Abstract
It could be argued that the greatest transformative aspect of the Human Genome Project has been not the sequencing of the genome itself, but the resultant development of new technologies. A host of new approaches has fundamentally changed the way we approach problems in basic and translational research. Now, a new generation of high-throughput sequencing technologies promises to again transform the scientific enterprise, potentially supplanting array-based technologies and opening up many new possibilities. By allowing DNA/RNA to be assayed more rapidly than previously possible, these next-generation platforms promise a deeper understanding of genome regulation and biology. Significantly enhancing sequencing throughput will allow us to follow the evolution of viral and bacterial resistance in real time, to uncover the huge diversity of novel genes that are currently inaccessible, to understand nucleic acid therapeutics, to better integrate biological information for a complete picture of health and disease at a personalized level and to move to advances that we cannot yet imagine.
What would you do if you could sequence everything? What if sample preparation was simple and unbiased? What types of experiments could you envision? Although we have not quite reached the point where cost and technology are no object, new sequencing techniques1–5 have not simply changed the landscape but have placed basic, clinical and translational research scientists into a new and unfamiliar world in which entirely different types of questions can be addressed. Sequence data are providing both a level of precision and a breadth of scope that were unimaginable only a few years ago. The rapidity with which this change has occurred has left many unaware of the opportunities that these new technologies provide. However, even those aware of the opportunities are faced with the issues of the up-front costs and infrastructure that limit access to these technologies.
After the completion of the Human Genome Project, the oft-asked question was how could the enormous capacity of genome centers be used productively once the most interesting sampling of other species was complete? Implicit in this question were two assumptions: first, that additional sequence data would not be particularly useful in the future and second, that genome sequencing would remain a costly endeavor that could be practiced only in ‘industrial’ centers. Instead, the better question is, how can cheap and accessible sequencing, delivered by new and emerging technologies, be leveraged with insightful approaches to biology and medicine to maximize the benefits to all? DNA sequence is no longer just an end in itself, but it is rapidly becoming the digital substrate replacing analog chip-based hybridization signals; it is the barcode tracking of enormous numbers of samples; and it is the readout indicating a host of chemical modifications and intermolecular interactions. Sequence data allow one to count mRNAs or other species of nucleic acids precisely, to determine sharp boundaries for interactions with proteins or positions of translocations and to identify novel variants and splice sites, all in one experiment with digital accuracy.
The brief history of molecular biology and genomic technologies has been marked by the introduction of new technologies, their rapid uptake and then a steady state or slow decline in use as newer techniques are developed that supersede them. For example, as measured by publications listed in PubMed, gel-based hybridization techniques for analyzing DNA and RNA that were introduced in the late 1970s reached their zenith in the early 1990s and then entered a decline when microarray technologies burst onto the scene (Fig. 1). Microarrays achieved a rapid uptake, displacing the less powerful blotting techniques. Similarly, DNA footprinting for determining protein-DNA interactions became widely used in the early 1980s, peaked in the mid-1990s and then began declining as approaches based on chromatin immunoprecipitation (ChIP) supplanted footprinting. Publications on ultra-high-throughput, second-generation or third-generation sequencing first appeared in the late 1990s, but have not yet attained the rapid rise in publications characteristic of the other technologies. This is because the technology is complex and it has been out of reach to the majority of researchers up until the present. Now, access to the technology is growing and one can predict a huge surge in usage as the new sequencing technologies supplant many aspects of not just the older sequencing methods, but also various methods for assessing gene expression, protein binding and other biological information.
Figure 1.
The number of publications with keywords for nucleic acid detection and sequencing technologies. PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez) was searched in two-year increments for key words and the number of hits plotted over time. For 2007–2008, results from January 1–March 31, 2008 were multiplied by four and added to those for 2007. Key words used were those listed in the legend except for new sequencing technologies (‘next-generation sequencing’ or ‘high-throughput sequencing’), ChIP (‘chromatin immunoprecipitation’ or ‘ChIP-Chip’ or ‘ChIP-PCR’ or ‘ChIP-Seq’), qPCR (TaqMan or qPCR or ‘real-time PCR’) and SNP analysis (SNPs or ‘single-nucleotide polymorphisms’ and not nitroprusside (nitroprusside is excluded because sodium nitroprusside is sometimes abbreviated as ‘SNP’ but is generally unrelated to genetics)).
The Human Genome Project gave scientists a nearly complete reference map of DNA sequences6. In addition to providing a scaffold on which to place sequence data, this map also revealed the gaping holes in our understanding of protein coding sequences and the diversity of RNAs and their multiple roles. All too glaring were the vast expanses of sequence with no recognizable features but very strong evolutionary conservation, implying novel, yet unknown, functions. Recent studies have demonstrated that much more of the genome is functionally active than previously imagined and that genomic structural diversity is greater than anticipated. Our understanding of genome biology and its involvement in disease etiology must be supplemented to include the whole genome and functional elements beyond just the protein coding regions.
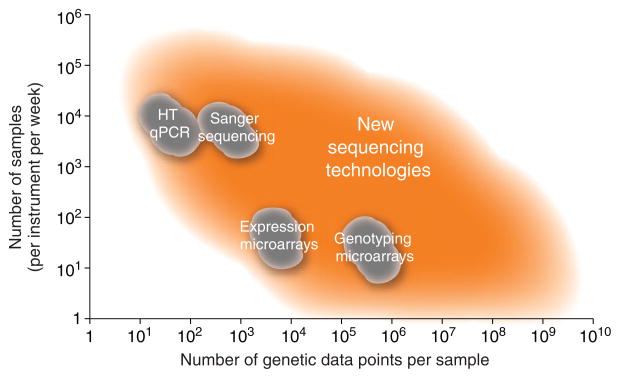
True understanding of how genes function requires knowledge of their expression patterns, their impact on all other genes and their effects on DNA structure and modifications. These data will have to be obtained across large numbers of cell types, individuals, environments and time points. The scale and precision of information required to even begin to approach these questions was unattainable without highly parallel sequencing technologies that are only now entering into use. New genetic analysis technologies not only are flexible, but also are of sufficient throughput and low enough cost for processing the large number of samples needed to generate statistically meaningful information (Fig. 2).
Figure 2.
Relative sample and data throughputs for different nucleic acid detection and sequencing technologies. A rough estimate of the number of samples that can be run on a single instrument in one week with the resultant data points is shown on a logarithmic scale for different technologies. This is intended for comparative scale and is not exact.
A door on a new era of genomic and biological insight has just opened, as demonstrated by the wide variety of large-scale, multi-dimensional studies recently reported. But these are just a taste of what will soon be possible. Here, we provide a review of genomic discoveries that both highlight the usefulness of integrative, high-throughput genomic technologies and paint a vision for the future of high-definition, sequence-driven biomedical research. The benefits of integrating information from various genomic assays, of increasing both the temporal and spatial resolution of experiments, and of applying these methods to much larger numbers of individuals will be explored and coupled with the expanding opportunities of new sequencing technologies that are making this kind of transformative experimentation possible.
Cataloging sequences and their variation
The most direct and obvious result of enhanced sequencing capabilities has been the simple accumulation of much more sequence data, and hence, many sequences from different species that allow more informative analyses of phylogeny and evolution. The wide variety of completed and draft genome sequences currently available is testament to the power of classical sequencing and brute force approaches. The rapid resequencing of multiple strains of Drosophila7, Caenorhabditis elegans8 and even humans9 provides a taste of things to come with the new techniques that are supplanting the earlier methods. Indeed, even DNA from long-extinct species is now being sequenced on a regular basis (reviewed by Millar et al.10). With the much larger range of sequences becoming available, it will now be possible to study the effects of selective forces in evolution at an individual level rather than aggregating many effects from population studies11. Individual variation has been used to great effect in recent whole-genome studies for understanding disease associations in humans with dozens of novel genes implicated in various phenotypes12.
Single-nucleotide polymorphisms
The search for genetic determinants of disease has depended greatly upon the discovery of ever better molecular markers. These markers are informative signposts distributed throughout the genome at the highest resolution feasible at the time. Initially, restriction fragment length polymorphisms, and then satellite tandem repeats, and more recently, single-nucleotide polymorphisms (SNPs) have represented the most commonly used genetic markers for disease-association studies. A high-resolution, genome-wide map of common SNPs is available and is amenable to high-throughput analysis. Classic sequencing provided the impetus for the SNP Consortium that delivered the first glimpses into the rich diversity of human DNA variation13. This was followed by the international HapMap project, which has catalogued the common patterns of genetic variation in humans14,15. The HapMap has expanded the usefulness of many individual SNPs by allowing the use of tagging SNPs as surrogates for other SNPs that are in linkage disequilibrium, or correlated, with the tagging SNPs. Availability of these data increased the effective coverage of genome-wide scans by allowing more efficient interrogation of known variation. The benefits of this understanding are unquestioned as results of whole-genome association studies reported in 2007 and 2008 have illuminated in incredible detail the role that simple variation plays in disease etiology and progression16. These studies have been possible because of the genome-wide nature of projects such as the Human Genome Project, the SNP database (dbSNPs) and HapMap, as well as the ease with which scientists could access these data.
Although the number of new disease-associated loci has been an impressive feat, allowing novel insights into many diseases, the results have also revealed issues that show the limitations of how far SNP-based genome-wide association studies with common variation can take us. Whereas some associations are located in well-characterized genes and generate easily predictable outcomes on protein function, many, in fact, land in poorly characterized regions of the genome that require extensive additional study before functional insight can occur17. Because these SNPs are potentially only markers of functional polymorphism, another round of studies is then required to fine-map the region of interest, either by studying additional SNPs or by sequencing the region in many individuals. Furthermore, SNP studies are limited to known and common variants. Although the current standard of 500,000–1,000,000 SNPs per assay is a lot of SNPs, it still only scratches the surface of variation by assessing less than 0.1% of DNA positions, thus addressing only known and common variation. Examination of haplotypes does not solve this problem completely, as they are limited in their ability to describe the genome, especially for uncommon variants. There is a large number of SNPs, referred to as singleton SNPs, which can be detected only by direct examination and not by other SNPs in linkage disequilibrium with them18. Only a tiny fraction of the genetic basis of common diseases can currently be explained by associations with common variants. This clearly indicates that uncommon and rare variants play a major role in many phenotypes, as has also been found by direct examination19,20. These rare variants are not readily addressed by current genotyping technologies. Furthermore, somatic mutations that may be causative for development of diseases such as cancer cannot be approached though haplotype analysis. Substantially expanded sequencing in large populations of individuals will be necessary for a deeper understanding of the genetic basis of disease. At present, sequencing is generally restricted to candidate genes, and provides only a narrow window on disease. Without question, if sequencing were as easy and cheap as genotyping, high-throughput genotyping would become a technique of the past.
Copy number variable regions
SNPs are not the only class of variation associated with important phenotypic consequences. From the early days of cytogenetics, structural genomic aberrations have been known to play a role in human disease. Most notably, chromosomal gains or losses have been associated with developmental defects and cancer. Until recently, only relatively large-scale rearrangements such as chromosomal aneuploidies and megabase-sized deletions, duplications, translocations, or inversions, detectable by traditional karyotyping techniques, were studied and used as diagnostic markers. With the advent of high-resolution genome-wide technologies and techniques21, previously undetected submicroscopic structural variations have been shown to exist, both in the genomes of diseased individuals, as well as in the genomes of normal populations22–25.
Using both genotyping SNP arrays and comparative genomic hybridization on large-insert clone arrays, all 270 HapMap samples have been analyzed for genomic gains or losses25. More than 1,400 copy number variable regions (CNVRs) were found, representing over 12% of the genome, greater than half of which were present in more than one individual. The unexpectedly high level of CNVRs in these HapMap individuals was further confirmed by the resequencing of individual genomes9,26. Also, by comparing these data to the latest reference genome assembly, almost half of the gaps in the reference assembly were found to be flanked by or contain CNVRs. Taken together, these findings have served as an impetus for the inclusion of CNVR assessment in population association studies, and highlight the fact that the reference human genome is, in fact, only an approximation of any individual genome. Like SNPs, CNVRs account for the modulation of many complex phenotypic traits and disease susceptibility in humans, and may play a role in environmental adaptation25,27,28. CNVRs, even more so than SNPs, will benefit from high-throughput sequence data, not only because their presence will be detectable, but also because the boundaries of insertions and deletions can be established with respect to other sequences.
Dynamic DNA and mixed genomes
Analysis of SNPs and CNVRs has been extraordinarily powerful when examining the static germline genome. However, there are situations in which the genome can deviate from its original sequence, and resequencing data are uniquely able to detect such alterations. In tumor cells, structural variation or preexisting SNPs may predispose a cell to uncontrolled growth and metastasis, but it is frequently de novo somatic mutations and rearrangements that have the greatest effect on tumor growth and disease progression29,30. Genomic alterations may vary even within different parts of the tumor because of variations in growth and selective pressure, so that the tumor genome may be a complex mixture of many genomes that defies simple SNP or CNVR classification. Resequencing of the coding regions in tumor genomes has already reinforced the importance of de novo mutations. These data have shown that many different mutations in many genes, and not simply mutations in the well-studied oncogenes, contribute to the process of tumorigenesis. Sequencing of a small number of samples from two classes of tumors across exons in thousands of genes demonstrated the power of searching broadly for mutations29,30. This allowed discovery of a far larger number of mutations than had been anticipated, many of these affecting pathways known to be involved in tumor development and progression31, providing an important window into the systems-level processes involved in cancer. However, it also indirectly showed that classic sequencing is simply impractical for analyzing large numbers of patient-based samples and complete cancer genomes, both of which would undoubtedly provide important therapeutic insights.
In addition to de novo mutations, DNA translocations are also important in the etiology of many cancers and discovery of new translocations benefits from a genome-wide approach. Already, deep sequencing has identified novel translocations in some cancers, providing information about their etiology and insight into effective therapeutic approaches32. The potential value of such data will be immense when comprehensive identification of translocations is possible in a clinically meaningful time frame. Deep sequencing of tumor samples also allows one to follow the mutation profile of a target and assess whether changes in the cell population may adversely affect the treatment paradigm33.
The Cancer Genome Atlas project (http://cancergenome.nih.gov/) aims to provide high-resolution molecular profiles of cancer. Cancer Genome Characterization Centers have been established to use the latest technologies to comprehensively detect genomic, epigenomic and transcriptomic aberrations that may play a role in cancer. All of the whole-genome analyses discussed above will be used to generate a broad vision of cancer, with the ultimate goal of developing strategies to better prevent and treat it. The use of high-throughput, novel sequencing technologies promises to provide far more complete profiles than would be possible with less comprehensive approaches.
Similarly, the rapidly changing genomes of some viruses (such as HIV) can be analyzed by deep resequencing of samples from multiple patients over time and with treatment34,35, allowing treatment protocols to be adapted to the evolving disease profile. As pathogens evolve drug resistance, quickly understanding how the resistance occurred is critical for both the diagnosis of the drug-resistant phenotype as well as for generating novel therapeutics for combating such resistance. For example, drug-resistant tuberculosis is a serious public health issue and, as a result, multiple tuberculosis genomes have been sequenced with potential benefits for enabling new treatments (reviewed by Loman and Pellen36). Similarly, 19 isolates of Salmonella enterica were sequenced to better understand its rate of evolution and its effective population size37. This information has improved understanding of how Salmonella interacts with its human hosts and thus provides insight into how the bacteria might be eradicated.
In addition to the direct approach just described, high-throughput sequencing can also identify viruses, bacteria and other organisms present in a complex biological sample by identifying their genomic signatures. When an infection, tumor or other undesirable outcome is caused by an unknown organism, sequencing all DNA and subtracting out what should be there from the total sequence profile leaves the signature of the contaminating or infecting organism. Such an approach has been used to identify viral sequences in tumor samples, thus implicating a virus in tumor growth38. In the same fashion, an arenavirus was linked with deaths after transplant surgery39 and two dicistroviruses were associated with honeybee colony collapse disorder40 through subtractive approaches. The deeper the sequence coverage, the more clearly such signals stand out.
When it is difficult or impossible to isolate different organisms for individual analysis, metagenomic studies can be carried out to identify the species present and to determine their representation in a particular sample41. Traditionally, 16S ribosomal RNA or other highly conserved genes have been used to characterize the variety of organisms in a given sample to minimize the amount of sequencing required, but that approach limits the amount of information that can be derived from a sample. The composition of a mixture could be determined as well as how that composition changes over time, but a deeper characterization of the genomes represented in those mixtures was not generally possible because of limiting sequencing capacity.
The ability to generate deep sequence data without the need for culturing organisms has opened another window on biology. Initial success with unculturable organisms using classic sequencing provided insight into bacteria that had been isolated as single cells42, but higher throughput sequencing can take it a step further with the ability to characterize complex systems in detail. The unexpected diversity of populations, such as the mixture of syntrophic microbes that utilize methane from deep sea vents, is causing a rethinking of how those complex ecosystems work and evolve43. A variety of other ecosystems is now just beginning to be examined with millions of sequences used to characterize the tremendous genetic diversity present in extreme environments44. The tremendous potential for the use of this diversity as substrate for novel industrial enzymes and processes can only be speculated on at this point.
Simply characterizing the organisms in an ecosystem can be extended to using genomic technologies for assessing the health of those ecosystems. Traditional means of assaying pollution or environmental stress have involved measuring macroscopic variables, such as growth and reproduction. More recently, measuring changes in transcription have proved very sensitive for detecting sublethal doses of multiple pollutants45,46. Because some studies have been done with microarrays, tester organisms required the generation of databases and construction of specialized arrays before use. Next-generation sequencing would have the advantage of not requiring the use of well-characterized species and would be able to detect novel transcripts that might not be expressed in unstressed organisms and hence not on arrays.
In addition to extreme environments, attention is also being directed at the variety of ecosystems present on and within the human body. The human microbiome project (http://nihroadmap.nih.gov/hmp/)47 is aimed at characterizing and understanding the diversity of microbes that inhabit different parts of the human body and how they may affect health and disease. Such studies will require deep sequencing of organisms to fully appreciate the complexity of interactions among microbes and between them and their host. These advances across the breadth of metagenomics will provide new insight into the role that infectious agents play in disease development and progression as well as provide an understanding of the diversity of species across many environments.
Epigenome: DNA information that simple sequencing misses
Increasingly, we are coming to recognize that the message encoded within the DNA sequence is regulated in a variety of complex ways, including modifications of the genome itself. One example, alterations in the pattern of DNA methylation in human DNA, has been associated with the level of transcription, a variety of disease states and a host of other phenotypes48,49. The most common variation in human DNA, DNA methylation at the 5′-position in cytosine, can be detected by several methods50. New technologies that allow us to see details of DNA methylation across the genome and to analyze a large number of patient samples may provide insight into how these epigenetic DNA modifications affect development and disease. However, technical limitations still hamper progress in examining genome-wide gene silencing and imprinting by methylation. Sequencing of bisulfite-treated DNA, a method that allows methylated and unmethylated positions to be distinguished, has been laborious and costly. Because of the resource-intensive nature of the methodology, it has traditionally been reserved for the analysis of candidate genes or gene regions and thus has provided only a limited view of the genome.
More recently, methylation has been studied on methylation-specific oligoarrays48,51 or using methylation-specific restriction enzymes52. Differences in DNA methylation patterns between tumor and adjacent normal tissue were identified, suggesting that tumor progression may rely not only on the introduction of somatic mutations, but also on changes in gene methylation. This may, in turn, result in the silencing of tumor suppressor genes or the activation of oncogenes. When the methylation results were overlaid with genome-wide expression data, there was a high degree of correlation between high methylation rates and low levels of gene expression. The expression changes were presumed to be a regulatory consequence of the detected changes in epistatic state52. Evidence has also been mounting for the role of methylation in development, aging and gene-environment interactions, emphasizing the need to study methylation in a wider variety of samples and conditions53,54. For example, comparing global and specific differences in methylation in a cohort of monozygotic twins revealed the accumulation of epigenetic differences with age55. Changes in methylation over time and upon exposure to different environmental factors may account for the onset of disease with age. These discoveries have recently led to calls for a comprehensive study of epigenetic modifications throughout the genome in the form of a Human Epigenome Project56–58. Initial studies have already provided data on three human chromosomes59, as well as Arabidopsis thaliana60. Nevertheless, the throughput of current technologies limits the analysis of multiple individuals or samples.
In addition to methylation patterns, it is possible to detect other types of structural variation as well. Numerous enzymes and chemical agents are differentially sensitive to various forms of DNA strain, structure, accessibility and other features. With sufficient sequencing capacity, the ability to see breakpoints induced by nucleases or modifications induced by chemical agents becomes practical at the whole-genome level. Hypersensitive regions are frequently associated with regulatory and protein binding phenomena, so the ability to detect them at high resolution will provide further understanding of many processes. For example, digestion of the nuclease-sensitive spacers between nucleosomes has been used as a means to map them at a genome-wide scale for both Caenorhabditis elegans and Drosophila61,62.
Interactome: the direct partners and first line of function
DNA sequence, structure and modifications are recognized by a wide variety of proteins like histones, which bind nonspecifically to DNA and help to compact it, and transcription factors, which bind to specific sequence motifs and activate or repress transcription. ChIP was developed to identify novel protein binding sites across the genome. This allows researchers to identify protein-binding sites within living cells and has been expanded to a genome-wide assay with the improvement of microarray and sequencing technologies63–65. The method allows researchers to compare protein-DNA binding profiles between tissue types, developmental stages or differentially treated cells. The ChIP technique has been broken down into broad categories, ChIP-Chip, ChIP-PCR and ChIP-Seq, depending on the platform used to characterize DNA fragments selected during the immunoprecipitation process, whether microarray hybridization, PCR or DNA sequencing, respectively66–68. As discussed in more detail below, the hybridization technologies have traditionally had the benefit of low cost, but have been limited by incomplete coverage of the genome, difficulties in distinguishing closely related sequences and poor resolution of sequence boundaries. As sequencing technologies achieve lower cost and higher throughput, ChIP-Seq approaches are yielding more sensitive and complete coverage of the entire genome, providing more accurate pictures of the complex regulatory landscape within the genome.
ChIP techniques are also well suited for detecting differences in modification patterns on DNA-binding proteins, like histones, which have a variety of different post-translational modifications thought to affect transcription. Although histones bind in a largely nonspecific manner, they are modified in specific ways that tag regions of the genome for selective action by transcription factors and polymerases. A genome-wide scan of histone methylation patterns was superimposed on transcriptional start site and gene expression data to determine the effects of differential modification on gene expression. Monomethylation of histones 2, 3 and 4 are all linked with gene activation, whereas trimethylation of histone 3 has been associated with deactivation69. Similarly, genome-wide patterns of histone acetylation and ubiquitination have also been linked with regulation of expression70,71 and differentiation status of embryonic stem cells and other cells72,73. Although the depth of sequencing data provided detailed information about such modifications, even deeper sequencing could greatly increase the resolution and aid in interpretation of the observed patterns. Understanding where the binding and modifications occur and how these interactions span the genome will provide further insight into gene expression.
As histones act on regions of the genome in a broad way, enhancers and promoters act on specific genes or regions. Enhancer sequences increase the expression of a gene or set of genes, but unlike promoters, enhancers are not always found near the genes they control. They are believed to physically interact with distal genomic targets via DNA interacting proteins or transcription factors, thus making their localization more problematic than that of simple promoters. Locating and profiling the physical interactions of these proteins with the genome in different tissue types throughout development or in response to external perturbations can be a powerful means of dissecting genome biology. Although the transcription factors bind specifically to particular DNA sequences, the consensus sequences for many are complex and cannot be deduced without direct experimental evidence for binding within a living cell.
The ChIP techniques have had a substantial impact on the understanding of how differential protein binding can affect the regulation of many processes. Transcription factors, such as OCT4, SOX2 and NANOG, and repressor proteins, such as the polycomb proteins, are known to play a role in stem cell development. Correlating protein-DNA interaction data with expression information has provided a rich insight into mechanisms of action74. Although generally activators of transcription, OCT4, SOX2 and NANOG ChIP-chip analysis has shown these factors also bind to and inhibit the expression of other lineage-specific transcription factor–encoding genes74,75.
Cost and throughput often limit the number of points used in time-course experiments, preventing the collection of the frequent measurements required for high temporal resolution. Rapid changes in genomic regulation are known to occur in cells that respond to external stimuli, such as macrophages reacting to bacterial lipopolysaccharide (LPS). ChIP and quantitative (q)PCR analysis have been used to identify early events responsible for macrophage activation76. After stimulating macrophages with LPS, a cluster of transcription factor genes were activated within one hour of treatment. Mining protein-protein interaction maps and searching for transcription factor–binding motifs in the genome revealed that two of the transcription factors identified in this early cluster, ATF3 and nuclear factor (NF)-κB, had binding sites upstream of cytokine-encoding genes IL6 and IL12b. Subsequent ChIP time-course experiments after LPS stimulation demonstrated that both of these transcription factors bound to the promoter of IL6, with an initial spike in NF-κB binding and a gradual sustained binding of ATF3 thereafter76. Carrying out similar but more complete kinetic experiments genome-wide will yield powerful information regarding the dynamics of genome regulatory pathways.
Transcriptome: more variants and greater precision for measuring RNA
The ultimate goal of understanding epigenomics, transcription factors and histone modifications is deducing how the genome responds to the complex collection of factors that influence cellular physiology. Measuring RNA expression has been accomplished by increasingly sophisticated tools of greater specificity and higher throughput that have allowed detection of increased numbers of RNA species in larger sample sets over a wider dynamic range. Although studies with the recently developed technologies of qPCR, DNA microarrays and serial analysis of gene expression (SAGE) have provided tremendous insight into biological processes, each suffers from its own biases and limitations. The most accurate techniques with the greatest dynamic range, such as qPCR, can be used to measure only a small subset of RNAs in any given sample. If one wishes to characterize a substantial fraction of RNAs in a sample with an oligonucleotide or cDNA microarray, it is at the expense of precision and dynamic range. With hybridization arrays, low-frequency and highly homologous sequences suffer the most in terms of the lower limit of quantification and reproducibility. Nonspecific binding of related or high-frequency sequences can obscure real signals. Neither qPCR nor a traditional array is able to discover novel transcripts and variants.
Initially, microarrays used oligonucleotides or cDNAs directed only to known transcripts and arrays were also limited by a small dynamic range and cross-hybridization of related transcripts. When different platforms were analyzed, comparison yielded a correlation of r = 0.7–0.8 and this was considered satisfactory77. The limitation to known transcripts has been overcome in a small number of instances by using much denser tiling arrays that can sample the entire genome of the species of interest. Using tiling arrays and other approaches, the Encyclopedia of DNA Elements (ENCODE) Project Consortium has embarked on a systematic effort to survey transcription from a subset of the genome78. A combination of genomic assays, comparative genomics and computational prediction algorithms were used to discover new exons and new transcription start sites for many genes, novel nonprotein-coding transcripts and chimeric transcripts for a number of tandem genes. It was possible to see that much more of the genome is transcribed than previously thought, demonstrating the existence of a large number of novel RNA species79–81.
Non-mRNA RNAs have been poorly characterized in the past, but recent research has highlighted the microRNA (miRNA) class of small RNAs that hybridize specifically to target mRNAs and inhibit their expression (either by inducing degradation or inhibiting translation). These miRNA expression profiles have been associated with developmental regulation, tissue differentiation and cancer82–88. Some studies have demonstrated that specific miRNAs are directly implicated in cancer pathways, underscoring the importance of including noncoding transcript profiling in disease research. Indeed, the small size and high degree of similarity across different classes of miRNA molecules make them particularly well suited to new sequencing approaches. In addition to miRNAs, the expression of larger noncoding transcripts has also been found to be altered in cancer89 and in patients at high cardiac disease and diabetes risk17. The roles of these new, nonprotein-coding transcripts are not yet understood, but they are believed to play fundamental roles in the regulation of gene expression and disease progression.
In addition to arrays, hybridization to RNA within cells in situ can also yield valuable information, although not in as quantitative a manner. Using an automated in situ hybridization platform, The Allen Institute for Brain Science (Seattle, WA) has created The Allen Brain Atlas, a web-based resource with a three-dimensional gene expression map for 20,000 mRNAs in 400 mouse brain structures. Among the Institute’s findings, 25% of the genes analyzed are expressed in a tissue-specific manner, and expression patterns can clearly differentiate substructures within known anatomical regions90. Because of technological constraints, these mRNA measurements could only be binned into large groupings of expression levels. The mouse atlas is serving as the basis for a much larger project recently announced and aimed at mapping gene expression throughout the human brain (http://humancortex.alleninstitute.org/has/). Another study using microarrays analyzed expression differences between neurons that were microdissected from disparate regions of the brain using laser-capture methods91. A study of 193 human brains for both gene expression and genetic variation revealed that over 20% of identified transcripts were genetically determined, but this study was limited by the sensitivity of the tools available92. Deeper sequencing of specific cell types would provide more detailed genetic information as well as expression data.
Microarray platforms for assessing expression levels have not generally been used for detecting genetic variation or splice variants, but some have recently been designed to distinguish between the expression of different splice variants93,94. However, array content is still usually limited to known alleles or splice junctions. As with transcription, alternative splicing is known to occur differentially during specific developmental stages, in specific tissues and in various disease states95.
To circumvent the inherent issues of hybridization, researchers have attempted to generate digital gene expression profiles in which every transcript is counted. Thus far, most published articles reporting digital expression patterns have employed SAGE (serial analysis of gene expression)96,97. This technique relies on sequencing short tags from each transcript, and could potentially address the issue of rare transcripts, but the costs associated with SAGE have limited the number of tags that are sequenced to only a fraction of what is typically estimated to be the transcript count in a single cell. Nonetheless, comparison of tag profiles of more than 200,000 tags per cell line has been useful in identifying cell lineage–specific transcripts and differences98. This quantitative limitation leads to reduced accuracy. Additionally, potential bias in transcript capture introduced by the complex sample preparation protocols raises concerns about the overall accuracy of the results. Capturing a wide range of expression levels, including RNAs that may occur at only a few copies per cell (as with many critical receptors and transcription factors), is important for a complete picture of the transcriptome. Greater depth is now achieved with more direct sequencing methodologies, which permit digital measurement of transcripts that provides data as detailed as one wishes, at a resolution that is unattainable by hybridization methodologies. Over 40 million mRNA reads from mouse brain, muscle and liver have been generated allowing not just quantification of the mRNA but insight into splicing and transcriptional starts and stops99. Similarly, nearly 100 million reads were obtained from mouse stem cells and embryoid bodies100 and other researchers have delved deeply into C. elegans101, HeLa cells102, yeast103 and fission yeast104. The availability of technologies that provide sequence data without the need for PCR and other manipulations that could lead to bias in detection will further add to the value of such data. In contrast to hybridization, sequencing can accrue the extra benefits of obtaining transcription start and stop sites, alternative splice sites and genetic variants at the same time as expression data.
Integrating ’omics: the values of synergy
Each of the techniques described thus far provides valuable data for understanding biological systems but this value can be greatly enhanced when multiple types of data are generated on the same system, a property inherent to many sequencing approaches. The recent availability of genome-wide expression data coupled with genotype data has already shown the value of integrating genetic and transcriptional approaches. RNA expression levels can be treated as quantitative traits and combined with genotyping or sequencing data to identify quantitative trait loci105. Mapping gene expression quantitative trait loci (eQTL) in conjunction with phenotypic data has allowed systems analysis and the dissection of regulatory relationships and their implications for disease106,107. Some of the eQTLs identified were known polymorphisms that have well- described effects on the transcription, splicing and mRNA turnover of their respective target genes, whereas other genes were novel loci associated with obesity and metabolic disease traits for the first time.
The feasibility of whole-genome approaches using multiple technologies has only emerged recently, but such approaches have been used previously with small regions or a single gene. For example, the study of the α-globin gene and its involvement in α-thalassemia108 was addressed by superimposing DNase hypersensitivity, phylogenetic footprinting, ChIP, expression and sequence data. A gain-of-function regulatory SNP involved in the creation of a ‘false’ transcription binding site was found upstream of the normal α-globin promoter, resulting in the underexpression of the α-globin genes in α-thalassemia patients108. This one-gene, multi-dimensional study demonstrates the type of biology that can be discovered only with an integrative approach, and makes the case for expanding this approach to genome-scale studies. Such studies are now emerging as integrated approaches, as evidenced by the simultaneous mapping of cytosine methylation, the mRNA transcriptome and the small RNA transcriptome in various Arabidopsis strains109. Some of the wide varieties of applications that can be envisioned are shown schematically in Figure 3.
Figure 3.
What can high-throughput sequencing do for you? The breadth of information that can be generated with high-throughput sequencing and the variety of sample sources is illustrated.
Current limitations and future possibilities
Our ability to undertake genome-wide integrative studies, at a scale that would include multiple technologies to be used across many samples, has been hampered by the technical, financial and throughput limitations imposed by the current genetic analysis technologies. Microarrays rely on hybridization probes of known sequence and are limited by probe density, thermodynamic constraints on probe design and cross-hybridization artifacts. This prevents a full analysis of the genome and leads to a reduction in the types of genomic or genetic changes that can be detected110. The throughput of current array platforms cannot accommodate experiments with 104–106 samples for a reasonable cost and within a reasonable time frame. Although the use of genome tiling arrays provides solutions to some of these challenges, a complete picture of the transcriptome remains a technical and algorithmic challenge. The increased content of tiling arrays makes cost of array manufacture and processing an issue that has limited their use for large numbers of samples. Microarrays will continue to be widely used for the foreseeable future because of their extensive legacy data and installed instrument base. However, depending on how deeply one wishes to sequence, digital gene expression has now matched or exceeded microarrays in terms of reagent and disposable costs per sample111. The different sequencing platforms can generate anywhere from 500,000 to 500,000,000 reads per run and those are distributed across up to 50 channels, making it possible to analyze that many samples simultaneously. The costs of both microarrays and sequencing are, in many cases, <$400 per sample, so the choice of platform becomes dependent on the type of experiment and the available instrumentation, with recurring costs becoming less of a factor.
Similarly, for genetic analysis, various types of genotyping arrays have provided a wealth of data on many phenotypes with the ability to readily analyze thousands of samples, but remain fundamentally limited by the requirements for known variations and the current inability to cost effectively include all rare variants and singleton SNPs not covered by linkage disequilibrium. Both genotyping and expression arrays are also limited by sequence differences across genomes, requiring new sets of arrays as different species are examined. If our understanding of genome variation is limited to human samples, the full benefits that are attainable by studying other species, including disease and drug safety models, will be lost.
Capillary electrophoresis–based sequencing technologies are also unable to pierce the price and performance barriers to enable high- content, genome-wide experiments requiring thousands of genomic samples. To make genome-scale studies tractable, researchers have applied complexity-reduction techniques, or have relied on biological inference to select genes or gene regions of interest, thus looking under the lamp-post rather than opening the genome for a full inspection. Without the availability of true whole-genome sequencing technologies, many regions of the genome will remain refractory to analysis, and many rare variants will remain undiscovered, limiting our understanding of genomic variation and disease.
There are now multiple high-throughput sequencing technologies that can address the present limitations of both hybridization-based technologies and classic sequencing. These technologies vary in their sequencing throughput in terms of samples and sequences, their complexity of sample preparation requirements and their output in terms of read-lengths1–5. Additionally, some of these techniques allow single-molecule sequencing, instead of the traditional sequencing of amplified sets of molecules, and this capability will further add to the value of these data. The details of a given application will determine which technology is best suited for a particular situation. For example, read-length is more important for de novo sequencing and metagenomics with unknown organisms. For digital gene expression, read-length is much less important and the number of reads becomes paramount. Thus, the absolute number of Mb/h is a useful metric but tells only part of the story with the number of reads sometimes a more important indicator. Whether interested in the number of reads or the number of bases, researchers and healthcare providers should be preparing now for what they will do with orders of magnitude more sequence data.
High-throughput sequencing is not without issues. Even if sequencing were entirely free (which is not likely to happen), there are other costs that will limit the benefits derived from very cheap data generation. The huge quantity of sequences will shift the bottleneck from the generation of data to its analysis. There are important challenges112 for the analysis of such data that will require changes to the programs used to align and assemble sequence. The various sequencing technologies generate different read-lengths, different error rates and different error profiles relative to traditional data and to each other. In addition to simply analyzing the sequence data, new methods will be needed to analyze and integrate the massive data sets and then apply those results to various types of biological information. The new technologies will no doubt raise issues with many aspects of the current research and diagnostic infrastructure, and those issues should be considered now. The complexity of analysis will rise markedly, but the opportunities for an immensely deeper understanding of disease, its causes and personalization of treatment will be even greater.
Overcoming these obstacles will be critical for taking full advantage of our new sequencing horsepower. We will be able to analyze bacterial and viral evolution in real time so that new and appropriate therapeutics can be directed at infectious agents intent on evading the current generation of drugs. We will be able to take advantage of the huge genetic diversity now hidden from view in microbes in extreme environments and apply novel enzymes and systems to ameliorate problems of disease, pollution, energy production and industrial processes. We will be able to better understand and track novel nucleic acid therapeutics and gene therapies so that they can be optimized more quickly for everyone’s benefit. We will better understand individual disease risk and be able to take personalized preventative measures.
We are now poised to take advantage of the huge advances in sequencing that the various new technologies have already attained and promise to achieve in the future. The widespread desire to accomplish the goal of cheap, accessible sequencing is clear from programs, such as the US National Institutes of Health’s (Bethesda, MD) $1,000 genome grants that seek to catalyze the rapid attainment of technological advances (http://www.genome.gov/15015208), and the Archon X Genomics prize in which $10 million will go to the first team able to sequence 100 genomes in 10 days for less than $10,000 per genome (http://genomics.xprize.org/). With the recent passage of the Genetic Information Non-discrimination Act (GINA) in the United States, many of the legal hurdles for the generation of data have been removed. We will soon be limited only by our imagination, as the new sequencing technologies remove the financial and technical barriers, and we move into an era of unexpected applications and discoveries, driven by access to massive quantities of DNA sequence data.
Conducting the larger and more complex experiments required to decode the biology of the genome and its role in disease would be impossible with the genomic technologies and budgets available up until now. As the above examples illustrate, researchers have been forced to make trade-offs in their experimental designs in terms of the number of samples they analyze, the extent of genomic coverage they extract, the level of genomic resolution they examine and the type of genomic data they obtain. To allow a truly integrative, hypothesis-free and statistically powerful survey of genomes, the new versatile, high-throughput, cost-effective nucleic acid sequencing technologies that are now emerging are required. This surge of new data can be received as a flood, overwhelming the unsuspecting researcher, or as a tremendous wave that can be surfed to new horizons. Those who are able to take advantage of the new technologies, whether researcher or patient, will benefit substantially. Sequencing everything is only the first step, as we then need to process that data into information that can be used broadly to benefit human health and productivity.
Acknowledgments
We thank Jennifer MacArthur (Helicos BioSciences) for assistance and critical reading of the manuscript and Stephen Quake (Stanford University) for critical reading and comments.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Church GM. Genomes for all. Sci Am. 2006;294:46–54. doi: 10.1038/scientificamerican0106-46. [DOI] [PubMed] [Google Scholar]
- 3.Zwolak M, DiVentra M. Physical approaches to DNA sequencing and detection. Rev Mod Phys. 2008;80:141–165. [Google Scholar]
- 4.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Harris TD, et al. Single-Molecule DNA Sequencing of a Viral Genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 8.Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 9.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 10.Millar CD, Huynen L, Subramanian S, Mohandesan E, Lamber DM. New developments in ancient genomics. Trends Ecol Evol. 2008;23:386–393. doi: 10.1016/j.tree.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Ellegren H, Sheldon BC. Genetic basis of fitness differences in natural populations. Nature. 2008;452:169–175. doi: 10.1038/nature06737. [DOI] [PubMed] [Google Scholar]
- 12.Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 13.Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 14.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellcome Trust Case Control Consortium. Genome-wide association study of 14, 000 cases of seven common diseases 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broadbent HM, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 18.Ke X, Taylor MS, Cardon LR. Singleton SNPs in the human genome and implications for genome-wide association studies. Eur J Hum Genet. 2008;16:506–515. doi: 10.1038/sj.ejhg.5201987. [DOI] [PubMed] [Google Scholar]
- 19.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 20.McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychol. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 21.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 22.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 23.Freeman JL, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 24.Komura D, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet. 2006;15(Spec No 1):R57–R66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- 28.McCarroll SA, Althshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2008;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 29.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 30.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Chittenden T, et al. Functional Classification Analysis of Somatically Mutated Genes in Human Breast and Colorectal Cancers. Genomics. 2008;91:508–511. doi: 10.1016/j.ygeno.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 33.Nardi V, et al. Quantitative monitoring by polymerase colony assay of known mutations resistant to ABL kinase inhibitors. Oncogene. 2008;27:775–782. doi: 10.1038/sj.onc.1210698. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann C, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loman NJ, Pallen MJ. XDR-TB genome sequencing: a glimpse of the microbiology of the future. Future Microbiol. 2008;3:111–113. doi: 10.2217/17460913.3.2.111. [DOI] [PubMed] [Google Scholar]
- 37.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2007;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palacios G, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 40.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 41.Woyke T, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 42.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pernthaler A, et al. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 45.Bundy JG, et al. ‘Systems toxicology’ approach identifies coordinated metabolic responses to copper in a terrestrial non-model invertebrate, the earthworm Lumbricus rubellus. BMC Biol. 2008;6:25. doi: 10.1186/1741-7007-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Straalen NM, Roelofs D. Genomics technology for assessing soil pollution. J Biol. 2008;7:19. doi: 10.1186/jbiol80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauch TA, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2007;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasui DH, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu M, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 53.Agrelo R, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci USA. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brena RM, Huang TH, Plass C. Quantitative assessment of DNA methylation: potential applications for disease diagnosis, classification, and prognosis in clinical settings. J Mol Med. 2006;84:365–377. doi: 10.1007/s00109-005-0034-0. [DOI] [PubMed] [Google Scholar]
- 55.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones PA, Martienssen R. A blueprint for a Human Epigenome Project: the AACR Human Epigenome Workshop. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- 57.Garber K. Momentum building for human epigenome project. J Natl Cancer Inst. 2006;98:84–86. doi: 10.1093/jnci/djj042. [DOI] [PubMed] [Google Scholar]
- 58.Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–1125. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- 59.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valouev A, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- 64.Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep. 2008;9:337–343. doi: 10.1038/embor.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 67.Euskirchen GM, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 69.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 70.Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minsky N, et al. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 72.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meissner A, et al. Genome-scale DNA methylation maps of pluirpotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 77.Petersen D, et al. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics. 2005;6:63. doi: 10.1186/1471-2164-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Encode Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 80.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 81.Kapranov P, et al. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Palatnik JF, et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 83.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 84.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 86.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 87.Huang Q, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 88.Morin RD, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 91.Liang WS, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2006;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 93.Fehlbaum P, Guihal C, Bracco L, Cochet O. A microarray configuration to quantify expression levels and relative abundance of splice variants. Nucleic Acids Res. 2005;33:e47. doi: 10.1093/nar/gni047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 95.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 96.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 97.Sun M, et al. SAGE is far more sensitive than EST for detecting low-abundance transcripts. BMC Genomics. 2004;5:1–4. doi: 10.1186/1471-2164-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirst M, et al. LongSAGE profiling of nine human embryonic stem cell lines. Genome Biol. 2007;8:R113. doi: 10.1186/gb-2007-8-6-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 100.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 101.Shin H, et al. Transcriptome analysis for Caenorhabditis elegans based on novel expressed sequence tags. BMC Biol. 2008;6:30. doi: 10.1186/1741-7007-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 103.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 105.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 106.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 107.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.De Gobbi M, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 109.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chou CC, Chen CH, Lee TT, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shendure J. The beginning of the end for microarrays? Nat Genet. 2008;5:585–586. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 112.Pop M, Salzburg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142–149. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]