Abstract
Humans as diurnal beings are active during the day and rest at night. This daily oscillation of behavior and physiology is driven by an endogenous circadian clock not environmental cues. In modern societies, changes in lifestyle have led to a frequent disruption of the endogenous circadian homeostasis leading to increased risk of various diseases including cancer. The clock is operated by the feedback loops of circadian genes and controls daily physiology by coupling cell proliferation and metabolism, DNA damage repair, and apoptosis in peripheral tissues with physical activity, energy homeostasis, immune and neuroendocrine functions at the organismal level. Recent studies have revealed that defects in circadian genes due to targeted gene ablation in animal models or single nucleotide polymorphism, deletion, deregulation and/or epigenetic silencing in humans are closely associated with increased risk of cancer. In addition, disruption of circadian rhythm can disrupt the molecular clock in peripheral tissues in the absence of circadian gene mutations. Circadian disruption has recently been recognized as an independent cancer risk factor. Further study of the mechanism of clock-controlled tumor suppression will have a significant impact on human health by improving the efficiencies of cancer prevention and treatment.
Keywords: Aging, cancer risk factors, cell cycle, cellular senescence, circadian rhythm, DNA damage response, metabolism, molecular clock, social jet lag, tumor suppression
Introduction
The evolutionary adaptation dictates that most, if not all, physiological processes in mammals follow a circadian rhythm. Circadian rhythm is generated by an endogenous circadian clock and coupled to the diurnal oscillation of environmental cues. Changes in lifestyles due to industrialization and economic globalization in the modern world has led to frequent disruption of endogenous circadian homeostasis, which is coupled with a dramatic increase in the modern-day diseases including psychological disorders, cardiovascular diseases, metabolic syndromes, aging, immune deficiencies, reproductive and neuroendocrine dysfunction, as well as cancer (1–4).
The circadian control of mammalian physiology
The mammalian circadian clock is composed of a central clock in the hypothalamic suprachiasmatic nucleus (SCN), peripheral oscillators in all peripheral tissues studied, and circadian input and output pathways. The SCN consists of multiple, self-sustained single-cell circadian oscillators that constantly synchronize to environmental cues to generate coordinated circadian outputs. The most potent environmental circadian cue is the ambient light received by a subset of melanopsin-expressing retinal ganglion neurons and transmitted directly to the SCN via the retinohypothalamic tract (RHT) (5). The SCN clock targets other hypothalamic centers to generate the circadian rhythm in the neuroendocrine and autonomic systems (NES and ANS) via direct neuronal connections and secreting diffusible molecules. The NES and ANS are circadian output pathways that generate a coupled circadian rhythm in diverse peripheral tissues by controlling cell signaling in a tissue-specific manner (Figure 1A) (5–7).
Figure 1.
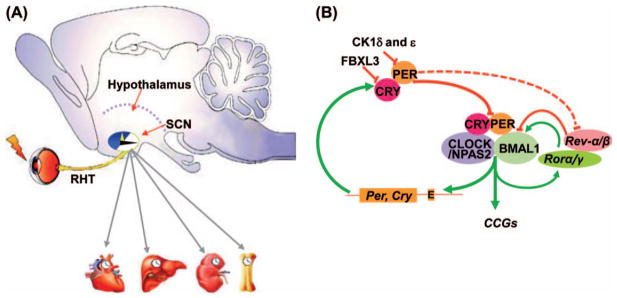
The mammalian circadian clock. (A) The mammalian circadian clock is comprised of circadian input pathways, such as the light input pathway via the retinohypothalamic tract (RHT), the central clock located in the hypothalamic suprachiasmatic nucleus (SCN), the circadian output pathways including the neuroendocrine and autonomic nervous systems (grey arrows) and peripheral clocks in all tissues studied. (B) A simplified model of the molecular clock. Solid lines show direct regulation of the positive and negative feedback loops by core circadian genes Bmal1, Clock, Ck1δ and ε, Cry, Naps2, Per, Rev-erbα and β, as well as Rorα and γ. The dashed lines show indirect regulation of Rev-erbα and β by PER. The molecular clock also targets clock-controlled genes (CCGs) that regulate diverse cellular processes. The first-order CCGs are controlled by the molecular clock directly at the transcriptional level.
The molecular clock is operated by interacting feedback loops of core circadian genes in all cells in the body. At the transcriptional level, the clock is driven by heterodimers of bHLHPAS transcription factors BMAL1/CLOCK or BMAL1/NPAS2 that activate core circadian genes Cryptochrome (Cry1,2) and Period (Per1–3) via E-boxes in gene promoters at the beginning of a subjective day. The PER and CRY proteins then form a transcriptional repressor complex that enters the nucleus at the beginning of a subjective night to inhibit the heterodimer activity by protein–protein interactions and/or recruitment of transcriptional termination complexes. Bmal1 is also rhythmically regulated by its own transcriptional targets Rorα, Rev-erbα and β encoding nuclear receptors RORα, REV-ERBα and β, respectively. Upon activation, RORα stimulates Bmal1 expression, while REV-ERBα and β suppress Bmal1 transcription (8,9). The molecular feedback loops are also controlled by post-translational mechanisms. The stability of PER and CRY, controlled by casein kinase 1ε and δ (CK1ε/δ) and the Skp1-cullin-F-box protein (SCF) E3 ubiquitin ligase complexes, respectively, determines the time of the PER/CRY repressor nuclear entry (10,11). The cell-autonomous oscillation of multiple interlocked feedback loops of circadian genes defines the intrinsic circadian rhythmicity of the molecular clock (Figure 1B) (8,9).
The clock targets clock-controlled genes (CCGs) to control diverse cellular processes in peripheral tissues. System-level approaches have identified a large number of first-order CCGs controlled by the clock at the transcriptional level. The majority of CCGs encode tissue-specific expressed mRNAs to control key tissue functions. A small group of ubiquitously expressed CCGs encode proteins supporting basic cellular functions (12,13). The rhythmic expression of these CCGs is controlled by mechanisms including direct transcriptional regulation by heterodimers via E-box sequences in gene promoters, indirect regulation by clock-controlled gene-specific transcriptional regulators, and circadian oscillation in chromatin remodeling (9,12,14).
The molecular clock constantly responds to daily entrainment signals to maintain the synchrony with the environment. In SCN neurons, light stimuli phase-shifts the molecular clock via activating immediate early responsive genes such as Ap1, Per1, and Per2 in a time-dependent manner via signal transduction pathways including the calcium/calmodulin-dependent protein kinases II (CaM kinases II), c-Jun N-terminal kinase (JNK), c-AMP-protein kinase A (PKA), extracellular signal-regulated kinases (ERK), mitogen-activated protein kinases (MAPK), nitric oxide (NO)/c-GMP, or protein kinase C alpha (PKCα) (15,16). In peripheral tissues, the circadian output pathways generate cyclic changes in the levels of neurotransmitters, growth factors, cytokines, and blood-borne hormones in the tissue microenvironment, which rhythmically entrain the molecular clock via intracellular signaling controlled by pathways including those mediating the light response in SCN neurons (4,7,17).
The homeostasis of internal physiology is maintained by coordinated activities of the central and peripheral clocks. Disruption of external light cues phase-shifts the SCN clock leading to a phase-shift in circadian output pathways, which then phase-shifts peripheral clocks via phase-shifting intracellular signaling in a tissue-specific manner. The consecutive phase-shifts in the hierarchical circadian timing system temporarily disrupts the homeostasis of physiology due to various rates of phase-shifts of peripheral tissues resulting from their differential innervation by circadian output pathways. The time needed for re-establishing internal circadian homeostasis is determined by when the circadian disruption occurs during a day and how many hours of phase-shift in the SCN clock it initially induces. Therefore, human rotating working schedules or frequent rapid long-distance transmeridian travel leads to chronic misalignment of internal physiology from environmental cues, which has been shown by recent studies to increase the risk of cancer significantly (1,2,4,18).
The mechanism of clock-controlled tumor suppression
The clock plays a key role in controlling tissue-specific functions. Tissues normally supported by cellular processes frequently deregulated in cancers are most likely sensitive to circadian dysfunction-induced tumor development. Recent studies have revealed that circadian disruption specially increases the risk of cancers in the immune, skeletal, digestive and reproductive organs that need cell proliferation, metabolism, and DNA damage repair to maintain daily function and are prone to cell death, senescence, and inflammatory response induced by adverse physiological conditions (1–4,19).
Circadian control of cell proliferation
Cell cycle progression follows a circadian rhythm in all rapidly renewing mammalian tissues studied, but is arrhythmic or displays ultradian rhythms in metastatic cancers (4, 20). The ubiquitously expressed CCGs include key cell cycle and proto-oncogenes as well as tumor suppressors (12,13). The existing evidence suggests that the molecular clock likely suppresses proto-oncogenes but stimulates tumor suppressors at transcriptional level. For example, the binding of BMAL1/CLOCK and BMAL1/NPAS2 heterodimers at E-boxes in gene promoters negatively regulates proto-oncogene c-Myc but stimulates the tumor suppressor Wee1 in response to extracellular mitogenic signals (19, 21–23). Disruption of the molecular clock leads to deregulation of these CCGs, which is coupled with increased risk of neoplastic growth and cancer in various circadian gene mutant mouse models (19, 22–27).
At the post-translational level, the molecular clock may modulate both positive and negative loops of the cell cycle clock. For example, PER1 and CRY2 were reported to regulate p53-controlled checkpoint function by interacting with the tumor suppressor ataxia-telangiectasia mutated (ATM) and Rad3-related (ATR), respectively (28,29), whereas CK1ε promotes β-catenin-mediated activation of T-cell-specific transcription factor/lymphoid enhancer factor-1 (TCF/LEF) family that stimulates c-Myc and Cyclin D1-mediated cell cycle progression (30). Thus, the molecular clock acts to maintain the homeostasis but not inhibition of cell proliferation at the cellular levels (4).
The molecular clock alone is unable to generate the circadian rhythm of cell proliferation because G1 cell cycle progression is controlled by extracellular mitogenic signals (31). In vivo, these signals include neurotransmitters, steroid and peptide hormones, chemokines, growth factors, or cytokines either directly released from the circadian output pathways or rhythmically produced by peripheral tissue via paracrine and autocrine signaling. The tissue-specific interaction of these signals with their targets, such as G-protein coupled receptors, tyrosine kinase receptors, integrins, and nuclear receptors, simultaneously entrains the molecular and cell cycle clocks by activation of early responsive genes (4).
One pathway for the central clock to control cell proliferation in peripheral tissues is via the sympathetic nervous system (SNS). The SNS innervates all peripheral organs to control diverse physiological functions (32). The direct neuronal targeting of hypothalamic paraventricular pre-sympathetic neurons by the SCN clock generates a robust circadian variation in the basal level of the SNS tone, which then drives the circadian rhythm of diverse cellular processes in peripheral tissues by controlling peripheral clocks and intracellular signaling (6). The SNS signaling is deregulated or arrhythmic in human night-shift workers and jet-lagged mouse models (19,33). Surgical ablation of SNS innervation abolishes circadian oscillation of immune function via inhibition of β-adrenergic receptor (ADRβ)-mediated hematopoietic stem cell proliferation, differentiation, and trafficking, which is a key mechanism of immune suppression that promotes cancer development (4). Uncontrolled SNS signaling promotes tumor initiation by stimulating cell proliferation, while sympathectomy inhibits tumor growth. Thus, β-blockers are suggested as novel anti-cancer drugs for future anticancer therapies (34, 35). The mechanism of SNS-mediated cell cycle progression can be explained by its ability to activate Ap1, Per1 and 2, and ATM (19). AP1 activation leads to Myc-dependent G1 cell cycle progression. Per induction stimulates BMAL1/CLOCK-mediated Myc transcriptional repression, whereas the peripheral clock-dependent induction of ATM leads to p53 activation. Thus, the rhythmic SNS signaling is a circadian cue for both cell cycle and peripheral clocks in vivo. Loss of function in the peripheral clock abolishes the activation of BMAL1/CLOCK and p53 in response to SNS signaling, but has no effect on AP1-controlled Myc activation, leading to Myc oncogenic activation, uncontrolled cell proliferation, and neoplastic growth in mice (19, 23). Together, these findings described above explain why disruption of the circadian homeostasis increases the risk of cancer in humans and rodent models (19, 36–58) and highlight the role of circadian disruption as one of the independent risk factors for the dramatic increase in the rate of sporadic cancers in the modern societies.
Circadian control of DNA damage response and cellular senescence
The activities of cell proliferation and metabolism generate a large amount of lesions in genomic DNA each day, which activates DNA repair machinery and cell cycle checkpoints to repair damaged DNA or induces cell death or cellular senescence when the damage exceeds the capacity of repair (32, 59). In mammals, ATM and ATR are two master regulators of DNA damage. The clock not only couples ATM activation with daily cell cycle progression in peripheral tissues but also activates ATM and ATR in response to acute DNA damage (19,22,28), leading to protein kinases CHK1/2 and p53-mediated cell cycle arrest and/or p53-mediated apoptosis (60). The clock also controls the expression of key DNA replication, recombination, and repair enzymes (12, 13). Loss of function in the molecular clock deregulates these CCGs leading to accumulation of DNA damage and increased risk of neoplastic growth in mice. For example, the gene encoding xeroderma pigmentosum A (XPA), an essential enzyme for nucleotide excision repair, is deregulated in mice lacking both Cry1 and 2, which is coupled with a dampened circadian rhythm in nucleotide excision repair after UVB irradiation in epidermis (61, 62). Keratinocyte-specific deletion of Bmal1 in mice results in deregulation of cell proliferation, DNA repair, and oxidative phosphorylation genes, leading to increased intracellular reactive oxygen species (ROS), elevated and arrhythmic cell proliferation, dampened UVB-induced DNA damage response, and accumulation of DNA lesions in epidermis (25). The circadian regulators may also directly participate in DNA damage response. After γ-radiation, CLOCK translocates to the sites of DNA double-strand breaks (63). PER1 was reported to interact directly with ATM and CHK2, and BMAL1 is required to activate the p53-p21WAF1/CIP1 pathway (28,64), whereas CRY2 is involved in intra-S checkpoint activation after UV radiation by interacting with ATR and CHK1 via TIMELESS (29). In mice, the activation of all circadian genes studied is required for a time-dependent γ-radiation response in thymus. Loss of function in Per2 abolishes the response of all circadian genes to γ-radiation, leading to radiation resistance and increased tumor incidence (19, 22).
The deregulation of DNA damage response and accumulation of DNA lesions are associated with premature aging phenotypes frequently observed among circadian gene mutant mice. Bmal1−/− mice display aggressive aging phenotypes leading to a significantly reduced lifespan (65). Mice carrying a mutated Clock or lacking Per or Cry also display premature aging phenotypes that are more evident after being exposed to γ-radiation (19, 22, 66, 67).
Aging in various tissues shares a common mechanism of replicative cellular senescence, which refers to a state of permanent withdrawal from the cell cycle due to accumulation of genetic alterations beyond the capacity of repair (68, 69). Since metastasizing tumors show unlimited capacity of cell proliferation, cellular senescence has been widely considered as a mechanism of tumor suppression (70). However, recent studies have revealed that senescent cells are still metabolically active but show deregulation of chromatin organization and gene expression, and increased secretion of proinflammatory cytokines, proteases, and growth factors that stimulate tumor growth and metastasizing (68). In addition, senescent cells can regain the ability of proliferation in response to changes in internal physiology (71). The uncontrolled Myc and Ras/MAPK oncogenic signaling, activation of p53/p21WAF1/CIP1 and pRB/p 16INK4a tumor suppressors, and loss of function in Sirt1, which encodes the NAD-dependent deacetylase sirtuin-1, a class III histone deacetylase, are established molecular mechanisms promoting cellular senescence (68, 71–73). Especially, SIRT1 may bridge aging and cancer prone phenotypes found in circadian gene mutant mice by directly deacetylating BMAL1, PER, p53, β-catenin, and DNA repair protein KU70 (11, 72–78). In mice, the early onset of cellular senescence is, at least in part, due to AKT-dependent vascular senescence in Per2 mutants, and via a p53-independent mechanism in Bmal1 nulls (79,80). Thus, cancer and aging share a common mechanism of DNA damage accumulation in vivo.
Circadian control of metabolic homeostasis and inflammatory response
Among various DNA damaging agents, ROS can induce oxidative DNA damage including double-strand DNA breaks to promote genomic instability (81). The exogenous sources of ROS include pollutants, tobacco, xenobiotics, and radiation. The endogenous ROS are natural by-products of cell metabolism mainly produced by NADPH oxidase complexes located in cell membranes, mitochondria, peroxisomes, and endoplasmic reticulum (82). ROS normally function in various signaling pathways to maintain the homeostasis of cellular function. Their intracellular levels are tightly controlled by multiple mechanisms (83). However, under pathological conditions, such as viral infection, chronic inflammation, and metabolic dysfunction, intracellular ROS levels can increase dramatically, leading to significant oxidative damages to cells including double-strand DNA breaks (84, 85).
The clock is a master regulator of cell metabolism. It controls the expression and activities of rate-limiting enzymes for ROS production and cellular antioxidative response including the NADPH oxidase complexes that produce ROS and superoxide dismutases, glutathione peroxidases, and peroxiredoxins that mediate antioxidative response (86–88). Loss of function in the molecular clock has been shown to deregulate cell metabolism leading to excess accumulation of ROS in vivo (25). Circadian dysfunction also promotes metabolic adaptations in favor of cancer progression. One example of such adaptations is the Warburg effect (89), which describes that, in contrast to normal somatic cells metabolizing glucose to CO2 and H2O via a low rate of glycolysis followed by oxidative phosphorylation, cancer cells predominantly use glucose to produce energy by a high rate of anaerobic glycolysis in the cytosol even in the presence of oxygen, leading to a dramatic increase in intracellular ROS and synthesis of purines, pyrimidines, non-essential amino-acids, and free fatty acids essential for cell growth and division. Recent findings suggest that the Warburg effect is driven by oncogene activation and, therefore, is considered as the result rather than the cause of neoplastic growth (85). In vivo, the clock not only controls the expression of proto-oncogenes and tumor suppressors but also the key metabolic genes involved in the Warburg effect, such as glucose-6-phosphatase, pyruvate kinase, and glucose transporter 2 (GLUT) (86,90). Thus, disruption of circadian homeostasis may promote the Warburg effect in the absence of any gene mutations by promoting oncogenic activation and metabolic dysfunction.
Recent studies have established a strong correlation between circadian disruption and the development of metabolic syndromes (1, 91). The mechanisms linking metabolic syndromes with cancer include deregulation of extracellular signaling, induction of angiogenesis, and chronic inflammation, a key event promoting cancer (92, 93). Chronic inflammation can be induced by excess ROS that activates NF-κB-mediated proinflammatory and pro-proliferation pathways (94, 95). Together, these events lead to tissue damage and cell death as well as the following tissue regeneration supported by active cell proliferation. However, the intrinsic deficiencies in cell cycle control, metabolic balance, cancer immuno-editing (reviewed in (4)), and the accumulation of DNA damage due to chronic circadian disruption would significantly increase the risk of neoplastic growth and cancer development.
The circadian genes in cancer
Recent studies have revealed that genes operating both positive and negative loops of the molecular clock are important for tumor suppression in vivo, and that the mechanism of clock-controlled tumor suppression is conserved among humans and rodents (Tables I and II).
Table I.
Circadian genes in human diseases.
| Clock genes | Cancer types | Other diseases | References |
|---|---|---|---|
| Bmal1 | Breast, colorectal, head and neck, ovarian, pancreatic, and prostate cancers, B-cell lymphoma, pleural mesothelioma, ALL, AML, and CML | Aging, hypertension, immune deficiency, mood disorders, neurodegeneration, and T2D | (45, 96–108) |
| CK1δ/ε | Breast, colorectal, ovarian, pancreatic, and prostate cancers | Aging, chronic inflammation, hypertension, insulin resistance, neurodegeneration, and sleep disorders | (102, 120–135) |
| Clock | Breast, colorectal, lung, ovarian, pancreatic, prostate and skin cancers, B-cell lymphoma, and CLL | Bipolar disorder, hypertension, obesity, neurodegeneration, NAFLD, and T2D | (50, 102–104, 106, 139–144) |
| Cry1/2 | Breast, colorectal, endometrial, head and neck, ovarian, pancreatic, prostate and skin cancers, glioma, pleural mesothelioma, CLL, CML, HCC, and NHL | Acute inflammation, hyperglycemia, mood disorders, obesity, Parkinson’s, and T2D | (102–105, 125, 126, 134, 139–140, 153–171) |
| Dec1/2 | Breast, endometrial, gastric, head and neck, lung, pancreatic, and renal cancers, and HCC | Autoimmune disease, chronic inflammation, and sleep disorders | (182–194) |
| Npas2 | Breast and prostate cancers, pleural mesothelioma, glioma, and NHL | Aging, chronic fatigue, chronic inflammation, hypertension, mood disorders, and neurodegeneration | (100, 102, 134, 161, 169, 197–204) |
| Per1/2/3 | Breast, colorectal, endometrial, head and neck, lung, ovarian, pancreatic, prostate, skin and thyroid cancers, B-cell lymphoma, glioma, pleural mesothelioma, AML, CLL, CML, and HCC | Aging, cardiovascular disease, chronic inflammation, insulin resistance, neurodegeneration, obesity, eating, mood and sleep disorders, and T2D | (98, 99, 104, 105, 126, 134, 139, 140, 157, 158, 160, 161, 164, 169, 170, 213, 223, 229, 240) |
| Rorα/γ | Breast, colorectal, pituitary, prostate, and thyroid cancers | Immune deficiency, insulin resistance, and obesity | (122, 255–264) |
| Rev-erbα/β | Breast and thyroid cancers and pleural mesothelioma | Autoimmune disease, chronic inflammation, and obesity | (162, 256, 273–280) |
| βTrCP1/2 | Breast, colorectal, gastric, pancreatic, and prostate cancers, and HCC | N/A | (284–289) |
ALL = acute lymphocytic leukemia; AML = acute myeloid leukemia; CLL = chronic lymphocytic leukemia; CML = chronic myeloid leukemia; HCC = hepatocellular carcinoma; NAFLD = non-alcoholic fatty liver disease; NHL = non-Hodgkin’s lymphoma; T2D = type 2 diabetes.
Table II.
Circadian gene mutant mouse models.
| Genetic model | Mouse strain | Mouse phenotype | References |
|---|---|---|---|
| Bmal1+/− | C57BL/6J | Premature aging, cancer prone | (19, 65) |
| Bmal1−/− | C57BL/6J | Arrhythmic, aggressive aging, hypoglycemia, redox deregulation, immune deficiency cellular senescence, cancer prone, metabolic syndromes, neurological disorders | (19, 23, 65, 111–113, 115, 116) |
| Bmal1−/− islets or adipocytes | C57BL/6J × 129 × ICR | Hyperglycemia, hypoinsulinemia, adipocyte hyperplasia, redox deregulation | (115, 116) |
| Bmal1−/− keratinocytes | C57BL/6J | Genomic instability, cellular senescence, deregulation of cell proliferation and metabolism, redox deregulation | (25, 65) |
| CK1δ or CK1ε | C57/BL6J | Lengthened period, autoimmune diseases, mammary gland neoplasms | (136–138) |
| ClockΔ19/Δ19 | C57BL/6J & C57BL6 × BALB/c | Arrhythmic in constant darkness, metabolic syndromes, obesity, premature aging, NAFLD | (21, 66, 116, 145–152) |
| Clock−/− | C57BL/6J | Aging, chronic inflammation | (150) |
| Clock−/− cardiomyocytes | FVB/NJ | Cardiac dysfunction, fatty acid dysregulation | (149) |
| Cry1−/− or Cry2−/− | C57BL/6J | Abnormal behavioral rhythm, premature aging, hypertension, chronic inflammation, sleep disorder, cancer prone, metabolic syndromes | (21, 61, 173–175) |
| Cry1−/−; Cry2−/− | C57BL/6J | Arrhythmic, premature aging, chronic inflammation, hypertension, impaired tissue regeneration, sleep disorder, cancer prone, metabolic syndromes | (19, 23, 62, 67, 175–178) |
| Dec1−/−; Dec2−/− | C57BL/6J | Lengthened circadian period | (195, 196) |
| Npas2−/− | C57BL/6J × 129Sv | Metabolic syndromes, behavioral abnormalities, sleep disorder | (205–208) |
| Per1−/− | C57BL/6J | Immune deficiencies, cancer prone | (242, 243, 252) |
| Per2−/− or Per2 tm1Brd/tm1Brd | C57BL/6J & C57BL/6J × 129Sv | Arrhythmic inconstant darkness, premature aging, cancer prone, metabolic syndromes, vascular diseases, immune deficiency | (19, 22, 23, 173, 241, 250, 253) |
| Per2S662G or Per2S662D | C57BL/6J | Shortened period, premature aging, deregulation of apoptosis, cancer prone | (24, 27) |
| Per1−/−; Per2 tm1Brd/tm1Brd | C57BL/6-Tyrc-2J | Arrhythmic, premature aging and neurological disorders, cancer prone, metabolic syndromes, neurological disorder | (19, 242, 243, 251) |
| Per3−/− | C57BL/6 × 129Sv | Metabolic syndromes, arteriosclerotic disease | (249, 250) |
| Rorα −/− or Rorγ −/− | C57/BL6J | Premature aging, adipocyte hyperplasia, immune-deficient, neurodegeneration | (265–271) |
| Rev-Erbα−/− | C57BL/6J × 129Sv | Metabolic syndromes, immune deficiencies | (272, 281, 282) |
| Rev-Erbα−/−; Rev-erbβ −/− | C57BL/6J × 129Sv | Arrhythmic, metabolic syndromes | (272) |
| βTrCP1m/m | C57BL/6J | Tissue-specific expression of dominant negative mutants promotes tumor development | (298–300, 302) |
Brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (Bmal1)
Bmal1 variants due to single nucleotide polymorphisms (SNPs) in humans are associated with various diseases, such as type 2 diabetes, hypertension, mood and sleep disorders, aging, neurodegeneration, and immune deficiencies (96–101), as well as cancers including breast, colorectal, prostate, pancreatic, and ovarian cancers, head and neck squamous cell carcinoma, B-cell lymphoma, pleural mesothelioma, acute lymphocytic leukemia (ALL), acute myeloid leukemia (AML), and chronic myeloid leukemia (CML) (45, 102–108). Suppression of Bmal1 expression in human prostate, lung, or glioma cancer cells significantly increases their metastasizing potential (109). The role for Bmal1 in tumor suppression include but are not limited to the suppression of uncontrolled PI3K-AKT-MMP2 signaling essential for tumor invasion and the activation of p53-mediated apoptosis and cell cycle arrest (64, 110). Ablation of Bmal1 in mice abolishes circadian behavioral homeostasis even under entrained conditions (111), which is coupled with phenotypes of aggressive aging (65, 112), cognitive deficits (113), chronic inflammation (65), cancer (19), and deregulated response to anti-cancer drugs such as docetaxel, etoposide, oxaliplatin, and cyclophosphamide (109, 114). Tissue-specific ablation of Bmal1 increases the risk of abnormal adipogenesis (115), insulin resistance (116), toxic ROS accumulation, genomic instability, senescence, and uncontrolled cell proliferation in epidermis (25).
Casein kinase 1δ and ε (Ck1δ and ε)
CK1ε plays a critical role in MYC-driven cancers in humans (117). It also regulates gluconeogenic genes in response to PGC-1α activation and modulates lipoprotein metabolism by phosphorylating PGC-1β (118, 119). Mutations and/or deregulation of Ck1δ and ε are associated with colorectal, pancreatic, prostate, breast, and ovarian cancers (102, 120–126), neurodegeneration and sleep disorders (127–131), chronic inflammation and aging (132, 133), as well as metabolic syndromes (134, 135). Disruption or deregulation of Ck1δ or ε in mice deregulates circadian homeostasis (136) and increases the risk of mammary carcinogenesis (137) and autoimmune diseases (138).
The circadian locomotor output cycles kaput (Clock)
The SNPs in the human Clock gene are associated with increased susceptibility to breast, colorectal, lung, ovarian, skin, pancreatic, and prostate cancers, B-cell lymphoma, and chronic lymphocytic leukemia (CLL) (50, 102–104, 106, 139–141), as well as bipolar disorder, neurodegeneration, and metabolic syndromes including obesity and obesity-related non-alcoholic fatty liver disease (NAFLD) (142–144). Mice homozygous in dominant negative Clock mutation (ClockΔ19/Δ19) are unable to maintain circadian behavior rhythm after prolonged exposure to constant darkness (145) and show increased risk of cardiac and immune malfunctions, premature aging, deregulation of DNA damage response, and various metabolic syndromes including obesity, adipocyte hypertrophy, NAFLD, diabetes, hypercholesterolemia, and hypertriglyceridemia (66, 116, 146–150). In peripheral tissues, ClockΔ19/Δ19 mice show deregulation of genes controlling glycolysis, mitochondrial oxidative phosphorylation, lipid metabolism, inflammatory response, fatty acid oxidation, chromatin remodeling, redox synthesis, cellular senescence DNA damage response, and tumor suppression (74, 86, 148, 151, 152).
Cryptochrome (Cry)
The SNPs or deregulation of Cry1 and/or 2 are associated with increased susceptibility and mortality to breast, colorectal, endometrial, prostate, skin, thyroid, and prostate cancers, hepatocellular carcinoma (HCC), pancreatic ductal adenocarcinoma, head and neck squamous cell carcinoma, glioma, CLL, CML pleural mesothelioma, and non-Hodgkin’s lymphoma (NHL) (102–105, 125, 126, 139–140, 153–161), Parkinson’s disease (162), mood disorders (163), acute inflammation (164), and metabolic syndromes including obesity and type 2 diabetes (134, 165–171). Inhibition of Cry2 expression in MCF-7 cells leads to dysregulation of genes important for proliferation, apoptosis, angiogenesis, inflammation, and tumor migration and invasion (172). Cry mutant mice are deficient in tissue regeneration (21) and also show increased risk of neoplastic growth (23, 173), chronic inflammation, high-fat diet-induced metabolic syndromes (174–176), premature aging (19, 67), deregulation of DNA damage repair (61, 62), hypertension (177), and sleep disorders (178). Although previously reported as lack of evidence of increased cancer risk (67), a recent study shows that mice lacking Cry1 and/or 2 also display increased risk of spontaneous and radiation-induced tumor development (19).
Deleted in esophageal cancer 1 and 2 (Dec1 and 2)
Dec1 and 2 are activated by BMAL1/CLOCK and encode bHLH transcription factors that suppress Per and Cry transcription (179–181). In humans, SNPs, deletion and/or deregulation of Dec1 or 2 are associated with gastric, non-small-cell lung, pancreatic, endometrial, renal, and breast cancers, HCC, head and neck esophageal squamous cell carcinoma and lymph node metastasis (182–191), sleep disorders (192), and inflammation and autoimmune disease (193, 194). Mice lacking Dec1 or 2 (Dec1−/− or Dec2 −/−) maintain normal circadian rhythmicity. However, mice lacking both Dec1 and 2 (Dec1−/−; Dec2−/−) have a lengthened circadian period (195, 196).
Neuronal PAS domain protein 2 (Npas2)
SNPs and deregulation of Npas2 in humans increase the risk of metabolic syndromes, aging, neurodegeneration, chronic fatigue syndrome, mood and sleep disorders, chronic inflammation (100, 134, 169, 197, 198), and breast and prostate cancers, pleural mesothelioma, glioma, and possibly NHL (102, 161, 199–204). The role of Npas2 in tumor suppression has not been vigorously studied using mouse models. However, Npas2 mice display a variety of pathological changes that may directly or indirectly promote tumor development, such as impaired responses to food entraining signals, metabolic syndromes (205, 206), and lack of sleep homeostasis and behavioral adaptability (207, 208). At the cellular level, Npas2 has been found to control redox levels, oxidative and DNA-damage response, and the expression of cell cycle genes and tumor suppressors (209–212).
Period (Per)
The SNPs and deregulation of Per1, 2, and/or 3 in humans are associated with increased risk of thyroid, breast, prostate, ovarian, endometrial, pancreatic, colorectal, skin, and non-small-cell lung cancers (NSCLC), HCC, colorectal carcinoma, diffusible large B-cell lymphomas, malignant pleural mesothelioma, head and neck squamous cell carcinoma, glioma, CML, AML, and CLL (104, 105, 126, 139, 140, 157, 158, 160, 161, 213–223), which often show deregulation of inflammatory cytokines, oncogenic and tumor suppressors including p38, ERα, G1 and S-phase cyclins, c-Myc, NF-κB, Bcl-XL, PKA, telomerase, ATM, p53, p21, and Wee1 (28, 106, 175, 219, 224, 228). The SNPs and deregulation of human Per genes are also linked to obesity, metabolic syndromes, type 2 diabetes, eating, mood and sleep disorders, cardiovascular diseases, aging, depression, chronic inflammation, and neurodegeneration (98, 99, 134, 164, 169, 170, 197, 229–240).
Various research teams have independently demonstrated that mouse models deficient in Per1, Per2, or both Per1 and 2 show increased risk of spontaneous and radiation-induced tumor development in immune, digestive, skeletal, and reproductive organs (19, 22, 24, 26, 27), neoplastic growth in bone (23, 173), premature aging and vascular senescence (19, 80, 241, 242), mood disorders (243), immune deficiencies (244, 255), and metabolic syndromes including diabetes, liver cholestatic diseases, hypoglycemia, and hyperinsulinemia (246–251). Per mutant mice show deregulation of key genes controlling cell proliferation, metabolism, senescence, inflammatory response, and death such as c-Myc, p53, Gadd45α, Atm, Ap1, Cyclin D1, Cyclin A, β-catenin, APC, PPARγ, Mdm2, Akt, Cyp7A1, interferon gamma (IFNγ), and nuclear receptors Fxr and Car (22, 23, 26, 27, 244, 247, 251–254).
Retinoic acid-related orphan receptor (Rorα and γ)
The deregulation and/or SNPs of Rorα and/or γ in humans have been linked to an increased risk of breast and prostate cancers, colorectal adenocarcinomas, pituitary and thyroid tumors (122, 255–260), immune deficiency (261), obesity, insulin resistance, and adipogenesis (262–264). The cancer risk of mice lacking Rorα or γ (Rorα−/− or Rorγ−/−) has not been carefully studied. However, these mice are immune-deficient (265, 266) and show adipocyte hyperplasia (267–269). Deregulation of Rorα, and/or γ is also associated with accelerated aging and neurodegeneration in mice (270, 271).
Reverse viral erythroblastosis oncogene products alpha and beta (Rev-erbα and β)
Rev-erbα and β are also known as the nuclear receptor subfamily 1, group D, member 1 and 2 (Nr1d1 and 2). The direct targets of REV-ERBα and β transcription also include key metabolic genes controlling lipid and energy homoeostasis (272). Rev-erbα is the only nuclear receptor gene frequently amplified in human breast cancers, which is associated with poor clinical outcomes and survival (273–276). The SNPs and/or deregulation of human Reverbα are also associated with thyroid tumors, pleural mesothelioma, obesity and metabolic syndromes, chronic inflammation, and autoimmune diseases (162, 256, 277–280). Mice lacking Rev-erbα (Nr1d1−/−) do not have circadian homeostasis and show immune deficiencies, metabolic syndromes, and diet-induced obesity (272, 281, 282).
Beta-transducin repeat-containing protein1 and 2 (βTrCP1 and 2)
βTrCP is also known as F-box/WD repeat containing protein (Fbxw) that belongs to one of the four components of the ubiquitin protein ligase complex. βTrCP-mediated PER cytoplasmic degradation controls the activity of the major negative loop in the molecular clock (283). SNPs, mutations, and/or deregulation of βTrCP in humans are associated with prostate, breast, colorectal, gastric, and pancreatic cancers as well as HCC (284–289). βTrCP plays important roles in cell cycle checkpoints and DNA damage response (290, 291), protein synthesis, cell growth, survival, and metabolism (292–294). It also regulates the pro-apoptotic protein BimEL, NF-κB, and WNT pathways to control immune response and cell survival (77, 295, 296). In vitro, loss of βTrCP stimulates angiogenesis and migration of human cervical and thyroid cancer cells (297).
Mice lacking βTrCP (βTrCP1−/− and βTrCP2−/−) do not show overt abnormalities. However, tissue-specific expression of a dominant negative mutant βTrCP1 in mice promotes tumor development in targeted tissues (298, 299). The βTrCP transgenic mice show deregulation of cell adhesion, migration, and proliferation, as well as suppression of p53 via activation of oncogene Mdm2 (290, 300–302).
Conclusion
The compelling evidence provided in this review supports the notion that the mammalian circadian clock suppresses tumor development in vivo. The fact that circadian disruption induces similar pathophysiological changes in the same organ systems in humans and animal models via deregulation of the same molecular pathways suggests that the mechanism of clock-controlled tumor suppression is conserved during evolution. The observation that circadian dysfunction induces a coupled increase in the risk of cancer and other modern-day diseases in affected individuals, such as accelerated aging, neurodegeneration, neuroendocrine dysfunction, metabolic syndromes, and immune deficiencies, indicates that the manifestations of various abnormal physiological conditions but not a single molecular pathway promote cancer development in vivo. These findings have opened an exciting new research direction to investigate the molecular mechanisms of cancer initiation and progression as a consequence of chronic malfunction in mammalian physiology. Such study will have important impact on human health by developing novel strategies for cancer prevention and treatment as, in industrialized societies, the majority of the population experiences chronic misalignment of endogenous circadian systems throughout their lifespan due to personal, social, and professional demands (1, 2, 303).
Key messages.
Circadian dysfunction is an independent risk factor for cancers in modern societies.
The circadian clock suppresses tumor development by maintaining homeostasis of physiology.
The mechanism of circadian control of tumor suppression is conserved during evolution.
Acknowledgments
This work is supported by the grants from USDA/ARS (6250–51000-055) and NIH/NCI (R01 CA137019-01A) to L. Fu.
Footnotes
Declaration of interest: The authors report no conflicts of interest.
References
- 1.Evans JA, Davidson AJ. Health consequences 1. of circadian disruption in humans and animal models. Prog Mol Biol Transl Sci. 2013;119:283–323. doi: 10.1016/B978-0-12-396971-2.00010-5. [DOI] [PubMed] [Google Scholar]
- 2.Foster RG, Peirson SN, Wulff K, Winnebeck E, Vetter C, Roenneberg T. Sleep and circadian rhythm disruption in social jetlag and mental illness. Prog Mol Biol Transl Sci. 2013;119:325–46. doi: 10.1016/B978-0-12-396971-2.00011-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Ptacek LJ, Fu YH. Diversity of human clock genotypes and consequences. Prog Mol Biol Transl Sci. 2013;119:51–81. doi: 10.1016/B978-0-12-396971-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013;119:221–82. doi: 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore RY. The suprachiasmatic nucleus and the circadian timing system. Prog Mol Biol Transl Sci. 2013;119:1–28. doi: 10.1016/B978-0-12-396971-2.00001-4. [DOI] [PubMed] [Google Scholar]
- 6.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nat Rev Neurosci. 2001;2:521–6. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 7.Buijs R, Salgado R, Sabath E, Escobar C. Peripheral circadian oscillators: time and food. Prog Mol Biol Transl Sci. 2013;119:83–103. doi: 10.1016/B978-0-12-396971-2.00004-X. [DOI] [PubMed] [Google Scholar]
- 8.Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 2013;(217):3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 10.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:2139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 11.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–20. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–54. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueda HR. Systems biology of mammalian circadian clocks. Cold Spring Harb Symp Quant Biol. 2007;72:365–80. doi: 10.1101/sqb.2007.72.047. [DOI] [PubMed] [Google Scholar]
- 14.Eckel-Mahan K, Sassone-Corsi P. Epigenetic regulation of the molecular clockwork. Prog Mol Biol Transl Sci. 2013;119:29–50. doi: 10.1016/B978-0-12-396971-2.00002-6. [DOI] [PubMed] [Google Scholar]
- 15.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21:359–68. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Vadigepalli R, Rafferty R, Gonye GE, Weaver DR, Schwaber JS. Integrative gene regulatory network analysis reveals light-induced regional gene expression phase shift programs in the mouse suprachiasmatic nucleus. PLoS One. 2012;7:e37833. doi: 10.1371/journal.pone.0037833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U. Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell. 2013;152:492–503. doi: 10.1016/j.cell.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 18.Barclay JL, Tsang AH, Oster H. Interaction of central and peripheral clocks in physiological regulation. Prog Brain Res. 2012;199:163–81. doi: 10.1016/B978-0-444-59427-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 22.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 23.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–15. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 24.Gu X, Xing L, Shi G, Liu Z, Wang X, Qu Z, et al. The circadian mutation PER2(S662G) is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012;19:397–405. doi: 10.1038/cdd.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci USA. 2012;109:11758–63. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X, Wood PA, Ansell CM, Ohmori M, Oh EY, Xiong Y, et al. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–97. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 27.Wood PA, Yang X, Taber A, Oh EY, Ansell C, Ayers SE, et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786–93. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–82. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 29.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 31.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 32.Brodal P. The central nervous system: structure and function. New York: Oxford University Press; 2010. [Google Scholar]
- 33.Adams SL, Roxe DM, Weiss J, Zhang F, Rosenthal JE. Ambulatory blood pressure and Holter monitoring of emergency physicians before, during, and after a night shift. Acad Emerg Med. 1998;5:871–7. doi: 10.1111/j.1553-2712.1998.tb02816.x. [DOI] [PubMed] [Google Scholar]
- 34.Raju B, Haug SR, Ibrahim SO, Heyeraas KJ. Sympathectomy decreases size and invasiveness of tongue cancer in rats. Neuroscience. 2007;149:715–25. doi: 10.1016/j.neuroscience.2007.07.048. [DOI] [PubMed] [Google Scholar]
- 35.Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, et al. Autonomic nerve development contributes to prostate cancer progression. Science. 2013;341:1236361. doi: 10.1126/science.1236361. [DOI] [PubMed] [Google Scholar]
- 36.Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–7. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 37.Li JC, Xu F. Influences of light-dark shifting on the immune system, tumor growth and life span of rats, mice and fruit flies as well as on the counteraction of melatonin. Biol Signals. 1997;6:77–89. doi: 10.1159/000109112. [DOI] [PubMed] [Google Scholar]
- 38.Wu M, Zeng J, Chen Y, Zeng Z, Zhang J, Cai Y, et al. Experimental chronic jet lag promotes growth and lung metastasis of Lewis lung carcinoma in C57BL/6 mice. Oncol Rep. 2012;27:1417–28. doi: 10.3892/or.2012.1688. [DOI] [PubMed] [Google Scholar]
- 39.Filipski E, Subramanian P, Carriere J, Guettier C, Barbason H, Levi F. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009;680:95–105. doi: 10.1016/j.mrgentox.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Logan RW, Zhang C, Murugan S, O’Connell S, Levitt D, Rosenwasser AM, et al. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. J Immunol. 2012;188:2583–91. doi: 10.4049/jimmunol.1102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yasuniwa Y, Izumi H, Wang KY, Shimajiri S, Sasaguri Y, Kawai K, et al. Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS One. 2010;5:e15330. doi: 10.1371/journal.pone.0015330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubo T, Ozasa K, Mikami K, Wakai K, Fujino Y, Watanabe Y, et al. Prospective cohort study of the risk of prostate cancer among rotating-shift workers: findings from the Japan collaborative cohort study. Am J Epidemiol. 2006;164:549–55. doi: 10.1093/aje/kwj232. [DOI] [PubMed] [Google Scholar]
- 43.Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res. 2007;67:10618–22. doi: 10.1158/0008-5472.CAN-07-2485. [DOI] [PubMed] [Google Scholar]
- 44.Lahti TA, Partonen T, Kyyronen P, Kauppinen T, Pukkala E. Night-time work predisposes to non-Hodgkin lymphoma. Int J Cancer. 2008;123:2148–51. doi: 10.1002/ijc.23566. [DOI] [PubMed] [Google Scholar]
- 45.Stevens RG. Working against our endogenous circadian clock: breast cancer and electric lighting in the modern world. Mutat Res. 2009;680:106–8. doi: 10.1016/j.mrgentox.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Touitou Y, Bogdan A, Levi F, Benavides M, Auzeby A. Disruption of the circadian patterns of serum cortisol in breast and ovarian cancer patients: relationships with tumour marker antigens. Br J Cancer. 1996;74:1248–52. doi: 10.1038/bjc.1996.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panzer A. Melatonin in osteosarcoma: an effective drug? Med Hypotheses. 1997;48:523–5. doi: 10.1016/s0306-9877(97)90123-7. [DOI] [PubMed] [Google Scholar]
- 48.Skibola CF, Holly EA, Forrest MS, Hubbard A, Bracci PM, Skibola DR, et al. Body mass index, leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2004;13:779–86. [PubMed] [Google Scholar]
- 49.Viswanathan AN, Schernhammer ES. Circulating melatonin and the risk of breast and endometrial cancer in women. Cancer Lett. 2009;281:1–7. doi: 10.1016/j.canlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloog I, Haim A, Stevens RG, Portnov BA. Global co-distribution of light at night (LAN) and cancers of prostate, colon, and lung in men. Chronobiol Int. 2009;26:108–25. doi: 10.1080/07420520802694020. [DOI] [PubMed] [Google Scholar]
- 51.Knutsson A, Alfredsson L, Karlsson B, Akerstedt T, Fransson EI, Westerholm P, et al. Breast cancer among shift workers: results of the WOLF longitudinal cohort study. Scand J Work Environ Health. 2013;39:170–7. doi: 10.5271/sjweh.3323. [DOI] [PubMed] [Google Scholar]
- 52.Bhatti P, Mirick DK, Davis S. Invited commentary: shift work and cancer. Am J Epidemiol. 2012;176:760–3. doi: 10.1093/aje/kws311. discussion 764–5. [DOI] [PubMed] [Google Scholar]
- 53.Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev. 2013;17:273–84. doi: 10.1016/j.smrv.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Buzzelli G, Dattolo P, Pinzani M, Brocchi A, Romano S, Gentilini P. Circulating growth hormone and insulin-like growth factor-I in nonalcoholic liver cirrhosis with or without superimposed hepatocarcinoma: evidence of an altered circadian rhythm. Am J Gastroenterol. 1993;88:1744–8. [PubMed] [Google Scholar]
- 55.Davis S, Mirick DK, Stevens RG. Night shift work, 55. light at night, and risk of breast cancer. J Natl Cancer Inst. 2001;93:1557–62. doi: 10.1093/jnci/93.20.1557. [DOI] [PubMed] [Google Scholar]
- 56.Rafnsson V, Tulinius H, Jonasson JG, Hrafnkelsson J. Risk of breast cancer in female flight attendants: a population-based study (Iceland) Cancer Causes Control. 2001;12:95–101. doi: 10.1023/a:1008983416836. [DOI] [PubMed] [Google Scholar]
- 57.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 58.Pukkala E, Aspholm R, Auvinen A, Eliasch H, Gundestrup M, Haldorsen T, et al. Cancer incidence among 10,211 airline pilots: a Nordic study. Aviat Space Environ Med. 2003;74:699–706. [PubMed] [Google Scholar]
- 59.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 60.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–8. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 61.Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci USA. 2010;107:4890–5. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci USA. 2011;108:18790–5. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cotta-Ramusino C, McDonald ER, 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–17. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mullenders J, Fabius AW, Madiredjo M, Bernards R, Beijersbergen RL. A large scale shRNA barcode screen identifies the circadian clock component ARNTL as putative regulator of the p53 tumor suppressor pathway. PLoS One. 2009;4:e4798. doi: 10.1371/journal.pone.0004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component-of the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–34. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 68.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Best BP. Nuclear DNA damage as a direct cause of aging. Rejuvenation Res. 2009;12:199–208. doi: 10.1089/rej.2009.0847. [DOI] [PubMed] [Google Scholar]
- 70.Sager R. Senescence as a mode of tumor suppression. Environ Health Perspect. 1991;93:59–62. doi: 10.1289/ehp.919359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;32:5129–43. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 72.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–68. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 73.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 74.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD +-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 76.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–60. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cho IR, Koh SS, Malilas W, Srisuttee R, Moon J, Choi YW, et al. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of beta-catenin. Biochem Biophys Res Commun. 2012;423:270–5. doi: 10.1016/j.bbrc.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 78.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 79.Khapre RV, Kondratova AA, Susova O, Kondratov RV. Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle. 2011;10:4162–9. doi: 10.4161/cc.10.23.18381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang CY, Wen MS, Wang HW, Hsieh IC, Li Y, Liu PY, et al. Increased vascular senescence and impaired endothelial progenitor cell function mediated by mutation of circadian gene Per2. Circulation. 2008;118:2166–73. doi: 10.1161/CIRCULATIONAHA.108.790469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 82.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 83.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 84.Gupta SC, Hevia D, Patchva S, Park B, Koh W, Aggarwal BB. Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid Redox Signal. 2012;16:1295–322. doi: 10.1089/ars.2011.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 87.Cipolla-Neto J, Abdalla DS, Markus RP, Campa A. Circadian variations of superoxide dismutase activity in the rat pineal gland. J Neural Transm Gen Sect. 1993;92:117–23. doi: 10.1007/BF01244871. [DOI] [PubMed] [Google Scholar]
- 88.Anea CB, Zhang M, Chen F, Ali MI, Hart CM, Stepp DW, et al. Circadian clock control of Nox4 and reactive oxygen species in the vasculature. PLoS One. 2013;8:e78626. doi: 10.1371/journal.pone.0078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 90.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–7. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 91.Bass J, Turek FW. Sleepless in America: a pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165:15–16. doi: 10.1001/archinte.165.1.15. [DOI] [PubMed] [Google Scholar]
- 92.Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol Cancer Res. 2006;4:221–33. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- 93.Okwan-Duodu D, Umpierrez GE, Brawley OW, Diaz R. Obesity-driven inflammation and cancer risk: role of myeloid derived suppressor cells and alternately activated macrophages. Am J Cancer Res. 2013;3:21–33. [PMC free article] [PubMed] [Google Scholar]
- 94.Hursting SD, Dunlap SM. Obesity, metabolic dysregulation, and cancer: a growing concern and an inflammatory (and microenvironmental) issue. Ann NY Acad Sci. 2012;1271:82–7. doi: 10.1111/j.1749-6632.2012.06737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 96.Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, et al. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci USA. 2007;104:14412–7. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Popa-Wagner A, Catalin B, Buga AM. Novel putative mechanisms to link circadian clocks to healthy aging. J Neural Transm. 2013 Dec 3; doi: 10.1007/s00702-013-1128-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 98.Milagro FI, Gomez-Abellan P, Campion J, Martinez JA, Ordovas JM, Garaulet M. CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol Int. 2012;29:1180–94. doi: 10.3109/07420528.2012.719967. [DOI] [PubMed] [Google Scholar]
- 99.Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med. 2007;39:229–38. doi: 10.1080/07853890701278795. [DOI] [PubMed] [Google Scholar]
- 100.Evans DS, Parimi N, Nievergelt CM, Blackwell T, Redline S, Ancoli-Israel S, et al. Common genetic variants in ARNTL and NPAS2 and at chromosome 12p13 are associated with objectively measured sleep traits in the elderly. Sleep. 2013;36:431–46. doi: 10.5665/sleep.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miao CG, Yang YY, He X, Huang C, Huang Y, Zhang L, et al. The emerging role of microRNAs in the pathogenesis of systemic lupus erythematosus. Cell Signal. 2013;25:1828–36. doi: 10.1016/j.cellsig.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 102.Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–22. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Relles D, Sendecki J, Chipitsyna G, Hyslop T, Yeo CJ, Arafat HA. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J Gastrointest Surg. 2013;17:443–50. doi: 10.1007/s11605-012-2112-2. [DOI] [PubMed] [Google Scholar]
- 104.Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87:1060–70. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]
- 105.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–55. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 106.Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69:8447–54. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- 107.Yang MY, Chang JG, Lin PM, Tang KP, Chen YH, Lin HY, et al. Down-regulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Elshazley M, Sato M, Hase T, Yamashita R, Yoshida K, Toyokuni S, et al. The circadian clock gene BMAL1 is a novel therapeutic target for malignant pleural mesothelioma. Int J Cancer. 2012;131:2820–31. doi: 10.1002/ijc.27598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, Huang YJ, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem. 2010;148:319–26. doi: 10.1093/jb/mvq069. [DOI] [PubMed] [Google Scholar]
- 110.Jung CH, Kim EM, Park JK, Hwang SG, Moon SK, Kim WJ, et al. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol Rep. 2013;29:2109–13. doi: 10.3892/or.2013.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–17. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bunger MK, Walisser JA, Sullivan R, Manley PA, Moran SM, Kalscheur VL, et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–32. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 113.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–35. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci USA. 2005;102:3407–12. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–77. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci USA. 2012;109:9545–50. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li S, Chen XW, Yu L, Saltiel AR, Lin JD. Circadian metabolic regulation through crosstalk between casein kinase 1delta and transcriptional coactivator PGC-1alpha. Mol Endocrinol. 2011;25:2084–93. doi: 10.1210/me.2011-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 120.Tsai IC, Woolf M, Neklason DW, Branford WW, Yost HJ, Burt RW, et al. Disease-associated casein kinase I delta mutation may promote adenomatous polyps formation via a Wnt/beta-catenin independent mechanism. Int J Cancer. 2007;120:1005–12. doi: 10.1002/ijc.22368. [DOI] [PubMed] [Google Scholar]
- 121.Rodriguez N, Yang J, Hasselblatt K, Liu S, Zhou Y, Rauh-Hain JA, et al. Casein kinase I epsilon interacts with mitochondrial proteins for the growth and survival of human ovarian cancer cells. EMBO Mol Med. 2012;4:952–63. doi: 10.1002/emmm.201101094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One. 2012;7:e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim SY, Dunn IF, Firestein R, Gupta P, Wardwell L, Repich K, et al. CK1epsilon is required for breast cancers dependent on beta-catenin activity. PLoS One. 2010;5:e8979. doi: 10.1371/journal.pone.0008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brockschmidt C, Hirner H, Huber N, Eismann T, Hillenbrand A, Giamas G, et al. Anti-apoptotic and growth-stimulatory functions of CK1 delta and epsilon in ductal adenocarcinoma of the pancreas are inhibited by IC261 in vitro and in vivo. Gut. 2008;57:799–806. doi: 10.1136/gut.2007.123695. [DOI] [PubMed] [Google Scholar]
- 125.Chu LW, Zhu Y, Yu K, Zheng T, Yu H, Zhang Y, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–8. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 126.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–51. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 127.Li L, Zhao D, Wei H, Yao L, Dang Y, Amjad A, et al. REGgamma deficiency promotes premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci USA. 2013;110:11005–10. doi: 10.1073/pnas.1308497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schwab C, DeMaggio AJ, Ghoshal N, Binder LI, Kuret J, McGeer PL. Casein kinase 1 delta is associated with pathological accumulation of tau in several neurodegenerative diseases. Neurobiol Aging. 2000;21:503–10. doi: 10.1016/s0197-4580(00)00110-x. [DOI] [PubMed] [Google Scholar]
- 129.Brennan KC, Bates EA, Shapiro RE, Zyuzin J, Hallows WC, Huang Y, et al. Casein kinase idelta mutations in familial migraine and advanced sleep phase. Sci Transl Med. 2013;5:183ra56, 1–11. doi: 10.1126/scitranslmed.3005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev. 2011;31:924–54. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- 131.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–4. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 132.Miao CG, Yang YY, He X, Li XF, Huang C, Huang Y, et al. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. 2013;25:2069–78. doi: 10.1016/j.cellsig.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 133.Garcia-Ibarbia C, Delgado-Calle J, Casafont I, Velasco J, Arozamena J, Perez-Nunez MI, et al. Contribution of genetic and epigenetic mechanisms to Wnt pathway activity in prevalent skeletal disorders. Gene. 2013;532:165–72. doi: 10.1016/j.gene.2013.09.080. [DOI] [PubMed] [Google Scholar]
- 134.Hernandez-Morante JJ, Gomez-Santos C, Margareto J, Formiguera X, Martinez CM, Gonzalez R, et al. Influence of menopause on adipose tissue clock gene genotype and its relationship with metabolic syndrome in morbidly obese women. Age. 2012;34:1369–80. doi: 10.1007/s11357-011-9309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Soliman NA, Zineldeen DH, El-Khadrawy OH. Effect of NUCKS-1 overexpression on cytokine profiling in obese women with breast cancer. Asian Pac J Cancer Prev. 2014;15:837–45. doi: 10.7314/apjcp.2014.15.2.837. [DOI] [PubMed] [Google Scholar]
- 136.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–66. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hirner H, Gunes C, Bischof J, Wolff S, Grothey A, Kuhl M, et al. Impaired CK1 delta activity attenuates SV40-induced cellular transformation in vitro and mouse mammary carcinogenesis in vivo. PLoS One. 2012;7:e29709. doi: 10.1371/journal.pone.0029709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ghosh S, Koralov SB, Stevanovic I, Sundrud MS, Sasaki Y, Rajewsky K, et al. Hyperactivation of nuclear factor of activated T cells 1 (NFAT1) in T cells attenuates severity of murine autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2010;107:15169–74. doi: 10.1073/pnas.1009193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. 2011;127:531–40. doi: 10.1007/s10549-010-1231-2. [DOI] [PubMed] [Google Scholar]
- 140.Lengyel Z, Lovig C, Kommedal S, Keszthelyi R, Szekeres G, Battyani Z, et al. Altered expression patterns of clock gene mRNAs and clock proteins in human skin tumors. Tumour Biol. 2013;34:811–19. doi: 10.1007/s13277-012-0611-0. [DOI] [PubMed] [Google Scholar]
- 141.Rana S, Munawar M, Shahid A, Malik M, Ullah H, Fatima W, et al. Deregulated expression of circadian clock and clock-controlled cell cycle genes in chronic lymphocytic leukemia. Mol Biol Rep. 2014;41:95–103. doi: 10.1007/s11033-013-2841-7. [DOI] [PubMed] [Google Scholar]
- 142.Gomez-Abellan P, Gomez-Santos C, Madrid JA, Milagro FI, Campion J, Martinez JA, et al. Circadian expression of adiponectin and its receptors in human adipose tissue. Endocrinology. 2010;151:115–22. doi: 10.1210/en.2009-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garaulet M, Lee YC, Shen J, Parnell LD, Arnett DK, Tsai MY, et al. CLOCK genetic variation and metabolic syndrome risk: modulation by monounsaturated fatty acids. Am J Clin Nutr. 2009;90:1466–75. doi: 10.3945/ajcn.2009.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sookoian S, Castano G, Gemma C, Gianotti TF, Pirola CJ. Common genetic variations in CLOCK transcription factor are associated with nonalcoholic fatty liver disease. World J Gastroenterol. 2007;13:4242–8. doi: 10.3748/wjg.v13.i31.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, et al. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci USA. 2012;109:E2457–65. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294:H1036–47. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- 150.Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010;2:936–44. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–27. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 153.Lin DW, FitzGerald LM, Fu R, Kwon EM, Zheng SL, Kolb S, et al. Genetic variants in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes are prognostic markers of prostate cancer-specific mortality. Cancer Epidemiol Biomarkers Prev. 2011;20:1928–36. doi: 10.1158/1055-9965.EPI-11-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hoffman AE, Zheng T, Stevens RG, Ba Y, Zhang Y, Leaderer D, et al. Clock-cancer connection in non-Hodgkin’s lymphoma: a genetic association study and pathway analysis of the circadian gene cryptochrome 2. Cancer Res. 2009;69:3605–13. doi: 10.1158/0008-5472.CAN-08-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70:1459–68. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Luo Y, Wang F, Chen LA, Chen XW, Chen ZJ, Liu PF, et al. Deregulated expression of cry1 and cry2 in human gliomas. Asian Pac J Cancer Prev. 2012;13:5725–8. doi: 10.7314/apjcp.2012.13.11.5725. [DOI] [PubMed] [Google Scholar]
- 157.Yang MY, Yang WC, Lin PM, Hsu JF, Hsiao HH, Liu YC, et al. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythms. 2011;26:136–48. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 158.Eisele L, Prinz R, Klein-Hitpass L, Nuckel H, Lowinski K, Thomale J, et al. Combined PER2 and CRY1 expression predicts outcome in chronic lymphocytic leukemia. Eur J Haematol. 2009;83:320–7. doi: 10.1111/j.1600-0609.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 159.Mannic T, Meyer P, Triponez F, Pusztaszeri M, Le Martelot G, Mariani O, et al. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab. 2013;98:4446–56. doi: 10.1210/jc.2013-2568. [DOI] [PubMed] [Google Scholar]
- 160.Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–33. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 161.Roe OD, Anderssen E, Helge E, Pettersen CH, Olsen KS, Sandeck H, et al. Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: the emerging gene portrait of the mesothelioma phenotype. PLoS One. 2009;4:e6554. doi: 10.1371/journal.pone.0006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin Q, Ding H, Zheng Z, Gu Z, Ma J, Chen L, et al. Promoter methylation analysis of seven clock genes in Parkinson’s disease. Neurosci Lett. 2012;507:147–50. doi: 10.1016/j.neulet.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 163.Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–89. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Haimovich B, Calvano J, Haimovich AD, Calvano SE, Coyle SM, Lowry SF. In vivo endotoxin synchronizes and suppresses clock gene expression in human peripheral blood leukocytes. Crit Care Med. 2010;38:751–8. doi: 10.1097/CCM.0b013e3181cd131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu C, Zhang R, Wang C, Wang J, Ma X, Hou X, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One. 2010;5:e15542. doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Barker A, Sharp SJ, Timpson NJ, Bouatia-Naji N, Warrington NM, Kanoni S, et al. Association of genetic Loci with glucose levels in childhood and adolescence: a meta-analysis of over 6,000 children. Diabetes. 2011;60:1805–12. doi: 10.2337/db10-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gomez-Abellan P, Hernandez-Morante JJ, Lujan JA, Madrid JA, Garaulet M. Clock genes are implicated in the human metabolic syndrome. Int J Obes (Lond) 2008;32:121–8. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 169.Pappa KI, Gazouli M, Anastasiou E, Iliodromiti Z, Antsaklis A, Anagnou NP. Circadian clock gene expression is impaired in gestational diabetes mellitus. Gynecological Endocrinology: The Official Journal Of The International Society Of Gynecological Endocrinology. 2013;29:331–5. doi: 10.3109/09513590.2012.743018. [DOI] [PubMed] [Google Scholar]
- 170.Tahira K, Ueno T, Fukuda N, Aoyama T, Tsunemi A, Matsumoto S, et al. Obesity alters the expression profile of clock genes in peripheral blood mononuclear cells. Arch Med Sci. 2011;7:933–40. doi: 10.5114/aoms.2011.26603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kelly MA, Rees SD, Hydrie MZ, Shera AS, Bellary S, O’Hare JP, et al. Circadian gene variants and susceptibility to type 2 diabetes: a pilot study. PLoS One. 2012;7:e32670. doi: 10.1371/journal.pone.0032670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hoffman AE, Zheng T, Ba Y, Stevens RG, Yi CH, Leaderer D, et al. Phenotypic effects of the circadian gene Cryptochrome 2 on cancer-related pathways. BMC Cancer. 2010;10:110. doi: 10.1186/1471-2407-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Maronde E, Schilling AF, Seitz S, Schinke T, Schmutz I, van der Horst G, et al. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS One. 2010;5:e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. J Biol Chem. 2012;287:25280–91. doi: 10.1074/jbc.M112.340786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662–7. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Barclay JL, Shostak A, Leliavski A, Tsang AH, Johren O, Muller-Fielitz H, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304:E1053–63. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 177.Doi M, Takahashi Y, Komatsu R, Yamazaki F, Yamada H, Haraguchi S, et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat Med. 2010;16:67–74. doi: 10.1038/nm.2061. [DOI] [PubMed] [Google Scholar]
- 178.Wisor JP, O’Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci. 2002;3:20. doi: 10.1186/1471-2202-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kawamoto T, Noshiro M, Sato F, Maemura K, Takeda N, Nagai R, et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem Biophys Res Commun. 2004;313:117–24. doi: 10.1016/j.bbrc.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 180.Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J. 2004;382(Pt 1):43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–4. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 182.Nishiwaki T, Daigo Y, Kawasoe T, Nakamura Y. Isolation and mutational analysis of a novel human cDNA, DEC1 (deleted in esophageal cancer 1), derived from the tumor suppressor locus in 9q32. Genes Chromosomes Cancer. 2000;27:169–76. [PubMed] [Google Scholar]
- 183.Miura K, Suzuki K, Tokino T, Isomura M, Inazawa J, Matsuno S, et al. Detailed deletion mapping in squamous cell carcinomas of the esophagus narrows a region containing a putative tumor suppressor gene to about 200 kilobases on distal chromosome 9q. Cancer Res. 1996;56:1629–34. [PubMed] [Google Scholar]
- 184.Wong VC, Ko JM, Qi RZ, Li PJ, Wang LD, Li JL, et al. Abrogated expression of DEC1 during oesophageal squamous cell carcinoma progression is age-and family history-related and significantly associated with lymph node metastasis. Br J Cancer. 2011;104:841–9. doi: 10.1038/bjc.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jia YF, Xiao DJ, Ma XL, Song YY, Hu R, Kong Y, et al. Differentiated embryonic chondrocyte-expressed gene 1 is associated with hypoxia-inducible factor 1alpha and Ki67 in human gastric cancer. Diagn Pathol. 2013;8:37. doi: 10.1186/1746-1596-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu Y, Miao Y, Wang J, Lin X, Wang L, Xu HT, et al. DEC1 is positively associated with the malignant phenotype of invasive breast cancers and negatively correlated with the expression of claudin-1. Int J Mol Med. 2013;31:855–60. doi: 10.3892/ijmm.2013.1279. [DOI] [PubMed] [Google Scholar]
- 187.Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y, et al. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol. 2013;34:1641–50. doi: 10.1007/s13277-013-0697-z. [DOI] [PubMed] [Google Scholar]
- 188.Wu Y, Sato F, Yamada T, Bhawal UK, Kawamoto T, Fujimoto K, et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol. 2012;41:1337–46. doi: 10.3892/ijo.2012.1559. [DOI] [PubMed] [Google Scholar]
- 189.Wei H, Ke HL, Lin J, Shete S, Wood CG, Hildebrandt MA. MicroRNA target site polymorphisms in the VHL-HIF1alpha pathway predict renal cell carcinoma risk. Mol Carcinog. 2014;53:1–7. doi: 10.1002/mc.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shi XH, Zheng Y, Sun Q, Cui J, Liu QH, Qu F, et al. DEC1 nuclear expression: a marker of differentiation grade in hepatocellular carcinoma. World J Gastroenterology. 2011;17:2037–43. doi: 10.3748/wjg.v17.i15.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yunokawa M, Tanimoto K, Nakamura H, Nagai N, Kudo Y, Kawamoto T, et al. Differential regulation of DEC2 among hypoxia-inducible genes in endometrial carcinomas. Oncol Rep. 2007;17:871–8. [PubMed] [Google Scholar]
- 192.Moller-Levet CS, Archer SN, Bucca G, Laing EE, Slak A, Kabiljo R, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110:E1132–41. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Martinez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J, et al. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4 + T cell response. J Exp Med. 2013;210:1603–19. doi: 10.1084/jem.20122387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bhawal UK, Ito Y, Tanimoto K, Sato F, Fujimoto K, Kawamoto T, et al. IL-1beta-mediated up-regulation of DEC1 in human gingiva cells via the Akt pathway. J Cell Biochem. 2012;113:3246–53. doi: 10.1002/jcb.24205. [DOI] [PubMed] [Google Scholar]
- 195.Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J, et al. Disturbed clockwork resetting in Sharp-1 and Sharp-2 single and double mutant mice. PLoS One. 2008;3:e2762. doi: 10.1371/journal.pone.0002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol. 2008;28:4080–92. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Englund A, Kovanen L, Saarikoski ST, Haukka J, Reunanen A, Aromaa A, et al. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J Circadian Rhythms. 2009;7:5. doi: 10.1186/1740-3391-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Smith AK, Fang H, Whistler T, Unger ER, Rajeevan MS. Convergent genomic studies identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology. 2011;64:183–94. doi: 10.1159/000326692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Monsees GM, Kraft P, Hankinson SE, Hunter DJ, Schernhammer ES. Circadian genes and breast cancer susceptibility in rotating shift workers. Int J Cancer. 2012;131:2547–52. doi: 10.1002/ijc.27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, Boyle P, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–5. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yi C, Mu L, de la Longrais IA, Sochirca O, Arisio R, Yu H, et al. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res Treat. 2010;120:663–9. doi: 10.1007/s10549-009-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu Y, Zhang X, Liu J, Hou G, Zhang S, Zhang J. Everolimus in combination with letrozole inhibit human breast cancer MCF-7/Aro stem cells via PI3K/mTOR pathway: an experimental study. Tumour Biol. 2014;35:1275–86. doi: 10.1007/s13277-013-1170-8. [DOI] [PubMed] [Google Scholar]
- 204.Madden MH, Anic GM, Thompson RC, Nabors LB, Olson JJ, Browning JE, et al. Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control. 2014;25:25–32. doi: 10.1007/s10552-013-0305-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wu X, Wiater MF, Ritter S. NPAS2 deletion impairs responses to restricted feeding but not to metabolic challenges. Physiol Behav. 2010;99:466–71. doi: 10.1016/j.physbeh.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.O’Neil D, Mendez-Figueroa H, Mistretta TA, Su C, Lane RH, Aagaard KM. Dysregulation of Npas2 leads to altered metabolic pathways in a murine knockout model. Mol Genet Metab. 2013;110:378–87. doi: 10.1016/j.ymgme.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O’Hara BF, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: genotype and sex interactions. Proc Natl Acad Sci USA. 2006;103:7118–23. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, et al. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301:379–83. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 209.Anantharam V, Lehrmann E, Kanthasamy A, Yang Y, Banerjee P, Becker KG, et al. Microarray analysis of oxidative stress regulated genes in mesencephalic dopaminergic neuronal cells: relevance to oxidative damage in Parkinson’s disease. Neurochem Int. 2007;50:834–47. doi: 10.1016/j.neuint.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–14. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 211.Yi CH, Zheng T, Leaderer D, Hoffman A, Zhu Y. Cancer-related transcriptional targets of the circadian gene NPAS2 identified by genome-wide ChIP-on-chip analysis. Cancer Lett. 2009;284:149–56. doi: 10.1016/j.canlet.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6:1461–8. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mostafaie N, Kallay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48:642–7. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 214.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xia HC, Niu ZF, Ma H, Cao SZ, Hao SC, Liu ZT, et al. Deregulated expression of the Per1 and Per2 in human gliomas. Can J Neurol Sci. 2010;37:365–70. doi: 10.1017/s031716710001026x. [DOI] [PubMed] [Google Scholar]
- 216.Fujioka A, Takashima N, Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochem Biophys Res Commun. 2006;346:169–74. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 217.Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 218.Jung-Hynes B, Huang W, Reiter RJ, Ahmad N. Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J Pineal Res. 2010;49:60–8. doi: 10.1111/j.1600-079X.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Oda A, Katayose Y, Yabuuchi S, Yamamoto K, Mizuma M, Shirasou S, et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 2009;29:1201–9. [PubMed] [Google Scholar]
- 220.Pogue-Geile KL, Lyons-Weiler J, Whitcomb DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Lett. 2006;243:55–7. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 221.Gery S, Komatsu N, Kawamata N, Miller CW, Desmond J, Virk RK, et al. Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin Cancer Res. 2007;13:1399–404. doi: 10.1158/1078-0432.CCR-06-1730. [DOI] [PubMed] [Google Scholar]
- 222.Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, et al. A role for the clock gene per1 in prostate cancer. Cancer Res. 2009;69:7619–25. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Thoennissen NH, Thoennissen GB, Abbassi S, Nabavi-Nouis S, Sauer T, Doan NB, et al. Transcription factor CCAAT/enhancer-binding protein alpha and critical circadian clock downstream target gene PER2 are highly deregulated in diffuse large B-cell lymphoma. Leuk Lymphoma. 2012;53:1577–85. doi: 10.3109/10428194.2012.658792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Blask DE, Hill SM, Dauchy RT, Xiang S, Yuan L, Duplessis T, et al. Circadian regulation of molecular, dietary, and metabolic signaling mechanisms of human breast cancer growth by the nocturnal melatonin signal and the consequences of its disruption by light at night. J Pineal Res. 2011;51:259–69. doi: 10.1111/j.1600-079X.2011.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sun CM, Huang SF, Zeng JM, Liu DB, Xiao Q, Tian WJ, et al. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res. 2010;16:403–11. doi: 10.1007/s12253-009-9227-0. [DOI] [PubMed] [Google Scholar]
- 226.Blask DE. Melatonin, sleep disturbance and cancer risk. Sleep Med Rev. 2009;13:257–64. doi: 10.1016/j.smrv.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 227.Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms. 2008;6:4. doi: 10.1186/1740-3391-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–20. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 229.Chu LW, Zhu Y, Yu K, Zheng T, Chokkalingam AP, Stanczyk FZ, et al. Correlation between circadian gene variants and serum levels of sex steroids and insulin-like growth factor-I. Cancer Epidemiol Biomarkers Prev. 2008;17:3268–73. doi: 10.1158/1055-9965.EPI-08-0073. [DOI] [PubMed] [Google Scholar]
- 230.Garaulet M, Corbalan-Tutau MD, Madrid JA, Baraza JC, Parnell LD, Lee YC, et al. PERIOD2 variants are associated with abdominal obesity, psycho-behavioral factors, and attrition in the dietary treatment of obesity. J Am Diet Assoc. 2010;110:917–21. doi: 10.1016/j.jada.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Garcia-Rios A, Perez-Martinez P, Delgado-Lista J, Phillips CM, Gjelstad IM, Wright JW, et al. A Period 2 genetic variant interacts with plasma SFA to modify plasma lipid concentrations in adults with metabolic syndrome. J Nutr. 2012;142:1213–18. doi: 10.3945/jn.111.156968. [DOI] [PubMed] [Google Scholar]
- 232.Lazar AS, Santhi N, Hasan S, Lo JC, Johnston JD, Von Schantz M, et al. Circadian period and the timing of melatonin onset in men and women: predictors of sleep during the weekend and in the laboratory. J Sleep Res. 2013;22:155–9. doi: 10.1111/jsr.12001. [DOI] [PubMed] [Google Scholar]
- 233.Below JE, Gamazon ER, Morrison JV, Konkashbaev A, Pluzhnikov A, McKeigue PM, et al. Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia. 2011;54:2047–55. doi: 10.1007/s00125-011-2188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guess J, Burch JB, Ogoussan K, Armstead CA, Zhang H, Wagner S, et al. Circadian disruption, Per3, and human cytokine secretion. Integr Cancer Ther. 2009;8:329–36. doi: 10.1177/1534735409352029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hida A, Kusanagi H, Satoh K, Kato T, Matsumoto Y, Echizenya M, et al. Expression profiles of PERIOD1, 2, and 3 in peripheral blood mononuclear cells from older subjects. Life Sci. 2009;84:33–7. doi: 10.1016/j.lfs.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 236.Jud C, Chappuis S, Revell VL, Sletten TL, Saaltink DJ, Cajochen C, et al. Age-dependent alterations in human PER2 levels after early morning blue light exposure. Chronobiol Int. 2009;26:1462–9. doi: 10.3109/07420520903385564. [DOI] [PubMed] [Google Scholar]
- 237.Lin C, Tang X, Zhu Z, Liao X, Zhao R, Fu W, et al. The rhythmic expression of clock genes attenuated in human plaque-derived vascular smooth muscle cells. Lipids Health Dis. 2014;13:14. doi: 10.1186/1476-511X-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shiino Y, Nakajima S, Ozeki Y, Isono T, Yamada N. Mutation screening of the human period 2 gene in bipolar disorder. Neurosci Lett. 2003;338:82–4. doi: 10.1016/s0304-3940(02)01290-9. [DOI] [PubMed] [Google Scholar]
- 239.Yoshida K, Hashiramoto A, Okano T, Yamane T, Shibanuma N, Shiozawa S. TNF-alpha modulates expression of the circadian clock gene Per2 in rheumatoid synovial cells. Scand J Rheumatol. 2013;42:276–80. doi: 10.3109/03009742.2013.765031. [DOI] [PubMed] [Google Scholar]
- 240.Ojeda DA, Perea CS, Nino CL, Gutierrez RM, Lopez-Leon S, Arboleda H, et al. A novel association of two non-synonymous polymorphisms in PER2 and PER3 genes with specific diurnal preference subscales. Neurosci Lett. 2013;553:52–6. doi: 10.1016/j.neulet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 241.Nakamura Y, Harama D, Shimokawa N, Hara M, Suzuki R, Tahara Y, et al. Circadian clock gene Period2 regulates a time-of-day-dependent variation in cutaneous anaphylactic reaction. J Allergy Clin Immunol. 2011;127:1038–45. e1–3. doi: 10.1016/j.jaci.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 242.Pilorz V, Steinlechner S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction. 2008;135:559–68. doi: 10.1530/REP-07-0434. [DOI] [PubMed] [Google Scholar]
- 243.Spencer S, Falcon E, Kumar J, Krishnan V, Mukherjee S, Birnbaum SG, et al. Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur J Neurosci. 2013;37:242–50. doi: 10.1111/ejn.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liu J, Malkani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, et al. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–6. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Bonney S, Kominsky D, Brodsky K, Eltzschig H, Walker L, Eckle T. Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS One. 2013;8:e71493. doi: 10.1371/journal.pone.0071493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, et al. Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat Med. 2012;18:774–82. doi: 10.1038/nm.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, et al. PER2 controls lipid metabolism by direct regulation of PPAR-gamma. Cell Metab. 2010;12:509–20. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bhatwadekar AD, Yan Y, Qi X, Thinschmidt JS, Neu MB, Li Calzi S, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes. 2013;62:273–82. doi: 10.2337/db12-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Cheng B, Anea CB, Yao L, Chen F, Patel V, Merloiu A, et al. Tissue-intrinsic dysfunction of circadian clock confers transplant arteriosclerosis. Proc Natl Acad Sci USA. 2011;108:17147–52. doi: 10.1073/pnas.1112998108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Carvas JM, Vukolic A, Yepuri G, Xiong Y, Popp K, Schmutz I, et al. Period2 gene mutant mice show compromised insulin-mediated endothelial nitric oxide release and altered glucose homeostasis. Front Physiol. 2012;3:337. doi: 10.3389/fphys.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ma K, Xiao R, Tseng HT, Shan L, Fu L, Moore DD. Circadian dysregulation disrupts bile acid homeostasis. PLoS One. 2009;4:e6843. doi: 10.1371/journal.pone.0006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Logan RW, Wynne O, Levitt D, Price D, Sarkar DK. Altered circadian expression of cytokines and cytolytic factors in splenic natural killer cells of Per1(−/−) mutant mice. J Interferon Cytokine Res. 2013;33:108–14. doi: 10.1089/jir.2012.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Arjona A, Sarkar DK. The circadian gene mPer2 regulates the daily rhythm of IFN-gamma. J Interferon Cytokine Res. 2006;26:645–9. doi: 10.1089/jir.2006.26.645. [DOI] [PubMed] [Google Scholar]
- 254.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469–76. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 255.Knower KC, Chand AL, Eriksson N, Takagi K, Miki Y, Sasano H, et al. Distinct nuclear receptor expression in stroma adjacent to breast tumors. Breast Cancer Res Treat. 2013;142:211–23. doi: 10.1007/s10549-013-2716-6. [DOI] [PubMed] [Google Scholar]
- 256.Mond M, Alexiadis M, Eriksson N, Davis M, Muscat G, Fuller P, et al. Nuclear receptor expression in human differentiated thyroid tumors. Thyroid. 2014 Feb 21; doi: 10.1089/thy.2013.0509. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 257.Xiong G, Wang C, Evers BM, Zhou BP, Xu R. RORalpha suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012;72:1728–39. doi: 10.1158/0008-5472.CAN-11-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Moretti RM, Montagnani Marelli M, Sala A, Motta M, Limonta P. Activation of the orphan nuclear receptor RORalpha counteracts the proliferative effect of fatty acids on prostate cancer cells: crucial role of 5-lipoxygenase. Int J Cancer. 2004;112:87–93. doi: 10.1002/ijc.20387. [DOI] [PubMed] [Google Scholar]
- 259.Karasek M, Gruszka A, Lawnicka H, Kunert-Radek J, Pawlikowski M. Melatonin inhibits growth of diethylstilbestrol-induced prolactin-secreting pituitary tumor in vitro: possible involvement of nuclear RZR/ROR receptors. J Pineal Res. 2003;34:294–6. doi: 10.1034/j.1600-079x.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- 260.Kottorou AE, Antonacopoulou AG, Dimitrakopoulos FI, Tsamandas AC, Scopa CD, Petsas T, et al. Altered expression of NFY-C and RORA in colorectal adenocarcinomas. Acta Histochem. 2012;114:553–61. doi: 10.1016/j.acthis.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 261.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN, et al. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS One. 2012;7:e44008. doi: 10.1371/journal.pone.0044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Klar J, Asling B, Carlsson B, Ulvsback M, Dellsen A, Strom C, et al. RAR-related orphan receptor A isoform 1 (RORa1) is disrupted by a balanced translocation t(4;15)(q22.3;q21.3) associated with severe obesity. European Journal Of Human Genetics: EJHG. 2005;13:928–34. doi: 10.1038/sj.ejhg.5201433. [DOI] [PubMed] [Google Scholar]
- 263.Gamboa-Melendez MA, Huerta-Chagoya A, Moreno-Macias H, Vazquez-Cardenas P, Ordonez-Sanchez ML, Rodriguez-Guillen R, et al. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61:3314–21. doi: 10.2337/db11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tinahones FJ, Moreno-Santos I, Vendrell J, Chacon MR, Garrido-Sanchez L, Garcia-Fuentes E, et al. The retinoic acid receptor-related orphan nuclear receptor gamma1 (RORgamma1): a novel player determinant of insulin sensitivity in morbid obesity. Obesity. 2012;20:488–97. doi: 10.1038/oby.2011.267. [DOI] [PubMed] [Google Scholar]
- 265.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, et al. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci USA. 2000;97:10132–7. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–73. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 267.Jetten AM, Kang HS, Takeda Y. Retinoic acid-r 267. elated orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Meissburger B, Ukropec J, Roeder E, Beaton N, Geiger M, Teupser D, et al. Adipogenesis and insulin sensitivity in obesity are regulated by retinoid-related orphan receptor gamma. EMBO Mol Med. 2011;3:637–51. doi: 10.1002/emmm.201100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Crumbley C, Wang Y, Banerjee S, Burris TP. Regulation of expression of citrate synthase by the retinoic acid receptor-related orphan receptor alpha (RORalpha) PLoS One. 2012;7:e33804. doi: 10.1371/journal.pone.0033804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Gohil K, Godzdanker R, O’Roark E, Schock BC, Kaini RR, Packer L, et al. Alpha-tocopherol transfer protein deficiency in mice causes multi-organ deregulation of gene networks and behavioral deficits with age. Ann NY Acad Sci. 2004;1031:109–26. doi: 10.1196/annals.1331.012. [DOI] [PubMed] [Google Scholar]
- 271.Caballero B, Vega-Naredo I, Sierra V, Huidobro-Fernandez C, Soria-Valles C, De Gonzalo-Calvo D, et al. Favorable effects of a prolonged treatment with melatonin on the level of oxidative damage and neurodegeneration in senescence-accelerated mice. J Pineal Res. 2008;45:302–11. doi: 10.1111/j.1600-079X.2008.00591.x. [DOI] [PubMed] [Google Scholar]
- 272.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REVERB-beta. Nature. 2012;485:123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Mol Endocrinol. 2013;27:350–65. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Davis LM, Harris C, Tang L, Doherty P, Hraber P, Sakai Y, et al. Amplification patterns of three genomic regions predict distant recurrence in breast carcinoma. J Mol Diagn. 2007;9:327–36. doi: 10.2353/jmoldx.2007.060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 276.Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res. 2010;70:1783–92. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Goumidi L, Grechez A, Dumont J, Cottel D, Kafatos A, Moreno LA, et al. Impact of REV-ERB alpha gene polymorphisms on obesity phenotypes in adult and adolescent samples. Int J Obes (Lond) 2013;37:666–72. doi: 10.1038/ijo.2012.117. [DOI] [PubMed] [Google Scholar]
- 278.Garaulet M, Smith CE, Gomez-Abellan P, Ordovas-Montanes M, Lee YC, Parnell LD, et al. REV-ERB-ALPHA circadian gene variant associates with obesity in two independent populations: Mediterranean and North American. Mol Nutr Food Res. 2014;58:821–9. doi: 10.1002/mnfr.201300361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Thessen Hedreul M, Moller S, Stridh P, Gupta Y, Gillett A, Daniel Beyeen A, et al. Combining genetic mapping with genome-wide expression in experimental autoimmune encephalomyelitis highlights a gene network enriched for T cell functions and candidate genes regulating autoimmunity. Hum Mol Genet. 2013;22:4952–66. doi: 10.1093/hmg/ddt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, et al. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFalpha. Gut. 2013;62:985–94. doi: 10.1136/gutjnl-2011-301213. [DOI] [PubMed] [Google Scholar]
- 281.Delezie J, Dumont S, Dardente H, Oudart H, Grechez-Cassiau A, Klosen P, et al. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–35. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 282.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–7. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ohsaki K, Oishi K, Kozono Y, Nakayama K, Nakayama KI, Ishida N. The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J Biochem. 2008;144:609–18. doi: 10.1093/jb/mvn112. [DOI] [PubMed] [Google Scholar]
- 284.Kim CJ, Song JH, Cho YG, Kim YS, Kim SY, Nam SW, et al. Somatic mutations of the beta-TrCP gene in gastric cancer. APMIS. 2007;115:127–33. doi: 10.1111/j.1600-0463.2007.apm_562.x. [DOI] [PubMed] [Google Scholar]
- 285.Gerstein AV, Almeida TA, Zhao G, Chess E, Shih Ie M, Buhler K, et al. APC/CTNNB1 (beta-catenin) pathway alterations in human prostate cancers. Genes Chromosomes Cancer. 2002;34:9–16. doi: 10.1002/gcc.10037. [DOI] [PubMed] [Google Scholar]
- 286.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 287.Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96:1161–70. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 288.Muerkoster S, Arlt A, Sipos B, Witt M, Grossmann M, Kloppel G, et al. Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65:1316–24. doi: 10.1158/0008-5472.CAN-04-1626. [DOI] [PubMed] [Google Scholar]
- 289.Chen S, He Y, Ding J, Jiang Y, Jia S, Xia W, et al. An insertion/deletion polymorphism in the 3′ untranslated region of beta-transducin repeat-containing protein (betaTrCP) is associated with susceptibility for hepatocellular carcinoma in Chinese. Biochem Biophys Res Commun. 2010;391:552–6. doi: 10.1016/j.bbrc.2009.11.096. [DOI] [PubMed] [Google Scholar]
- 290.Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 291.Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 292.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1-and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–71. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 293.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, et al. mTOR generates an auto-amplification loop by triggering the betaTrCP-and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–24. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–16. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–83. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, et al. betaTrCP-and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33:109–16. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Shaik S, Nucera C, Inuzuka H, Gao D, Garnaas M, Frechette G, et al. SCF(beta-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J Exp Med. 2012;209:1289–307. doi: 10.1084/jem.20112446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, et al. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–94. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Belaidouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guerin C. Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 2005;24:2271–6. doi: 10.1038/sj.onc.1208418. [DOI] [PubMed] [Google Scholar]
- 300.Huang C. Roles of E3 ubiquitin ligases in cell adhesion and migration. Cell Adh Migr. 2010;4:10–18. doi: 10.4161/cam.4.1.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci USA. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–59. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Roenneberg T. Chronobiology: the human sleep project. Nature. 2013;498:427–8. doi: 10.1038/498427a. [DOI] [PubMed] [Google Scholar]


