Abstract
Mitochondrial diseases are commonly caused by mutations in the mitochondrial DNA (mtDNA), which in most cases co–exists with the wild–type (mtDNA heteroplasmy). We have engineered TAL–effector nucleases (TALENs) to localize to mitochondria and cleave different classes of pathogenic mtDNA mutations. MitoTALEN expression led to permanent reductions in deletion or point mutant mtDNA in patient–derived cells, raising the possibility that they can be curative to some of these diseases.
Most often, pathogenic mtDNA mutations are heteroplasmic, and residual wild–type mtDNA can partially compensate for the mutated mtDNA1–3, averting a bioenergetic crisis. The levels of mutated mtDNA in affected tissues have to reach a high threshold, usually above 80%, for biochemical and clinical manifestations 1. Therefore, several elegant approaches to reduce the levels of mutant mtDNA have been tried 4–7. However, previous attempts have either not worked in living cells, lacked the flexibility to target clinically relevant mutations or did not produce stable changes in heteroplasmy to be readily translatable to animals.
To circumvent these limitations, we re–engineered TAL effector Nucleases (TALEN) 8,9 as a potentially flexible platform to reduce the levels of specific mtDNA mutations. A particular feature of the system is that TALENs work as heterodimers, requiring two TALEN monomers to bind closely spaced DNA sequences, allowing their FokI nuclease domains to dimerize and cleave DNA. We started testing this approach by designing mitochondrial–targeted TALEN (mitoTALEN) monomers to bind to the breakpoint region of a mtDNA with a large 5 Kb deletion, m.8483_13459del4977. This mutation is known as the “common deletion” because it is present in approximately 30% of all patients with mtDNA deletions 10, as well as in normal aging tissues 11,12. The design concept of the mitoTALEN for eliminating the common deletion (Δ5–mitoTALEN) places each monomer in specific wild–type sequences. However, only in the deletion mutant mtDNA are the monomers close enough to promote FokI dimerization and cleavage (Fig. 1a). No perfect homology of the selected target region was detected in the nuclear DNA (Supplementary Table 1).
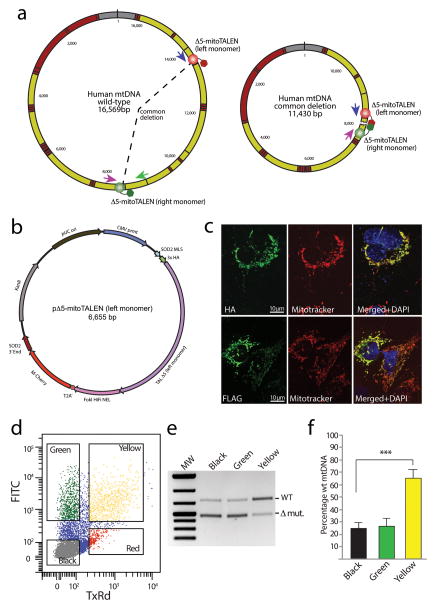
Figure 1. Using mitoTALEN to eliminate mtDNAs with the common deletion.
(a) We designed mitoTALEN monomers that bind wild–type sequence adjacent to the breakpoint present in the mtDNA carrying the “common deletion”. In the wild–type mtDNA, these sites are far apart, blocking FokI dimerization. (b) Plasmids express one of the two monomers of the mitoTALEN, and a fluorescent marker (eGFP or mCherry) from the same transcriptional unit through the use of a T2A translation stuttering sequence. The plasmids also contained additional elements to optimize expression and mitochondrial targeting, such as 3′UTRs from nuclear encoded mitochondrial genes. (c) Anti–FLAG or anti–HA antibodies were used in immunocytochemistry, showing strict mitochondrial localization of Δ5–mitoTALEN monomers in 24 hrs–transfected COS7 cells. (d) Cells were sorted for eGFP and mCherry 48 hours after transfection with two individual plasmids coding for the Δ5–mitoTALEN monomers. DNA isolated from the cell fractions that were not expressing fluorescent markers (“Black”), only one marker (“Green”) and both eGFP and mCherry (“Yellow”) were subjected to the 3–primer PCR analysis to quantify the relative levels of deleted mtDNA (e). PCR was performed with the primers depicted by colored arrows in Fig. 1a. Panel f shows the quantitation of 3 independent experiments (t–Test unpaired between “black” and “yellow” values; n=3; ***P = <0.001). Error bars correspond to SD of the mean.
We have made a series of modifications to rationally design mitoTALENs (Fig. 1b, and supplementary material). Each TALEN monomer had a unique epitope tag and a mitochondrial localization sequence at the N–terminus. The basic structure of the protein is shown (Supplementary Fig. 1a). Western blots showed that monomers of the expected size were synthesized after transient transfection (Supplementary Fig. 1b). Likewise, immunocytochemistry showed that they co–localized with the mitochondrial marker Mitotracker (Fig. 1c). Within the transcriptional unit of each mitoTALEN monomer we added a fluorescent marker, so that transfected cells could be FACS sorted, mCherry for the left monomer and eGFP for the right monomer (Fig 1b).
We tested the Δ5–mitoTALEN in human osteosarcoma cells heteroplasmic for the mtDNA common deletion (BH10.9 cells)13. We transfected cells with two independent plasmids, each coding for one mitoTALEN monomer. Two days after transfection cells were FACS sorted for mCherry and eGFP expression (Fig. 1d). Cells expressing both markers were labeled as “yellow” and were expected to also express both Δ5–mitoTALEN monomers. We also isolated cells not expressing any of the fluorescent markers (“black”) and cells expressing only eGFP (“green”). MtDNA heteroplasmy was analyzed by a previously described 3–primer PCR technique (Supplementary Methods), using the primers depicted in Fig. 1a. This analysis showed that the Δ5–mitoTALEN was effective in reducing the mtDNA deletion load and changing mtDNA heteroplasmy towards a predominance of wild–type mtDNA (Fig. 1e–f). We then measured the levels of the different mtDNA species by qPCR and found that the change in heteroplasmy was primarily due to a reduction in the absolute levels of deletion mutant (Supplementary Fig. 2). There was a trend towards a reduction in the total mtDNA levels after 2 days, and a small reduction was also observed after 14 days. A significant increase in wild–type mtDNA was observed at 14 days (Supplementary Fig. 2). Transfection with the Δ5–mitoTALEN in the homoplasmic parental osteosarcoma line (143B) did not change total mtDNA levels at either 2 or 14 days (Supplementary Fig. 2 b–c), confirming the specificity of the mitoTALEN.
We next tested the approach for the Leber’s optic neuropathy–dystonia point mutation m.14459G>A in the MT–ND6 gene 14. The rationale, is that one of the monomers of the14459A–mitoTALEN would bind to wild–type sequence adjacent to the mutation but cleavage would be dictated by binding of the right monomer to a recognition sequence harboring m.14459A (Fig. 2a). A TALEN was designed to differentiate these molecules and initially tested by yeast Single–Strand Annealing assay (Supplementary Fig. 3). This test showed that the TALE domain was able to differentially bind wild–type and mutant sites. We then re–engineered the construct and built the 14459A–mitoTALEN as described for the Δ5–mitoTALEN. Although nuclear pseudogenes of the MT–ND6 gene exist, we did not detect a complete target site with the m.14459A sequence (Supplementary Table 1).
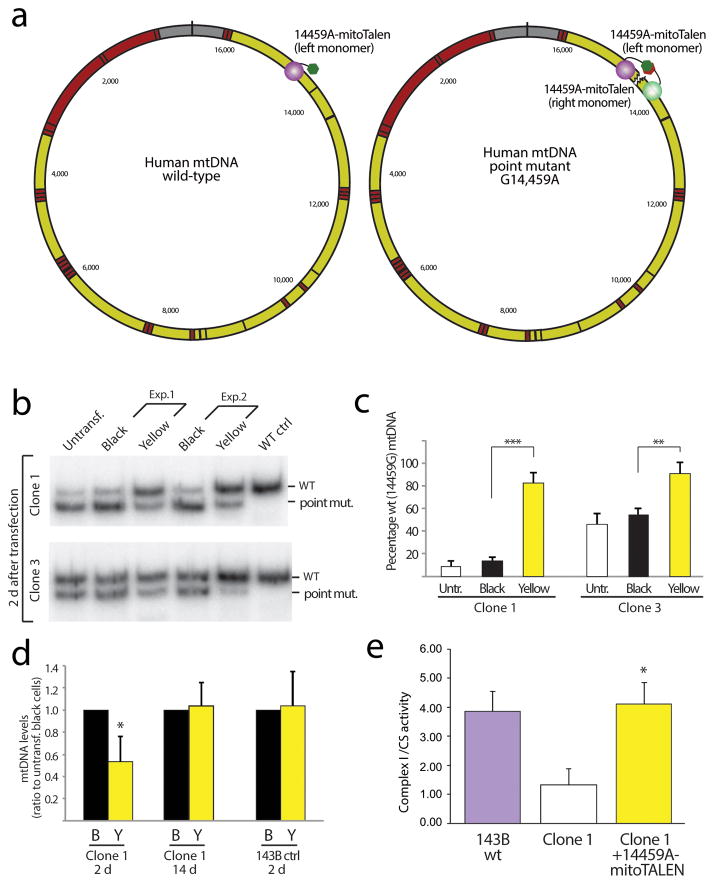
Figure 2. Using mitoTALEN to reduce the levels of the pathogenic m.14459A mtDNA point mutation.
(a) Approach for the elimination of a point mutant (m.14459G>A). In this case, one of the mitoTALEN monomers binds to wild–type sequence (left monomer) and the other to sequence containing the point mutation within the MT–ND6 gene (right monomer). (b) Cybrids harboring the Leber’s–dystonia m.14,459A mtDNA mutation (Clones 1 and 3) were transiently transfected with 14459A–mitoTALEN expressing plasmids (Exp.1 and 2). Clone 1 harbors approximately 90% m.14459A whereas clone 3 harbors approximately 55%. The panel shows the RFLP analysis of the mutation load in the “black” and “yellow” cells 48 hours after transfection FACS sorted as shown in Fig. 1d. Quantification of n=3 independent experiments is shown (c). Untr=untransfected cells. (d) Quantitation of the total mtDNA levels by qPCR showed a decrease in clone 1 two days after transfection, but not at 14 d. Likewise, wild–type control 143B cells did not show a decrease in mtDNA levels, even after 2 d. (e) Enzyme activity (complex I/citrate synthase ratio) in control, mutant and mutant cells transfected with the 14459A mitoTALEN (14 d after transfection). (t–Test unpaired between “black” and “yellow” values; n=3; * P = <0.005, ** P = <0.002, *** P = <0.001). Error bars correspond to SD of the mean.
We tested the 14459A–mitoTALEN in osteosarcoma cybrids harboring heteroplasmic levels of the m.14459A mtDNA (Clones 1 and 3). Transfections with the mitoTALEN were followed by FACS sorting and isolation of “black” and “yellow” cells. Heteroplasmy analyses of amplicons spanning m.14459 showed that the mtDNA heteroplasmy once again shifted in the predicted direction with a marked relative increase of WT mtDNA (Fig. 2b–c). The change was stable at 14 days after transfection (Supplementary Fig. 4), even though at this time point the mitoTALEN could not be detected in the cells by western blot (Supplementary Fig. 5). Analyses by qPCR showed that one of the tested clones (clone 1) had a transient decrease in total mtDNA levels, which is not surprising considering that it had close to 90% mutant mtDNA (Fig. 2d). However, this decrease was neither observed after 14 days, nor in transfected parental 143B cells, showing that the 14459A–mitoTALEN is specific towards m.14459A and that associated mtDNA depletion was due to the elimination of target mtDNA.
In contrast to the cells with heteroplasmic mtDNA deletions, which had enough wild–type genomes to show a normal biochemical phenotype, complex I activity was partially decreased in the clone with high levels of m.14459A mutant mtDNA (Clone 1). This partial defect (approximately 35–40% residual activity) was similar to the activity previously reported for cells harboring high levels of this mtDNA mutation 14. As expected, transfection with the 14459A–mitoTALEN not only reduced the mutant mtDNA load, but also increased complex I activity (Fig. 2e).
Although animal experimentation may be required to ensure the safety of mitoTALENs, our previous experience with mitochondrial–targeted restriction endonucleases showed that transient endonucleases activity in mitochondria is well tolerated in mouse muscle 15, heart and liver 4. Although there was transient mtDNA depletion, mtDNA levels were rapidly normalized due to a well–recognized, but yet uncharacterized “mtDNA copy number control” mechanism 16,17. Nevertheless, caution will have to be exercised in situations when the levels of the mtDNA haplotype targeted for elimination is close to 100%, as mtDNAs that carry mutations may contribute to the functional mitochondrial gene pool in some cases.
The delivery of therapeutic genes and proteins remains a barrier to the fast implementation of genetic therapies, particularly of large genes, such as TALEN. However, it is important to keep in mind that in comparison to gene therapy of the nuclear genome, where lifelong expression of the corrective factor is the goal, transient expression of mitoTALEN should be sufficient to produce lasting changes in mtDNA heteroplasmy 4,15. It is therefore reasonable to expect that permanent correction of heteroplasmy levels, potentially rescuing the OXPHOS deficiency in affected tissues, might be achieved after one or a small number of administrations of mitoTALEN, either as genetic or protein agents.
ONLINE METHODS
MitoTALEN construct
Custom made TALE domains were developed by Cellectis Bioresearch Company (http://www.cellectis-bioresearch.com) according to the mtDNA sequence provided by the authors. The TALEN binds to the following mtDNA sequences (TAL binding underlined). The TAL–domain amino acid sequences are also listed.
Common deletion breakpoint (Δ5 mitoTALEN)
AAAATATTAAACACAAACTACCACCTACCTCCCTCACCATTGGCAGCCTAGCATTAG
Left Δ5 TAL–Domain
DIADLRTLGYSQQQQEKIKPKVRSTVAQHHEALVGHGFTHAHIVALSQHPAALGTVAV KYQDMIAALPEATHEAIVGVGKQWSGARALEALLTVAGELRGPPLQLDTGQLLKIAKR GGVTAVEAVHAWRNALTGAPLNLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLT PQQVVAIASNGGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNGGGKQALETVQRLL PVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPEQVVAIASNIGGKQ ALETVQALLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPEQVVA IASHDGGKQALETVQRLLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAH GLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPEQVVAIASNIGGKQALETVQA LLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPEQVVAIASNIGGK QALETVQALLPVLCQAHGLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPQQV VAIASNGGGKQALETVQRLLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQ AHGLTPQQVVAIASNGGGRPALESIVAQLSRPDPALAALTNDHLVALACLGGRPALDAV KKGLG
Right Δ5Tal–Domain
DIADLRTLGYSQQQQEKIKPKVRSTVAQHHEALVGHGFTHAHIVALSQHPAALGTVAV KYQDMIAALPEATHEAIVGVGKQWSGARALEALLTVAGELRGPPLQLDTGQLLKIAKR GGVTAVEAVHAWRNALTGAPLNLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGL TPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNGGGKQALETVQRL LPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPQQVVAIASNNGGK QALETVQRLLPVLCQAHGLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGLTPEQV VAIASHDGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNGGGKQALETVQRLLPVLC QAHGLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGLTPEQVVAIASHDGGKQALE TVQRLLPVLCQAHGLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPEQVVAIAS NIGGKQALETVQALLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQAHGLT PQQVVAIASNGGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNNGGKQALETVQRLL PVLCQAHGLTPQQVVAIASNGGGRPALESIVAQLSRPDPALAALTNDHLVALACLGGRP ALDAVKKGLG
Point mutant m.14459A (14459A mitoTALEN)
CCCCTGACCCCCATGCCTCAGGATACTCCTCAATAGCCATCaCTGTA
Left (m.14459A Tal–Domain)
DIADLRTLGYSQQQQEKIKPKVRSTVAQHHEALVGHGFTHAHIVALSQHPAALGTVAV KYQDMIAALPEATHEAIVGVGKQWSGARALEALLTVAGELRGPPLQLDTGQLLKIAKR GGVTAVEAVHAWRNALTGAPLNLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGL TPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPEQVVAIASHDGGKQALETVQRLL PVLCQAHGLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPEQVVAIASHDGGK QALETVQRLLPVLCQAHGLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPEQV VAIASHDGGKQALETVQRLLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQ AHGLTPQQVVAIASNGGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNGGGRPALESI VAQLSRPDPALAALTNDHLVALACLGGRPALDAVKKGLG
Right (m.14459A Tal–Domain)
DIADLRTLGYSQQQQEKIKPKVRSTVAQHHEALVGHGFTHAHIVALSQHPAALGTVAV KYQDMIAALPEATHEAIVGVGKQWSGARALEALLTVAGELRGPPLQLDTGQLLKIAKR GGVTAVEAVHAWRNALTGAPLNLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGL TPEQVVAIASNIGGKQALETVQALLPVLCQAHGLTPQQVVAIASNGGGKQALETVQRLL PVLCQAHGLTPQQVVAIASNNGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNNGGK QALETVQRLLPVLCQAHGLTPEQVVAIASHDGGKQALETVQRLLPVLCQAHGLTPQQV VAIASNGGGKQALETVQRLLPVLCQAHGLTPEQVVAIASNIGGKQALETVQALLPVLCQ AHGLTPQQVVAIASNGGGKQALETVQRLLPVLCQAHGLTPQQVVAIASNGGGKQALET VQRLLPVLCQAHGLTPQQVVAIASNGGGRPALESIVAQLSRPDPALAALTNDHLVALAC LGGRPALDAVKKGLG
We modified plasmids constructs for mitoTALEN monomers using the In–Fusion HD cloning kit from Clontech Laboratories. Modifications included: Removal of the stock nuclear localization signal; inclusion of a mitochondrial localization signal derived from SOD2 or COX8A; inclusion of a tag in the N–terminus of the mature protein (Hemmaglutinin–HA or Flag); inclusion of a 3′ UTR from a mitochondrial gene, inclusion of an independent fluorescent marker to select for double transfected cells (eGFP in one monomer and mCherry in the other); inclusion of a picornaviral 2A–like sequence (T2A) between the mitoTALEN and the fluorescent marker 18. Besides using the two plasmids for co–transfection (see figure 1), both left and right monomers were fused in a tandem plasmid as shown in Supplementary Fig. 6a. This single plasmid produced more “yellow” cells after sorting (Supplementary Fig. 6b), although the results related to heteroplasmy changes in “yellow” cells were identical whether we used two plasmids or the combined one (not shown).
Single–Strand Annealing Assay
Single–Strand Annealing in Saccharomyces cerevisiae was performed as described in The Journal of Visualized Experiments (JoVE) article 19.
Cells, transfection and sorting
Cybrids harboring heteroplasmic common deletion were derived from fusions of an osteosarcoma cell line devoid of mtDNA (143B/206 line 20) and dermal fibroblast from a patient harboring the “common deletion”, m.8483_13459del4977 13,21. We used clone BH10.9, which was previously characterized 13. Cybrid clones for the point mutation model were derived in a similar way by fusing dermal fibroblast from a patient harboring the heteroplasmic missense mutation m.14,459A in MT–ND6 with 143B/206 cells22 and manually picking and screening colonies for heteroplasmy levels of m.14459A. The m.14,459A point mutation has been reported and characterized in other patients 14,23. Cells were transfected with GenJet DNA in vitro transfection reagent version II (#SL100489; SignaGen Laboratories), following the protocol suggested by the manufacturers. After 48hrs, cells were sorted using a FACSAria IIu by gating on single cell fluorescence using 561 nm laser and 600LP; 610/20 filter set for mCherry and 488nm laser and 505LP; 530/30 filter set for eGFP.
Western–blot
We subjected 40 μg of total proteins from cellular homogenates of Cos 7 (48 hours post transfection) to electrophoresis in 4–20% Tris–HCl polyacrylamide gels (#4561094; Bio–Rad Laboratories) and transferred the separated proteins to nitrocellulose membranes (#162–0115; Bio–Rad) as described 4. Antibodies used for blotting were: Rat anti–HA monoclonal antibody (#1867423; Roche Biochemical), mouse monoclonal anti–Flag antibody (#F3165; Sigma–Aldrich) and a mouse monoclonal anti–tubulin antibody (Cat# T9026; Sigma–Aldrich); Secondary antibodies: IRDye 800 conjugated anti–rat IgG (#612–732–120), IRDye 800 conjugated anti–mouse IgG (#610–732–124) and IRDye 700 conjugated anti–mouse IgG (#610–430–002) from Rockland. An Odyssey Infrared Imaging System (LI–COR) was used to scan the blots.
Immunocytochemical studies
We plated Cos 7 cells onto coverslips and then transfected them with mitoTALEN plasmids for 24hs. Cells were incubated for 30 min at 37°C with 200nM Mito Tracker Red CMXRos (Invitrogen) and fixed with 2% paraformaldehyde in PBS for 20 min. After a brief treatment with methanol (5 min), primary antibodies, anti–HA (Roche) or anti–FLAG (Sigma–Aldrich) in 2% bovine serum albumin /PBS were incubated overnight at 4°C in a humid chamber. The next day coverslips were incubated for 2 hrs at room temperature with Alexa Fluor 488–goat anti–rat IgG (#A–11006; Molecular Probes) or Alexa Fluor 488–goat anti–mouse IgG (A–11001; Molecular Probes) as previously described 4. Images were recorded using a confocal microscope Zeiss LSM510.
DNA extraction
We extracted DNA of the sorted cells with the NucleoSpin Tissue XS kit (#740901.50; Macherey–Nagel, Clontech) according to manufacturer’s instructions.
3–Primer PCR to quantify the ratios of deleted/WT molecules (Δ/WT)
We applied this 3–primer PCR approach as previously described 21. Primers were: F–m.8273_8289, B1–m.9028_9008 and B2–m.13720_13705. Primer B1 corresponds to a mtDNA region inside the common deletion, whereas primers F and B2 flank the deleted region. Primers F and B1 only amplify wt–mtDNAs, and primers F and B2 amplify Δ–mtDNAs. PCR products were cleaned using spin columns (#740609.250; Macherey–Nagel, Clontech) and quantified with a bio–analyzer system (Bioanalyzer 2100; Agilent).
Quantification of m.14,459A by “Last–cycle hot” PCR
We determine the levels of mutation by “Last–cycle hot” PCR 24,25, which only visualizes nascent amplicons and removes interference from heteroduplexes formed during melting and annealing cycles. Total DNA from sorted cells was used as template and PCR was performed with the following mtDNA primers in which the mutant allele m.14459A completes a BclI half–site allowing RFLP:
F–BclI–mut–F: 5′–CCCCCATGCCTCAGGATACTCCTCAATAGTGATC–3′
14579B: 5′–TGATTGTTAGCGGTGTGGTCGGGTGTGT–3′.
PCR products were digested with BclI and resolved in a 12% polyacrylamide gel. Radioactive signal was quantified using a Cyclone phosphor imaging system (Perkin–Elmer).
Quantitative PCR
We performed quantitative PCR reactions using SYBR/ROX chemistry (#172–5264; BioRad, SsoAdvanced SYBR Green) 15 on a BioRad CFX96/C1000 qPCR machine using manufacturers’ software to calculate ΔΔCT values. We determined the levels of different mtDNA species between sorted cell populations by quantifying the levels of deletion breakpoint/actin, wild–type/actin and total mtDNA/actin. We used the following primers:
Inside the “common deletion” (detects only wt mtDNA):
8537F: 5′–ATCTGTTCGCTTCATTCATTGC–3′
8661B: 5′–GGTGGTGATTAGTCGGTTGT–3′
Breakpoint of the “common deletion” (detects only Δ5 deleted mtDNA):
8410F: 5′–CATACTCCTTACACTATTCCTCAT–3′
13479B: 5′–TGCTAATGCTAGGCTGCCA–3′
Outside the “common deletion” (detects total mtDNA):
12s rRNA–F: 5′–CTCACCACCTCTTGCTCAG–3′
12s rRNA–B: 5′–GGCTACACCTTGACCTAACG–3′
Cytb–F: 5′–AATCACCACAGGACTATT–3′
Cytb–B: 5′–GTAGGAAGAGGCAGATAA–3′
Nuclear DNA primers:
β–actin Exon 6–F: 5′–GCGCAAGTACTCTGTGTGGA–3′
β–actin Exon 6–B: 5′–CATCGTACTCCTGCT–3′
Search for mitoTALEN target sequences in the nuclear genome
We created a BLAST database containing all the chromosomes from HG19 (GRCh37.p4) except mtDNA (chrM) using CLCBio genomics work bench. This was aligned with complete mitoTALEN target sites, including spacer sequence, to create a list of similar sequences. We screened each list using a motif search algorithm to count identical occurrences of individual mitoTALEN monomer recognition sites.
Enzyme Activities
For spectrophotometric analysis, we subjected 2.5 × 107 exponentially growing cells to centrifugation and washed them once with PBS. The pellets were resuspended in 1 ml of 10 mM HEPES buffer pH 7.4, 0.5 mM EDTA, 0.5 mM EGTA and 250 mM Sucrose, and then sonicated twice in ice–cold water for 10 seconds with 30 seconds intervals. To eliminate cellular debris, samples were centrifuged at 500 × g and the supernatant was used as reaction sample for spectrometric analysis as previously described 26. Complex I (NADH CoQ oxidoreductase) reaction was started with coenzyme Q1 (100 μM) and inhibited with rotenone (10 μM). Citrate synthase reaction was started with Oxaloacetate (0.5 mM). Assay results were normalized to protein concentration obtained by the Bradford method, and subsequently Complex I activity was normalized to Citrate synthase activity. Assays were performed in a BioTek Synergy H1 hybrid plate reader.
Supplementary Material
Acknowledgments
This work is supported by the US National Institutes of Health Grants 5R01EY010804, 1R01AG036871, 1R01NS079965, the Muscular Dystrophy Association and The JDM Fund. The authors declare no competing financial interests. We are grateful to Dr. F. Delacote (Cellectis bioresearch) for expert assistance in designing TALENs, and Dr. G. Zhai for critically reading the manuscript and Dr. A. Pickrell for scientific suggestions.
Footnotes
Author Contributions
S.R.B. and S.L.W. designed and conducted most experiments and assisted in writing the manuscript; M.P. assisted in the quantitation of mtDNA deletions and assisted in writing the manuscript; S.P. conducted the complex I activity experiments and assisted in writing the manuscript; C.T.M. designed and supervised the project and wrote the manuscript.
References
- 1.Schon EA, DiMauro S, Hirano M. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vafai SB, Mootha VK. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 4.Bacman SR, Williams SL, Garcia S, Moraes CT. Gene Ther. 2010;17:713–720. doi: 10.1038/gt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comte C, et al. Nucleic Acids Res. 2013;41:418–433. doi: 10.1093/nar/gks965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Proc Natl Acad Sci U S A. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Nat Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 8.Hockemeyer D, et al. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung YH, et al. Nat Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 10.Schon EA, et al. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- 11.Corral-Debrinski M, et al. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 12.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Nat Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 13.Diaz F, et al. Nucleic Acids Res. 2002;30:4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jun AS, Trounce IA, Brown MD, Shoffner JM, Wallace DC. Mol Cell Biol. 1996;16:771–777. doi: 10.1128/mcb.16.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacman SR, Williams SL, Duan D, Moraes CT. Gene Ther. 2012;19:1101–1106. doi: 10.1038/gt.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carling PJ, Cree LM, Chinnery PF. Mitochondrion. 2011;11:686–692. doi: 10.1016/j.mito.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Moraes CT. Trends Genet. 2001;17:199–205. doi: 10.1016/s0168-9525(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 18.Szymczak AL, et al. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 19.Liddell L, Manthey G, Pannunzio N, Bailis A. J Vis Exp. 2011 doi: 10.3791/3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King MP, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 21.Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Hum Mol Genet. 1994;3:13–19. doi: 10.1093/hmg/3.1.13. [DOI] [PubMed] [Google Scholar]
- 22.Bacman SR, Moraes CT. Methods Cell Biol. 2007;80:503–524. doi: 10.1016/S0091-679X(06)80025-7. [DOI] [PubMed] [Google Scholar]
- 23.Jun AS, Brown MD, Wallace DC. Proc Natl Acad Sci U S A. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Proc Natl Acad Sci U S A. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA. Am J Hum Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez B, et al. J Neurochem. 2001;78:1054–1063. doi: 10.1046/j.1471-4159.2001.00487.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.