the plant, opt3-2 seedlings were grown hydroponically for 6 weeks, exposed to 20 μM CdCl2 for 72h and the metal concentration of roots and leaves was measured by ICP–OES (Figure 2). The roots of opt3-2 over-accumulated Cd compared to wild-type; however, unexpectedly, Cd concentrations in leaves were almost five-fold less than those of wild-type plants (Figure 2). Conversely, seeds of opt3-2 plants show a large increase in Cd levels compared to wild-type seeds (Figure 2).
Figure 1.
opt3-2 Over-Accumulates Cd in Seeds and Is Cd-Hypersensitive.
(A) Ionomic profile of opt3-2 seeds grown on soil supplemented with heavy metals. Metal concentrations were determined by ICP–OES, normalized against Mg, and plotted as standard deviation from the wild-type mean (Z-value) (Lahner et al., 2003). Each line represents seeds from independent plants grown on heavy metal-laden soil. Z-values are considered significant when |z| > 1.96 (p < 0.05). (B) opt3-2 seedlings are hypersensitive to Cd. Wild-type and opt3-2 seeds were grown on ¼ MS media with or without 50 μM CdCl2 for 2 weeks.
Figure 2.
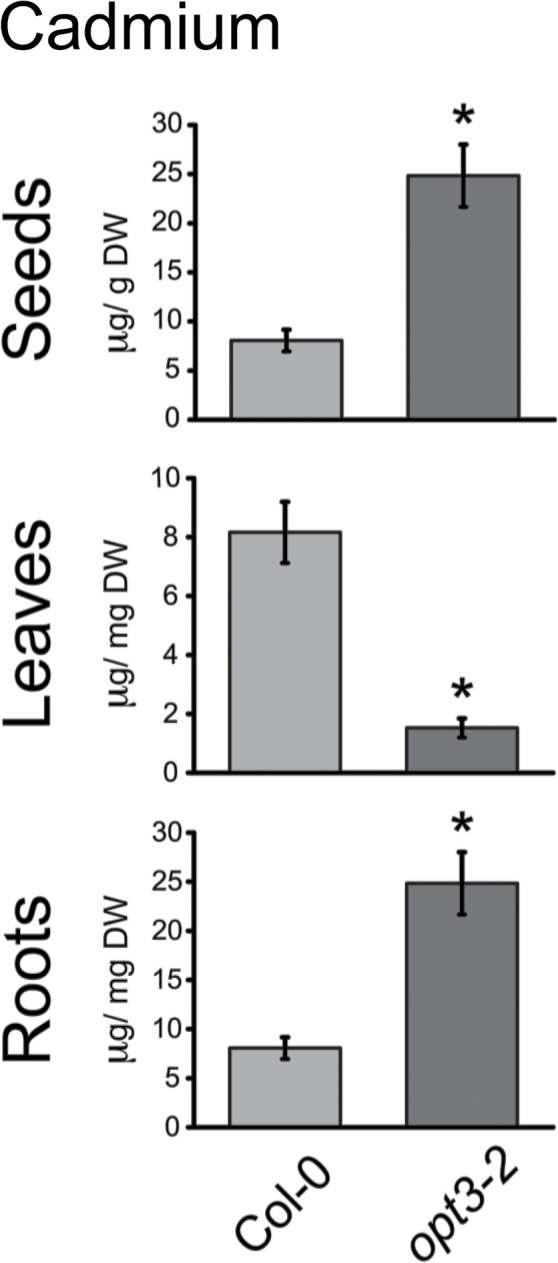
Cadmium Distribution between Tissues Is Altered in opt3-2 Plants.
Cd concentration was measured in roots (n = 5) and rosette leaves (n = 10) of 6-week-old hydroponically grown plants exposed to 20 μM CdCl2 for 72h and dried seeds of plants (n = 18) grown on soil containing a defined content of heavy metals (Lahner et al., 2003). Data represent mean ± SE (* p < 0.05).
Cadmium Distribution in opt3-2 Shoots Is Different from Essential Metals
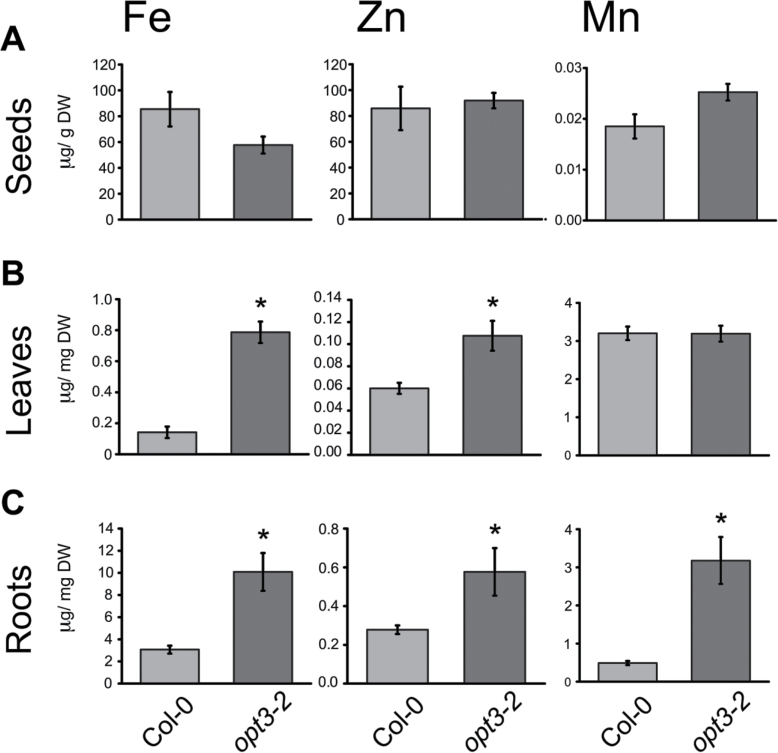
To determine whether the altered distribution of Cd in opt3-2 correlated with the distribution of essential metals in plant tissues, the levels of Zn, Fe, and Mn in opt3-2 were also measured and compared to wild-type plants (Figure 3). No dramatic differences in the concentration of Zn and Mn in seeds were found between wild-type and opt3-2 (Figure 3A). However, in contrast to Cd accumulation, opt3-2 over-accumulated significant levels of Zn and Fe in leaves compared to wild-type (Figure 3B). In roots, the concentration of Fe, Zn, and Mn was increased in opt3-2 compared to wild-type (Figure 3C). The different distribution of Cd in aerial parts of the plants (leaves and seeds) (Figure 2) suggests that the mechanisms mediating accumulation of metals in opt3-2 leaves is different for Cd compared to the essential metals Fe, Zn, and Mn.
Figure 3.
The Distribution of Iron, Zinc, and Manganese Is Different from Cd in opt3-2.
Metal concentration in roots, leaves, and seeds was determined as in Figures 1 and 2.
(A) Concentration of Fe, Zn, and Mn in opt3-2 seeds was similar to wild-type (n = 18).
(B) In leaves, only Zn and Fe were over-accumulated while Mn concentration was unaffected (n = 10).
(C) In roots, opt3-2 plants exhibited over-accumulation of Zn, Fe, and Mn compared to wild-type plants (n = 5). Data represent mean ± SE (* p < 0.05).
Ectopically Expressed OPT3 Complements opt3-2
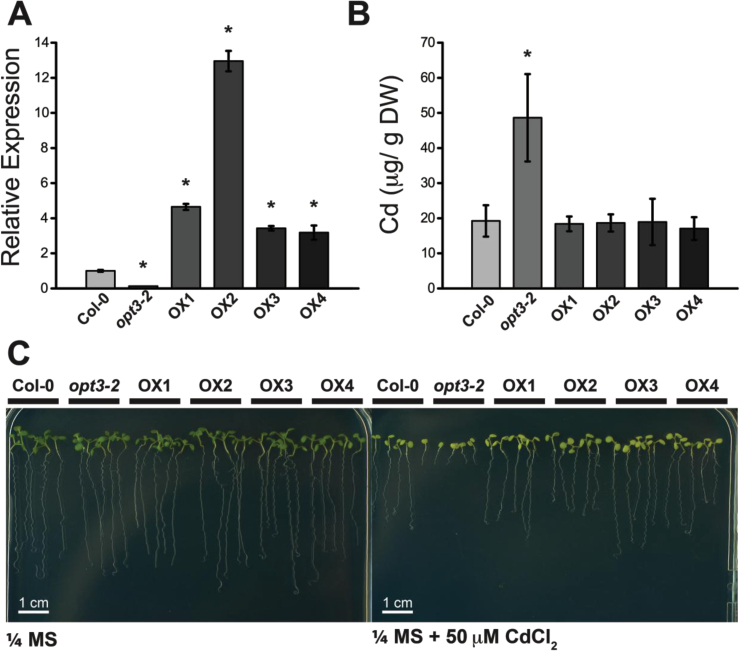
To evaluate whether the observed metal accumulation phenotypes were due to impaired expression of the OPT3 gene, the coding sequence of OPT3 was expressed in opt3-2 under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Overexpression of OPT3 in four independent lines was confirmed by qPCR (Figure 4A). opt3-2 contains a T-DNA insertion in the 5’ UTR of OPT3 (Stacey et al., 2008). Therefore, the residual OPT3 transcript observed in opt3-2 is expected in this knockdown line. OPT3 complementation lines were grown on heavy metal-containing soil, and the metal concentration of their seeds was determined by ICP–OES. Cd accumulation in seeds of the four complemented lines was reduced to wild-type levels (Figure 4B). Overexpression also rescued the seedling sensitivity of opt3-2 to Cd (Figure 4C), indicating that ectopic expression of OPT3 is sufficient to complement the sensitivity and metal accumulation phenotypes of opt3-2.
Figure 4.
Ectopic Overexpression of OPT3 in opt3-2 Reduces Cadmium Concentration in Seeds and Rescues the Seedling Hypersensitivity to Cd.
(A) Relative OPT3 expression levels of four representative 35S pro:OPT3 overexpression lines. Wild-type, opt3-2, and four OPT3 overexpression lines were grown on ¼ MS for 2 weeks, and OPT3 expression was determined by qPCR and normalized against wild-type OPT3 expression levels. Data represent mean ± SE (n = 3).
(B) OPT3 overexpression reduces the over-accumulation of Cd in seeds and (C) the Cd hypersensitivity of opt3-2 seedlings. Wild-type, opt3-2, and four complemented lines were grown on ¼ MS with or without 50 μM CdCl2 for 2 weeks. Data represent mean ± SE (n = 6; * p < 0.05).
OPT3 Is a Plasma Membrane Transporter Preferentially Expressed in the Phloem
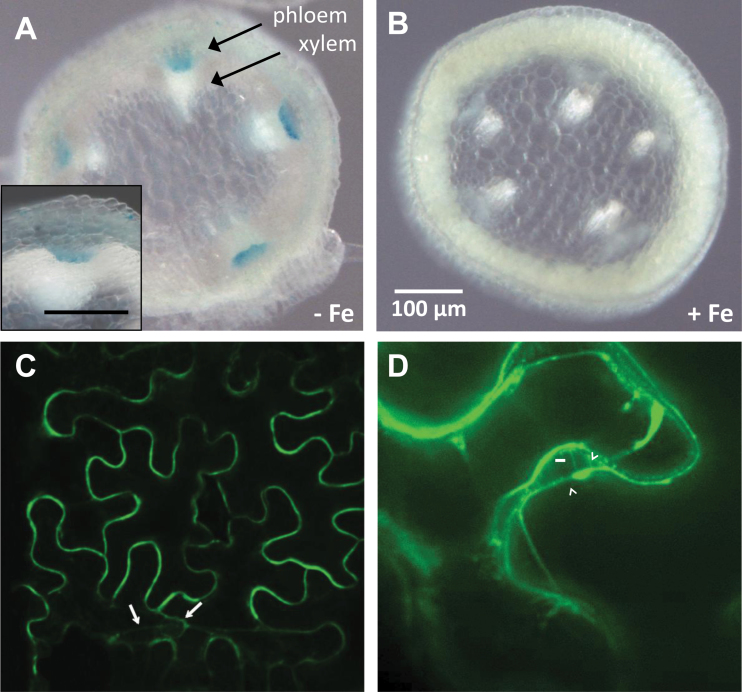
Previous GUS staining experiments have shown that OPT3 is expressed throughout the vasculature; however, localization at a higher resolution has not been evaluated (Stacey et al., 2006). To identify where in the vasculature OPT3 is preferentially expressed, β-glucuronidase (GUS) was expressed under the control of the native OPT3 promoter. Under standard growth conditions, GUS staining was negligible; however, under Fe-limiting conditions (under which OPT3 expression is induced), staining was clearly observed in the phloem, but not in the pith or endodermis (Figure 5A and 5B). Consistent with our findings, cell-type-specific microarray data sets show the highest intensity values of OPT3 in the phloem, comparable to the phloem sucrose transporter SUC2 (Supplemental Figure 1) (Mustroph et al., 2009). Thus, two independent approaches show preferential expression of OPT3 in the phloem. To gain insight into the subcellular localization of OPT3, an N-terminal YFP–OPT3 translational fusion was infiltrated into Nicotiana benthamiana leaves. Fluorescence was detected along the cell periphery, indicative of plasma membrane localization (Figure 5C). A weaker perinuclear fluorescence and transvacuolar strands were also observed in some cells (see arrows in Figure 5C), indicating that a fraction of the YFP–OPT3 localizes to the endoplasmic reticulum (ER). The ER fluorescence pattern, however, was not present in all cells. Furthermore, Hechtian strands were clearly present connecting the cell wall to the plasma membrane of plasmolyzed leaf cells (Figure 5D). A previous large-scale proteomics study in Arabidopsis also found OPT3 at the plasma membrane (Li et al., 2012). These results suggest that OPT3 is a plasma membrane transporter preferentially expressed in the phloem.
Figure 5.
OPT3 Is a Plasma Membrane Transporter Expressed in the Phloem.
(A, B) GUS staining was performed in fixed and sectioned stems of OPT3 pro:GUS plants under (A) iron-deficient conditions and (B) Fe-sufficient conditions.
(C) OPT3 localizes to the plasma membrane. N. benthamiana epidermal cells were infiltrated with Agrobacterium carrying 35S pro:YFP–OPT3 and imaged using confocal microscopy 3 d later. Fluorescence in the cell perimeter is indicative of plasma membrane localization. ER fluorescence is also present (arrows) surrounding the nucleus and as strands traversing the cytoplasm.
(D) Leaves of N. benthamiana epidermal cells transiently expressing YFP–OPT3 as in the panel were plasmolyzed with 4% NaCl. Note the Hechtian strands (arrows) connecting the cell wall to the plasmolyzed protoplast (double arrowheads), indicative of plasma membrane localization.
Shoot-Specific Expression of OPT3 Is Sufficient to Restore Metal Homeostasis
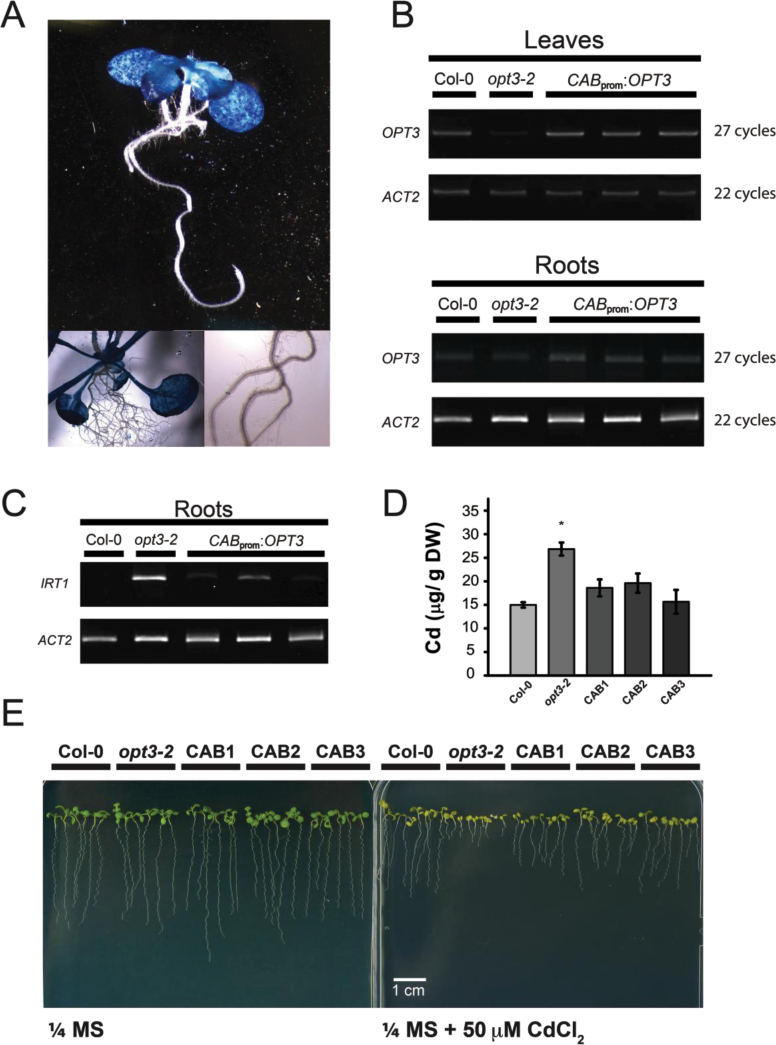
The Arabidopsis mutant opt3-2 shows a constitutive Fe-deficiency response in roots including the up-regulation of the Fe/Zn/Mn transporter IRT1 (Stacey et al., 2008). Despite this Fe-deficiency response, Fe sensing in shoots remains intact (Stacey et al., 2008). The molecular mechanisms mediating shoot-to-root signaling of iron status in plants remain largely unknown. The impaired iron sensing in roots but not shoots of opt3-2, in conjunction with phloem localization, suggests a possible role of OPT3 in shoot-to-root transport of a signal reporting metal status. To test this hypothesis, the OPT3 coding sequence was expressed in opt3-2 under the control of the shoot-specific chlorophyll a/b binding protein promoter (CAB2 pro:OPT3) (Chen et al., 2006). Shoot specificity of the CAB2 promoter was determined by GUS staining (Figure 6A). RT–PCR analyses confirmed that OPT3 is preferentially expressed in the shoots of three independent transgenic lines (Figure 6B). The residual OPT3 transcript in opt3-2 roots expressing CAB2 pro:OPT3 plants is consistent with the knockdown nature of the opt3-2 allele. Thus, the low level of OPT3 transcript in roots is not sufficient to properly regulate metal homeostasis in roots (Stacey et al., 2008).
Figure 6.
Shoot-Specific Expression of OPT3 Is Sufficient to Complement the Fe-Deficiency Response in opt3-2 Roots.
(A) CAB2 pro is preferentially active in shoots and is not active in roots. GUS staining in a whole seedling expressing CAB2 pro:GUS is evident only in shoots.
(B) RT–PCR confirmed the shoot specificity of CAB2 pro:OPT3. Wild-type, opt3-2, and three independent CAB2 pro:OPT3 lines were grown vertically on ¼ MS plates for 2 weeks, and cDNA was prepared separately from root and leaf RNA. OPT3 expression was determined in roots and shoots of wild-type, opt3-2, and three independent CAB2 pro:OPT3 lines. ACT2 was used a loading control, and the number of PCR cycles is shown to the right of each gel image. Note that complete knockout of OPT3 causes embryo lethality (Stacey et al., 2002), and opt3-2 shows reduced expression of OPT3 transcript.
(C) CAB2 pro:OPT3 successfully restores regulation of IRT1 in opt3-2. IRT1 expression in roots of wild-type, opt3-2, and CAB2 pro:OPT3 was determined by RT–PCR as in panel (A). RT–PCR was performed for 22 cycles, and ACT2 was used as a loading control.
(D) opt3-2 plants expressing CAB2 pro:OPT3 accumulate wild-type levels of Cd in seeds. Wild-type, opt3-2, and three CAB2 pro:OPT3 lines were grown on heavy metal-laden soil, and their seed metal concentration was determined by ICP–OES as in Figure 1. Data represent mean ± SE (n = 6; * p < 0.05).
(E) CAB2 pro:OPT3 complements seedling sensitivity to Cd in opt3-2. Wild-type, opt3-2, and three CAB2 pro:OPT3 lines were grown on ¼ MS with or without 50 μM CdCl2 for 2 weeks.
Two of the major phenotypes described in opt3-2 are the constitutive iron-deficiency response in roots, as illustrated by high IRT1 expression (Figure 6C), and the over-accumulation of Cd in seeds (Figure 6D). Thus, we tested whether shoot-specific expression of OPT3 was able to complement both phenotypes. As shown by RT–PCR, IRT1 transcript levels were greatly reduced in the roots of CAB2 pro:OPT3-expressing plants compared to the opt3-2 mutant (Figure 6D). These results show that shoot-specific expression of OPT3 is sufficient for proper regulation of metal homeostasis, including communication between leaves and roots. Furthermore, the Cd accumulation in CAB2 pro:OPT3 seeds was reduced to wild-type levels (Figure 6D). Seedling hypersensitivity to Cd was also rescued in the three independent CAB2 pro:OPT3-expressing lines (Figure 6E). Collectively, these results demonstrate that shoot-specific OPT3 expression is sufficient to complement opt3-2 root phenotypes, suggesting that OPT3 may mediate the long-distance transport of a signaling molecule from leaves to relay information about metal status, thus contributing to whole-plant metal homeostasis.
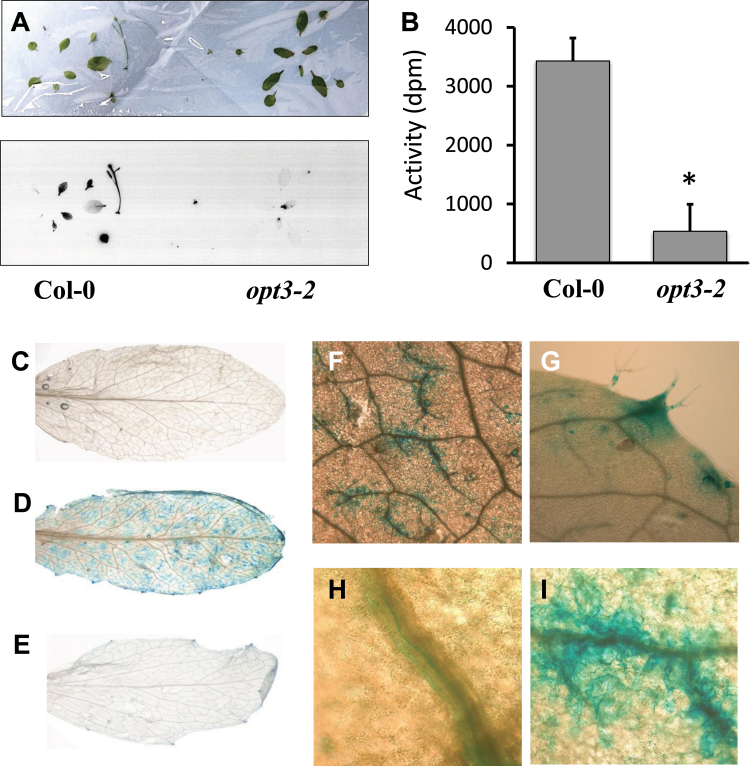
Leaf-to-Leaf Transport of Fe Is Impaired in opt3-2
To test whether OPT3 functions in the mobilization of Fe or other molecules, we first assessed the capacity of wild-type and opt3-2 to remobilize Fe from one leaf to other leaves using the radiotracer 59Fe. In these experiments, 59Fe was loaded into a mature leaf as Fe2+ at a slightly acidic pH (pH 6.2) to resemble the apoplastic pH. The addition of ascorbic acid was used to reduce Fe3+ to Fe2+ and maintain it in the reduced form. Figure 7A shows that Fe can be re-mobilized from one leaf to adjacent leaves in wild-type. In contrast, opt3-2 shows negligible movement of 59Fe between leaves. Figure 7B shows the 59Fe activity (dpm) in the four leaves adjacent to the leaf where the 59Fe was originally applied. Compared to wild-type, opt3-2 shows a severe reduction in the quantity of 59Fe mobilized from one leaf to the adjacent leaves (Figure 7B), suggesting that OPT3 is required for the reallocation of Fe between plant tissues. In fact, opt3-2 plants over-accumulate Fe in mature leaves compared to wild-type, as visualized by Perls’ staining (Figure 7C and 7D). Interestingly, over-accumulation of Fe in opt3-2 occurs only in mature leaves but not in young leaves (Figure 7D and 7E). Moreover, accumulation of Fe in opt3-2 is more evident at the base of the trichomes and near the vasculature in the minor veins but not in the main vasculature, suggesting that, in opt3-2, the reallocation of Fe between leaves is impaired, particularly at advanced stages of leaf development (Figure 7F–7I).
Figure 7.
Mobilization of Iron between Leaves Is Impaired in opt3-2.
(A) 59Fe was applied to a mature wild-type or opt3-2 leaf and the distribution of 59Fe was monitored after a 12-h incubation period. Lower panel: Signal coming from 59Fe was detected in wild-type leaves adjacent to the leaf where the 59Fe was applied while only a fraction of the signal was detected in opt3-2 leaves.
(B) The specific activity measured in the four adjacent leaves to which the 59Fe was originally applied show that the movement of 59Fe in opt3-2 was marginal. Data represent mean ± SE (n = 4, * p < 0.05).
(C–I) Visualization of Fe using Perls’ stain shows that, compared to a mature wild-type leaf (C), opt3-2 contains substantial amounts of stainable Fe (D). (E) Over-accumulation of Fe in opt3-2 leaves is less evident in younger leaves. Accumulation of Fe in opt3-2 leaves is more evident close to (F) secondary veins, (G) the base of trhichomes, and (H, I) surrounding the vasculature.
To test whether OPT3 functions as a Fe2+ transporter similarly to IRT1, we expressed OPT3 in a yeast strain deficient in Fe2+ uptake (fet3fet4). As previously shown, IRT1 expression in yeast allows the fet3fet4 strain to grow on minimal media without the addition of extra Fe (Supplemental Figure 2). OPT3 was unable to rescue the fet3fet4 strain (Supplemental Figure 2), suggesting that, in yeast, OPT3 does not mediate the uptake of Fe2+ like IRT1. Subcellular localization studies, however, show that OPT3–YFP protein fusions do not localize to the plasma membrane in yeast (Supplemental Figure 3) in contrast to in planta (Figure 5C). This mislocalization of OPT3 in yeast precluded further characterization of OPT3 using yeast as a heterologous system. Note that, if fet3fet4 yeast cells were not sufficiently pre-starved of iron, growth of the fet3fet4 mutant was observed, and therefore long-term starvation of yeast was required for these complementation tests. We attempted complementation with different yeast promoters, starting with the strong GAL promoter (Supplemental Figure 2A). Using the phosphoglycerate kinase (PGK) yeast promoter, OPT3 also did not complement the pre-iron-starved fet3fet4 yeast mutant, consistently with previous studies showing that OPT3 does not complement this yeast mutant (Supplemental Figure 2A). Nevertheless, 59Fe re-mobilization studies suggest that OPT3 is essential for the remobilization of Fe within plant tissues; whether this transport occurs as Fe2+ or as a Fe-ligand complex remains to be determined.
OPT3 is a member of the oligopeptide transporter family and some members of this family have been found to have broad substrate specificity for peptides of different length and amino acid composition (Osawa et al., 2006; Pike et al., 2009). To test whether OPT3 mediates the long-distance transport of GSH in planta, we pursued radiotracer experiments to assess the movement of 35S-GSH from one leaf to adjacent leaves (Supplemental Figure 4). No differences were found between wild-type and opt3-2, suggesting that OPT3 does not participate in the mobilization of GSH between plant tissues. Interestingly, opt3-2 rosette leaves from plants exposed to 20 μM CdCl2 (in the hydroponic solution) and supplemented with 0.5mM GSH (foliar application) accumulated Cd, but no other metals, to wild-type levels. These results suggest that GSH is required for Cd retention in leaves. On the other hand, GSH supplemented to the roots reduced Cd in the leaves of both opt3-2 and wild-type plants (Supplemental Figure 5) likely because GSH trapped Cd in roots of both opt3-2 and wild-type plants (Supplemental Figure 5).
Glutathione has recently been shown to play a critical role in Fe signaling in yeast by stabilizing FeS clusters in the cytosol (Rutherford et al., 2005; Li et al., 2009). In Arabidopsis, GSH is also important to maintain proper homeostasis and crosstalk between Zn and Fe metabolism (Shanmugam et al., 2012). To test whether long-distance transport of GSH is important for proper shoot-to-root signaling and homeostasis of trace metals in roots, we measured the constitutive high activity of the root ferric reductase in opt3-2 after foliar application of GSH (Supplemental Figure 6A). In all cases, including the application of foliar GSH, GSH applied to roots, or foliar application of Fe, the activity of the root ferric reductase remained constitutively high in opt3-2. We also tested whether iron applied directly to roots or complexed with GSH, citrate, or nicotianamine is sufficient to repress the high activity of the Fe chelate reductase in opt3-2 roots. Supplemental Figure 6B shows that iron alone (Fe2+), or in complex with GSH, nicotianamine, or citrate, cannot down-regulate the constitutive iron-deficiency response in opt3-2 back to wild-type levels.
DISCUSSION
opt3-2 Shows an Altered Phloem-Mediated Cd Distribution
We have identified an Arabidopsis mutant, opt3-2, that over-accumulates Cd in seeds and roots (Figure 1) but, unexpectedly, under-accumulates Cd in leaves (Figure 2). Cadmium distribution throughout the plant is an orchestrated process dictated by root uptake, root-to-shoot translocation through the xylem, and redistribution of Cd from leaves to sink tissues (i.e. seeds, younger leaves, and roots) via the phloem. opt3-2 displays constitutive up-regulation of IRT1, a root transporter with broad specificity for heavy metals including Cd (Eide et al., 1996; Rogers et al., 2000). Over-accumulation of Cd, Zn, Fe, and Mn in roots (Figures 2 and 3) may be explained by the constitutively high expression of IRT1. However, under-accumulation of Cd in leaves and over-accumulation of Cd in seeds, which is different from essential metals (Figures 1 and 2), is inconsistent with the high expression of IRT1 (Stacey et al., 2008). Nutrients, water, and heavy metals are mobilized from leaves into seeds through the phloem (Turgeon and Wolf, 2009). Accumulation of metabolites in sink tissues (roots and seeds) and under-accumulation in source tissues (leaves) is best described as an increased redistribution process, likely through the phloem (Nour-Eldin et al., 2012). Notably, of the analyzed metals, only Cd under-accumulates in leaves (Figure 2). These results suggest that, in contrast to the broad specificity of heavy metal uptake at the root level, metal-specific mechanisms mediate the remobilization of heavy metals from leaves to sink tissues.
In addition to the altered distribution of heavy metals within the plant leading to over-accumulation of Cd in seeds, opt3-2 also shows hypersensitivity to Cd at the seedling stage (Figures 1–3). Both the increased accumulation of Cd in seeds and the Cd hypersensitivity of seedling growth are restored to wild-type levels by ectopically expressing OPT3, demonstrating that the altered redistribution of Cd through the plant is the result of the reduced expression of OPT3 in opt3-2 (Figure 4).
OPT3 Mediates Shoot-to-Root Signaling of Iron Status
The opt3-2 mutant displays a constitutive iron-deficiency response in roots, while the leaves properly respond to iron levels as indicated by wild-type levels of ferritin expression (Stacey et al., 2008), suggesting that the iron status response is mainly disrupted in roots. In plants, the root iron-deficiency response is regulated by local signals within the root and also by unknown systemic signals originating from aerial tissues (Vert et al., 2003; Hindt and Guerinot, 2012). OPT3 is a plasma membrane transporter preferentially expressed in phloem cells during iron starvation (Figure 5). Cell-specific microarrays (Mustroph et al., 2009) (Supplemental Figure 1) and OPT3 pro:GUS analysis under Fe-limiting conditions (Figure 5A) show preferential expression of OPT3 in phloem cells, suggesting a role of OPT3 in long-distance transport processes. Notably, shoot-specific expression of OPT3 (CAB2 pro:OPT3) in the opt3-2 background rescued the constitutively high expression of IRT1 in roots, the seed Cd over-accumulation phenotype, and the seedling sensitivity to Cd (Figure 6). These results suggest that the impaired metal homeostasis in opt3-2 roots is caused by a disruption of the shoot-to-root signaling of the leaf metal status. Thus, OPT3 is the first shoot-expressed gene required for proper communication from leaves to roots to maintain metal homeostasis at the whole-plant level.
Several Arabidopsis and tomato mutants displaying an Fe-deficiency response in roots can be rescued by foliar application of Fe (Garcia et al., 2013); these experiments suggest that shoot-to-root Fe signaling plays an important role in Fe homeostasis (Garcia et al., 2013) which in turn could also impact the uptake and accumulation of other transition metals such as Zn, Mn, and Cd, as seen in opt3-2 (Figures 1 and 3). Foliar application of Fe does not repress the Fe-deficiency response in opt3-2 roots to wild-type levels (Garcia et al., 2013) (Supplemental Figure 6), suggesting that source-to-sink transport of Fe, or a molecule mediating Fe signaling, is impaired in opt3-2. Radiotracer experiments using 59Fe demonstrate that the movement of Fe between leaves is impaired in opt3-2 (Figure 7A); whether this leaf-to-leaf transport occurs as Fe2+ or as an Fe–ligand complex remains to be determined. OPT3 is a member of the oligopeptide transporter family and members of this family have been shown to mediate the transport of a broad range of peptides (Osawa et al., 2006; Pike et al., 2009). Arabidopsis OPT3 has also been reported to rescue the ability of yeast mutants defective in Cu and Mn transport to grow on low concentrations of these transition metals (Wintz et al., 2003). However, so far there is no direct evidence to suggest that OPT3 mediates the transport of transition metals, in the ionic form or complexed with a ligand, or whether OPT3 mediates the transport of a ligand that facilitates the uptake and accumulation of transition metals into the cell. In fact, our OPT3 localization experiments in yeast show that OPT3–YFP fusions are unable to transit out of the ER to the plasma membrane (Supplemental Figure 3). This intracellular localization of OPT3 makes it difficult to interpret the ability of yeast strains defective in transition metal transport to grow on minimal media when expressing OPT3.
Glutathione is a small peptide that has also gained recent attention in metal-status signaling via GSH-coordinated intermediaries of the iron–sulfur cluster assembly machinery (Rutherford et al., 2005; Rouhier et al., 2007; Bandyopadhyay et al., 2008). Regulation of GSH levels is also essential for regulating the iron-deficiency response in fungi (Li et al., 2009; Kumar et al., 2011). Our radiotracer experiments using 35S-GSH (Supplemental Figure 4) and the ferric reductase assay in opt3-2 roots (Supplemental Figure 6) show that leaf-to-leaf movement of GSH was unaffected and that foliar application of GSH does not suppress the constitutive Fe-deficiency response in opt3-2. These results suggest that shoot-to-root transport of GSH alone has little effect on the long-distance signaling of the Fe status in Arabidopsis.
opt3-2 as a Model for Long-Distance Cd and Nutrient Transport
Phloem transport plays a key role in delivering nutrients, including metals, to developing seeds (Turgeon and Wolf, 2009; Mendoza-Cozatl et al., 2011). However, the mechanisms of toxic heavy metal loading into seeds are largely unknown. Nicotianamine, GSH, and PCs are the main metal-chelating molecules found in phloem sap (Mendoza-Cozatl et al., 2011). Nicotianamine has been shown to form complexes with Fe, Cu, Zn, and Mn, while GSH and PCs preferentially bind to Cd (Dorčák and Krężel, 2003). The differential partitioning of Cd among roots, leaves, and seeds in opt3-2 relative to the essential metals Fe, Zn, and Mn suggests that independent mechanisms mediate the partitioning of essential and non-essential metals, likely as specific metal–chelate complexes. Understanding phloem-mediated transport and seed-loading mechanisms of individual metals and metal–ligand complexes will be important to restrict accumulation of toxic metals in seeds while ensuring the accumulation of essential metals.
In summary, we show that Arabidopsis OPT3 is expressed in the phloem and functions in the long-distance shoot-to-root signaling of Fe/Zn/Mn status. When OPT3 expression is compromised, there is a misregulation of genes mediating uptake and mobilization of trace metals leading to an over-accumulation of cadmium, but not other metals, in seeds. We further show that mobilization of Fe2+ between leaves is impaired in opt3-2 and that targeted OPT3 expression in leaves is sufficient to restore Fe/Zn/Mn status signaling to roots providing molecular information on shoot-to-root Fe-status signaling. Sensing and regulation of trace-metal homeostasis in plants have been long-standing questions in plant biology and the results presented here offer new insights and avenues to advance our understanding of how essential and non-essential metals are accumulated and distributed within plant tissues.
METHODS
Plant Materials and Growth Conditions
Wild-type (Col-0) and opt3-2 seeds were surface-sterilized, stratified at 4°C for 48h in the dark, and germinated under a 16-h light/8-h dark photoperiod. For Cd sensitivity experiments, ¼ MS plates were supplemented with 50 μM CdCl2 and allowed to grow vertically for 14 d.
For metal determination in seeds, 2-week-old seedlings were transferred to Sunshine Basic Mix 2 soil supplemented with heavy metals as described (McDowell et al., 2013). For metal determination in roots and leaves, plants were grown hydroponically as described previously (Chen et al., 2006). Elemental analyses were performed by ICP–OES at the UCSD/Scripps Institution of Oceanography analytical facility using dried rosette leaves, roots, or seeds digested overnight in trace-metal grade 70% HNO3 as described previously (Chen et al., 2006).
Plasmid Construction
All primers used for PCR amplification for cloning are listed in Supplemental Table 1. For OPT3 expression driven by the CaMV 35S promoter (35S pro:OPT3), the OPT3 genomic DNA fragment was amplified using the primers OPT3-A and OPT3-B. The amplified OPT3 DNA was cloned as an AvrII/BstEII fragment into a modified pCambia 1391Z binary vector encoding the CaMV 35S promoter derived from pRT101 (Topfer et al., 1987). For confocal localization studies, the OPT3 coding sequence was amplified from Col-0 cDNA to create pENTR–OPT3 using OPT3-C and OPT3-D, and cloned into pENTR/D-TOPO® (Invitrogen, Carlsbad, CA, USA). The YFP–OPT3 fusion was obtained by recombining the OPT3 coding sequence into pH35YG (Nishimura et al., 2010) using LR Clonase II® (Invitrogen, Carlsbad, CA, USA).
For shoot-specific expression, the OPT3 coding sequence was recombined into a Gateway® compatible pGreenII plasmid (Hellens et al., 2000) containing the CAB2 pro and the NOS terminator (CAB2 pro:GW–NOS ter). The CAB2 promoter was amplified from Col-0 genomic DNA using the primers CABP-A and CABP-C, and cloned into the KpnI/HindIII sites of pGreenII upstream of the Gateway® cassette. For CAB2 pro GUS staining, the 2-kb promoter fragment was amplified from genomic DNA using CABP-B and CABP-C, inserted into pENTR/D-TOPO®, and recombined into pBGGUS (Kubo et al., 2005). For OPT3 pro:YFP expression studies, a 2-kb fragment upstream of the start codon of OPT3 was amplified from Col-0 genomic DNA using the primers OPT3P-A and OPT3P-B, and introduced into pDONRZeo® using BP Clonase II® (Invitrogen, Carlsbad, CA, USA). OPT3 pro was then recombined into a Gateway®-compatible pGreen II plasmid containing the coding sequence of YFP and the NOS terminator. For localization of OPT3 in Saccharomyces cerevisiae, the N-terminal YFP fusion YFP–OPT3 was amplified from pH35YG–OPT3 with YFP-A and OPT3-D, cloned into pENTR/D-TOPO®, and inserted into pYES–DEST52 via recombination. The C-terminal YFP fusion OPT3–YFP was created by amplifying OPT3 with OPT3-E and OPT3-F, and amplifying YFP with YFP-B and YFP-C. The two PCR fragments were combined with the USER® system (New England Biolabs, MA, USA), inserted into pDONRZeo® using BP Clonase II®, and then recombined into pYES–DEST52.
Plant Transformation
A. thaliana was transformed using the floral dip method (Clough and Bent, 1998), and N. benthamiana was Agro-infiltrated as previously described (Wydro et al., 2006). Agrobacterium tumefaciens strain GV3101 was used for all transformations, and pSoup was used as the helper plasmid for pGreenII-carrying strains (Hellens et al., 2000). OPT3 pro:YFP was transformed into Col-0 plants, and CAB2 pro:OPT3 and 35S pro:OPT3 were transformed into the opt3-2 background.
Reverse Transcription PCR and Quantitative PCR
For RT–PCR of CAB2 pro:OPT3 transgenic lines, plants were grown vertically on ¼ MS media for 14 d. Leaves and roots were then separated, and RNA was prepared using the RNEasy Plant Mini Kit® (Qiagen, Hilden, Germany). The RNA was DNase treated and reverse-transcribed with SuperscriptIII® (Invitrogen, Carlsbad, CA, USA). RT–PCR was performed for the indicated number of cycles and normalized against ACT2 using the primers listed in Supplemental Table 2. RT–PCR was performed for additional cycles to confirm that amplification at the indicated cycles were in the logarithmic phase.
For qPCR analysis of 35S pro:OPT3 lines, plants were grown on ¼ MS media for 14 d. cDNA was then prepared from RNA of whole seedlings as for RT–PCRs. Transcript abundance was then determined with SYBR® Green (Sigma-Aldrich, St. Louis, MO, USA) using a LightCycler® 1.5 Real-Time PCR System (Roche Diagnostics, Indianapolis, IN, USA). OPT3 transcript levels were normalized to ACT2 transcript levels and relative OPT3 expression levels were determined using the comparative Ct method (Schmittgen and Livak, 2008). Primers used for qPCR are listed in Supplemental Table 3.
Yeast Transformation and Growth
S. cerevisiae BY4741 was transformed with pYES–DEST52 plasmids harboring YFP–OPT3 or OPT3–YFP via the lithium acetate method (Schiestl and Gietz, 1989), and transformants were selected on glucose-containing YNB-Ura media (Mäser et al., 2002). Expression of YFP–OPT3 and OPT3–YFP was induced by culturing yeast overnight in YNB-Ura supplemented with 2% galactose and 1% raffinose.
GUS Staining and Fluorescence Microscopy
For GUS staining, transgenic plants carrying the OPT3 pro:GUS fusion were grown on ½ MS medium for 20 d before being transferred to Fe-sufficient or Fe-deficient medium (Yi and Guerinot, 1996) and grown for an additional 10 d. Inflorescence stems were isolated, hand-sectioned, and stained for GUS as previously described (Stacey et al., 2002). CAB2 pro:GUS staining was performed on 3-week-old seedlings grown on ¼ MS medium. Staining patterns were observed and documented using a Nikon SMZ1500 stereomicroscope. Fluorescence microscopy was performed on a Nikon TE-200U microscope equipped with a Yokogawa Nipkow spinning disc confocal head and a Roper Cascadell 512b EM CCD camera. YFP was excited with a Chroma HQ480/40 band-pass emission filter. Fluorescence images were captured using Metamorph v.5.0 (Universal Imaging, Sunnyvale, CA, USA) and edited using NIH ImageJ (http://imagej.nih.gov/ij/). For plasmolysis experiments, N. benthamiana leaf sections were incubated in 4% NaCl for 15min prior to imaging.
Leaf-to-Leaf Transport of 59Fe and 35S-GSH
Plants grown for at least 3 weeks under 16-h/8-h light/dark cycles were used for radiotracer experiments. For 59Fe experiments, a fully developed leaf was immersed in a buffer containing 50mM MES (pH 6.2), 15 μM FeCl3, 1mM ascorbic acid, and 30 μCi ml–1 of 59Fe (Perkin Elmer, USA). Wild-type and opt3-2 leaves were incubated with the radiotracer for 12h before detaching the load leaf, which was placed into a separate 20-ml scintillation vial to determine its specific activity. Following the incubation, the remaining rosette leaves were dissected onto a platform for autoradiography using a phosphorimaging plate (Fujifilm 20 cm × 40 cm phosphorimaging plate, BAS-IP MS 2040, or Fujifilm 20 cm × 25 cm phosphorimaging plate, BAS-IP MS 2025). The phosphorimaging plate was exposed for 2h and then scanned on a Typhoon FLA 9000 (GE Healthcare Lifesciences) using the phosphorimaging settings with 100-μm resolution (approximately 15-min scan). After scanning, the dissected leaves were placed in 20-ml scintillation vials. Opti-Fluor (10 ml, PerkinElmer) was added to each sample and the specific activity was determined using a liquid scintillation counter (TriCarb Liquid Scintillation Counter, PerkinElmer). To reduce the excitation between samples with higher activity, two spaces in the racks were left empty between samples. A background sample was analyzed and its count rate subtracted from each sample.
Remobilization of 35S-GSH between leaves was performed as described for 59Fe with some modifications. A mature leaf from wild-type or opt3-2 was immersed in a solution containing 25mM GSH, Tris 50mM (pH 7.5) supplemented with 35S-GSH (30 μCi ml–1, Perkin Elmer, USA). After a 12-h incubation under continuous light, the immersed leaf was removed and the remaining rosette leaves were dissected and placed on the phosphorimaging plate which was exposed for 5h and then scanned using a Typhoon FLA 9000 (GE Healthcare Lifesciences).
Perls’ Staining of Ferric Iron
The Perls’ staining method was used as described (Green and Rogers, 2004; Schuler et al., 2012). Mature leaves, young leaves, and siliques were harvested freshly from 7–8-week-old plants and incubated with fixative solution (methanol/chloroform/pure acetic acid, 6:3:1) for 1h at room temperature. After incubation, the fixative solution was removed, and the samples were washed by exchanging distilled water three times. Perls’ staining solution (4% HCl and 4% K-ferrocyanide, 1:1; Green and Rogers, 2004) was added for 1h at room temperature. The reaction was terminated by washing three times with distilled water.
Cd Treatment Together with Foliar and Root Application of GSH
Plants were grown on soil until right before bolting (5–6 weeks old) and then transferred to 0.5 Hoagland’s nutrient hydroponic solution pH 5.9 (Heeg et al., 2008) for 1 week and exposed to defined treatments for 72h as follows: (A) Cd: 0.5 Hoagland’s nutrient hydroponic solution + 20 μM Cd; (B) Cd+Shoot GSH: same as Cd treatment with (0.5mM GSH + 0.125%Tween20 (added as surfactant) application to leaves; (C) Cd+Root GSH: same as Cd treatment hydroponic media with 0.5mM GSH added to the hydroponic solution. Tissue digestion and elemental concentrations were determined as described above.
Ferric Reductase Activity Determinations
Plants were grown on soil right until before bolting and then transferred to 0.5 Hoagland’s hydroponic nutrient solution pH 5.9 described above for 1 week and then transferred to Fe-limited hydroponic solution (0.5 Hoagland’s nutrient solution pH 5.9 without Fe-EDTA) for 7 d. Plants were then transferred to the following treatments: –Fe + Foliar Fe: hydroponic nutrient solution without Fe and with (0.05% FeSO4 + 0.125%Tween20 application to leaves; –Fe + Foliar Fe + Shoot GSH: same as –Fe + Foliar Fe, with 0.5mM GSH application to leaves; –Fe + Foliar Fe + Root GSH: same as –Fe + Foliar Fe but adding 0.5mM GSH to the Fe-limited hydroponic solution. For foliar Fe and GSH spray treatments, leaves were sprayed once a day until they were completely moistened. After 5 d of foliar treatment, root ferric reductase activity was determined as previously described (Lucena et al., 2006).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This research was supported by a National Institute of Environmental Health Sciences (Grant No. P42 ES010337), OPT3 transport analyses were funded by the Chemical Sciences, Geosciences, and Biosciences Division of the Office of Basic Energy Sciences at the US Department of Energy (DE-FG02-03ER15449). Research was supported by a US National Science Foundation Arabidopsis 2010 grant (IOB 0419695 to J.I.S.) and a US National Science Foundation CAREER grant (IOS-1252706 to D.G.M.C.). D.G.M.C. also received support from the University of Missouri Research Board Grant (Project CB000519). 59Fe and 35S-glutathione experiments were supported by the Department of Energy (Projects for Interrogations of Biological Systems, DE-SC0002040 to S.S J.). T.O.J. was supported by the UCSD-Salk IGERT Plant Systems Biology Interdisciplinary Graduate Training Program (Grant No. 0504645) and D.W.D. by the NIH-NIBIB Training Grant (5 T32 EB004822).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Olena Vatamaniuk for helpful discussions regarding possible substrates for OPT3. No conflict of interest declared.
REFERENCES
- Bandyopadhyay S., Gama F., Molina-Navarro M.M., Gualberto J.M., Claxton R., Naik S.G., Huynh B.H., Herrero E., Jacquot J.P., Johnson M.K, et al. (2008). Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe–2S] clusters. EMBO J. 27, 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Komives E.A., Schroeder J.I. (2006). An improved grafting technique for mature Arabidopsis plants demonstrates long-distance shoot-to-root transport of phytochelatins in Arabidopsis . Plant Physiol. 141, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Aarts M.G., Thomine S., Verbruggen N. (2013). Plant science: the key to preventing slow cadmium poisoning. Trends Plant Sci. 18, 92–99 [DOI] [PubMed] [Google Scholar]
- Clemens S., Antosiewicz D.M., Ward J.M., Schachtman D.P., Schroeder J.I. (1998). The plant cDNA LCT1 mediates the uptake of calcium and cadmium in yeast. Proc. Natl Acad. Sci. U S A. 95, 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Dorčák V., Krężel A. (2003). Correlation of acid-base chemistry of phytochelatin PC2 with its coordination properties towards the toxic metal ion Cd(II). Dalton Transactions. 2253 [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl Acad. Sci. U S A. 93, 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz E., Romkens P., van Raamsdonk L., van der Fels-Klerx I. (2008). A chain modeling approach to estimate the impact of soil cadmium pollution on human dietary exposure. J. Food Prot. 71, 2504–2513 [DOI] [PubMed] [Google Scholar]
- Garcia M.J., Romera F.J., Stacey M.G., Stacey G., Villar E., Alcantara E., Perez-Vicente R. (2013). Shoot to root communication is necessary to control the expression of iron-acquisition genes in Strategy I plants. Planta. 237, 65–75 [DOI] [PubMed] [Google Scholar]
- Green L.S., Rogers E.E. (2004). FRD3 controls iron localization in6Arabidopsis . Plant Physiol. 136, 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeg C., Kruse C., Jost R., Gutensohn M., Ruppert T., Wirtz M., Hell R. (2008). Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell. 20, 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- Hendrick D.J. (1996). Occupational and chronic obstructive pulmonary disease (COPD). Thorax. 51, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt M.N., Guerinot M.L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta. 1823, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il’yasova D., Schwartz G.G. (2005). Cadmium and renal cancer. Toxicol. Appl. Pharmacol. 207, 179–186 [DOI] [PubMed] [Google Scholar]
- Koh S., Wiles A.M., Sharp J.S., Naider F.R., Becker J.M., Stacey G. (2002). An oligopeptide transporter gene family in Arabidopsis . Plant Physiol. 128, 21–29 [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C., Igbaria A., D’Autreaux B., Planson A.G., Junot C., Godat E., Bachhawat A.K., Delaunay-Moisan A., Toledano M.B. (2011). Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 30, 2044–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E., Abid K., Rogers E., Guerinot M.L., Harper F.J., Ward J., McIntyre L, et al. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana . Nat. Biotechnol. 21, 1215–1221 [DOI] [PubMed] [Google Scholar]
- Li B., Takahashi D., Kawamura Y., Uemura M. (2012). Comparison of plasma membrane proteomic changes of Arabidopsis suspension-cultured cells (T87 Line) after cold and ABA treatment in association with freezing tolerance development. Plant Cell Physiol. 53, 543–554 [DOI] [PubMed] [Google Scholar]
- Li H., Mapolelo D.T., Dingra N.N., Naik S.G., Lees N.S., Hoffman B.M., Riggs-Gelasco P.J., Huynh B.H., Johnson M.K., Outten C.E. (2009). The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe–2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 48, 9569–9581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.A., Tsukagoshi H., Busch W., Lahner B., Salt D.E., Benfey P.N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 22, 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C., Waters B.M., Romera F.J., Garcia M.J., Morales M., Alcantara E., Perez-Vicente R. (2006). Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J. Exp. Bot. 57, 4145–4154 [DOI] [PubMed] [Google Scholar]
- Mäser P., Hosoo Y., Goshima S., Horie T., Eckelman B., Yamada K., Yoshida E., Bakker A., Shinmyo S., Schroeder J.I, et al. (2002). Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl Acad. Sci. U S A. 99, 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell S.C., Akmakjian G., Sladek C., Mendoza-Cozatl D., Morrissey J.B., Saini N., Mittler R., Baxter I., Salt D.E., Ward J.M, et al. (2013). Elemental concentrations in the seed of mutants and natural variants of Arabidopsis thaliana grown under varying soil conditions. PloS One. 8, e63014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M.J., Bell M.J., Wright G.C., Cruickshank A. (1997). Inter- and intra-specific variation in accumulation of cadmium by peanut, soybean, and navybean. Aust. J. Agric. Res. 48, 1151 [Google Scholar]
- Mendoza-Cozatl D.G., Butko E., Springer F., Torpey J.W., Komives E.A., Kehr J., Schroeder J.I. (2008). Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus: a role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 54, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cozatl D.G., Jobe T.O., Hauser F., Schroeder J.I. (2011). Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic. Curr. Opin. Plant Biol. 14, 554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis . Proc. Natl Acad. Sci. U S A. 106, 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrot T., Plusquin M., Hogervorst J., Roels H.A., Celis H., Thijs L., Vangronsveld J., Van Hecke E., Staessen J.A. (2006). Environmental exposure to cadmium and risk of cancer: a prospective population-based study. Lancet Oncol. 7, 119–126 [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J, et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis . Plant J. 61, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Andersen T.G., Burow M., Madsen S.R., Jorgensen M.E., Olsen C.E., Dreyer I., Hedrich R., Geiger D., Halkier B.A. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature. 488, 531–534 [DOI] [PubMed] [Google Scholar]
- Osawa H., Stacey G., Gassmann W. (2006). ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem. J. 393, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.M., Guerinot M.L. (2009). Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike S., Patel A., Stacey G., Gassmann W. (2009). Arabidopsis OPT6 is an oligopeptide transporter with exceptionally broad substrate specificity. Plant Cell Physiol. 50, 1923–1932 [DOI] [PubMed] [Google Scholar]
- Rogers E.E., Eide D.J., Guerinot M.L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl Acad. Sci. U S A. 97, 12356–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N., Unno H., Bandyopadhyay S., Masip L., Kim S.K., Hirasawa M., Gualberto J.M., Lattard V., Kusunoki M., Knaff D.B, et al. (2007). Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe–2S] cluster in poplar glutaredoxin C1. Proc. Natl Acad. Sci. U S A. 104, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford J.C., Ojeda L., Balk J., Muhlenhoff U., Lill R., Winge D.R. (2005). Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron–sulfur protein biogenesis. J. Biol. Chem. 280, 10135–10140 [DOI] [PubMed] [Google Scholar]
- Satarug S., Garrett S.H., Sens M.A., Sens D.A. (2010). Cadmium, environmental exposure, and health outcomes. Environ. Health Perspect. 118, 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R.H., Gietz R.D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schuler M., Rellan-Alvarez R., Fink-Straube C., Abadia J., Bauer P. (2012). Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis . Plant Cell. 24, 2380–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam V., Tsednee M., Yeh K.C. (2012). ZINC TOLERANCE INDUCED BY IRON 1 reveals the importance of glutathione in the cross-homeostasis between zinc and iron in Arabidopsis thaliana . Plant J. 69, 1006–1017 [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Koh S., Becker J., Stacey G. (2002). AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis . Plant Cell. 14, 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M.G., Osawa H., Patel A., Gassmann W., Stacey G. (2006). Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta. 223, 291–305 [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Patel A., McClain W.E., Mathieu M., Remley M., Rogers E.E., Gassmann W., Blevins D.G., Stacey G. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topfer R., Matzeit V., Gronenborn B., Schell J., Steinbiss H.H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R., Wolf S. (2009). Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60, 207–221 [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hermans C., Schat H. (2009). Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 181, 759–776 [DOI] [PubMed] [Google Scholar]
- Vert G.A., Briat J.F., Curie C. (2003). Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 132, 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E.L., Connolly E.L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11, 530–535 [DOI] [PubMed] [Google Scholar]
- Wintz H., Fox T., Wu Y.Y., Feng V., Chen W., Chang H.S., Zhu T., Vulpe C. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278, 47644–47653 [DOI] [PubMed] [Google Scholar]
- Wydro M., Kozubek E., Lehmann P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana . Acta. Biochim. Pol. 53, 289–298 [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10, 835–844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.