Abstract
This article describes the development of a reliable CZE-ESI-MS method to simultaneously separate and quantitate three specific metabolites (3-hydroxyanthranilic acid (3-HAA), quinolinc acid (QA), and picolinic acid (PA)) of the kynurenine pathway (KP) of tryptophan catabolism. Using a covalently bonded sulfonated capillary. the parameters such as pH, type of background electrolyte, type of organic solvent, nebulizer pressure as well as both negative and positive ESI-MS modes were optimized to achieve the best Rs and S/N of three KP metabolites. The developed CZE-ESI-MS assay provided high resolution of PA/QA, high specificity, a total analysis time of 10 min with satisfactory intra-day and inter-day repeatability of migration time and peak areas. Under optimized CZE-ESI-MS conditions, the calibration curves over a concentration range of 19–300 μM for 3-HAA and QA, and 75–300 μM for PA were simultaneously generated. The method was successfully applied for the first time to profile the concentrations of initial substrate, 3-HAA, and its eventual products, PA and QA, formed in the complex multienzyme system. As the ratio of two enzymes, 3-hydroxyanthranilate 3,4-dioxygenase (HAO) and α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) decreases, the concentration of QA approaches essentially zero indicating that all ACMS formed by the action of HAO is consumed by ACMSD rather than its spontaneous decay to QA.
Keywords: Capillary zone electrophoresis-mass spectrometry, Enzyme, 3-Hydroxyanthranilic acid, Picolinic acid, Quinolinic acid
1 Introduction
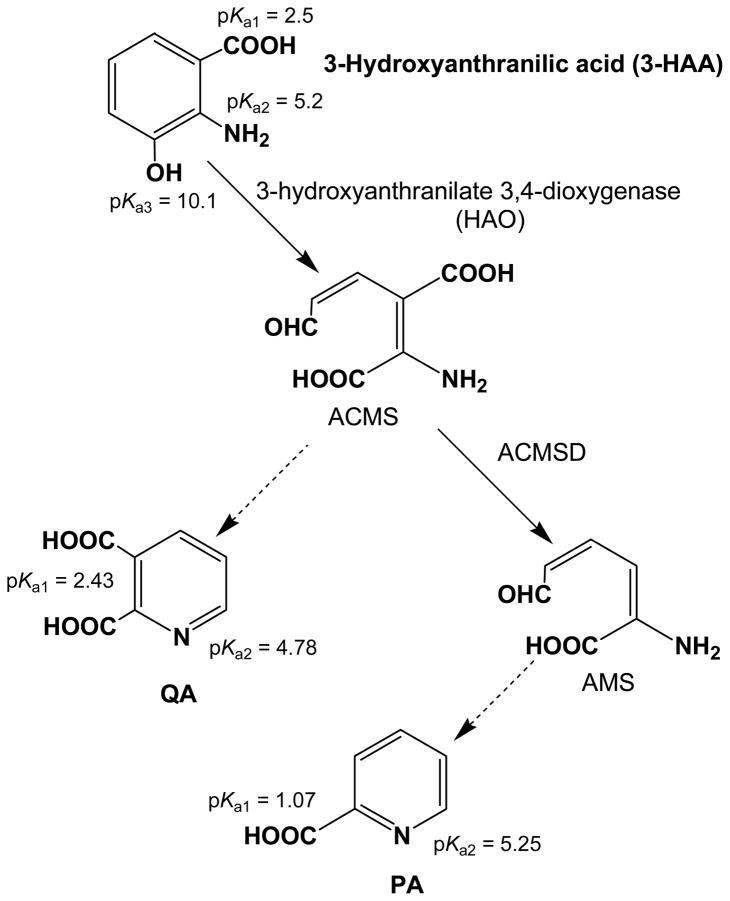
Quinolinic acid (QA), a metabolite of tryptophan catabolism in the kynurenine pathway (KP) and a precursor for mammalian biosynthesis of nicotinamide adenine dinucleotide (NAD+), has been identified as a neurotoxin when its concentrations in the cerebrospinal fluid (CSF) and blood are sufficiently elevated [1–3]. The high level of QA has been observed in many neurodegenerative diseases: Huntington’s disease, Alzheimer’s disease, Parkinson’s disease, epilepsy, AIDS-dementia complex, schizophrenia, and traumatic brain injuries [4]. The QA is formed as the result of a spontaneous, non-enzymatic, dehydrative cyclization of α-amino-β-carboxymuconate-ε-semialdehyde (ACMS), a metabolic intermediate in the KP (Figure 1) [5]. However, in the presence of α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD), the substrate ACMS is converted enzymatically to α-aminomuconate-ε-semialdehyde (AMS). This unstable metabolite, AMS undergoes a non-enzymatic cyclization reaction with the elimination of water, to form picolinic acid (PA). Although PA has not been shown to be neuroactive in physiological relevant conditions, measured concentrations in CSF have been seen to increase with age [6]. Studying the formation of the QA and PA is therefore of interest. Hence a reliable analytical method should be developed to quantitate PA and QA concentrations as disease diagnostic markers and in mechanistic studies of the enzymes competing with the non-enzymatic side reactions.
Figure 1.
Schematic representation of 3-HAA, PA and QA in the kynurenine (KP) pathway. The KP is very complex and the pathway shows only a few compounds relevant to this paper.
Numerous high performance liquid chromatography (HPLC) methods for separation and detection of tryptophan metabolites have been reported with different detectors [7, 8], including UV and fluorescence [9–11], electrochemical [12] as well as mass spectrometric (MS) detection [7, 8, 13–15]. Most of the aforementioned HPLC techniques are time consuming, require gradient elution or multistep detection and separation systems. Gas chromatography-mass spectrometry (GC/MS) has been described for the analysis of tryptophan metabolites [16, 17]. However, the method requires sample derivatization to yield volatile and thermally stable analytes. This is not only a time-consuming procedure, but may involve risk for sample contamination and alteration of the content.
The coupling of CZE with ESI-MS represents a powerful combination due to high separation efficiency, high resolution, and high throughput of CZE combined with high detection sensitivity, and high specificity provided by ESI-MS [18, 19]. In contrast to HPLC-MS, CE-MS is particularly suitable for resolving a wide range of polar and charged metabolites [18]. Another attractive feature associated with CZE compared to HPLC is its ultra-small (i.e., nanoliters) injection, making it particularly well-suited for samples that are volume-limited [20]. To-date there are several CZE methods, which have been described to analyze tryptophan metabolites by coupling to UV detection [21, 22] but only one by electrochemical detection [23] and one by ESI-MS detection [24]. However, variability of migration time and peak area on bare fused silica capillary (BFS) is a major concern in CZE and CZE-MS. Although there are some procedures to correct for migration time shifts and aligning electropherograms [25, 26], still, for the studies on enzyme samples, reproducibility of migration times and peak areas is of utmost importance to observe small changes in product composition when optimizing the assay. Using CZE with BFS capillaries, the analysis of small molecules in enzyme samples or body fluids with minimal sample pretreatment is often not possible due to adsorption of proteins or other matrix components to the capillary wall causing irreproducible electroosmotic flows (EOF) and migration times. A promising approach to minimize this problem is the use of chemically modified covalently coated capillaries [27, 28], which have a longer lifetime and better reproducibility than non-covalently coated capillary, i.e. physically coating the BFS capillary. In addition, the covalently coated capillaries are easy to prepare and operate, and do not require the arduous fabrication of any frits for packed formats or blending of monomeric reagents with suitable porogens in precise proportions typically used in the preparation of monolithic column [28].
In this study, a covalently coated sulfonated capillary was prepared to develop a CZE-MS assay for the simultaneous analysis of the tryptophan metabolites (HAA, PA and QA) of the KP. To find the best conditions, several parameters, including the pH, type of background electrolyte (BGE), type of organic modifier, nebulizer gas pressure as well as the negative and positive ESI-MS mode were tested to find the best settings for the simultaneous separation of HAA with PA and QA. Apart from the optimization and quantitation of KP metabolites, the intra-day and inter-day reproducibility were performed. The ultimate goal was to study the enzymes indirectly responsible for the formation of QA and PA, which have been associated with neurodegenerative diseases and ageing. This presents a unique problem because QA and PA metabolites of the KP are formed spontaneously by non-catalytic decay of intermediates 2-amino-3-carboxymuconic semialdehyde (ACMS) and 2-aminomuconic semialdehyde (AMS), respectively. Therefore, the feasibility of CZE-ESI-MS assay was investigated for profiling the enzyme catalyzed reaction of the substrate, 3-HAA, to generate PA and QA. To our knowledge, this is the first report in which CZE-ESI-MS was developed and applied for the mechanistic studies of the enzymes competing with the non-enzymatic side reactions. This report opens up the possibility for studying KP enzymes in vitro by providing a highly repeatable and facile CZE-ESI-MS method for measuring the product distribution of the multi-enzyme system. In addition, the method has a potential to measure relevant concentration of PA and QA in biological samples (e.g., serum and cereberospinal fluid).
2 Materials and methods
2.1 Chemicals and reagents
2,2′-azobis (2-methylpropionitrile) (AIBN), 3-(trimethoxysilyl) propyl methacrylate (γ-MAPS) and 2-acrylamido-2-methyl-1-propanesulphonic acid (AMPS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (ACN), methanol (MeOH), proline and analytical-grade ammonium acetate (as 7.5 M NH4OAc solution) was also purchased from Sigma-Aldrich (St. Louis, MO, USA). Ammonium hydroxide (NH4OH), acetic acid (HOAc), and 3-hydroxyanthranilic acid, 97% (3-HAA) were obtained from Fisher Scientific (Springfield, NJ, USA). Quinolinic acid (QA), >98%, picolinic acid (PA), >99.9%, and N-2-hydroxy-ethylpiperazine-N-2-ethanesulfonic acid (HEPES), >99.5% were purchased from VWR International (Atlanta, GA, USA). Water used in all experiments was triply deionized and obtained from Barnstead Nanopure II water system (Barnstead International, Dubuque, IA, USA). The BFS capillary (375 μm od, 50 μm id) was obtained from Polymicro Technologies (Phoenix, AZ, USA).
2.2 Buffer and sheath liquid preparation
The background electrolyte (BGE) was prepared by diluting the stock 7.5 M NH4OAc solution and then adjusting to the desired pH values with HOAc or NH4OH using an Orion 420A pH meter (Beverly, MA, USA). The HEPES buffer was prepared at 25 mM with 5% glycerol by volume and the pH adjusted to 7.4 as measured by a Fisher Scientific AP115 pH meter (Springfield, NJ, USA) with ammonium hydroxide. The sheath liquids (80:20 (v/v) of MeOH/H2O containing 20 mM HOAc or NH4OH) were prepared by mixing aqueous solutions with appropriate volume ratio of MeOH. The running buffer and sheath liquid were filtered with 0.45 μm PTFE membranes and degassed for 20 min before use.
2.3 Enzyme preparation
3-Hydroxyanthranilate 3,4-dioxygenase (HAO) and α-amino-β-carboxy-ε-semialdehyde decarboxylases (ACMSD) were prepared as previously described with slightly modified procedures [29]. Briefly, cells were lysed by two passes through a microfluidizer processor, Microfluidics M-110P (Newton, MA USA). Crude lysate was clarified by centrifugation and cell free extract purified by affinity and size exclusion chromatography with an ÄKTA FPLC, GE Healthcare (Piscataway, NJ USA). Enzymes were concentrated with Amicon Ultra Centrifugal Filters, Ultra-15, pore size 10 kDa NMWCO, Millipore (Billerica, MA USA), and stored at −80°C until use.
2.4 Standard solutions and enzymes samples preparation
The stock solutions of PA, QA and proline (used as the internal standard (IS)) were prepared at 20 mM in 15 mM HEPES pH 7.4. Because 3-HAA is very poorly soluble in aqueous solution, a very concentrated stock solution (500 mM) was made in DMSO, a solvent in which 3-HAA is highly soluble. A mixture containing all analytes was prepared by taking certain amount of each stock solution and diluting to the desired concentrations at levels of 3, 6, 9, 19, 38, 75, 150 and 300 μM for 3-HAA and QA, whereas 10, 20, 50, 75, 150, 200, 250 and 300 μM concentrations were used for PA. The IS concentration was kept constant at 0.1 mM using 15 mM HEPES pH 7.4. For each of the enzyme samples, total enzyme concentration, the concentration of substrate, 3-HAA, and total sample volume were fixed at 100 nM, 300 μM, and 500 μL respectively. Five parallel enzyme samples were made by varying the ratio of HAO to ACMSD: 15:1, 10:1, 5:1, 2:1, and 1:1 respectively. Samples were prepared by diluting concentrated enzyme and substrate to the desired concentration in oxygen saturated HEPES buffer. The enzyme and the substrate were allowed to react for 12 h, after which the enzymes were removed by filtration with Amicon Ultra centrifugal filters, Ultracel – 0.5 mL 10 kDa MWCO, Millipore (Billerica, MA, USA).
2.5 Preparation of sulfonated capillary
First, a rehydroxylation process to maximize the number of silanol groups of the BFS capillary was performed. In brief, the capillary was flushed under vacuum with water, 1.0 M sodium hydroxide, water, 0.1 M hydrochloric acid, water, and MeOH for 0.5, 3, 0.5, 0.5, 0.5, and 0.5 h respectively. Second, silanization of the capillary inner wall with γ-MAPS was performed as described elsewhere [30]. The γ-MAPS was dissolved in methanol at 1:1 v/v, then filled through the pretreated capillary under vacuum. The filled capillary was sealed with rubber septa and kept at 60°C for 20 h in a GC oven. The capillary was rinsed with MeOH to flush out the residual reagents. Next, it was dried by nitrogen gas at 60°C for 3 h. Finally, the pretreated capillary was modified with the AMPS by the following procedures. The reaction mixture consisting of 20.0 mg AIBN, 40.0 mg AMPS, and 5.0 mL MeOH was sonicated for 20 min to obtain a homogeneous solution and then filled into the pretreated capillary. After the pretreated capillary was completely filled with the mixture, it was sealed at both ends with rubber septa. The sealed capillary was reacted at 60°C for 24 h and then flushed with MeOH to remove the unreacted materials.
2.6 Instrumentation
All CZE-ESI-MS experiments were carried out on an Agilent CE system interfaced to an Agilent 1100 series MSD quadrupole mass spectrometer (Agilent Technologies, Palo Alto, CA) equipped with an Agilent CE-MS adapter kit (G1603A), and an Agilent CE-ESI-MS sprayer kit (G1607). The sheath liquid was delivered using an Agilent 1100 series isocratic HPLC pump with a 1:100 splitter. Nitrogen was used as both nebulizing gas and drying gas. The Agilent ChemStation and CE-MSD add-on software were employed for instrument control and data analysis.
2.7 CZE-ESI-MS conditions
The method development was carried out with a BFS capillary (60 cm total length) or a sulfonated capillary (60 cm total length). For the BFS capillary, before the capillary was installed into the ESI-MS sprayer, it was flushed with 1 M NH4OH for 30 min and water for 10 min at 45°C. After installation of the capillary into the nebulizer, it was flushed with 1 M NH4OH and water for 3 min each before injection. As a preconditioning step, the capillary was rinsed with the running buffer for 4 min. After each run, the capillary was flushed with water (3 min), 1M NH4OH (4 min) and water (3 min). For the sulfonated capillary, the capillary was rinsed with the running buffer for 30 min before its first use. The capillary was flushed with the running buffer for 4 min after each CE-MS run. The separation voltage for both the BFS and sulfonated capillaries was set at +15 kV employing a voltage ramping of 3 kV/s. To ensure a good run-to-run reproducibility, a fresh buffer vial was always used for each run. The ESI-MS detection was performed in the selective ion monitoring (SIM) mode. The fragmentor voltage was optimized by direct infusion method, which involved continuously flushing 1 mg/mL analyte through a 60 cm long, 75 μm id BFS capillary at 50 mbar to the ESI interface. Unless otherwise stated, the following ESI-MS conditions were used: fragmentor voltage, 52 V; capillary voltage, 3500 V; drying gas flow rate, 5.0 L/min; drying gas temperature, 200°C. Resolution (Rs), noise level and S/N ratio was calculated with Agilent Chemstation software (V 9.0). The S/N ratio was calculated by taking the ratio of the peak height and noise, where the noise was measured as six times the standard deviation of the linear regression. The value for noise used in this work was determined in the time range closest to the peak. The peak symmetry was calculated by taking the ratio of the back and front half-widths at 10% of the peak height [31]. The positive [M+H]+ ions were monitored at 154 m/z for 3-HAA, 124 m/z for PA, 168 m/z for QA and 116 m/z for IS, and the negative [M-H]− ions were monitored at 152 m/z for 3-HAA, 122 m/z for PA and 166 m/z for QA. The calibration curves for 3-HAA, PA and QA were obtained by plotting the normalized peak area ratios of each analyte/(IS) versus concentrations of the analytes. The normalized peak area is defined as the ratio of the peak area to retention time.
3. Results and Discussion
3.1 Optimization of CZE-ESI-MS
3.1.1 Effect of BGE pH
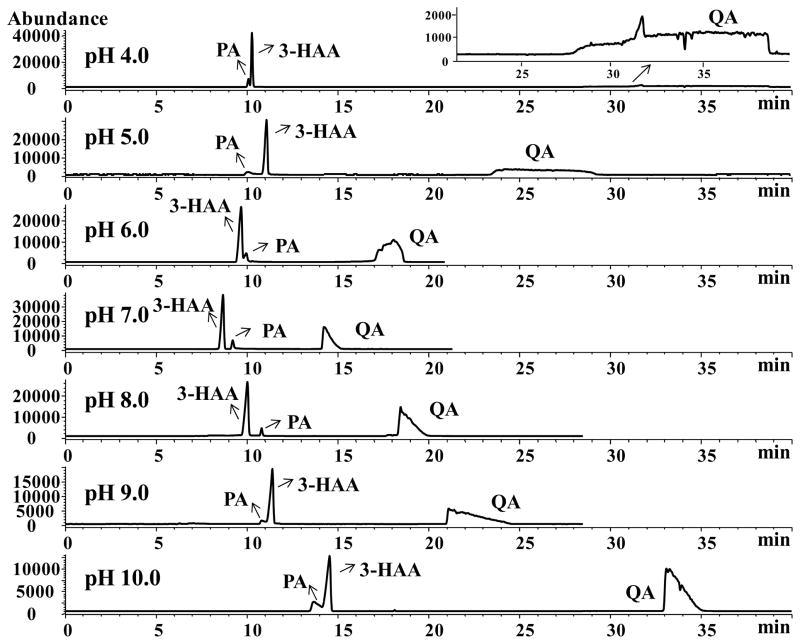
The substrate 3-HAA contains three substituted functional groups (primary amine, carboxylate and hydroxyl) on the benzene ring with a range of pKa, whereas the products (PA and QA) contain carboxylate and a heterocyclic nitrogen. Hence both substrate and product may acts as zwitterionic compounds at a certain pH (Figure 1). A series of BGEs containing 15 mM NH4OAc were studied over the range of pH from 4.0 to 10.0 in 1.0 unit increment. At pH lower than 4.0, the silanol groups on the surface of BFS capillary are partially protonated. Thus, the electroosmotic flow (EOF) is minimized, as a result, anionic QA was not eluted in 60 min at pH 3.0 (data not shown). As demonstrated in Figure 2, QA has a significantly longer migration time than 3-HAA and PA. This is because QA contains two ionizable carboxyl groups and one heterocylic nitrogen resulting in doubly negative charge at pH 7.0 or higher due to complete deprotonation of heterocyclic nitrogen and carboxyl groups. However, at lower pH values (e.g., pH 4–6), the peak shapes of QA is severely distorted due to electrostatic interaction of protonated pyridine nitrogen with capillary. It is not too surprising that QA acquires more effective negative charge than 3-HAA or PA over the studied pH range. Therefore, its electrophoretic mobility towards the anodic end (inlet) will be larger than 3-HAA and PA, thus leading to the longest migration time of the three. An interesting trend in the reversal of migration order of PA and 3-HAA was observed first upon increasing the pH from 5.0 to 6.0, and then a second reversal was observed in the pH range of 8.0 to 9.0 (Figure 2). This behavior is associated with different electrophoretic mobility of the two analytes, which is dependent on the effective negative charge of PA and 3-HAA. Speculating from the molar mass and pKa values of PA (123 g/mol, pKa1 = 1.07, pKa2 = 5.25) and 3-HAA (153 g/mol, pKa1 = 2.5, pKa2 = 5.2, pKa3 = 10.1), PA may acquire relatively more effective negative charge-to-mass ratio than 3-HAA in the pH range 6.0–8.0, leading to larger electrophoretic mobility towards the anodic (injector) end and longer migration time towards the cathodic (detector) end. However, at pH 9.0, the hydroxyl group of HAA is partially ionized leading to increase in negative charge on 3-HAA. A BGE containing 15 mM NH4OAc at pH 7.0 was selected for the further study because it provided a reasonable compromise between resolution, migration time and peak shapes for the three compounds. Optimization of the types of other volatile BGEs (i.e., ammonium carbonate and ammonium formate) and the additions of organic solvent (i.e., ACN and MeOH) provided no improvement in peak shapes of QA (Supporting Information, Figure S1 and S2).
Figure 2.
Effect of the buffer pH values on the separation of 3-HAA, PA and QA. CZE-ESI-MS conditions: 60 cm long (50 μm I.D.) BFS capillary. Running buffer, 15 mmol/L NH4OAc. Applied voltage, +15 kV. Capillary temperature, 20°C. Injection, 5 mbar, 10 s; Spray champer parameters: nebulizer gas pressure at 3 psi; drying gas temperature at 200°C; dry gas flow at 5 L/min; capillary voltage at −3500 V; fragmentor at 65 V. Sheath liquid: 80:20 MeOH/water containing 20 mM NH4OH, sheath liquid flow rate at 0.5 mL/min. Analytes: 0.5 mM for 3-HAA, 0.2 mM for PA and 0.3 mM for QA.
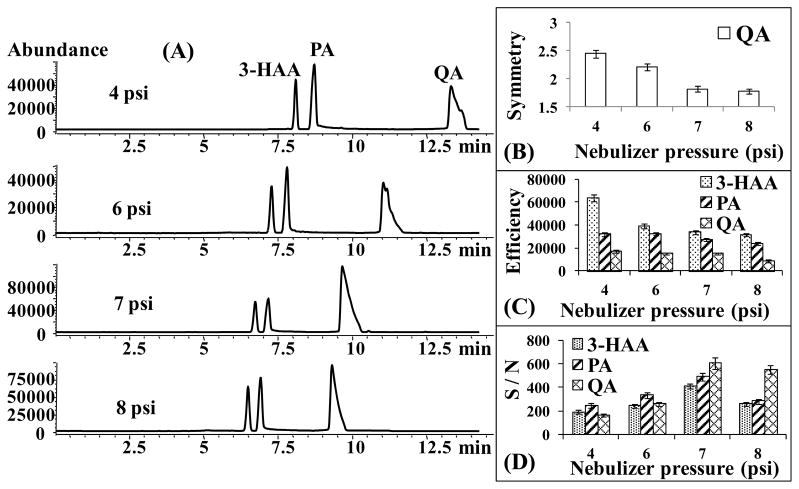
3.1.2 Effect of nebulizer gas pressure
The nebulizer gas pressure is an important parameter which needs to be optimized for CZE-ESI-MS separation. The effect of nebulizer gas pressure on the separation and detection of 3-HAA, PA and QA was investigated by varying from 4 to 8 psi while keeping all other parameters constant. One of the advantages of using higher nebulizer pressure is the improvement of the peak shape. As shown in Figure 3B, the peak symmetry of QA was apparently improved upon increasing the nebulizer gas pressure from 4 to 8 psi. However, the disadvantage is that the nebulizer gas pressure has the potential to generate a suction force at the capillary outlet [32]. Consequently, a laminar flow is formed inside the capillary, decreasing the separation efficiency (N) (Figures 3C). On the other hand, the plot shown in Figure 3D demonstrates that increasing the nebulizer gas pressure improves initially the detection sensitivity. More specifically, the maximum abundance of the analytes was improved ~2-fold as the nebulizer gas pressure increased from 4 to 8 psi (Figure 3A), but the S/N ratio only increased to a maximum at 7 psi (Figure 3D). Further increase of the nebulizer pressure to 8 psi increases the noise level, decreasing S/N. This trend indicates that high nebulizer gas pressure could influence the stability of the electrospray [33]. Therefore, at 7 psi a good compromise was achieved due to higher S/N and improved peak shape of QA.
Figure 3.
Effect of the nebulizer gas pressure on the separation and sensitivity of 3-HAA, PA and QA. CZE-ESI-MS conditions: Running buffer, 15 mmol/L NH4OAc pH 7.0. Injection, 5 mbar, 100 s. Analytes: 0.4 mM for 3-HAA, 1 mM for PA and 1 mM for QA. Other conditions are the same as in Figure 2.
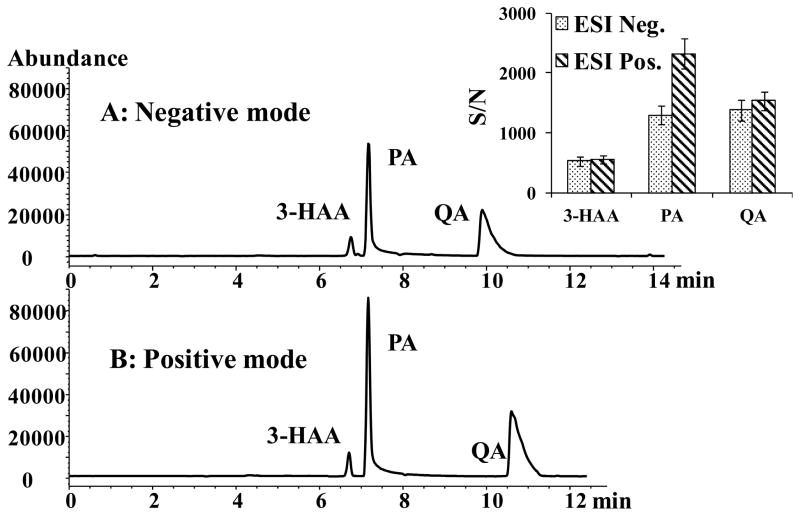
3.1.3 Comparison of positive and negative CZE-ESI-MS detection
As mentioned earlier, 3-HAA, PA and QA are polyprotic compounds containing acidic carboxyl or hydroxyl groups and basic primary amine or heterocyclic nitrogen groups. These functional groups can be easily ionized in ESI negative and positive mode, respectively. Comparison of TIC chromatograms generated in both negative and positive CZE-ESI-MS modes is shown in Figure 4. While the S/N for 3-HAA and QA are not significantly different in positive or negative ion mode, the S/N of PA is much higher in positive ion mode. One possible reason is that PA has a pyridinium nitrogen whose gas phase proton affinity is much higher compared to the carboxylate group, (which has a higher tendency to loose proton) resulting in overall highly intense protonated molecular ion [34]. In contrast, in case of 3-HAA and QA, the presence of multiple acidic residues enhances the negative ion ESI-MS signal. Thus, presence of one high affinity protonation sites and two deprotonation sites on 3-HAA and QA results in overall similar ability to gain or loss a proton generating similar S/N in the gas phase in both modes. Furthermore, when the enzyme sample with HAO:ACMSD of 15:1 was analyzed in ESI positive and negative mode, respectively the sample matrix effects was a problem in the latter mode. For example, the peak of QA was masked partially by a broad impurity peak from the enzyme sample in the negative ion mode whereas a clear background was obtained in the positive mode.
Figure 4.
Comparison of negative and positive ion mode of CZE-ESI-MS. CZE-ESI-MS conditions: Running buffer, 15 mmol/L NH4OAc pH 7.0. (A) Negative mode: sheath liquid: 80:20 MeOH/water containing 20 mM NH4OH, capillary voltage at −3500 V; (B) Positive mode: sheath liquid: 80:20 MeOH/water containing 20 mM HOAc, capillary voltage at +3500 V. Analytes: 0.1 mM for 3-HAA, 0.8 mM for PA and 0.3 mM for QA. Other conditions are the same as in Figure 2.
3.1.4 Repeatibility of migration time of sulfonated capillary
It is well known that variability of migration time and peak area is an important issue in CZE-MS. When preparing polymer-based monolithic columns [35–37], AMPS was usually chosen as a charged monomer to generate a strong and stable EOF to transport the mobile phase through the monolithic column. To obtain reproducible migration times and peak areas of analytes, sulfonated groups were chemically introduced to the inner wall of untreated BFS capillary by chemical modification with AMPS via a silanization reaction followed by an in situ graft polymerization reaction (see experimental section). This modification to produce sulfonated capillary yields a pH independent and relatively constant EOF [27, 38, 39]. Comparison of migration time and peak area repeatability on sulfonated capillary versus BFS is shown in Table 1. Clearly, the % RSDs of the migration times and peak areas of analytes with the sulfonated capillary were less than 0.94 and 7.5%, respectively which were significantly lower than those obtained with a BFS capillary (5.9 and 17.7%, respectively).
Table 1.
Reproducibility of the migration times and peak areas on the sulfonated and BFS capillaries.
| Compounds(a) | Sulfonated capillary | BFS capillary | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Intraday repeatability (%RSD) (n = 10) | Interday repeatability (%RSD) (n = 5) | Intraday repeatability (%RSD) (n = 10) | Interday repeatability (%RSD) (n = 5) | |||||
|
| ||||||||
| Time | Area | Time | Area | Time | Area | Time | Area | |
| 3-HAA | 0.51 | 5.7 | 0.63 | 7.5 | 3.5 | 14.3 | 5.1 | 17.1 |
| PA | 0.55 | 5.1 | 0.69 | 7.2 | 4.0 | 15.9 | 5.4 | 17.7 |
| QA | 0.83 | 4.9 | 0.94 | 7.0 | 4.7 | 14.8 | 5.9 | 16.6 |
All the conditions are the same as Figure 4B.
0.3 mM in 15 mM HEPS pH 7.4.
3.2 Comparison of CZE-UV and CZE-ESI-MS
The standard mixtures of 3-HAA, PA and QA were separated by CZE and detected by both UV and MS, respectively to compare the sensitivity of the two detection techniques (Supporting Information, Figure S3). As shown in Figure S3A, the CZE-UV at a concentration of 0.5 mM provided the S/N of 22 for all three analytes (3-HAA, PA and QA). However, the CZE-ESI-MS (Figure S3B) provided much higher S/N at the same injection size. For example, the S/N was ~128-fold higher for 3-HAA, and 70–75-fold higher for 0.2 mM PA and 0.3 mM QA. The results confirm that, CZE-ESI-MS is significantly more sensitive than CZE-UV.
3.3 Profiling substrate and products in enzyme samples
First, the calibration curves, using an internal standard (IS), proline, were set up to simultaneously quantitate 3-HAA, PA and QA. The normalized peak area ratios of each analyte/IS were plotted versus the concentrations of each analyte (data not shown). The calibration curves covered at least one order of magnitude with good linearity. Certainly, high concentrations can be analyzed, which may not be required for most studies. For the substrate, 3-HAA, the range of 19–300 μM (R2 = 0.9948) whereas for the products, 75–300 μM for PA (R2 = 0.9979) and 19–300 μM for QA (R2 = 0.9906) were obtained. The limit of quantitation (LOQ) of 3-HAA, PA and QA were found to be 19, 75 and 19 μM, respectively (based on S/N of 10:1). The limit of detection (LOD) of 3-HAA, PA and QA were found to be 3, 20 and 6 μM, respectively (based on S/N of 3:1).
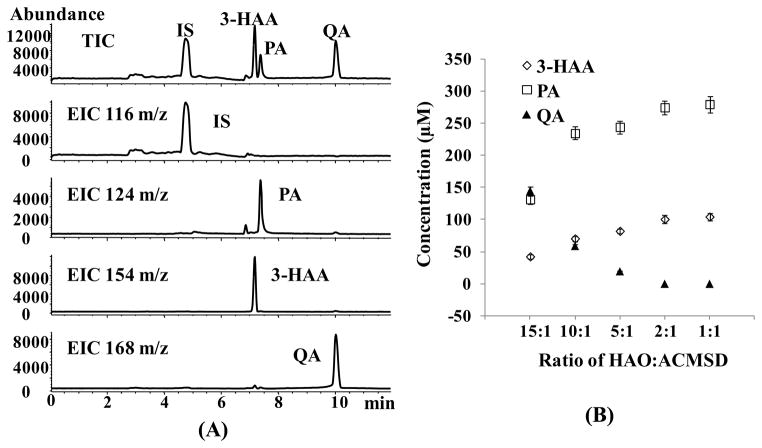
The developed CZE-ESI-MS method was applied to profile the initial substrate (3-HAA) as well as the formation of products (PA and QA) after incubation of 3-HAA with a pair of enzyme (HAO and ACMSD). Five parallel samples were prepared by varying the ratio of HAO:ACMSD at 15:1, 10:1, 5:1, 2:1, and 1:1, respectively, while keeping the total enzyme concentration fixed at 100 nM, substrate concentration at 300 μM, and reaction time at 12 hr. By applying the developed CZE-ESI-MS method, these enzyme/substrate samples with different ratio of HAO to ACMSD were analyzed. The results show that the relative amount of each enzyme present in the reaction system does affect the product distribution. A representative electrochromatogram of an enzyme sample with HAO:ACMSD of 15:1 (Figure 5A) showed good selective separation of HAA from PA and QA and clear background. In addition, Table 2 showed good run-to-run repeatibility for migration times and peak areas of the enzyme samples. The run-to-run migration times repeatability of <2.9% RSD (n = 15) suggest that the EOF was stable, with minimal protein adsorption to the capillary wall of the sulfonated coated capillary.
Figure 5.
CZE-ESI-MS of 3-HAA, PA and QA in the enzyme samples with different ratio of HAO:ACMSD. (A): Represented total ion chromatogram and extracted ion electrochromatograms of substrate 3-HAA incubated with the enzyme samples (HAO:ACMSD) in the ratio of 15:1. (B): Concentrations of substrate and products profile with different ratio of HAO:ACMSD. Other conditions are the same as Figure 4B.
Table 2.
Reproducibility of the migration times and peak areas of the enzyme samples.
| Samples with HAO:ACMSD | Run-to-run repeatability (% RSD) (n = 3)(a) | |||||
|---|---|---|---|---|---|---|
| 3-HAA | PA | QA | ||||
|
| ||||||
| Time | Area | Time | Area | Time | Area | |
| 15: 1 | 0.41 | 4.0 | 0.45 | 4.4 | 0.61 | 5.1 |
| 10: 1 | 0.48 | 3.9 | 0.53 | 4.1 | 0.69 | 5.8 |
| 5: 1 | 0.61 | 4.5 | 0.67 | 4.3 | 0.89 | 6.9 |
| 2: 1 | 0.57 | 4.9 | 0.67 | 5.3 | -(b) | - |
| 1: 1 | 0.63 | 5.6 | 0.72 | 5.7 | - | - |
|
| ||||||
| 2.1 (n = 15) | N (c) | 2.9 (n = 15) | N | 2.5 (n = 9) | N | |
All the conditions are the same as Figure 5.
Each enzyme sample was run in triplicate to calculate the run-to-run repeatability.
Not available because the peak was below LOQ.
Not available because the peak areas were different for the enzyme samples with variable HAO:ACMSD.
The profiles of the substrate and its products concentration after enzymatic reaction at five different ratio of HAO to ACMSD is shown in Figure 5B. Clearly, as the ratio of HAO to ACMSD decreased from 15:1 to 1:1, the concentration of 3-HAA and PA increased but the concentration of QA decreased. As shown in KP in Figure 1, HAO is known to oxidize 3-HAA to ACMS, from which QA is non-enzymatically derived. In addition, ACMS can be diverted by ACMSD to a benign catabolite, AMS, a much less stable molecule that rapidly converts to PA [6, 40]. Note that keeping the total enzyme concentration fixed at 100 nM, when the ratio of HAO to ACMSD decreased, the concentration of HAO decreased, and the concentration of ACMSD increased. Consequently, less substrate, 3-HAA, was oxidized to ACMS, which in turn decrease the concentration of non-enzymatically derived QA. Simultaneously more ACMSD would convert ACMS to AMS. As mentioned earlier, AMS is a much less stable molecule which rapidly converts to PA. The results showed that as the ratio of HAO:ACMSD decreased, the concentration of QA approached essentially zero, indicating that all ACMS formed by the action of HAO was consumed by ACMSD rather than decaying to QA. Therefore, the study demonstrates that the product distribution of PA and QA can be measured by varying the function of two enzyme ratio. Moreover, the developed CZE-ESI-MS method for profiling substrate and products may provide a potential method to study the effects of possible relevant enzyme-enzyme interactions which may take place in the in vitro multienzyme system.
4. Concluding remarks
A rapid, specific and repeatable CZE-ESI-MS method was developed for assaying the initial substrate, 3-HAA, and its products, PA and QA, in the multienzyme system. The combined use of a sulfonated capillary and a single quadrupole MS provided a simple hyphenation method, which produces high repeatability of migration time and peak area for selective MS detection. In addition, only a simple filtration of enzyme sample was required, which makes the sample preparation significantly less time-consuming. Next, the developed CZE-ESI-MS method was applied to profile the initial substrate, 3-HAA, as well as the formation of PA and QA after enzymatic reaction. As the ratio of HAO to ACMSD decreased from 15:1 to 1:1, the concentration of 3-HAA and PA increased, and the concentration of QA decreased. These trends of the concentrations of substrate and products versus enzyme ratio of HAO to ACMSD demonstrated the enzymes (HAO and ACMSD) compete with those of non-enzymatic side reactions to PA and QA.
The results shown in this study suggest that CZE-ESI-MS method has the potential to provide good repeatability and rapid separation in analyzing other polar and charged tryptophan metabolites involved in the KP. Through the use of the developed CZE-ESI-MS method with the sulfonated capillary it will be possible to study other relevant enzyme-enzyme interactions, which may take place in the in vitro multi-enzyme system for use in both diagnostics and mechanistic studies. While improve peak shape of QA was obtained at higher nebulizer pressure, in our future work we plan to change the modified functional groups on the surface of the coated capillary to further improve the peak shape of the analytes. Moreover, the improvement in sensitivity of the PA and QA by CZE-ESI-MS/MS should provide an efficient diagnostic tool to measure the concentration of these metabolites and help usher the next generation of highly reliable and sensitive CZE-ESI-MS method for the analysis of tryptophan metabolites of KP in biological samples.
Supplementary Material
Acknowledgments
This work was financially supported by the National Institutes of Health Grant (R01-GM-062314).
Abbreviations
- QA
quinolinic acid
- PA
picolinic acid
- 3-HAA
3-hydroxyanthranilic acid
- KP
kynurenine pathway
- NAD+
nicotinamide adenine dinucleotide
- ACMS
α-amino-β-carboxymuconate-ε-semialdehyde
- AMS
α-aminomuconate-ε-semialdehyde
- ACMSD
α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase
- BFS
bare fused silica
- γ-MAPS
3-(trimethoxysilyl) propyl methacrylate
- AMPS
2-acrylamido-2-methyl-1-propanesulphonic acid
- HAO
3-hydroxyanthranilate 3,4-dioxygenase
References
- 1.Stone TW, Darlington LG. Nat Rev Drug Discov. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 2.Schwarcz R. Curr Opin Pharmacol. 2004;4:12–17. doi: 10.1016/j.coph.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Mehler AH, Yano K, May EL. Science. 1964;145:817–819. doi: 10.1126/science.145.3634.817. [DOI] [PubMed] [Google Scholar]
- 4.Pérez-De La Cruz V, Köningsberg M, Santamaría A. CNS Neurol Disord Drug Targets. 2007;6:398–410. doi: 10.2174/187152707783399229. [DOI] [PubMed] [Google Scholar]
- 5.Henderson LM, Hirsch HM. J Biol Chem. 1949;181:667–675. [PubMed] [Google Scholar]
- 6.Grant RS, Coggan SE, Smythe GA. Int J Tryptophan Res. 2009;2:71–79. doi: 10.4137/ijtr.s2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jong WHA, Smit R, Bakker SJL, De Vries EGE, Kema IP. J Chromatogr B. 2009;87:603–609. doi: 10.1016/j.jchromb.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Möller M, Du Preez JL, Harvey BH. J Chromatogr B. 2012;898:121–129. doi: 10.1016/j.jchromb.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 9.Dazzi C, Candiano G, Massazza S, Ponzetto A, Varesio L. J Chromatogr B. 2001;751:61–68. doi: 10.1016/s0378-4347(00)00450-3. [DOI] [PubMed] [Google Scholar]
- 10.Herve C, Beyne P, Jamault H, Delacoux E. J Chromatogr B. 1996;675:157–161. doi: 10.1016/0378-4347(95)00341-x. [DOI] [PubMed] [Google Scholar]
- 11.Vignau J, Jacquemont MC, Lefort A, Imbenotte M, Lhermitte M. Biomed Chromatogr. 2004;18:872–874. doi: 10.1002/bmc.445. [DOI] [PubMed] [Google Scholar]
- 12.Vaarmann A, Kask A, Maeorg U. J Chromatogr B. 2002;769:145–153. doi: 10.1016/s1570-0232(01)00639-0. [DOI] [PubMed] [Google Scholar]
- 13.Amirkhani A, Rajda C, Arvidsson B, Bencsik K, Boda K, Seres E, Markides KE, Vecsei L, Bergquist J. Eur J Neurol. 2005;12:625–631. doi: 10.1111/j.1468-1331.2005.01041.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Miyazaki T, Shibata T, Hara N, Tsuchiya M. J Chromatogr B. 2008;867:57–61. doi: 10.1016/j.jchromb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Liao X, Zhu J, Rubab M, Feng Y, Poon R. J Chromatogr B. 2010;878:1003–1006. doi: 10.1016/j.jchromb.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Eckstein JA, Ammerman GM, Reveles JM, Ackermann BL. J Mass Spectrom. 2008;43:782–790. doi: 10.1002/jms.1376. [DOI] [PubMed] [Google Scholar]
- 17.Notarangelo FM, Wu H, Macherone A, Graham DR, Schwarcz R. Anal Biochem. 2012;421:573–581. doi: 10.1016/j.ab.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramautar R, Demirci A, De Jong GJ. Trends Anal Chem. 2006;25:455–466. [Google Scholar]
- 19.Shamsi SA, Miller BE. Electrophoresis. 2004;25:3927–3961. doi: 10.1002/elps.200406131. [DOI] [PubMed] [Google Scholar]
- 20.Castro-Puyana M, García-Cañas V, Simó C, Cifuentes A. Electrophoresis. 2012;33:147–167. doi: 10.1002/elps.201100385. [DOI] [PubMed] [Google Scholar]
- 21.Weber PL, Malis M, Palmer SD, Klein TL, Lunte SM. J Chromatogr A. 1997;697:263–268. doi: 10.1016/s0378-4347(97)00094-7. [DOI] [PubMed] [Google Scholar]
- 22.Tenorio-López FA, del Valle-Mondragón L, Martínez-Lazcano JC, Sánchez-Mendoza A, Ríos C, Pastelín-Hernández G, Pérez-Severiano F. Chromatographia. 2007;65:725–731. [Google Scholar]
- 23.Malone MA, Zuo H, Lunte SM, Smyth MR. J Chromatogr A. 1995;700:73–80. [Google Scholar]
- 24.Arvidsson B, Johannesson N, Citterio A, Righetti PG, Bergquist J. J Chromatogr A. 2007;1159:154–158. doi: 10.1016/j.chroma.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 25.Ullsten S, Danielsson R, Bäckström D, Sjöberg P, Bergquist J. J Chromatogr A. 2006;1117:87–93. doi: 10.1016/j.chroma.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 26.Guillo C, Barlow D, Perrett D, Hanna-Brown M. J Chromatogr A. 2004;1027:203–212. doi: 10.1016/j.chroma.2003.11.102. [DOI] [PubMed] [Google Scholar]
- 27.Lu M, Zhang L, Lu Q, Chi Y, Chen G. Electrophoresis. 2009;30:2273–2279. doi: 10.1002/elps.200800683. [DOI] [PubMed] [Google Scholar]
- 28.Dong X, Wu R, Dong J, Wu M, Zhu Y, Zou H. Electrophoresis. 2009;30:141–154. doi: 10.1002/elps.200800412. [DOI] [PubMed] [Google Scholar]
- 29.Li T, Iwaki H, Fu R, Hasegawa Y, Zhang H, Liu A. Biochemistry. 2006;45:6628–6634. doi: 10.1021/bi060108c. [DOI] [PubMed] [Google Scholar]
- 30.Dong J, Ou J, Dong X, Wu R, Ye M, Zou H. J Sep Sci. 2007;30:2986–2992. doi: 10.1002/jssc.200700402. [DOI] [PubMed] [Google Scholar]
- 31.Kanoatov M, Retif C, Cherney LT, Krylov SN. Anal Chem. 2012;84:149–154. doi: 10.1021/ac203129h. [DOI] [PubMed] [Google Scholar]
- 32.Shamsi SA. Anal Chem. 2001;73:5103–5108. doi: 10.1021/ac0105179. [DOI] [PubMed] [Google Scholar]
- 33.Feng HT, Yuan LL, Li SFY. J Chromatogr A. 2003;1014:83–91. doi: 10.1016/s0021-9673(03)00942-7. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann ED, Stroobant V. Mass Spectrometry. 3. John Wiley and Sons, Ltd; 2007. [Google Scholar]
- 35.Delaunay-Bertoncini N, Demesmay C, Rocca JL. Electrophoresis. 2004;25:3204–3215. doi: 10.1002/elps.200305975. [DOI] [PubMed] [Google Scholar]
- 36.Pucci V, Raggi MA, Svec F, Fréchet JMJ. J Sep Sci. 2004;27:779–788. doi: 10.1002/jssc.200401828. [DOI] [PubMed] [Google Scholar]
- 37.Bedair M, El Rassi Z. Electrophoresis. 2002;23:2938–2948. doi: 10.1002/1522-2683(200209)23:17<2938::AID-ELPS2938>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Kodama S, Yamamoto A, Terashima H, Honda Y, Taga A, Honda S. Electrophoresis. 2005;26:4070–4078. doi: 10.1002/elps.200500445. [DOI] [PubMed] [Google Scholar]
- 39.Xu L, Feng Y, Shi Z, Da S, Wei F. J Chromatogr A. 2004;1033:161–166. doi: 10.1016/j.chroma.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Fukuoka S, Ishiguro K, Yanagihara K, Tanabe A, Egashira Y, Sanada H, Shibata K. J Biol Chem. 2002;277:35162–35167. doi: 10.1074/jbc.M200819200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.