Abstract
Although small-molecule drug discovery efforts have focused largely on enzyme, receptor, and ion-channel targets, there has been an increase in such activities to search for protein-protein interaction (PPI) disruptors by applying high-throughout screening (HTS)–compatible protein-binding assays. However, a disadvantage of these assays is that many primary hits are frequent hitters regardless of the PPI being investigated. We have used the AlphaScreen technology to screen four different robust PPI assays each against 25,000 compounds. These activities led to the identification of 137 compounds that demonstrated repeated activity in all PPI assays. These compounds were subsequently evaluated in two AlphaScreen counter assays, leading to classification of compounds that either interfered with the AlphaScreen chemistry (60 compounds) or prevented the binding of the protein His-tag moiety to nickel chelate (Ni2+-NTA) beads of the AlphaScreen detection system (77 compounds). To further triage the 137 frequent hitters, we subsequently confirmed by a time-resolved fluorescence resonance energy transfer assay that most of these compounds were only frequent hitters in AlphaScreen assays. A chemoinformatics analysis of the apparent hits provided details of the compounds that can be flagged as frequent hitters of the AlphaScreen technology, and these data have broad applicability for users of these detection technologies.
Keywords: AlphaScreen, protein-protein interaction, assay development, frequent hitter, drug discovery, high-throughput screening
Introduction
Small-molecule drug discovery efforts have historically largely focused on the enzyme, receptor, and ion-channel target classes, although many others have been implicated in disease states.1 The protein-protein interaction (PPI) target class has been somewhat underexplored with respect to identifying small molecules that can disrupt them, especially when considering that they play an important role in many biological processes and are also implicated in many diseases, including cancer, bacterial infections, leukemia, and neurodegenerative disease.2,3 Consequently, the discovery and development of drugs targeting PPIs have become increasingly prominent from a drug discovery perspective. However, the potential for identifying small-molecule PPI disruptors remains largely untapped and thus they remain an attractive proposition from a drug discovery perspective, especially when searching for first-in-class drugs and identifying novel intellectual property. This is particularly poignant when considering that only a handful of small-molecule disruptors that target PPIs have been developed.4–6 However, a significant hurdle for a small-molecule drug discovery program to search for a disruptor of a PPI has been the availability of screening compatible assays and suitable libraries of compounds that can yield hits acting via the desired mechanism. Moreover, a bottleneck to identifying small-molecule disruptors of PPIs is that their contact surfaces are predominantly large and flat and devoid of clefts or pockets.7
The most incisive in vitro strategies to reliably identify and characterize PPI disruptors are low-throughput methods such as isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and microscale thermophoresis (MST).8–10 These assays are extensively employed at the post small-molecule high-throughput screening (HTS) stage, where a selection of apparent hits are evaluated to provide key parameters for their respective targets, such as their dissociation constant (Kd), stoichiometry, and the kinetics of binding (kon and koff). In contrast, in vitro HTS-compatible assays that can be performed in microplate format are employed extensively to identify apparent PPI disruptors, especially when there is insufficient structural information about the target.11 The activities of these apparent hits are subsequently confirmed in suitable assays, and often a large proportion of these are shown to act by nonspecific mechanisms (frequent hitters) and some of those have been reported.12,13 The work of Baell and Holloway13 is of particular interest in this respect as the authors initially identified frequent hitters in 6 AlphaScreen assays and then they ascertained that many also appeared in screens with other technologies. This led them to establish the Pan Assay INterference Substances (PAINS) filters, which could be used to identify problematic compounds from different small-molecule screening campaigns. The impact of these frequent hitters can also depend on the assays that are implemented, especially when looking for PPI disruptors where the problem of promiscuous compounds is confounded as the probability of finding real PPI disruptors is low compared with, for example, enzyme inhibitors.
We have used the outputs of four different and robust PPI AlphaScreen HTS campaigns (25,000 compounds each) to develop filters for identifying AlphaScreen frequent hitters and report them in this article. We analyzed the frequency and type of small molecules that were identified as apparent hits from the HTS campaigns. Our selection criteria highlighted 137 compounds yielding consistent activity regardless of the PPI being investigated. To initially confirm and evaluate these results, we screened the 137 compounds in one of the primary HTS assays (confirmation assay) and the AlphaScreen TruHits assay (counter assay). With this strategy, we identified 77 compounds exclusively acting as frequent hitters in assays employing His-tagged proteins (AlphaScreen-His-FH), with the other 60 compounds acting as frequent hitters without any specificity (AlphaScreen-FH). To characterize these compounds further, we evaluated the activity of the 137 frequent hitters in an additional assay with time-resolved fluorescence resonance energy transfer (TR-FRET) format and confirmed that most of the compounds were only apparent hits in the AlphaScreen assays. Using the chemoinformatics analysis of our screening data, we developed and validated new filters that can be used to flag compounds that have the potential to interfere with AlphaScreen technology.
Materials and Methods
Reagents
Biological reagents
Affinity-tagged proteins used for development of PPI assays were cloned, expressed, and purified at the Institute of Molecular Toxicology and Pharmacology, Helmholtz Zentrum München, Germany. Expression of recombinant proteins in Escherichia coli strain BL21 RIPL were induced when bacterial cultures reached an OD600 = 0.6 to 0.8 using 1 mM isopropyl-b-D-thiogalactopyranoside (IPTG). Fusion proteins were purified using affinity columns. Subsequently, size-exclusion chromatography was performed using an ÄKTA purifier system with a Superdex 75 column (Healthcare, Munich, Germany). The purity of each protein for assay development purposes was >95% as confirmed by Coomassie staining. The His-tagged glutathione-S-transferase (His-GST) protein used in the TR-FRET counter assay was purchased from Upstate Biotechnology (Placid, NY; product no. 12-523).
AlphaScreen reagents
The AlphaScreen detection system (PerkinElmer, Waltham, MA) made use of glutathione donor beads (product number 6765300), Strep-Tactin Alpha donor beads (product number AS106D), streptavidin donor beads (product number 6760002), nickel chelate (Ni2+-NTA) donor beads (product number AS101D), a Histidine (Nickel Chelate) Detection Kit (product number 6760619C), a C-Myc Detection Kit (product number 6760611C), and the TruHits Kit (product number 6760627D).
TR-FRET reagents
The TR-FRET detection system (Cisbio, Codolet, France) made use of anti–GST-XL665 (product number 61GSTXLB) and anti–His-K (product number 61HISKLB).
The 25,000-compound diverse small-molecule library
The diverse small-molecule library used in the HTS campaigns was composed of compounds acquired from three providers—namely, ChemDiv (San Diego, CA; 10,000 compounds), Enamine Ltd. (Princeton, NJ; 10,000 compounds), and ChemBridge (San Diego, CA; 5000 compounds). The following properties were used to select the 25,000 compounds from those that were available from each provider: molecular weight (MW) <600, diverse chemical scaffolds, satisfying Lipinski’s rule of 5,14 and predicted to be soluble in DMSO.15 Subsequent to clustering of the compounds, representatives with the highest solubility according to ALOGPS 2.116 and lowest probability of predicted AMES test mutagenicity were selected.17 In addition, several chemoinformatics filters were used to exclude reactive, unstable, and toxic chemical groups, which are implemented in ToxAlerts.18 The purity of the compounds was >90%, as reported by the providers of the compounds.
Instruments
Plate handling was performed using a Cell::Explorer HTS platform (PerkinElmer) system, Echo 550 (Labcyte, Sunnyvale, CA), Sciclone G3 with a Twister II robotic arm (PerkinElmer), Flexdrop (PerkinElmer), Multidrop (Thermo, Waltham, MA), and Mosquito (TTP Labtech, Cambridge, UK) liquid-handling systems. AlphaScreen and TR-FRET measurements were performed using an EnVision Multilabel Reader (PerkinElmer). Assays were performed in white 384 well polystyrene microplates (Greiner Bio-One, Monroe, NC; product number 784904) or a white 384-well OptiPlate (PerkinElmer; product number 6007290).
Other reagents
All other reagents not listed above (e.g., buffers) were purchased from Sigma-Aldrich (Taufkirchen, Germany) and Roth (Karlsruhe, Germany) and were of the highest quality.
Development of AlphaScreen Assays
Target proteins and their respective tags
The four HTS-compatible PPI assays selected for study are anonymized and implicated in different cellular signaling pathways. The combination of target proteins in each assay was as follows: PROTEIN(1)-GST/PROTEIN(2)-His, PROTEIN(3)-StrepTagII/PROTEIN(4)-His, PROTEIN(5)-His/PROTEIN(6)-Myc, PROTEIN(7)-Biotin/PROTEIN(8)-His.
Development and automation of the AlphaScreen primary assays
To identify the optimal protein concentration for each PPI assay (robust signal with minimal protein concentration), matrix titration experiments were performed in accordance with the manufacturer’s protocol (PerkinElmer). Dilutions of proteins and other reagents were made in an assay buffer containing 1× phosphate-buffered saline (PBS; pH 7.4), 0.5% bovine serum albumin (BSA), and 0.01% Tween-20. The reproducibility, signal stability, and robustness (Z′) were determined for each PPI assay to ensure they were HTS compatible. Prior to performing the HTS campaigns, the PPI assays were adapted to automation using a liquid handler and a compound transfer station (see Instruments). The final assay volume was 60 µL with AlphaScreen bead concentrations 3 to 5 µg/mL. As the diverse small-molecule library to be screened was stored in 100% v/v DMSO, it was possible to obtain a screening concentration of 10 µM for each test compounds with 1% v/v DMSO. In all screening campaigns, the negative control was based on the use of PPI binding mutant controls (single-point mutation) that would prevent the PPI from forming, and the positive control contained 1% v/v DMSO only. The quality and robustness of the assay, represented as Z′, were calculated.11
Execution of the AlphaScreen high-throughput primary screening campaigns against the 25,000 diverse small-molecule library
Each HTS campaign was performed in a 384-well microplate format (test compound concentration = 10 µM, n = 1) with a final volume of 60 µL per well as follows: (1) dispensation of 30 µL of 2× concentrated PROTEIN(1), PROTEIN(3), PROTEIN(6), or PROTEIN(8) into a microplate using a robotic liquid handler; (2) transfer of 0.6 µL of test compounds in DMSO (1-mM stock) into each well using a compound transfer station with a nanoliter head, yielding a final assay concentration of each compound of 10 µM; (3) dispensation of 10 µL of 6× concentrated PROTEIN(2), PROTEIN(4), PROTEIN(5), or PROTEIN(7) into the respective assay microplates using a robotic liquid handler; (4) incubation of the assay microplates for 1 h at room temperature; (5) addition of each 10 µL of the appropriate AlphaScreen beads (3–5 µg/mL) followed by a further incubation for 1 h at room temperature in the dark due to the photosensitivity of the beads; and (6) reading of the assay microplates using laser excitation at 680 nm, with emission detected at 520 to 620 nm using the EnVision 2102 Multilabel Reader (PerkinElmer). Each assay microplate included the following controls: 16 wells containing a PPI binding mutant protein as negative control and 16 wells containing 1% v/v DMSO as the positive control. All data were processed using Excel (Microsoft Corp., Redmond, WA) and visualized using Prism (GraphPad Software, La Jolla, CA).
AlphaScreen TruHits counter assay protocols
The TruHits kit contains streptavidin donor beads and biotinylated acceptor beads, which bind to each other without the addition of any further reagents. In total, 30 µL TruHits kit bead premix was dispensed into each well of a microplate and incubated for 30 min at room temperature. Subsequent to this, test compounds (final concentration of 10 µM) were transferred and the mixture was incubated for 1 h at room temperature. The assay microplates were read using the EnVision 2102 Multilabel Reader as described above (PerkinElmer). All data were processed using Excel (Microsoft Corp.) and visualized using Prism (GraphPad Software).
Development of TR-FRET Counter assays
TR-FRET counter assay development
The concentration of His-GST protein used in these experiments was determined from matrix titration experiments where linear TR-FRET signal was observed. The final concentrations of the components of the assay made use of 2 nM His-GST protein, 4.6 nM MAb anti–GST-XL665, and 7 nM MAb anti–His-K (containing the europium chelate). The sensitivity of the assay toward DMSO was determined up to 10% v/v DMSO.
The TR-FRET counter assay was performed in a 384-well microplate format as follows: (1) dispensation of 100 nL of test compounds in 100% v/v DMSO into microplates (final concentration 10 µM); (2) a single 10-µL per-well addition of all assay components containing 2 nM His-GST protein, 4.6 nM MAb anti–GST-XL665, and 7 nM MAb anti–His-K in 25 mM Tris/HCl (pH 7.5), 150 mM NaCl, 0.2 M KF (potassium fluoride), and 0.1% BSA into microplates; (3) the assay microplates incubated for 1 h at 25 °C; and (4) fluorescence at 615 nm and 665 nm measured simultaneously using an EnVision 2102 Multilabel Reader. All data were processed using Excel (Microsoft Corp.) and visualized using Prism (GraphPad Software). Assay buffer lacking the His-GST protein (16 wells) was used as the negative control, and 1% v/v DMSO was used as the positive control.
Analysis of the Output of the High-Throughput Primary Screening Campaigns against the 25,000 Diverse Small-Molecule Library
Primary HTS hit identification
The primary screening data from the four PPI AlphaScreen HTS campaigns (final test compound concentration 10 µM, n = 1) were processed as follows: (1) the Z′, signal window (SW), and coefficient of variation (CV) were calculated and then compared with the minimum pass criteria (Z′ >0.5, SW >2, CV < 20%); (2) a heat-map and a scatterplot for each assay microplate were generated to reveal any plate effects, including drift in assay signal, edge effects, and other systematic sources of variability; and (3) the primary hits for each of the PPI assays were classified as compounds that led to >50% disruption of a PPI complex (i.e., the normalized assay signal decreased >50%). All HTS data were processed using Excel (Microsoft Corp.) and visualized using Prism (GraphPad Software).
Primary HTS hit evaluation
The primary hits were confirmed in one of the primary PPI assays (i.e., the PROTEIN(7)-Biotin/PROTEIN(8)-His also being the confirmation assay) and the AlphaScreen TruHits counter assay (see above for protocols). The results of these screening activities were correlated in a variety of ways to identify those compounds that appeared to specifically interfere with the AlphaScreen chemistry and those that appeared to interfere with the interaction of the His-tag and the nickel chelate (Ni2+-NTA) donor bead. Further annotation of the primary hits was performed using a TR-FRET counter assay that made use of the His-GST protein to provide information regarding the general interference of compounds with these two commonly employed assays in drug discovery.
Chemoinformatics Analysis
Published frequent hitters filters
Previously developed filters for the identification of frequent hitters were uploaded using SMiles ARbitrary Target Specification (SMARTS) language19 and are available at ToxAlerts (http://ochem.eu/alerts).18 These include 178 promiscuous compounds filters (PCs)20 and 480 PAINS.13 For the PCs, the original SMARTS were available. The PAINS filters,13 which were provided as SYBYL line notations, were encoded into SMARTS.21
Rules to identify frequent hitters
The SMARTS language was also used for the development of new filters for the analysis of the experimental data reported herein. Each filter incorporated a single SMARTS string, and construction of substructural patterns involved extraction of common molecular scaffolds and comparative analysis of substituents in the scaffold nodes for active and inactive compounds as well as grouping chemicals based on structural similarity and proposed mode of action. Scaffold Hunter,22 ISIDA,23 and Silicos-IT scaffolds24 were analyzed using the SetCompare utility of the online chemical modeling environment (OCHEM) platform,21 and overrepresented structural elements in the set of promiscuous compounds were identified. The acceptability of each filter was assessed using the enrichment value (EV) according to Baell and Holloway.13 EV was defined as the percentage of compounds that were active in two or more PPI assays relative to the number of “clean” compounds that were inactive in four of the PPI assays using equation (1), where HITSi is number of compounds identified by the filter, which are active in i assays.
The EV threshold of 30%, as reported in a previous publication,13 was used for an acceptance of each filter. Filters with EV <30% were either refined or eliminated. The latter was done if we were able to extend (generalize) other filters in such a way that they covered the filter that was eliminated. The refinement of a filter included analysis of valence/connectivity of scaffold atoms, valence/connectivity of atoms adjacent to scaffold, a size and an atomic composition of aromatic fragments (if they existed), and so on. The refinement was performed to identify new restrictions, which could be incorporated in the filter. The introduction of new restrictions made the filters more specific and increased their EV values. After the values of EV were >30% threshold, additional restrictions to a filter were added only if some structural differences were obvious and easily recognizable and/or they were able to contribute to a mechanistic interpretation.
The PubChem BioAssay database25 was used to validate the developed filters. A query line <alphascreen assay AND (pcassay_protein_target[filt]) AND (screening[filt]) AND 1000:1000000[Total Sid Count]> retrieved five primary screening assays implemented using AlphaScreen technology. Three of these assays used His-tagged proteins and thus could be used for the assessment of quality of developed AlphaScreen-His-FH filters. A description of the assays can be found in Supplemental Table S1. Data from the assays were compiled, and only those substances (in total 208,000), which were tested in all assays, were kept for further consideration. There were also 2230 compounds, which coincided between the PubChem and Helmholtz Zentrum München für Gesundheit und Umwelt (HMGU) screening collection. These compounds were excluded from the PubChem data set, and the final collection included 205,974 substances, which were used to validate the results of the developed filters.
Results and Discussion
Overall Strategy in Identifying Frequent Hitters in AlphaScreen HTS Hit Populations
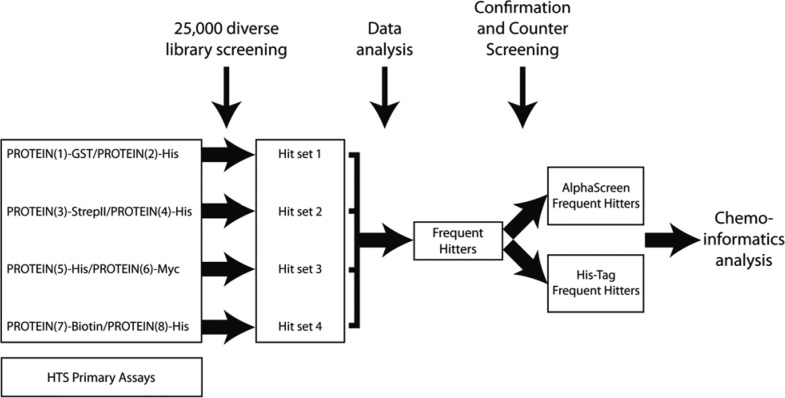
HTS is a popular method for discovering small molecules as starting points for drug discovery purposes.11 A major issue with this methodology is that many of the primary hits from HTS campaigns are artifacts and not direct modulators of the target being investigated. The overall strategy reported herein was to identify frequent hitters from AlphaScreen HTS hit populations that made use of PPI assays and to provide a detailed chemoinformatics analysis of the results obtained from the primary HTS campaigns and counter assays. An overview of the workflow that has been implemented herein is shown in Figure 1. This strategy included six steps: (1) development of primary assays, (2) HTS against 25,000 compounds for each assay, (3) data analysis and identification of frequent hitters that reduced the AlphaScreen signal in all assays irrespective of the PPI being investigated, (4) confirmation and counter screening of the frequent hitters with two AlphaScreen assays (i.e., the PROTEIN(7)-Biotin/PROTEIN(8)-His and the TruHits kit), (5) counter screening with a TR-FRET binding assay, and (6) chemoinformatics analysis.
Figure 1.
Pictorial overview of the workflow implemented in the study. Data for four independent AlphaScreen high-throughput screening primary assays were selected for analysis and identification of AlphaScreen-FH as well as AlphaScreen-His-FH. The specific anonymized protein-protein interaction (PPI) pairs (PROTEIN(1)-GST/PROTEIN(2)-His, PROTEIN(3)-StrepTagII/PROTEIN(4)-His, PROTEIN(5)-His/PROTEIN(6)-Myc, PROTEIN(7)-Biotin/PROTEIN(8)-His) were screened against a diverse library of 25,000 compounds (each PPI pair comprised one His-tagged protein). All small molecules that led to a decrease in the AlphaScreen signal >50% were classified as hits. Combinatorial analysis of all four screening campaigns allowed the identification of frequent hitters. With appropriate counter screening assays, frequent hitters were differentiated by AlphaScreen-FH and AlphaScreen-His-FH. These data formed the basis for new substructure filter tools to annotate promiscuous screening compounds within screening libraries.
AlphaScreen Primary HTS assays
Criteria for AlphaScreen primary assay development
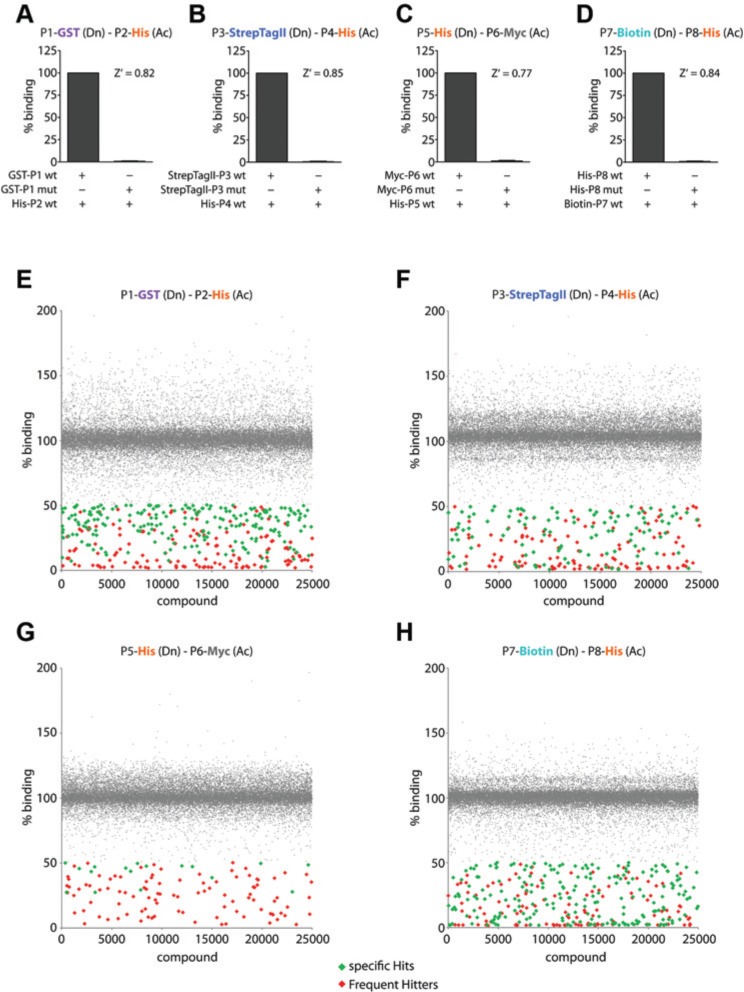
Many HTS assay technologies tend to provide primary hits that frequently appear independent of the target being screened, and these are termed frequent hitters. To identify these frequent hitters from the PPI HTS campaigns that were performed in the study reported herein, we analyzed the primary hit lists of four independent AlphaScreen HTS campaigns (Fig. 2). The principle of the AlphaScreen assay is illustrated in Supplemental Figure S1A. The four primary HTS-compatible assays selected for this study are anonymized and made use of PPI assays from unrelated signaling pathway targets. All PPI assays were independently optimized using the respective recombinant proteins. The proteins were expressed in E. coli and purified with purity >95% (data not shown) and contained various tags (His, GST, StrepTagII, Myc, or biotin tag) to facilitate their purification and/or detection in AlphaScreen assays. The specific PPI protein pairs were PROTEIN(1)-GST/PROTEIN(2)-His, PROTEIN(3)-StrepTagII/PROTEIN(4)-His, PROTEIN(5)-His/PROTEIN(6)-Myc, and PROTEIN(7)-Biotin/PROTEIN(8)-His. A common feature of all PPI pairs was the presence of the His-tag on one protein of the pair. To determine the optimal protein concentration for the AlphaScreen PPI assays, matrix titrations of both binding partners for each PPI pair were performed (data not shown). Using this approach, it was possible to develop robust HTS-compatible assays that made use of protein concentrations in the low nanomolar range. As no known small-molecule disruptors of the PPIs being investigated were available, it was possible to use binding mutant controls (single-point mutants) of one protein in the PPI as an alternative. Each point mutation was selected on the basis of structural data and was conclusively shown to prevent the PPI from forming (Fig. 2A–D) as well as in other in vitro binding assays (data not shown). All assays and HTS campaigns exceeded the minimum criteria (Z′ >0.5, SW >2, and CV <20%). In addition, the assays met the general requirements for HTS compatibility such as acceptable DMSO tolerance and signal stability (see Fig. 2). In all cases, the same assay buffer was used containing 1× PBS (pH 7.4), 0.5% BSA, and 0.01% Tween-20 as these components have been shown to reduce compound aggregation and minimize nonspecific inhibition.12
Figure 2.
The output of four AlphaScreen primary high-throughput screening (HTS) assays selected for this study. (A–D) The four protein-protein interaction (PPI) pairs produced excellent and robust signals (Z′ > 0.5). The first bar in each panel shows the AlphaScreen signal of the wild-type (wt) protein pairs. The second bar depicts the reduced signal coming from a protein-binding mutant (mut). (E–H) Scatterplots of the four HTS campaigns. The PPI pairs were screened against a diverse library of 25,000 compounds. Inactive compounds are marked in gray. Comparison of all four screening campaigns allowed the distinction of specific Hits (green) and frequent hitters (red). Dn, donor bead; Ac, acceptor bead; P, PROTEIN.
Output of the primary HTS campaigns against the 25,000-compound diverse library
A diverse set of 25,000 small molecules was screened in 384-well format with a final compound concentration of 10 µM (n = 1). The classification of a primary hit compound was based on compounds that yielded a decrease in the normalized assay signal >50% (Fig. 2E–H). However, this population would include false positives that arise for a multitude of reasons from optical interference,26 compound insolubility, aggregation,12 and compounds reported in the literature that are known to be problematic.13 More specifically, in the case of the Alpha–Screen technology, compound-mediated nonspecific effects include interference by way of singlet oxygen quenching, color quenching, autofluorescence, and disruption of the interaction between the tag of the protein and binding site of the detection system.
Compounds were classified as frequent hitters when they yielded a median decrease in the normalized signal >50% in all four PPI assays. Using this evaluation method, 137 compounds were classified as frequent hitters. Some compounds generally interfered with the AlphaScreen technology (AlphaScreen-FH), while others specifically prevented the binding of the His-tag of the protein in the PPI complex with the nickel chelate (Ni2+-NTA) of the beads (AlphaScreen-His-FH). The possibility that frequent hitters interfered with the other protein tags (GST, Myc, biotin, or StrepTagII) could be excluded as these were different in each of the four assays.
Post-HTS Confirmation and Counter assays
Output of the AlphaScreen confirmation and counter assays
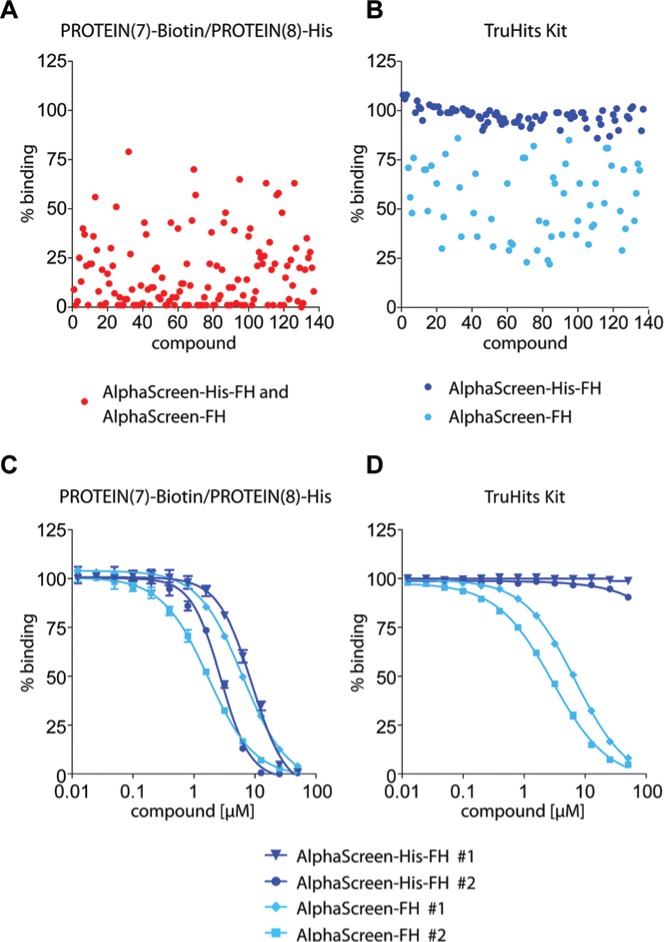
To discriminate between AlphaScreen-FH and AlphaScreen-His-FH, we selected the 137 frequent hitters and confirmed their activity in the PROTEIN(7)-Biotin/PROTEIN(8)-His Primary assay (n = 3) and the AlphaScreen TruHits counter assay (n = 3). The TruHits counter assay contained streptavidin donor and biotinylated acceptor beads and was designed to identify false-positive compounds that act by way of singlet oxygen quenching, redox reaction interference, or any general reaction with proteins.
To determine compound concentration-dependent effects, the primary hits were screened at 10 µM and 20 µM. In addition, 10 control compounds were included that were not classified as frequent hitters in the four PPI primary HTS campaigns, and these did not yield an effect in either the confirmation or counter assays (data not shown). All 137 frequent hits clearly showed AlphaScreen signal decrease in the PROTEIN(7)-Biotin/PROTEIN(8)-His confirmation assay (Fig. 3A). Moreover, 60 of the 137 compounds reduced AlphaScreen signal in the TruHits counter assay, indicating that these compounds are general AlphaScreen-FH (Fig. 3B). The remaining 77 compounds did not affect the TruHits counter assay (Fig. 3B). As these compounds were active in the PROTEIN(7)-Biotin/PROTEIN(8)-His assay but not in the TruHits assay, the data demonstrate that these compounds are histidine mimetics and/or nickel chelators and are referred to as AlphaScreen-His-FH.
Figure 3.
The output of the AlphaScreen confirmation and counter assays. (A) In total, 137 compounds were cherry-picked and reconfirmed as frequent hitters in the primary PROTEIN(7)-Biotin/PROTEIN(8)-His assay (red). Control treatment was set to 100%. (B) The identical compounds were subsequently investigated for their activity against the AlphaScreen technology using the TruHits counter assay. Combining the results from A and B enabled the classification of frequent hitters into AlphaScreen-FH (light blue) and AlphaScreen-His-FH (dark blue). (C, D) Two compounds from each of the classified groups were selected and retested in dose-response experiments (13-point dose-response, n = 3) using the assay shown in A and B.
The confirmation and counter assays were able to discriminate between the two types of frequent hitters—namely, general frequent hitters of the AlphaScreen technology and His-Tag/Ni2+-NTA frequent hitters in AlphaScreen assays. The streptavidin-biotin binding in the TruHits assay is one of the strongest interactions in biological systems and in theory is impossible to be disrupted by any compound.27 Thus, the TruHits assay reliably identifies frequent hitters of the AlphaScreen technology. However, this assay lacks the His-tag and is therefore not suitable for identifying His-Tag/Ni2+-NTA frequent hitters. For this reason, we used the PROTEIN(7)-Biotin/PROTEIN(8)-His assay to analyze His-Tag/Ni2+-NTA frequent hitters, because of the following reasons: (1) this assay contains the biotin-streptavidin binding, which is one of the strongest interactions; (2) interference of the compounds with the PROTEIN(7)-Biotin/PROTEIN(8)-His interaction can be excluded as the frequent hitters were also active in the other three PPIs from different pathways; and (3) consequently, all active compounds in this particular assay target the His-Tag/Ni2+-NTA binding or the AlphaScreen technology. Thus, a compound interfering with general AlphaScreen technology would generate a signal reduction in both assays, whereas histidine mimetics or nickel chelators should just appear as hits in the PROTEIN(7)-Biotin/PROTEIN(8)-His assay.
In addition, we exemplary tested two AlphaScreen-FH and two AlphaScreen-His-FH in 13-point dose-dependent titration experiments. The chemical structures of the four selected compounds are shown in Supplemental Figure S2. Again, we could see that the two AlphaScreen-FH were dose-dependently active in both AlphaScreen counter assays, while the two AlphaScreen-His-FH only appeared as hits in the PROTEIN(7)-Biotin/PROTEIN(8)-His assay (Fig. 3C, D).
In summary, of the 25,000 compounds available in our diversity library, we identified 60 frequent hitters of the AlphaScreen technology and 77 compounds interfering with the His-Tag/Ni2+-NTA binding used for protein detection.
Output of the TR-FRET counter assay
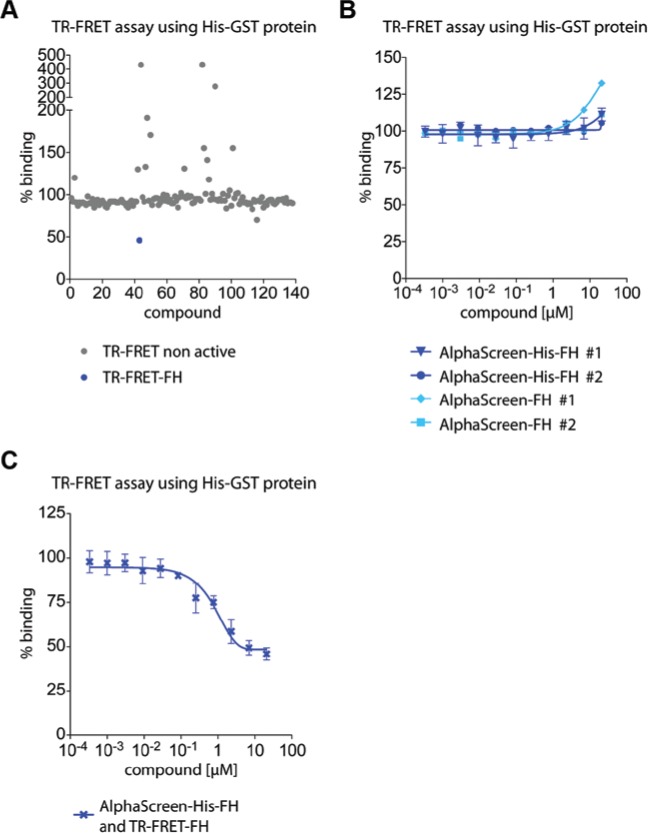
The 137 compounds identified as frequent hitters in the AlphaScreen Primary HTS screening campaigns were evaluated in a TR-FRET counter assay. The principle of the TR-FRET assay is shown in Supplemental Figure S1B. All compounds were screened using a commercial GST-His protein (n = 3) with the detection system being a MAb anti–GST-XL665 (acceptor) and a MAb anti–His-K (donor). As the GST-His protein was a single protein, any decrease in the TR-FRET signal in the presence of test compound would be due to the acceptor and donor moieties being either physically separated or more likely in the case of this assay, due to compound-mediated optical interference. It was possible to distinguish whether compounds were autofluorescent or quenching by monitoring the fluorescence intensities at 665 nm (acceptor) and at 615 nm (donor). A real hit compound would be expected to yield a reduction in 665-nm emission with the 615-nm emission being unaffected. In the case of the 137 compounds that were screened at a concentration of 10 µM, only 4 compounds were identified as being autofluorescent (2 compounds being autofluorescent at both 665 nm and 615 nm) and only 10 compounds being identified as quenchers at both 665 nm and 615 nm. One compound was identified as a disruptor of the PPI/detection system complex with a 54% ± 3% effect, which was also an Alpha–Screen-His-FH (Fig. 4A). A key difference of the AlphaScreen and TR-FRET assays was that the former made use of nickel chelate (Ni2+-NTA) beads to bind the His-tag of the proteins, whereas in the latter, the His-tag was bound to the detection system via an antibody, and this was less prone to compound-mediated interference. A dose-response with five compounds was also performed in TR-FRET assays (Fig. 4B, C). Only the TR-FRET active compound from Figure 4A showed a dose-dependent effect (Fig. 4C).
Figure 4.
Time-resolved fluorescence resonance energy transfer (TR-FRET) counter assay. (A) The TR-FRET counter assay was performed at optimal concentrations of all reagents. All 137 compounds from Figure 3 were analyzed for interference with the TR-FRET technology. A linear fusion protein (GST-His) produced a robust TR-FRET signal. Control treatment was set to 100%. One compound also causing a reduction of the TR-FRET signal is marked in blue and is classified as a TR-FRET frequent hitter (TR-FRET-FH). (B) The four compounds tested in AlphaScreen dose-response (Fig. 3C, D) were also evaluated in TR-FRET dose-response assays. (C) The TR-FRET-FH identified in A exhibited a sigmoidal dose-dependent reduction of TR-FRET signal.
Chemoinformatics Analysis of Frequent Hitters
The analysis of frequent hitters was performed separately for AlphaScreen-His-FH (77) and AlphaScreen-FH (60). Screening of both sets using PC filters20 identified only two hits. This result was expected since PC filters were not specifically developed for the AlphaScreen technology. The analysis of our data using PAINS filters13 correctly identified the majority (52 of 60) of the AlphaScreen-FHs. However, only five AlphaScreen-His-FHs were covered with the PAINS filters (Suppl. Table S2). The high percentage of detected AlphaScreen-FHs is expected considering that for the development of PAINS filters, results for six AlphaScreen assays were used. Lower sensitivity of the PAINS filters to AlphaScreen-His-FHs is also predicted because only 2 of their assays incorporated His-tagged proteins and the nickel chelate (Ni2+-NTA) donor beads. Even if a compound was detected as active in those two assays, it would not comply with a definition of frequent hitter proposed by Baell and Holloway13 and therefore was not considered for the filter development.
Development of filters to identify AlphaScreen-His-FH
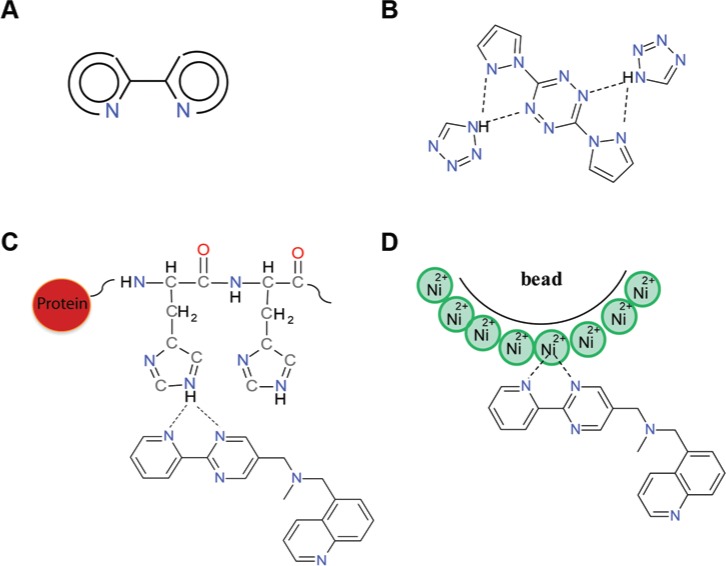
Preliminary structural analysis of the AlphaScreen-His-FHs showed that almost a quarter of them (18 compounds) comprised a common molecular fragment depicted in Figure 5A. Such a molecular moiety is a known synthon in supramolecular chemistry due to its ability to form bifurcated hydrogen bonds with XH-donating groups (where X = N, O)28,29 and coordination complexes with metal ions.30 Figure 5B shows an example of molecular trimers, detected in crystals and formed by compounds incorporating the fragment with NH-donating azoles. Taking the good NH-donor properties of imidazole rings into account, it could be assumed that similar bifurcated hydrogen-bonded complexes are formed between histidine residues of tagged proteins and the compounds (Fig. 5C). These complexes would prevent immobilization of the proteins on the bead surfaces. The same compounds may also act as chelating agents and adsorb on the bead surface, thus also preventing binding of the proteins to acceptor beads (Fig. 5D). In both cases, fluorescence would decrease and contribute to the identification of the false-positive hits.
Figure 5.
Possible mechanism of action of 18 AlphaScreen-His-FH sharing the same common molecular fragment. (A) The common molecular fragment for 18 AlphaScreen-His-FH. (B) Examples of molecular trimers formed by the fragment and NH-donating azoles according to Tetko et al.15 and SMARTS.19 (C) Hypothetical hydrogen bond interaction of the fragment with the PROTEIN-His-tag. (D) Hypothetical chelating complexes formed by the fragment with Ni2+-ions covering an acceptor bead surface (dashed lines show interactions).
Structural analysis of the remaining 59 AlphaScreen-His-FHs showed that most are known chelating agents such as 8-hydroxyquinolines,31 picolylamines,32,33 and pyridines.28,30,34 Part of them incorporates molecular moieties described in the Chelator Fragment Library (CFL-1), which was developed for the identification of compounds that would bind zinc(II) metalloproteins.35 The prevalence of chelating fragments in the AlphaScreen-His-FHs may be due to their coordination properties. Therefore, our strategy for filter development leading to the identification of potential chelating sites was successful as it led 19 of such filters to be established to identify AlphaScreen-HIS-FHs (filters 1–19 in Suppl. Table S3). It was also possible that some molecular fragments from the CFL-1 library, which were not present or were underrepresented in our screening library, could be important for the identification of AlphaScreen-His-FHs due to the same mechanism of action, and therefore, we also encoded structural fragments from CFL-1 as SMARTS patterns.
Filters for identification of AlphaScreen chemistry frequent hitters
In total, 60 frequent hitters were confirmed to interfere by way of AlphaScreen chemistry. It is important to mention that although these compounds were identified using different PPI assays, they will also appear as frequent hitters in other assays using the AlphaScreen technology (e.g., phosphatase assay, kinase assay, etc.). The PAINS filters did not recognize eight compounds, thus indicating their high coverage. These eight compounds predominantly comprised fused aromatic systems or quinone moieties in their structures and might quench excitation/emitted radiation or singlet oxygen. Since these compounds were structurally diverse, we developed an additional six filters to recognize them (see filters 20–25 in Suppl. Table S3).
Validation of filters
The results reported in Table 1 indicate that the filters developed in the work presented herein have high EV values and thus successfully identified frequent hitters in both HMGU and PubChem screening data sets. New filters developed for AlphaScreen chemistry compounds were rather specific, and only 23 compounds were flagged. This number included 9 compounds classified as frequent hitters (8 compounds used to develop filters and 1 compound active in three assays). The AlphaScreen-His filters flagged 312 compounds, including 92 classified as frequent hitters.
Table 1.
Numbers of Compounds and Enrichment Values Calculated with Different Filters Applied to Screen HMGU and PubChem Data Sets.
| HMGU |
PubChem |
|||||||
|---|---|---|---|---|---|---|---|---|
| FH Type | Reference | No. of Filters | Active in 2–4 Assays (FH) | Nonactive in All Assays | Enrichment Value, % | Active in 1–3a Assays (FH) | Nonactive in All Assays | Enrichment Value, % |
| PC | 20 | 178 | 6 | 286 | 2.1 | 349 | 13,114 | 2.6 |
| PAINS | 13 | 480 | 74 | 807 | 9.2 | 1217 | 14,270 | 8.5 |
| CFL-1 | 35 | 56 | 12 | 183 | 6.6 | 452 | 4891 | 9.2 |
| AlphaScreen-His-FH | This study | 19 | 92 | 220 | 42 | 534 | 1475 | 36 |
| AlphaScreen-FH | This study | 6 | 9 | 14 | 64 | 0 | 8 | NA |
FH, frequent hitter; NA, not available.
Due to the limited number of assays (n = 3) in PubChem, we also counted compounds active in only one assay as frequent hitters for the enrichment value (EV) calculation. The HMGU (Helmholtz Zentrum München für Gesundheit und Umwelt) data were the same four assays used for the development of the filters.
We also assessed EVs of other sets of filters. The PC filters had the lowest EV values for both collections, thus confirming that they are not applicable to the used screening technology. CFL-1 filters had 3- to 5-fold higher EV enrichments relative to PC filters for both data sets. These filters had EV even higher than that of PAINS filters for the PubChem data set. The high EV values calculated for these filters confirmed the importance of the chelating properties of compounds for their promiscuity when using AlphaScreen technology. The EVs of PAINS filters for both HMGU and PubChem data sets were below 30% reported in the original study, and the lower values were presumably due to the differences in the composition of both sets used in this study compared with the data sets analyzed by Baell and Holloway.13
To further validate filters, we analyzed 6239 compounds from DrugBank (http://www.drugbank.ca). The same set was used in our previous study.18 The PAINS and newly proposed filters flagged 281 (4.5%) and 49 (0.8%) compounds, respectively. This result indicates that both types of filters are specific and there is no danger of overfiltering of drug-like molecules.
The identification and exclusion of frequent hitters is important to avoid erroneous interpretation of results of HTS experiments. However, most HTS experiments are rather heterogeneous in that the different technologies may contribute different frequent hitter populations. The use of inappropriate filters may not allow correct identification of promiscuous compounds, as it was exemplified by a failure of filters developed by Pearce et al.20 PAINS filters,13 proposed for AlphaScreen technology, were able to successfully identify about half of 137 frequent hitters. The analysis of the remaining frequent hitters allowed us to establish a clear hypothesis of their mechanism of action (i.e., histidine mimetics or nickel chelators). Following analysis of these frequent hitters, we developed new filters, which are specific for the use of His-tag proteins in the AlphaScreen technology. Of note, the classification of these compounds into the two different subtypes was only possible by conducting appropriate counter assays. Although chemoinformatics filters are very useful for prediction of frequent hitters, this will not completely replace in vitro counter screening to experimentally confirm the frequent hitters.
The newly developed filters as well as those from previous studies were made publicly available at the OCHEM Web site (http://ochem.eu/alerts) and can be freely accessed by Web users to interpret results of their HTS screening campaigns.
Acknowledgments
We thank Scarlett Dornauer and Stefanie Brandner for excellent technical assistance.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This project was partially supported by FP7 MC ITN project “Environmental Chemo-informatics” (ECO), grant 238701, and GO-Bio BMBF project number 0315647 “iPRIOR—On-Line Platform for Toxicity Prediction and Prioritization of Chemical Compounds for Drug Discovery and REACH.”
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
References
- 1. Overington J. P., Al-Lazikani B., Hopkins A. L. How Many Drug Targets Are There? Nat. Rev. Drug Discov. 2006, 5, 993–996. [DOI] [PubMed] [Google Scholar]
- 2. Ryan D. P., Matthews J. M. Protein-Protein Interactions in Human Disease. Curr. Opin. Struct. Biol. 2005, 15, 441–446. [DOI] [PubMed] [Google Scholar]
- 3. Sperandio O., Reynes C. H., Camproux A. C., et al. Rationalizing the Chemical Space of Protein-Protein Interaction Inhibitors. Drug Discov. Today 2010, 15, 220–229. [DOI] [PubMed] [Google Scholar]
- 4. Whitty A., Kumaravel G. Between a Rock and a Hard Place? Nat. Chem. Biol. 2006, 2, 112–118. [DOI] [PubMed] [Google Scholar]
- 5. Cochran A. G. Antagonists of Protein-Protein Interactions. Chem. Biol. 2000, 7, R85–R94. [DOI] [PubMed] [Google Scholar]
- 6. Zinzalla G., Thurston D. E. Targeting Protein-Protein Interactions for therapeutic Intervention: A Challenge for the Future. Fut. Med. Chem. 2009, 1, 65–93. [DOI] [PubMed] [Google Scholar]
- 7. Wells J. A., McClendon C. L. Reaching for High-Hanging Fruit in Drug Discovery at Protein-Protein Interfaces. Nature 2007, 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 8. Jerabek-Willemsen M., Wienken C. J., Braun D., et al. Molecular interaction Studies Using Microscale Thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willander M., Al-Hilli S. Analysis of Biomolecules Using Surface Plasmons. Methods Mol. Biol. 2009, 544, 201–229. [DOI] [PubMed] [Google Scholar]
- 10. Ghai R., Falconer R. J., Collins B. M. Applications of Isothermal Titration Calorimetry in Pure and Applied Research—Survey of the Literature from 2010. J. Mol. Recogn. 2012, 25, 32–52. [DOI] [PubMed] [Google Scholar]
- 11. Macarron R., Banks M. N., Bojanic D., et al. Impact of High-Throughput Screening in Biomedical Research. Nat. Rev. Drug Discov. 2011, 10, 188–195. [DOI] [PubMed] [Google Scholar]
- 12. Coan K. E., Shoichet B. K. Stoichiometry and Physical Chemistry of Promiscuous Aggregate-Based Inhibitors. J. Am. Chem. Soc. 2008, 130, 9606–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baell J. B., Holloway G. A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- 14. Lipinski C. A., Lombardo F., Dominy B. W., et al. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [DOI] [PubMed] [Google Scholar]
- 15. Tetko I. V., Novotarskyi S., Sushko I., et al. Development of Dimethyl Sulfoxide Solubility Models Using 163 000 Molecules: Using a Domain Applicability Metric to Select More Reliable Predictions. J. Chem. Inf. Model. 2013, 53, 1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tetko I. V., Tanchuk V. Y., Kasheva T. N., et al. Estimation of Aqueous Solubility of Chemical Compounds Using E-State Indices. J. Chem. Inf. Comput. Sci. 2001, 41, 1488–1493. [DOI] [PubMed] [Google Scholar]
- 17. Sushko I., Novotarskyi S., Körner R., et al. Applicability Domain for In Silico Models to Achieve Accuracy of Experimental Measurements. J. Chemometrics 2010, 24, 202–208. [Google Scholar]
- 18. Sushko I., Salmina E., Potemkin V. A., et al. ToxAlerts: A Web Server of Structural Alerts for Toxic Chemicals and Compounds with Potential Adverse Reactions. J. Chem. Inf. Model. 2012, 52, 2310–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. SMARTS—A Language for Describing Molecular Patterns. http://www.daylight.com/dayhtml/doc/theory/theory.smarts.html
- 20. Pearce B. C., Sofia M. J., Good A. C., et al. An Empirical Process for the Design of High-Throughput Screening Deck Filters. J. Chem. Inf. Model. 2006, 46, 1060–1068. [DOI] [PubMed] [Google Scholar]
- 21. Sushko I., Novotarskyi S., Korner R., et al. Online Chemical Modeling Environment (OCHEM): Web Platform for Data Storage, Model Development and Publishing of Chemical Information. J. Comput. Aided. Mol. Des. 2011, 25, 533–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wetzel S., Klein K., Renner S., et al. Interactive Exploration of Chemical Space with Scaffold Hunter. Nat. Chem. Biol. 2009, 5, 581–583. [DOI] [PubMed] [Google Scholar]
- 23. Varnek A., Fourches D., Horvath D., et al. ISIDA—Platform for Virtual Screening Based on Fragment and Pharmacophoric Descriptors. Cur. Comp. Aid. Drug Des. 2008, 4, 191–198. [Google Scholar]
- 24. Strip-it. http://silicos-it.com/software/strip-it/1.0.2/strip-it.html Accessed July 8, 2013.
- 25. Wang Y., Xiao J., Suzek T. O., et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gul S., Gribbon P. Exemplification of the Challenges Associated with Utilising Fluorescence Intensity Based Assays in Discovery. Exp. Opin. Drug Discov. 2010, 5, 681–690. [DOI] [PubMed] [Google Scholar]
- 27. Green N. M. Avidin and Streptavidin. Methods Enzymol. 1990, 184, 51–67. [DOI] [PubMed] [Google Scholar]
- 28. Slepukhin P. A., Salmina E. S., Potemkin V. A., et al. Crystal and Electronic Structure of Heteromolecular Complexes of 3,6-bis-(3,5-dimethylpyrazol-1-yl)-1,2,4,5-tetrazine with Azoles. J. Struct. Chem. 2013, in press. [Google Scholar]
- 29. Salmina E.S., Rusinov G.L., Slepukhin P.A., et al. Intermolecular Interactions in Heteromolecular Crystals of Tetrazine Derivatives with Azoles. J. Struct. Chem. 2011, 52, 1134–1138. [Google Scholar]
- 30. Kaim W. The Coordination Chemistry of 1,2,4,5-Tetrazines. Coord. Chem. Rev. 2002, 230, 127–139. [Google Scholar]
- 31. Albrecht M., Fiege M., Osetska O. 8-Hydroxyquinolines in Metallosupramolecular Chemistry. Coord. Chem. Rev. 2008, 252, 812–824. [Google Scholar]
- 32. Chen M., Lv X., Liu Y., et al. An 2-(2′-Aminophenyl)Benzoxazole-Based OFF-ON Fluorescent Chemosensor for Zn2+ in Aqueous Solution. Org. Biomol. Chem. 2011, 9, 2345–2349. [DOI] [PubMed] [Google Scholar]
- 33. Bratsos I., Urankar D., Zangrando E., et al. 1-(2-Picolyl)-Substituted 1,2,3-Triazole as Novel Chelating Ligand for the Preparation of Ruthenium Complexes with Potential Anticancer Activity. Dalton Trans. 2011, 40, 5188–5199. [DOI] [PubMed] [Google Scholar]
- 34. Constable C.E., Morris D., Carr S. Functionalised 3,3[Prime or Minute]-Bipyridines—A New Class of Dinucleating Ligands. N J Chem. 1998, 22, 287–294. [Google Scholar]
- 35. Agrawal A., Johnson S. L., Jacobsen J. A., et al. Chelator Fragment Libraries for Targeting Metalloproteinases. ChemMedChem 2010, 5, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]