Abstract
The glassy-winged sharpshooter (GWSS) is an invasive insect species that transmits Xylella fastidiosa, the bacterium causing Pierce's disease of grapevine and other leaf scorch diseases. X. fastidiosa has been shown to colonize the anterior foregut (cibarium and precibarium) of sharpshooters, where it may interact with other naturally-occurring bacterial species. To evaluate such interactions, a comprehensive list of bacterial species associated with the sharpshooter cibarium and precibarium is needed. Here, a survey of microbiota associated with the GWSS anterior foregut was conducted. Ninety-six individual GWSS, 24 from each of 4 locations (Bakersfield, CA; Ojai, CA; Quincy, FL; and a laboratory colony), were characterized for bacteria in dissected sharpshooter cibaria and precibaria by amplification and sequencing of a portion of the 16S rRNA gene using Illumina MiSeq technology. An average of approximately 150,000 sequence reads were obtained per insect. The most common genus detected was Wolbachia; sequencing of the Wolbachia ftsZ gene placed this strain in supergroup B, one of two Wolbachia supergroups most commonly associated with arthropods. X. fastidiosa was detected in all 96 individuals examined. By multilocus sequence typing, both X. fastidiosa subspecies fastidiosa and subspecies sandyi were present in GWSS from California and the colony; only subspecies fastidiosa was detected in GWSS from Florida. In addition to Wolbachia and X. fastidiosa, 23 other bacterial genera were detected at or above an average incidence of 0.1%; these included plant-associated microbes (Methylobacterium, Sphingomonas, Agrobacterium, and Ralstonia) and soil- or water-associated microbes (Anoxybacillus, Novosphingobium, Caulobacter, and Luteimonas). Sequences belonging to species of the family Enterobacteriaceae also were detected but it was not possible to assign these to individual genera. Many of these species likely interact with X. fastidiosa in the cibarium and precibarium.
Introduction
Homalodisca vitripennis (Germar) (Hemiptera: Cicadellidae: Cicadellinae) or glassy-winged sharpshooter (GWSS) is a leafhopper that exclusively ingests plant xylem fluid from a wide range of host plants [1]. GWSS is considered an important pest in commercial agriculture because it, along with other sharpshooter species, transmits the phytopathogenic bacterium Xylella fastidiosa [2]. X. fastidiosa is the causative agent of a variety of scorch diseases including Pierce's disease (PD) of grapevine, almond leaf scorch disease (ALSD), and citrus variegated chlorosis (CVC). X. fastidiosa is unique as the only known arthropod-transmitted plant pathogen that is both propagative and non-circulative, multiplying solely in the insect's anterior foregut without becoming circulative in the insect's hemolymph [3].
Morphologically, the functional anterior foregut of a sharpshooter consists of: 1) the precibarium, a narrow channel that conveys fluid from the stylets (piercing-sucking mouth parts) into 2) the cibarium, the sucking pump, from which the fluid is swallowed into the rest of the gut. Studies of the cibarium and precibarium, using both light and electron microscopy, have shown patch-like colonies of bacteria interspersed with naked cuticle. The earliest microscopy study used both light and scanning electron microscopy (SEM) to show rod-shaped bacteria embedded in a gum-like matrix in inoculative, i.e. competent to inoculate pathogen, but not in non-inoculative, Graphocephala atropunctata (Signoret) (blue-green sharpshooter; BGSS) [4]; these bacteria were assumed to be the PD bacterium (now known as X. fastidiosa). Later SEM studies found similar bacteria in Oncometopia nigricans (Walker) and H. vitripennis individuals that had fed on plants colonized by X. fastidiosa [5]; sharpshooters fed on healthy plants were reported to be bacteria-free. A more recent SEM study of G. atropunctata again demonstrated that the presence of bacteria in the cibarium and precibarium was highly correlated with infectivity [6]. Three confocal light microscopy studies introducing and visualizing fluorescent protein-expressing bacteria (either X. fastidiosa or an Alcaligenes sp.) have shown single bacterial cells and small clumps or colonies of cells in the anterior foregut of inoculative sharpshooters [3], [7], [8].
Several small-scale studies of bacterial 16S sequences associated with sharpshooters have previously been published. In 2003, Moran et al. [9] dissected bacteriomes from several species of Cicadellinae and detected a novel symbiont they named “Candidatus Baumannia cicadellinicola.” Subsequent work from the same group identified another novel symbiont, “Candidatus Sulcia muelleri,” associated with various sharpshooters and other related insects [10], [11]. B. cicadellinicola, W. pipientis, as well as bacteria closely related to Pseudomonas, Stenotrophomonas and Acintobacter were identified in GWSS hemolymph in an independent study [12]. Lacava et al., 2007 [13] used a combination of denaturing gradient gel electrophoresis, culturing, and 16S sequencing to characterize bacteria associated with GWSS heads. In a larger 16S sequencing study, Hail et al., 2011 [14] detected predominantly Pectobacterium in hemolymph and Cardiobacterium associated with the alimentary canal and whole insect. In contrast to previous 16S sequencing studies, Wolbachia was not detected in the hemolymph, only in whole insects [14]. These previous studies have been limited in the number of sequences examined and have not compared insects from different locations; therefore, details about the extent of microbiome variation among GWSS individuals and geographic locations are unknown.
This study is the first to use next-generation sequencing (NGS) techniques to exhaustively catalog bacteria associated with the GWSS cibarium and precibarium. A short, diagnostic region of the bacterial 16S RNA gene was amplified by PCR from 96 individual insects (24 each from 4 different geographic locations). Illumina sequencing technology was used to pool PCR products and obtain at least 50,000 individual reads per insect. To more precisely identify a few common bacterial genera (Wolbachia, Xylella, Anoxybacillus, Caulobacter, and Novosphingobium), additional Sanger sequencing is presented. Cataloging foregut-associated bacteria is a necessary first step toward examining interactions among bacterial species in GWSS foregut and addressing the hypothesis that other bacterial species can affect the ability of a sharpshooter to acquire, retain, and/or inoculate X. fastidiosa.
Results
16S rRNA Sequencing
In total, more than 19 million paired-end reads were recovered using Illumina-based sequencing (Table 1). Approximately 92% of these reads passed Illumina's stringent quality control (QC) measures and of those, just over 84% could be classified as bacterial 16S amplicons (including 16S sequences from chloroplasts). Demultiplexing, where the dual-end indices are used to assign each read to an individual insect, was performed using Illumina's MiSeq Reporter software, giving between about 50,000 and one million high quality reads per insect sample (Table 1).
Table 1. Summary of Sequencing Results.
| All Sequences: | total | average | low | high |
| # raw reads retrieved: | 19,473,000 | |||
| # reads passing quality control: | 17,882,483 | 186,276 | 57,736 | 1,045,276 |
| unclassified (not 16S): | 2,838,274 | 29,565 | 5,343 | 105,435 |
| classified (bacterial 16S): | 15,044,209 | 156,711 | 49,833 | 943,506 |
The number of sequencing reads (total, average per insect, lowest value, and highest value) are presented.
Assignment of Bacterial Classifications
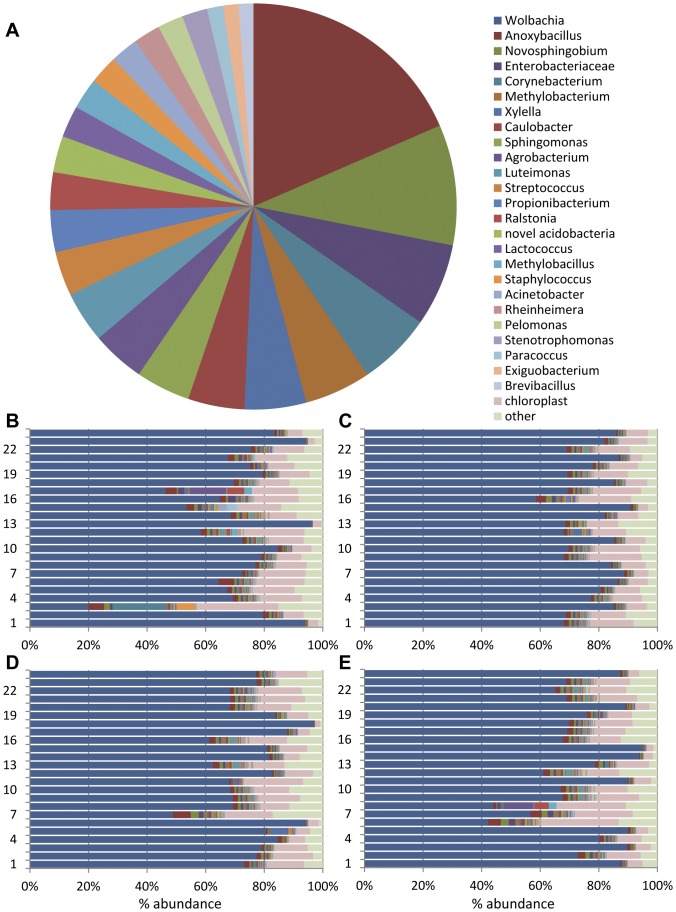
MiSeq Reporter automatically assigns taxonomic classifications to reads by comparing sequences to an Illumina-curated version of the Greengenes 16S rRNA gene database. In addition, FastQ files were exported from MiSeq Reporter and Genomics Workbench was used to align paired ends, remove primer sequence, and group reads into operational taxonomic units (OTUs) of at least 97% identity. Taxonomic classifications were assigned to OTUs using the BLAST function within Genomics Workbench. These two methods gave very similar results; the number of reads assigned to either Wolbachia or to X. fastidiosa varied by less than 2% for all 96 insects. To compare taxon and OTU prevalence between insects, percent abundance was calculated based on total number of classified reads. In addition, annotation of each genus was manually checked by BLAST (www.ncbi.nlm.nih.gov) against both the non-redundant and reference RNA sequence databases. Taxonomic classifications were identical except for the OTU listed as “novel acidobacteria” in Table 2. MiSeq Reporter assigned that group to the genus Candidatus Solibacter. However, manual BLAST searches revealed only 93% identity between the OTU and published Solibacter usitatus Ellin6076 16S sequence. Since this is below the 97% identity threshold for OTU formation, this group was relabeled “novel acidobacteria.” Its consensus 16S sequence is 99% identical to several uncultured bacterium clones from two separate unpublished studies, one of a constructed wetland in France and the other from Tibetan permafrost (i.e. accession numbers KC432438 and JQ684311). Many genera from the family Enterobacteriaceae have identical V6 regions of the 16S rRNA gene; therefore it was not possible to identify this OTU below the family level. The MiSeq Reporter results are presented in Table 2 as average % abundance over all 96 samples. The category “other/unclassified” represents taxa present at less than 0.1% or reads that were not sufficiently similar to known 16S genes to be mapped to a genus. Because Wolbachia is likely not present in the cibarium and precibarium, it, along with chloroplast and other/unclassified, were omitted from Figure 1A in order to better demonstrate the diversity and abundance of identified bacterial genera potentially present in precibaria and cibaria; all classifications are included in Figure 1B–E to convey inter-insect variability.
Table 2. Bacterial Genera Abundance.
| Classifications: | average (%) | SD (%) |
| Wolbachia | 74.55 | 12.98 |
| Anoxybacillus | 1.54 | 1.07 |
| Novosphingobium | 0.80 | 0.49 |
| Enterobacteriaceae | 0.55 | 0.44 |
| Corynebacterium | 0.49 | 1.85 |
| Methylobacterium | 0.44 | 0.32 |
| Xylella | 0.41 | 0.65 |
| Caulobacter | 0.37 | 0.20 |
| Sphingomonas | 0.36 | 0.23 |
| Agrobacterium | 0.35 | 1.63 |
| Luteimonas | 0.34 | 0.50 |
| Streptococcus | 0.29 | 0.20 |
| Propionibacterium | 0.28 | 0.20 |
| Ralstonia | 0.25 | 0.75 |
| novel acidobacteria | 0.24 | 0.14 |
| Lactococcus | 0.21 | 0.17 |
| Methylobacillus | 0.21 | 0.39 |
| Staphylococcus | 0.19 | 0.66 |
| Acinetobacter | 0.18 | 0.27 |
| Rheinheimera | 0.18 | 0.27 |
| Pelomonas | 0.17 | 0.23 |
| Stenotrophomonas | 0.17 | 0.15 |
| Paracoccus | 0.11 | 0.34 |
| Exiguobacterium | 0.10 | 0.08 |
| Brevibacillus | 0.10 | 0.09 |
| chloroplast | 10.15 | 5.23 |
| other/unclassified | 6.99 | 3.65 |
All bacterial genera present at or above 0.1% of the total are listed. Data are from MiSeq Reporter; classifications were verified by BLAST.
Figure 1. Genera most commonly associated with H. vitripennis cibarium and precibarium.
A) Average abundance of non-Wolbachia genera over all 96 insects; B–E) Abundance of all classifications from B) Bakersfield individuals; C) Ojai individuals; D) Florida individuals; E) colony individuals. Color legend in panel A applies to all panels (A–E); order of classifications in the legend is the same as the order around the pie graph in A (starting with the dark red Anoxybacillus in the upper right) and in the bar graphs B–E (starting with the dark blue Wolbachia on the left).
α- and β-Diversity
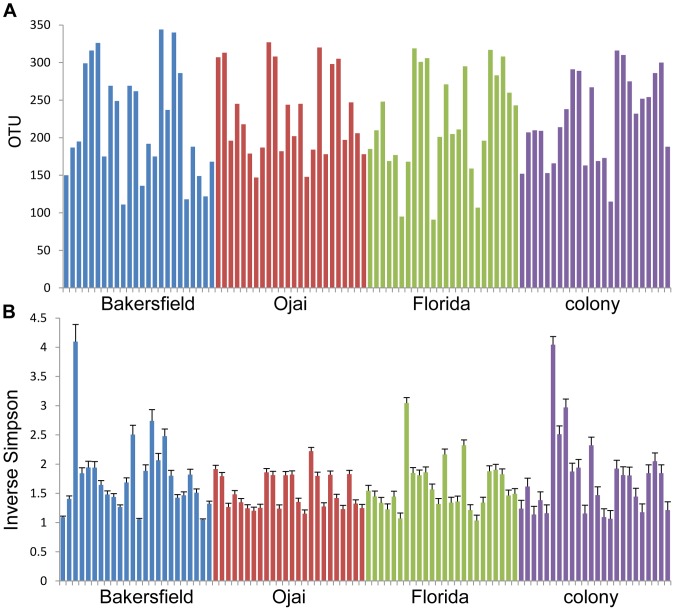
The total number of OTUs observed in each of the 96 insects is shown in Figure 2A; Figure 2B shows the inverse Simpson index, another measure of α-diversity or richness of each individual microbiome. Examining β-diversity, AMOVA (analysis of molecular variance) showed no significant differences among the bacterial profiles from each of the four locations (Fs = 1.14, df = 95, p-value = 0.337). To examine potential geographical differences, the California 72 insects (24 from Bakersfield, 24 from Ojai, and 24 colony) were grouped together and compared with the 24 Florida insects also using AMOVA. Again, no significant differences were observed (Fs = 0.438, df = 95, p-value = 0.579).
Figure 2. α Diversity Measures.
Total number of OTU observed per insect (A) and inverse Simpson index (B) for each of the 96 insects as calculated using mothur. Error bars in B are 95% confidence levels. Bars are color-coded: blue (Bakersfield), red (Ojai), green (Florida), and purple (colony).
Wolbachia ftsZ gene sequence
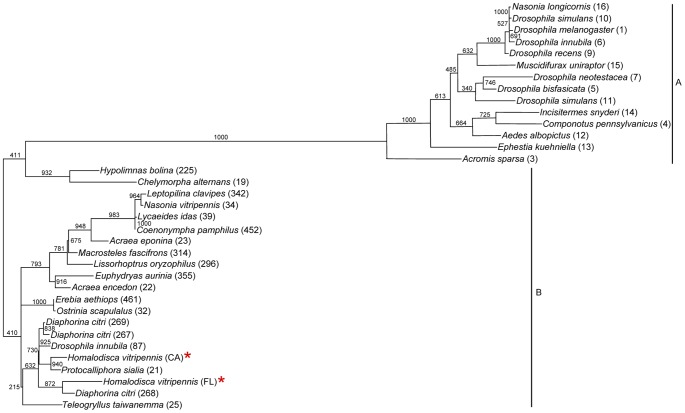
The 16S rRNA gene shows little to no divergence in many strains of Wolbachia isolated from disparate insect hosts and geographic locations. MLST (multilocus sequence typing) analysis been used to further characterize Wolbachia strains [15]. Sequences from the three California insect sources (Bakersfield, Ojai, and the colony, which was originally collected in Bakersfield) were identical and are grouped together in Table 3. However, there are quite a few polymorphisms between the California insects and the Florida ones, especially in the coxA and wsp loci. A larger, overlapping region of the ftsZ gene was also cloned by PCR and sequenced [16]. There were no differences in this longer ftsZ sequence among the four geographic locations. As shown in Figure 3, the MLST sequences determined here place the Wolbachia from H. vitripennis in supergroup B of Wolbachia pipientis [16], [17]. W. pipientis supergroups A and B are most commonly associated with arthropod species.
Table 3. Wolbachia MLST Results.
| loci | California | Florida | bp / aa differences between CA and FL |
| gatB | 9 | 227* | 3 |
| coxA | 212* | 221* | 14 |
| hcpA | 88 | 251* | 1 |
| ftsZ | 195* | 195* | 0 |
| fbpA | 27 | 403* | 2 |
| wsp | 378 | 675* | 59a |
| HVR1 | 150 | 233* | 4 |
| HVR2 | 165 | 266* | 10b |
| HVR3 | 24 | 8 | 7 |
| HVR4 | 3 | 7 | 10 |
59 differences include 56 substitutions and 3 indels (3 bp each).
10 differences include 8 substitutions and 2 indels (1 aa each).
Five individual cloned PCR products were sequenced for each locus from each insect population. Sequences from all three California populations (Bakersfield, Ojai, and colony) were identical and have been grouped together. Allele numbers are from http://pubmlst.org/wolbachia/ and novel alleles are marked (*). Differences are base pairs for genes (gatB, coxA, hcpA, ftsZ, fbpA, and wsp) and amino acids for peptides (HVR1, 2, 3 and 4).
Figure 3. Phylogeny of Wolbachia MLST genes from various insects.
A maximum likelihood tree (1000 bootstrap replicates) is based on multiple alignments of concatenated DNA sequences encoding the coxA, fbpB, ftsZ, gatB, hcpA, and wsp genes. Bootstrap values are shown for all nodes; red asterisks indicate sequences from this study. Sequence types from pubmlst.org/wolbachia are given in parentheses. Lines to the right indicate Wolbachia supergroups A and B.
X. fastidiosa MLST Analysis and Quantitative PCR
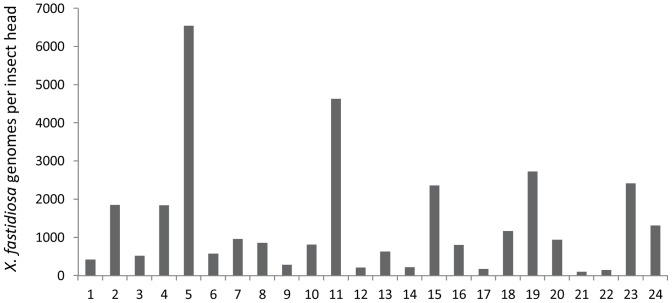
MLST has also been used to characterize X. fastidiosa strains and assign subspecies [18], [19]. Due to limited amounts of starting DNA, only 4 of the typical 8 loci were sequenced (pilU, petC, leuA, and gltT). As shown in Table 4, sequences corresponding to both the fastidiosa and sandyi subspecies were detected in GWSS from both California locations and from the colony; however, only the fastidiosa subspecies was detected in Florida GWSS. It is noteworthy that subspecies sandyi has not been found in Florida. X. fastidiosa levels were quantitated using real time PCR on DNA extracted from 24 GWSS from the colony. As shown in Figure 4, X. fastidiosa genomes per dissected insect head ranged from 100 to 6500 (average 790). Due to the limited amount of DNA in a dissected GWSS head, these are different insects than those used for 16S rRNA sequencing.
Table 4. Xylella fastidiosa MLST Results.
| Loci | Bakersfield | Ojai | Florida | Colony |
| pilU | fastidiosa (1) sandyi (4) | fastidiosa (5) | fastidiosa (5) | sandyi (5) |
| petC | fastidiosa (3) sandyi (2) | fastidiosa (2) sandyi (3) | fastidiosa (5) | fastidiosa (5) |
| leuA | fastidiosa (3) sandyi (2) | sandyi (5) | n.a.* | fastidiosa (1) sandyi (4) |
| gltT | fastidiosa (3) sandyi (2) | fastidiosa (1) sandyi (4) | fastidiosa (5) | n.a.* |
* n.a. = no amplification.
Five individual cloned PCR products were sequenced for each locus from each insect population. Numbers in parentheses are the number of times sequences of each subspecies were detected. Due to limited amounts of DNA, leuA in the Florida population and gltT from the colony were not amplified.
Figure 4. X. fastidiosa genomes per GWSS head.
Quantitative RT-PCR was used to measure the number of X. fastidiosa genomes associated with 24 different dissected GWSS heads from the colony.
Additional Sequencing of the 16S rRNA genes from Anoxybacillus, Caulobacter, and Novosphingobium
To further characterize the species present, additional sequencing of the 16S rRNA gene was performed for these three genera. Sequences were compared to the GENBANK non-redundant DNA database by BLAST (www.ncbi.nlm.nih.gov). For Anoxybacillus, the longer 16S sequences did not allow assignment to the species level. Sequences obtained here, which did not vary among the four geographic locations, are identical to 27 sequences; 10 of these are annotated as Anoxybacillus (A. flavithermus, A. kamchatkensis, or unspecified Anoxybacillus spp.). Many Anoxybacilli are found in hot springs but some are insect-associated (e.g. JQ894876 from spiraling whitefly or KC170396 from the aphid Cinara tujafilina) or plant-associated (e.g. KC478840 from Indian mulberry Morinda citrifolia).
The Caulobacter sequence obtained here is 100% identical to 49 sequences in GENBANK, all uncultured and annotated as Caulobacter spp. or C. henricii. These sequences are from such diverse habitats as endosphere of hybrid poplar, coral and coral reef water, sea floor, soil, the South American moth Rothschildia lebeau larva, citrus trees, rice seeds, and rice paddy soil. The Novosphingobium sequence obtained here is 100% identical to 6 sequences in GENBANK: one uncultured from Artic seawater; and 5 cultured bacterial strains from rice roots and paddy soil or sugarcane and associated soils. By comparison to the NCBI reference RNA database, the sequence obtained here is 99% identical to Novosphingobium resinovorum strain NCIMB 8767 originally isolated from soil.
Discussion
Bacteria have been found associated with the guts of many insect species [20]. Therefore it is not surprising that so many disparate bacterial genera were found associated with GWSS cibarium and precibarium. Wolbachia is a common insect pathogen and has been detected in GWSS and other sharpshooters previously [10], [11], [13], [14]. Because Wolbachia is an endosymbiont [17], the Wolbachia detected here most probably comes from cellular contents or hemolymph liberated during the dissection process.
Many of the non-Wolbachia genera found in this study have been detected previously in association with plants or soil: Novosphingobium, Sphingomonas, Methylobacterium, Methylobacillus, Agrobacterium, Ralstonia, and Lutemonas. The casparian strip is an impermeable barrier in a root that separates the bulk soil solution and vascular fluids. However, it is possible for solutes and bacterial cells present in soil to passively enter the root xylem stream through lateral root junctions, wounds or root tips, which lack a casparian strip [21]. Therefore, it is not surprising to find soil bacteria present in xylem or in an exclusively xylem-ingesting insect.
The presence of chloroplast DNA may indicate that cellular contents of non-xylem cells are taken up into GWSS precibarium and cibarium during plant feeding. It has been hypothesized [22] that sharpshooters take up fluid into the precibarium from non-xylem cells encountered along the stylet pathway prior to reaching a xylem vessel. Perhaps uptake of non-xylem cell contents is performed to taste chemical constituents using gustatory sensilla lining the precibarium [23], and may aid in orienting the stylets towards xylem. Chloroplast DNA also likely enters the xylem stream through wounds and as part of the xylem maturation process. It is possible that chloroplasts brought into the precibarium lodged in bacterial biofilm colonizing the cuticle and thus were retained.
Several of the bacterial genera detected here have been previously reported to be sharpshooter-associated: Exiguobacterium, Stenotrophomonas, Pelomonas, Acinetobacter, Ralstonia, and Methylobacterium [12]–[14]. Sequences consistent with the previously reported GWSS endosymbionts Baumannia cicadellinicola and Sulcia muelleri were detected here at levels below 0.1% average abundance [9], [10]. It is likely these endosymbionts originated in hemolymph liberated during dissection, but at very low levels because they normally reside intracellularly in abdominal bacteriomes [9]. The Enterobacteriaceae includes the human pathogens Escherichia, Salmonella, and Shigella. Therefore it is possible that sequences from this family represent contaminants introduced during insect handling, dissection, or DNA processing. Similarly, Proprionibacterium has been most commonly found associated with human skin and also may be a contaminant.
It is not possible to directly calculate numbers of bacterial cells per insect head from 16S sequencing data. However, there is good correlation between percent abundance and copy number as determined by quantitative PCR (qPCR) [24]. There was not sufficient DNA to perform qPCR on GWSS sampled here; however, qPCR performed on other GWSS from the laboratory colony typically detected approximately 800 X. fastidiosa per insect (Figure 4).
Several microscopy studies have described uniform “shag carpets” of bacteria lining the cibaria and precibaria in inoculative sharpshooters and have hypothesized these are pure cultures of X. fastidiosa. Data presented here suggests there may be other bacteria in these locations, including other gram-negative rods that would be very similar in appearance to X. fastidiosa. Novosphingobium, Sphingomonas, Caulobacter, Methylobacterium, Methylobacillus, Agrobacterium, Ralstonia, and Lutemonas are all gram-negative, rod-shaped bacteria. It cannot be stated with certainty that any non-X. fastidiosa bacteria detected here attached to or actively colonized the precibarial and cibarial cuticle. Consequently, microscopy studies of appropriately labeled bacteria would be needed to provide definitive proof of multiple bacterial species binding to the cuticle of sharpshooter cibarium and/or pre-cibarium.
Materials and Methods
Insect Collection and Dissection
H. vitripennis were field-collected from: 1) oleander (Nerium oleander (L.)) and photinia (Photinia spp.) plants in Bakersfield, CA on June 21, 2012 (Bakersfield), 2) citrus in Ojai, CA on July 24, 2012 (Ojai), and 3) wild sunflower (Helianthus annuus (L.)) in Quincy, FL on August 31, 2012 (Florida). Insects also were taken from 4) a H. vitripennis colony maintained in Parlier, CA on October 23, 2012 (colony). Colony insects were originally collected on oleander and photinia in Bakersfield, CA in 2003–2005, and maintained since then under artificial conditions (24–30°C, 16∶8 L∶D) without diapause, on a mixture of cowpea (Vigna unguiculata (L.)), sorghum (Sorghum bicolor (L.)), sweet basil (Ocimum basilicum (L.)), and sunflower plants in the same cage. H. vitripennis is not an endangered or protected species; permission to collect H. vitripennis was given by land owners in all locations.
Field-collected insects were frozen whole while colony insects were decapitated and only the heads were frozen; insect tissue was stored at −20°C until dissection. Twenty-four individuals from each geographic location were rapidly dissected on a cold plate to reduce thawing, using a Leica MZ16 stereoscan microscope, until only the precibarium and cibarium of the anterior foregut remained. Precautions were taken to maintain sterile technique and to avoid contamination of the sample. Dissected precibarium and cibarium from each individual insect was refrozen at −20°C until DNA isolation was performed according to a previously described protocol [25].
Construction of Libraries and DNA Sequencing
PCR primers were designed to amplify approximately 200 bp spanning V6 (the 6th variable region) of the 16S ribosomal RNA gene. In order to provide the nucleotide diversity needed by Illumina sequencing platforms, 6 random nucleotides were added to the 5′ end of each primer. Additional diversity was created using pools for 6 forward and 6 reverse primers where the start of each primer was shifted by one bp (Table 5). Ninety-six individual PCR reactions were performed using the Expand High Fidelity PCR System (Roche Applied Science) in an S1000 Thermal Cycler (BioRad) according to manufacturers' directions. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen). Libraries were constructed by ligating sequencing adapters and indices onto purified PCR products using the TruSeq DNA HT Sample Preparation Kit (Illumina). This kit adds one of 12 indices (unique 8 bp sequence tags) to the forward end and one of 8 different indices to the reverse end for 96 unique index combinations. Libraries were quantitated using a KAPA Library Quantification Kit (Kapa Biosystems) in a StepOnePlus Real Time PCR System (Applied Biosystems). Equimolar amounts of each of the 96 libraries were pooled and submitted for sequencing on a MiSeq Personal Sequencer using a 150-bp paired-end protocol at the University of Georgia Genomics Facility.
Table 5. Primers used to amplify 16S DNA.
| Forward primers | Reverse primers |
| NNNNNNAAACTCAAAGGAATTGACGG | NNNNNNGGGTTGCGCTCGTTGCGG |
| NNNNNNAACTCAAAGGAATTGACGGG | NNNNNNGGTTGCGCTCGTTGCGGG |
| NNNNNNACTCAAAGGAATTGACGGGG | NNNNNNGTTGCGCTCGTTGCGGGA |
| NNNNNNCTCAAAGGAATTGACGGGGR | NNNNNNTTGCGCTCGTTGCGGGAC |
| NNNNNNTCAAAGGAATTGACGGGGRC | NNNNNNTGCGCTCGTTGCGGGACT |
| NNNNNNCAAAGGAATTGACGGGGRCC | NNNNNNGCGCTCGTTGCGGGACTT |
Equimolar amounts of all 6 forward primers and all 6 reverse primers were pooled and used in PCR. Forward primers span nt 926–950; reverse primers span nt 1107–1129 (numbering relative to E. coli 16S DNA).
DNA Sequence Analysis
Sequence analysis, including demultiplexing and removal of indices, was performed using the bacterial metagenomics workflow in the MiSeq Reporter software (Illumina). FastQ sequence files (after demultiplexing and index removal) were also exported from MiSeq Reporter for further analysis using CLC Genomics Workbench (CLC Bio). The two analysis methods gave very similar results; discrepancies are noted in the Results. Inverse Simpson indices and AMOVAs (analysis of molecular variance) were calculated using the mothur package: www.mothur.org [26], [27]. To construct the phylogenetic tree, concatenated MLST sequences were downloaded from http://pubmlst.org/wolbachia and aligned with sequences from this study using ClustalW at Biology Workbench 3.2 (http://seqtool.sdsc.edu/CGI/BW.cgi) [28]. Maximum likelihood tree was drawn using T-REX (http://www.trex.uqam.ca) [29], [30]. The fastQ sequence files have been deposited at MG-RAST (http://metagenomics.anl.gov/metagenomics.cgi?page=Home) with ID # 4522303–4522398. A summary of the raw data is presented in Table S1.
Wolbachia ftsZ and MLST sequencing
All 24 original DNA samples from each location were pooled and PCR was performed using the previously described ftsZUNIF and ftsZUNIR primers [16] and MLST primers [15]. PCR products were cloned using a TOPO TA Cloning Kit with pCR2.1 (Life Technologies). Five individual clones from each location were sequenced using the M13 forward and reverse primers on a 3130×l Genetic Analyzer (Applied Biosystems). Sanger sequence data were analyzed using Sequencher 5.1 (Gene Codes Corporation). Final ftsZ sequence was deposited in GenBank (accession number KF636751) and final MLST sequences were uploaded to http://pubmlst.org/wolbachia/.
Additional sequencing of selected 16S genes
The Illumina sequences were used to design specific primers to the V6 region of the 16S gene for three common genera: Anoxybacillus (AB-F CCCCTGACAACCCGAGAGATCGGGCG), Caulobacter (CA-F GCCCGGACCGCCACAGAGATGTGGCTT), and Novosphingobium (NS-F CCCGCGCTACACAGAGAGATTTGTGG). These primers were used along with a 1544R universal 16S DNA primer (AAGGAGGTGATCCAGCC) to amplify approximately 600 bp spanning variable regions 7, 8, and 9 of selected 16S DNAs. As with the Wolbachia MLST, PCR products were cloned into pCR2.1 and five individual clones of each gene from each location were sequenced.
X. fastidiosa MLST sequencing and quantitative PCR
Previously reported primers were used to amplify portions of the pilU, petC, leuA, and gltT genes [19] from pooled DNA from each location. As with the Wolbachia MLST, PCR products were cloned into pCR2.1 and five individual clones of each gene from each location were sequenced. Subspecies were assigned using the Xylella fastidiosa MLST Databases at pubmlst.org/xfastidiosa/. Quantitative real-time PCR was performed on a StepOnePlus Real Time PCR System (Applied Biosystems) as previously described [25].
Supporting Information
OTU Table. Number of times each genera was detected in each of the 96 insects.
(XLSX)
Acknowledgments
The authors thank Dr. Russell Mizell for collecting insects from Florida, Dr. Drake C. Stenger for assistance with the tree in Figure 3, Dr. Mark S. Sisterson for help with statistical analyses, and Mark Schreiber and Holly Shugart for excellent technical assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. USDA is an equal opportunity provider and employer. This article is in the public domain and not copyrightable. It may be freely reprinted with customary crediting of the source.
Funding Statement
Funding for this research was from USDA-ARS appropriated project 5302-22000-010-00D. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Redak RA, Purcell AH, Lopes JRS, Blua MJ, Mizell RF III, et al. (2004) The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annual Review of Entomology 49: 243–270. [DOI] [PubMed] [Google Scholar]
- 2. Hill BL, Purcell AH (1995) Acquisition and retention of Xylella fastidiosa by an efficient vector, Graphocephala atropunctata . Phytopathology 85: 209–212. [Google Scholar]
- 3. Backus EA, Morgan DJW (2011) Spatiotemporal colonization of Xylella fastidiosa in its vector supports the role of egestion in the inoculation mechanism of foregut-borne plant pathogens. Phytopathology 101: 912–922. [DOI] [PubMed] [Google Scholar]
- 4. Purcell AH, Finlay AH, McLean DL (1979) Pierce's disease bacterium: mechanism of transmission by leafhopper vectors. Science 206: 839–841. [DOI] [PubMed] [Google Scholar]
- 5. Brlansky RH, Timmer LW, French WJ, McCoy RE (1983) Colonization of the sharpshooter vectors, Oncometopia nigricans and Homalodisca coagulata, by xylem-limited bacteria. Phytopathology 73: 530–535. [Google Scholar]
- 6. Almeida RPP, Purcell AH (2006) Patterns of Xylella fastidiosa colonization on the precibarium of sharpshooter vectors relative to transmission to plants. Annals of the Entomological Society of America 99: 884–890. [Google Scholar]
- 7. Newman KL, Almeida RPP, Purcell AH, Lindow SE (2003) Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera . Applied and Environmental Microbiology 69: 7319–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bextine B, Lauzon C, Potter S, Lampe D, Miller TA (2004) Delivery of a genetically marked Alcaligenes sp. to the glassy-winged sharpshooter for use in a paratransgenic control strategy. Current Microbiology 48: 327–331. [DOI] [PubMed] [Google Scholar]
- 9. Moran NA, Dale C, Dunbar H, Smith WA, Ochman H (2003) Intracellular symbionts of sharpshooters (Insecta: Hemiptera: Cicadellinae) form a distinct clade with a small genome. Environmental Microbiology 5: 116–126. [DOI] [PubMed] [Google Scholar]
- 10. Moran NA, Tran P, Gerardo NM (2005) Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Applied and Environmental Microbiology 71: 8802–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takiya DM, Tran PL, Dietrich CH, Moran NA (2006) Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Molecular Ecology 15: 4175–4191. [DOI] [PubMed] [Google Scholar]
- 12. Montllor Curley C, Brodie E, Lechner M, Purcell AH (2007) Exploration for facultative endosymbionts of glassy-winged sharptshooter (Hemiptera: Cicadellidae). Annals of the Entomological Society of America 100: 345–349. [Google Scholar]
- 13. Lacava PT, Parker J, Andreote FD, Dini-Andreote F, Ramirez JL, et al. (2007) Analysis of the bacterial community in glassy-winged sharpshooter heads. Entomological Research 37: 261–266. [Google Scholar]
- 14. Hail D, Lauziere I, Dowd SE, Bextine B (2011) Culture independent survey of the microbiota of the glassy-winged sharpshooter (Homalodisca vitripennis) using 454 pyrosequencing. Environmental Entomology 40: 23–29. [DOI] [PubMed] [Google Scholar]
- 15. Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, et al. (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis . Applied and Environmental Microbiology 72: 7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Casiraghi M, Anderson T, Bandi C, Bazzocchi C, Genchi C (2001) A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 122: 93–103. [DOI] [PubMed] [Google Scholar]
- 17. Casiraghi M (2005) Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151: 4015–4022. [DOI] [PubMed] [Google Scholar]
- 18. Scally M, Schuenzel EL, Stouthamer R, Nunney L (2005) Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Applied and Environmental Microbiology 71: 8491–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan X, Morano L, Bromley R, Spring-pearson S, Stouthamer R, et al. (2010) Multilocus sequence typing of Xylella fastidiosa causing Pierce's disease and oleander leaf scorch in the United States. Phytopathology 100: 601–611. [DOI] [PubMed] [Google Scholar]
- 20. Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiology Reviews 37: 699–735. [DOI] [PubMed] [Google Scholar]
- 21. Turner TR, James EK, Poole PS (2013) The plant microbiome. Genome Biology 14: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backus EA (in press) Sharpshooter feeding behavior in relation to transmission of Xylella fastidiosa: A model for foregut-borne transmission mechanisms. In: Brown JK, editor. Vector-Mediated Transmission of Plant Pathogens: American Phytopathological Society Press. [Google Scholar]
- 23. Backus EA, McLean DL (1982) The sensory systems and feeding behavior of leafhoppers. I. The aster leafhopper, Macrosteles fascifrons Stål (Homoptera: Cicadellidae). Journal of Morphology 172: 361–379. [DOI] [PubMed] [Google Scholar]
- 24. Fagen J, Giongo A, Brown C, Davis-Richardson A, Gano K, et al. (2012) Characterization of the relative abundance of the citrus pathogen Ca. Liberibacter asiaticus in the microbiome of its insect vector, Diaphorina citri, using high throughput 16S rRNA sequencing. The Open Microbiology Journal 6: 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ledbetter CA, Rogers EE (2009) Differential susceptibility of Prunus Germplasm (Subgenus Amygdalus) to a California Isolate of Xylella fastidiosa . HortScience 44: 1928–1931. [Google Scholar]
- 26. Kozich J, Westcott S, Baxter N, Highlander SK, Schloss P (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and Environmental Microbiology 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson J, Higgens D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gam pentalties, and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guindon S, Gascuel O (2003) A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Systematic Biology 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 30. Boc A, Diallo AB, Makarenkov V (2012) T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Research 40: W573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OTU Table. Number of times each genera was detected in each of the 96 insects.
(XLSX)