Summary
Cells acquire their ultimate identities by activating combinations of transcription factors that initiate and sustain expression of the appropriate cell-type specific genes. T-cell development depends on the progression of progenitor cells through three major phases associated with distinct transcription factor ensembles that control their recruitment to and proliferation in the thymus, their lineage commitment, and their responsiveness to T-cell receptor (TCR) signals, before the allocation of cells to particular effector programs. All three phases are essential for proper T cell development, as are the mechanisms that determine the boundaries between each phase: cells failing to shut off one set of regulators before the next gene network phase is activated are predisposed to leukemic transformation.
Introduction
The T-cell developmental program, initiated and sustained by the unique thymic environment, guides small numbers of pluripotent cells through multiple rounds of proliferation and differentiation, leading to T-lineage commitment, T-cell receptor (TCR) rearrangements, and generation of αβ TCR- or γδ TCR-expressing T-cells that function as killers, regulatory cells, or producers of specific cytokines 1-6. In the past five years, the transcriptional and epigenetic mechanisms that forge T-cell identity and suppress other developmental pathways have come into focus. It is not enough for cells to simply activate the set of transcription factors that maintain T-cell gene expression in mature T-cells; instead, the developmental program depends on the sequential operation of several distinct developmental gene networks.
From the time a lymphoid precursor arrives in the mouse thymus to the first expression of an αβTCR, it traverses at least 8 phenotypically distinct stages defined by expression of CD4, CD8 and other markers 1-6 — Flt3+ early thymic progenitor (ETP), ETP, double negative 2a (DN2a), DN2b, DN3a, DN3b, transitional DN4 and immature single-positive (ISP), and double positive (DP) (DN: CD4- CD8-, DP: CD4+ CD8+)(Fig. 1a). Most of these stages undergo proliferation, but the degree of proliferation and the time required to reach the DP αβ TCR+ stage vary between lymphoid precursor cohorts. It takes a little over a day for the first wave of lymphoid precursors that populate the fetal mouse thymus to generate DN2 cells (E12.5-E14) and only a total of four days for the first DP cells to appear (E16). In contrast, the lymphoid precursors that continuously trickle into the thymus throughout young adult life can take ten days to reach DN2 stages and two weeks to develop into DP cells, with the extra time providing the opportunity for much more extensive proliferation7, 8.
Figure 1. αβT-cell development: stages, surface markers, and transcription factor expression.
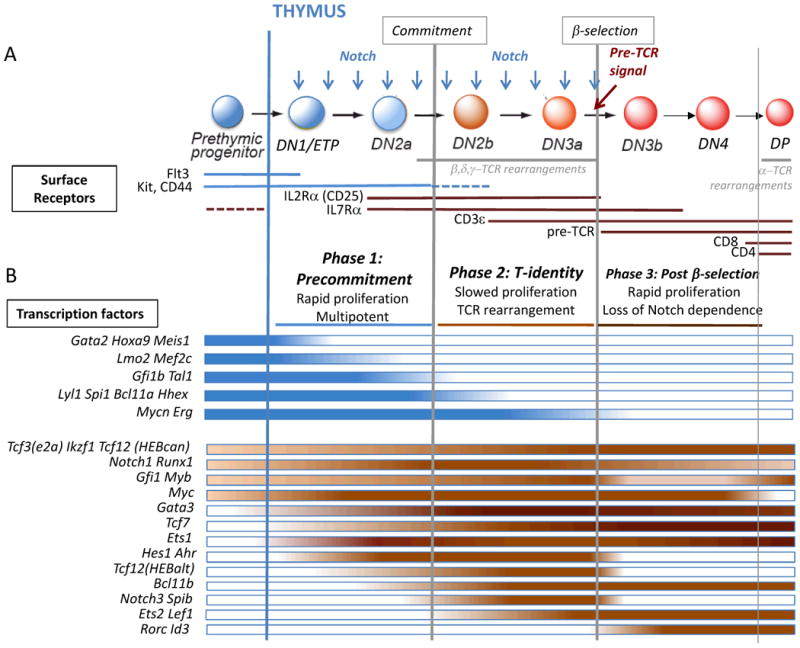
a. Adult mouse T-cell development begins in the bone marrow from lymphoid-primed prethymic progenitors that migrate to the thymus and begin differentiation in the thymic environment, which provides Notch ligands (blue arrows). Cells transit sequentially through DN1/ETP, DN2a, DN2b, DN3a, DN3b, DN4, and DP stages on the way to becoming αβT-cells (DN: CD4- CD8-; DP: CD4+ CD8+; ISP transitional-stage cells not shown). DN1 (CD44+ CD25-) cells include a subset with high Kit expression that contains the Early T-cell Precursors (ETP; CD44+Kit++CD25-), which contain essentially all the T-cell progenitor activity and are the only kind of DN1 cells that will be considered further here. ETPs lack or have downregulated IL7R, but as they differentiate to DN2, they turn on IL7R. Key cell surface receptors used to identify these stages are shown indicating the stages during which each receptor is expressed. Dotted lines indicate stages with lower expression levels. The stages during which TCR rearrangements occur are also marked. Development is divided by the commitment and β-selection checkpoints into three major regulatory phases (Phases 1, 2, and 3: post-β-selection), each with unique gene networks and cellular characteristics. Cells in phase 1 proliferate extensively and retain multipotentiality, while phase 2 cells are committed, slow their proliferation, and undergo TCR rearrangements. Only cells with a rearranged TCRβ that can combine with pre-Tα and transduce a signal can continue through the β-selection checkpoint into phase 3, a second highly proliferative but increasingly Notch-independent phase leading to CD4 and CD8 upregulation, then proliferative arrest, and TCRα rearrangement.
b. Stage-specific patterns of expression of important transcription factor genes are shown below the developmental stages. The color intensity variations provide an approximation of the dynamic changes in expression of the genes, grouped together based on similar expression patterns although not necessarily similar levels; for more accurate quantitation of gene expression, see sources of this figure in Zhang et al.31 and ImmGen32, 197 (www.immgen.org). The transcription factor genes are divided into legacy stem and progenitor genes that are mostly turned off in Phase 1 and are all off by β-selection (blue bars), and genes that are critical to different aspects of the T-cell specification and commitment programs (red bars). Additional information about each of these genes is provided in Tables 1 and 2.
Despite these kinetic differences, the gene expression patterns at given developmental stages of fetal and adult thymocytes are similar9. This similarity extends to the transcription factor genes that are characteristically expressed at each stage (Fig. 1b) as well as to the differentiation genes that these factors regulate. Thus, the transcriptional control of proliferation and of developmental progression is to some extent modular and may depend upon checkpoints to ensure orderly differentiation. This implies that distinct phases of T-cell development are governed not only by key transcription factors, but also by the coordination among such transcription factors, synchronized by gene regulatory network connections.
All the events that establish the T-cell identity of precursors are driven by Notch signaling10-13. Notch1 molecules on lymphoid precursors interact with Notch ligands in the thymic microenvironment, leading to activation of the T-cell-specific developmental program. During the first developmental stages, Notch signaling interacts with a “legacy” stem and progenitor-cell gene network inherited from multipotent precursors. Both legacy genes that will play ongoing roles in T cells and progenitor-specific legacy genes with roles confined to the earliest stages participate in this network that we term “phase 1” (Fig. 1b). Although still incompletely understood, the phase 1 network may support the extensive proliferative expansion of the ETP and DN2a cells, as well as impact upon the order, timing, and level of T-cell gene activation. Notch signaling also activates the first T-lineage specific transcription factors by its interaction with the phase 1 network, although the newly induced factors only express full T cell specification activity under the continuing influence of Notch signals in a second phase network.
T-cell specific transcription factors in the phase 2 network mediate commitment-linked functions that drive T-cell specific gene expression and open the TCR gene loci for rearrangement, as well as repress the expression of the progenitor-cell-specific phase 1 genes. The phase 2 network thus creates the distinctive T-cell identity. However, this phase 2 network also must be profoundly modified, once TCR gene rearrangement occurs and enables some cells to express either a pre-TCR (TCRβ with invariant pre-TCRα) or a γδ TCR. The resulting TCR signal transduction, again under the influence of Notch signaling, triggers another regulatory factor network shift, transitioning to phase 3. The phase 3 network includes newly activated genes that support the T cell phenotype but leads to the extinction of the expression of some factors that had played crucial roles in phase 2, reduces Notch target gene expression, and enables the cells to become Notch-independent.
The switching between the three phases must be correctly regulated to ensure the fidelity of T-cell development and to avoid leukemogenesis. Activation of new regulatory genes and repression of those from earlier phases must be accurately coordinated to insure proper passage through the developmental checkpoints. An important question is how homogeneous the precise trajectory of gene expression changes is between the surface receptor-defined stages, and this awaits single-cell transcriptional analysis14. However, a strong relationship has recently emerged between the generation of T-cell leukemias and thymocyte precursor competition and self-renewal as well as inappropriate persistence of gene expression from the phase 1 restricted genes of normal T-cell development 15-19. This relationship provides insights into the origins of certain forms of human T-cell acute lymphoblastic leukemias (T-ALL) as well as suggesting the roles of specific stem and progenitor cell genes in normal T-cell developmental regulatory networks.
Thus, a three-phase succession of gene networks processes multipotent precursors into T-lineage committed cells, and sets them on the paths to distinct functional roles. Expression of certain individual transcription factors can extend across the boundaries of these phases, but their functions in each phase are modulated according to their level and the overall gene network state. In this review we explore what these functions are and how they are interlinked.
Major features of early T-cell development: a framework
Progenitor interaction with the thymic environment
T-cell progenitors from bone marrow or fetal liver continuously seed the thymus via the blood. The major types of prethymic cells that appear competent to do this are multipotent precursors: primarily common lymphoid precursors, which express higher levels of the interleukin-7 receptor (IL-7R) and lower levels of the receptor tyrosine kinase Kit (CD117), and lymphoid-primed multipotent precursors, with lower IL-7R expression but expressing high levels of Kit20-23. Whereas the respective contributions of these precursors in vivo is an important question, once in the thymus the ETPs derived from them share a Kit-high, IL-7R-low phenotype and develop into Kit-high, IL-7R-high DN2a cells before commitment. In fact, both of the defining growth factor receptors are crucial for early pro-T cell expansion within the thymus. Both types of thymic immigrants also express the functionally important growth factor receptor Flt3 (FMS-related tyrosine kinase 3; Flk2), as well as Notch124. The unique thymic environment not only provides supportive cytokines, Kit ligand and IL-7, which maintain and expand the precursors, but it critically provides an environment rich in the Notch ligand, Delta-like 4 (DLL4)6. Engagement of Notch1 by environmental Notch ligands triggers proteolytic release of intracellular Notch1, which binds to DNA-bound RBPJ (or CSL) and recruits a transcriptional co-activator complex to activate Notch target genes25.
Stages of early T-cell development
The process of differentation from thymic entry to expression of a full heterodimeric TCR can be divided coarsely into Notch-dependent and TCR-dependent parts (Fig. 1a). The early cells do not express a TCR, and they proliferate and differentiate in a TCR-independent, Notch-dependent fashion. As soon as TCR gene rearrangement yields its first successful products, cell survival and differentiation become TCR-dependent, and Notch influence is downregulated. For αβ T-cells, TCRβ rearrangement normally occurs in the DN3a stage, with TCRα rearrangement delayed until the DP stage. DP αβTCR+ cells then await positive selection based on TCR recognition specificity, which is a prerequisite for further differentiation into effector T-cell subsets (such as CD4+ T cells, CD8+ T cells, natural killer T cells and regulatory T cells). Rearrangements of both TCRγ and TCRδ, leading to γδTCR-expressing cells, take place more synchronously in the DN2 or DN3a stages (Box 1).
Box 1. γδ T-Cell Development Overview.
Two classes of T cells expressing distinct TCR dimers, αβ and γδ, have been maintained in all jawed vertebrate taxa since TCR-expressing cells first evolved171. γδ T-cells share common intrathymic precursors with αβ T-cells, but undergo different differentiation programs following TCR expression172. Successful assembly of a γδTCR triggers signals that lead to down-regulation of CD25; however, γδTCR+ cells proliferate much less than pre-TCR+ cells and do not turn on CD4 or CD8. Specific characteristics of individual precursor cells at different stages may bias them towards the γδ- or αβ-lineage. DN2 cells in general, particularly those with high levels of IL7R and/or Sox13, can frequently adopt a γδ fate, but individual DN3a cells are more likely to produce αβ T-cells38, 173-175 (Fig. 2). Interplay of Notch signals with signals from the newly expressed pre-TCR or γδTCR is important for αβ vs. γδ divergence, although the outcomes of this interaction differ between mouse and human176. A γδTCR can produce a stronger signal than a pre-TCR, leading to more intense upregulation of Id3 and Erg, and initiation of a γδ rather than an αβ T-cell program177, 178.
Interestingly, γδTCR signaling triggers one of several different genetic programs available for γδTCR-expressing cells, differentially depending upon Id3, Sox13, Zbtb16 (encoding PLZF), and/or Bcl11b for further development and effector functions127, 130, 174, 179-181. The range of accessible γδ-subtype fates narrows with differentiation from DN2a to DN3a130. These γδ–lineage-specific programs mirror the range of Th1, Th2, NKT and Th17 effector programs that αβ T-cells acquire at later stages, either after positive selection or in the periphery. However, γδ T-cells can implement specific effector programs directly upon first TCR expression and signaling within the thymus. Unlike αβ T-cells, these functions correlate with particular TCRγ and δ segment usages179(see reviews172, 182, 183).
A major landmark in the Notch-dependent developmental process is T-lineage commitment, which occurs at the DN2a to DN2b transition (Fig. 1a). Commitment marks the final loss of factors and gene regulatory networks that allow alternative lineage fate choices under permissive conditions4, 26 (Box 2). Under normal conditions in the thymus Notch ligands push and retain progenitor cells in the T cell pathway and block alternative fate choices, but during commitment the accumulation of Notch-dependent gene regulatory changes makes these fates inaccessible regardless of the environment. The commitment transition was originally detected with an Lck-GFP transgene27 and is marked in non-transgenic animals by a deceptively subtle decrease in Kit expression1, 28. In fact, however, this transition is a major gene regulation upheaval which causes both substantial upregulation of many T-cell genes and the loss of progenitor-specific gene expression. Dendritic-cell, myeloid, NK (ILC1) and probably ILC2 and mast-cell potentials, indicative of sustained diverse and permissive regulatory states, are all terminated here4, 26, 29, 30 (Fig. 2). Thus, the Notch-dependent period of T-cell development is divided into two phases (phase 1: pre-commitment and phase 2: T-identity), which are profoundly distinct in their transcription factor gene network states and developmental properties (Fig. 1b).
Box 2. Mechanisms of Alternative Lineage Exclusion.
CLPs and LMPPs retain the potential to generate diverse lymphocyte types, as well as natural killer, dendritic, and myeloid cells under somewhat different ranges of permissive conditions 21-23, 52. Contact with the thymic environment efficiently blocks the cells’ access to these non-T cell lineage potentials through a variety of mechanisms, however, and under normal conditions the overwhelming majority of precursors only generate T cells in situ184, 185. Nevertheless, a gene network state providing latent competence to switch to other fates persists throughout the first few stages of T-cell development, which can be detected by experimentally removing the cells from the normal thymus. Alternative lineage potentials are lost at different times (Fig. 2) and by at least three different mechanisms. At the earliest thymic stage, as ETP cells turn off Flt3 expression, they unconditionally lose their B-cell potential100, 125, 126. The mechanism depends on Notch signals and GATA-397, 186, but this remains incompletely defined21, 99, 187.
Exclusion of myeloid and dendritic cell potentials is caused by removal of a positive regulator. Access to both options correlates with the expression profile of PU.1, and at early stages C/EBP factors, present at low levels, may also contribute188, 189. But while PU.1 is present Notch signaling keeps myeloid options under check. First, Notch-induced Hes1 represses Cebpa75. Notch signaling also modulates the spectrum of PU.1 transcriptional activities irrespective of C/EBPα66, 188, 190. PU.1 is silenced during DN2b, under the influence of Runx and possibly TCF-1 or GATA-329, 191-193 as well, closing the myeloid options.
The loss of access to NK and ILC2 fates may instead be due to activation of a repressor, i.e. activation of Bcl11b. Both innate lymphocyte programs require Id2 in order to neutralize E protein activity, and Bcl11b is implicated directly in keeping Id2 silent117, 118, 122.
Ironically, essential T-cell lineage regulatory inputs may themselves represent a threat for alternative lineage diversion during the uncommitted stages of T-cell development if their levels are dysregulated. Overexpression of GATA-3 can drive the cells toward a mast-cell fate29, antagonizing Notch-dependent survival systems. GATA-3 and TCF-1 can also work in a low-E protein regulatory state to promote development to ILC2s30, 194, 195 instead of T cells, and even excessive Notch signals may favor ILC fates in human precursors196. These alternatives too are removed during the commitment transition, through mechanisms that need to be clarified.
Figure 2. T-lineage commitment and alternative lineage potentials.
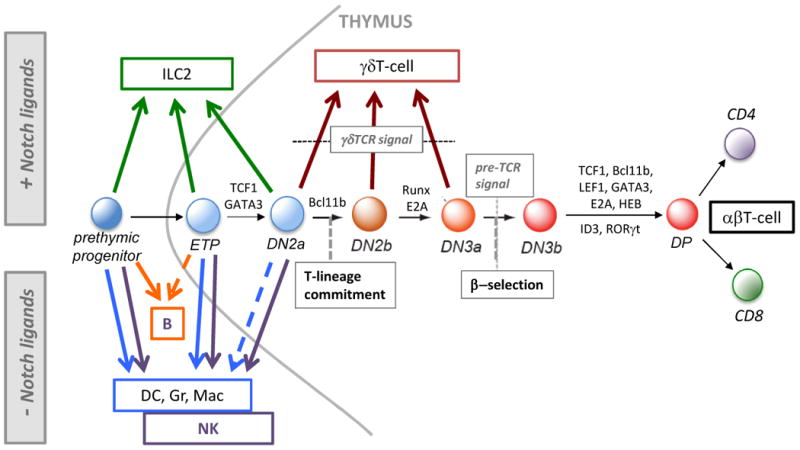
Diagram of early T-cell development for αβT-cells from bone marrow prethymic progenitors to entry into and development in the thymus, showing the stages from which alternative lineages can develop. The figure shows which fates can be accessed by cells in the presence of Notch signaling (upward pointing arrows), including γδT-lineage cells that can develop from DN2a/b and DN3 cells (brown arrows), as well as those fates that can only be accessed by experimental withdrawal from Notch signaling (downward pointing arrows). Arrows shown emerging from the thymus show conditional outcomes usually only observed if the cells are removed from the thymic environment. Alternative non-T-lineage innate lymphocytes (ILC2) can be generated from bone marrow precursors, as well as ETP and DN2 cells, but only in the presence of Notch ligands and specific cytokines. In the absence of Notch ligands, B-cells can be generated from bone marrow precursors and a subset of the earliest (Flt3+) ETP cells, while dendritic cells (DC) and granulocytes (Gr) or macrophages (Mac), can be derived from prethymic progenitors as well as from thymic ETP cells and to a lesser extent from DN2a cells, but not from later stage cells. Natural killer (NK) cells also develop well from prethymic, ETP and DN2a cells, but not from later stage cells. Broken-line arrows denote potentials seen in low-frequency precursors within a population. Shown also are some key transcription factors that are critical for specific transitions (see text and Fig. 3).
By the time commitment has occurred (DN2b–DN3a stages), most T-cell identity genes are fully turned on31, 32. Such genes include all of the invariant TCR components and mediators of TCR-dependent signaling, the expression of which prepares the cells for TCR-dependent survival. Proliferation slows in progression to the DN3a stage, and many cells enter G1 arrest. The cells of this stage then persist until they successfully rearrange TCR genes, or die. Growth and differentiation beyond the β-selection checkpoint at DN3a normally requires expression of a functional TCR, which activates the phase 3 gene network. This occurs when newly made TCRβ protein assembles at the cell membrane with TCR complex components that are already expressed, including pre-TCRα (Ptcra gene product), CD3γ, CD3δ, CD3ε and TCRζ. This pre-TCR complex assembly triggers cell enlargement, progression into rapid cell-cycling, termination of CD25 (IL-2RA) expression, and increased surface expression of CD27 (TNFRSF7) and the co-stimulatory molecule CD28, as the cells enter DN3b and then DN4/ISP stages33-37. Although Notch signals are required for passage through the β-selection checkpoint38-40, cells responding to this “pre-TCR” then free themselves from their Notch-dependence and turn off expression of Notch target genes. A somewhat different pathway is followed by cells that productively rearrange the TCRγ and TCRδ genes in the DN2-DN3 stages; these become one of a variety of TCRγδ cell types (Box1). They too transition from Notch-dependent to Notch-independent differentiation at this point.
Critical transcription factors in the induction and establishment of T cell regulatory networks
Expression of T-cell differentiation genes and many aspects of mature T-cell function eventually depends on a toolkit of transcription factors that includes TCF-1 (Tcf7 gene product) and its relative LEF-1, GATA-3, Bcl11b, E2A (Tcf3 gene product) and HEB (Tcf12 gene product), Ikaros (Ikzf1, or family members), Myb, Gfi1, and complexes of Runx1 (or family members) with CBFβ (Table 1) (see reviews 3, 4, 41-47). The process of T-cell programming, through phase 2 and beyond, must establish gene networks that stably maintain appropriate levels of these factors. However, these are only part of a much larger set of transcription factors that is differentially regulated during the T-cell specification process31, 48-50(Fig. 1b)(Table 1).
Table 1. Critical T-cell specification transcription factor genes in murine early T-cells and their progenitors.
| Gene | Gene Family | Knockout phenotype | Overexpression phenotype | Selected References |
|---|---|---|---|---|
| Notch1 | NOTCH, ankyrin repeat | Block in T development | T-ALL | 1-3 Reviewed in 4, 5 |
| Runx1 (AML1) | RUNT | Early stem cell defects; Block before DN3; Myeloproliferation | AML | 6 7 |
| Ets1 | ETS | Impaired DN3 to DP development and allelic exclusion; Defective NK and CD8 cell development | 8, 9 10 | |
| Ikzf1 (IKAROS) | IKAROS C2H2 ZnF | Loss of lymphoid cells and progenitors; T- and B-ALL | 11, 12 | |
| Tcf3 (E2A) | E protein basic helix-loop-helix (bHLH) | Defects in hematopoietic stem cell pools; DN2 defects; T-ALL | 13, 14 Reviewed in 15 | |
| Myb | MYB SANT (helix-turn-helix, HTH) | Multiple blocks from HSC to DP | 16-19 | |
| Myc (c-myc) | MYC bHLH | Defects in proliferation and survival of HSC, lymphoid progenitors, after β-selection | T-ALL | 20-22 |
| Tcf7 (TCF1) | TCF HMG | Almost complete loss of T cell development; T-ALL | Limited T development in the absence of Notch signals | 23-26 27 |
| Gata3 | GATA ZnF | No DN cells; if deleted later, β-selection block and loss of CD4 cells | Poor viability, diversion to mast cells | 28, 29 Reviewed in 30 31 |
| Hes1 | HAIRY bHLH | Defective T-cell development | 32, 33 34 | |
| Bcl11b (CTIP2) | ZnF, C2H2-like | No αβ T cells; Accumulation of DN2a and NK cells | 35, 36 37, 38 | |
| Tcf12 (HEBcan/alt) | E protein bHLH | Partial developmental blocks at DN1 and between DN3 and DP | Enhanced T-cell differentiation, reduced proliferation (esp. HEBcan) | 39 Reviewed in 40 |
| Lef1 | TCF HMG | No phenotype alone; ISP block with hypomorphic Tcf7 | T-ALL | 26, 41 |
| Gfi1 | SNAG, C2H2-like ZnF | Partial block between ETP and DN2 | Potentiates T-ALL | 42, 43 |
bHLH, basic helix-loop-helix; C2H2, Cys2 His2, a form of Zinc finger; ETS, E26 transformation-specific; HMG, high mobility group; HTH, helix turn helix; SNAG, Snail/Gfi1 domain, a repression domain; Znf, zinc finger
Radtke, F. et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10, 547-58 (1999).
Pui, J.C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299-308 (1999).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269-271 (2004).
Radtke, F., Macdonald, H.R. & Tacchini-Cottier, F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol 13, 427-37 (2013).
Aster, J.C., Blacklow, S.C. & Pear, W.S. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol 223, 262-73 (2011).
Growney, J.D. et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494-504 (2005).
Egawa, T., Tillman, R.E., Naoe, Y., Taniuchi, I. & Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med 204, 1945-57 (2007).
Clements, J.L., John, S.A. & Garrett-Sinha, L.A. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J Immunol 177, 905-12 (2006).
Zamisch, M. et al. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med 206, 2685-99 (2009).
Eyquem, S., Chemin, K., Fasseu, M. & Bories, J.C. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc Natl Acad Sci U S A 101, 15712-7 (2004).
Yoshida, T., Ng, S.Y., Zuniga-Pflucker, J.C. & Georgopoulos, K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7, 382-91 (2006).
Schjerven, H. et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol 14, 1073-83 (2013).
Yan, W. et al. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol 17, 7317-27 (1997).
Semerad, C.L., Mercer, E.M., Inlay, M.A., Weissman, I.L. & Murre, C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A 106, 1930-5 (2009).
De Pooter, R.F. & Kee, B.L. E proteins and the regulation of early lymphocyte development. Immunol.Rev. 238, 93-109 (2010).
Allen, R.D., 3rd, Bender, T.P. & Siu, G. c-Myb is essential for early T cell development. Genes Dev 13, 1073-8 (1999).
Lieu, Y.K., Kumar, A., Pajerowski, A.G., Rogers, T.J. & Reddy, E.P. Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A 101, 14853-8 (2004).
Bender, T.P., Kremer, C.S., Kraus, M., Buch, T. & Rajewsky, K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 5, 721-9 (2004).
Emambokus, N. et al. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J 22, 4478-88 (2003).
Douglas, N.C., Jacobs, H., Bothwell, A.L. & Hayday, A.C. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol 2, 307-15 (2001).
Smith, D.P., Bath, M.L., Metcalf, D., Harris, A.W. & Cory, S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood 108, 653-61 (2006).
Laurenti, E. et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611-24 (2008).
Verbeek, S. et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374, 70-4 (1995).
Germar, K. et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108, 20060-5 (2011).
Weber, B.N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63-8 (2011).
Yu, S. et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813-26 (2012).
Tiemessen, M.M. et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol 10, e1001430 (2012).
Ho, I.C. & Pai, S.Y. GATA-3 - not just for Th2 cells anymore. Cell Mol Immunol 4, 15-29 (2007).
Taghon, T., Yui, M.A. & Rothenberg, E.V. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 8, 845-55 (2007).
Ho, I.C., Tai, T.S. & Pai, S.Y. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat.Rev.Immunol 9, 125-135 (2009).
Hosoya, T., Maillard, I. & Engel, J.D. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev 238, 110-25 (2010).
Tomita, K. et al. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13, 1203-1210 (1999).
Wendorff, A.A. et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 33, 671-84 (2010).
De Obaldia, M.E. et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol 14, 1277-84 (2013).
Wakabayashi, Y. et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol 4, 533-9 (2003).
Ikawa, T. et al. An essential developmental checkpoint for production of the T cell lineage. Science 329, 93-6 (2010).
Li, L., Leid, M. & Rothenberg, E.V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89-93 (2010).
Li, P. et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329, 85-9 (2010).
Wojciechowski, J., Lai, A., Kondo, M. & Zhuang, Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol 178, 5717-26 (2007).
Braunstein, M. & Anderson, M.K. HEB in the spotlight: Transcriptional regulation of T-cell specification, commitment, and developmental plasticity. Clin Dev Immunol 2012, 678705 (2012).
Okamura, R.M. et al. Redundant regulation of T cell differentiation and TCRβ gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8, 11-20 (1998).
Yucel, R., Karsunky, H., Klein-Hitpass, L. & Moroy, T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med 197, 831-44 (2003).
Khandanpour, C. & Moroy, T. Growth factor independence 1 (Gfi1) as a regulator of p53 activity and a new therapeutical target for ALL. Oncotarget 4, 374-5 (2013).
The induction of critical T cell specification genes such as Tcf7, Gata3 and Bcl11b by thymic Notch ligands occurs in the context of previously established legacy stem/progenitor cell transcriptional networks. These legacy networks are not immediately dismantled and many of these progenitor-specific factors persist through multiple cell cycles and stages within the thymus (Figure 1), even though continued expression of these regulatory genes beyond the ETP and DN2a stages may be oncogenic. Furthermore, some of these progenitor genes are now known to play important roles during the precommitment phase, as described below. Understanding what these “phase 1-restricted factors” contribute to normal T-cell development is an important frontier in T-cell developmental homeostasis.
Phase 1-restricted transcription factors
Because development of each T-cell precursor cohort is coupled with proliferation, the steady-state percentage of thymocytes that are still in precommitment stages is tiny. However, in these early stages TCR-independent proliferation from any given immigrant can be extensive, spanning more than 10 cell cycles before commitment7, 51, and this proliferative expansion could be a major function supported by the legacy progenitor network . Many of the transcription factors that are expressed only in phase 1 ETP and/or DN2a cells have roles in the proliferation, survival, and self-renewal of other hematopoietic cells and leukemias, where the genes that encode them are functionally implicated as proto-oncogenes (see below; Table 2).
Table 2. Critical Phase 1-specific transcription factor genes in murine early T-cells and their progenitors.
| Gene | Gene Family | Knockout Phenotype | Overexpression Phenotype | Selected References |
|---|---|---|---|---|
| Lmo2 (Rbtn2) | LIM | Severe stem cell defects; No T cell effects if deleted after DN2 | T-ALL | 1-4 |
| Gata2 | GATA | Early stem cell defects | 5, 6 | |
| Mef2c | MADS-box | T, B and NK defects | AML, T-ALL association | 7 8 |
| Meis1 | MEIS Homeodomain | Early stem cell defects | AML | 9, 10 |
| Hoxa9 | HOX Homeobox | Defect in HSC proliferation; Block in DN, partial DN2 block | AML | 11, 12 |
| Tal1 (SCL) | TAL bHLH | Early stem cell defects | T-ALL | 13, 14 |
| Gfi1b | SNAG, C2H2-like ZnF | HSC expansion | 15 | |
| Lyl1 | TAL bHLH | Defects in LMPP, ETP, DN2a cells | ALL (B and T) | 16 17, 18 |
| Spi1 (Sfpi1, PU.1) | ETS | Absence of T-cell and NK development; AML | Diversion to DC or myeloid lineages | 19-22 Reviewed in23 |
| Bcl11a (Evi9, Ctip1) | ZnF, C2H2-like | Required for B, T, NK development | Myeloid, B, T oncogene | 6, 24 25 |
| Hhex (Prhx) | Homeobox | Impaired monocyte development | T-ALL | 2 26 |
| Mycn (N-myc) | MYC bHLH | Defects in HSC survival, proliferation (with c-myc) | AML | 27, 28 |
| Erg | ETS | Early stem cell defects | T-ALL and other leukemias | 29 30 |
bHLH, basic helix-loop-helix; C2H2, Cys2 His2, a form of Zinc finger; ETS, E26 transformation-specific; SNAG, Snail/Gfi1 domain, a repression domain; Znf, zinc finger
Yamada, Y. et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc Natl Acad Sci U S A 95, 3890-5 (1998).
McCormack, M.P. et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879-83 (2010).
McCormack, M.P., Forster, A., Drynan, L., Pannell, R. & Rabbitts, T.H. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol Cell Biol 23, 9003-13 (2003).
Cleveland, S.M. et al. LIM domain only-2 (Lmo2) induces T-cell leukemia with epigenetic deregulation of CD4. Exp Hematol (2014).
Johnson, K.D. et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest 122, 3692-704 (2012).
Lim, K.C. et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest 122, 3705-17 (2012).
Stehling-Sun, S., Dade, J., Nutt, S.L., DeKoter, R.P. & Camargo, F.D. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat Immunol 10, 289-96 (2009).
Homminga, I. et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 19, 484-97 (2011).
Hisa, T. et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J 23, 450-9 (2004).
Azcoitia, V., Aracil, M., Martinez, A.C. & Torres, M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol 280, 307-20 (2005).
Lawrence, H.J. et al. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89, 1922-30 (1997).
Izon, D.J. et al. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood 92, 383-93 (1998).
Robb, L. et al. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J 15, 4123-9 (1996).
Porcher, C. et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86, 47-57 (1996).
Khandanpour, C. et al. Evidence that growth factor independence 1b regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood 116, 5149-61 (2010).
Zohren, F. et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol 13, 761-9 (2012).
Zhong, Y., Jiang, L., Hiai, H., Toyokuni, S. & Yamada, Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene 26, 6937-47 (2007).
McCormack, M.P. et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood 122, 2093-103 (2013).
Dakic, A. et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med 201, 1487-502 (2005).
Iwasaki, H. et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106, 1590-600 (2005).
Franco, C.B. et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A 103, 11993-8 (2006).
Laiosa, C.V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L. & Graf, T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 25, 731-44 (2006).
Carotta, S., Wu, L. & Nutt, S.L. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev 238, 63-75 (2010).
24. Liu, P. et al. Bcl11a is essential for normal lymphoid development. Nat Immunol 4, 525-32 (2003).
Yu, Y. et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med 209, 2467-83 (2012).
Keng, V.W. et al. Homeobox gene Hex is essential for onset of mouse embryonic liver development and differentiation of the monocyte lineage. Biochem Biophys Res Commun 276, 1155-61 (2000).
Kawagoe, H., Kandilci, A., Kranenburg, T.A. & Grosveld, G.C. Overexpression of N-Myc rapidly causes acute myeloid leukemia in mice. Cancer Res 67, 10677-85 (2007).
Laurenti, E. et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611-24 (2008).
Tsuzuki, S., Taguchi, O. & Seto, M. Promotion and maintenance of leukemia by ERG. Blood 117, 3858-68 (2011).
Thoms, J.A. et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood 117, 7079-89 (2011).
From thymic entry to commitment the cells progress through several variants of the phase 1 regulatory state, due to the staggered losses of legacy phase 1 restricted genes and gain of Notch target genes (Fig. 1b;Fig. 3a-c). The most restricted subset of phase 1 regulators, including Gata2, Meis1, and Hoxa9, is expressed solely or predominantly in ETPs. Some of these genes may be co-expressed with Flt3 in a pattern restricted to the most primitive ETPs, as these genes are known to be involved in regulatory networks in Flt3+ prethymic progenitors52, 53. The possibilities for their functions in prethymic progenitors are intriguing, but most likely they are important for supporting engraftment in the thymus. Mef2c and Lmo2 are turned off a little later, persisting into the DN2a stage, especially in adult thymocytes, with their protracted period of expansion31, 32. The phase 1 factors Lmo2, Meis1 and N-myc (see below) have a direct connection to “stem-ness”: they have been identified recently as potent components of a transcription factor “cocktail” that can reprogram differentiated blood cells back to hematopoietic stem cells54.
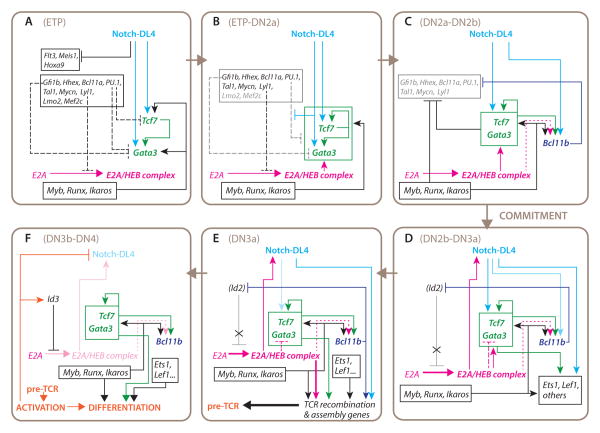
Figure 3. Progression of gene regulatory network states in T-cell commitment: current model.
Panels a-f (clockwise progression) show a model of the sequential changes in regulatory relationships between the genes, based on evidence discussed in the text, from the early Flt3+ ETP stage (a) through commitment (c-d) to β-selection (f). Passage through the indicated changes in gene network states (a-f) occurs during periods that correspond approximately to developmental stages, as defined by cell-surface markers, which are indicated at the top of each panel, but the regulatory states may not be homogeneously equivalent to these stages. Phase 1: panels a, b. Transition to phase 2: panel c. Phase 2: panels d, e, in which panel d emphasizes the regulatory gene expression changes and panel E emphasizes the consequences of those changes. Phase 3, post β-selection (for αβT lineage): panel f. Boxes enclose genes that operate coordinately at a given stage even if the details of regulation that link them are not known. Arrows indicate activation. Lines ending in bars indicate repression. Dashed lines indicate “soft repression” which limits the maximal activity or deployment of a transcription factor but does not silence its expression. Dotted lines: likely but undemonstrated linkages. Faded colors: decreasing expression or functional impact of a given gene or gene group as a result of regulatory effects shown elsewhere in the panel. (Id2), potential for activation of Id2 in response to cytokine signaling or other stimuli, which is held in check by Bcl11b. Note that E2A/HEB heterodimers can play a positive role in Gata3 and Tcf7 induction as well as exerting restraint on maximum levels of Gata3 expression. TCR recombination and assembly genes steeply increase in expression from DN2a to DN3a, but are most functionally significant in DN3a. As revealed by T-ALL phenotypes, cells that take a “short cut” from the states represented in panels a-b to the activation events in panel f, bypassing the commitment process, are susceptible to oncogenic transformation. For details, see text.
The most characteristic group of phase 1 transcription factors is expressed throughout the precommitment stages. These include Spi1 (PU.1, also called Sfpi1), Hhex, Bcl11a, Gfi1b, Erg, Mycn (N-myc) and Lyl1 (and its relative Tal1, expressed at much lower levels)(Figure 1b; Table 2). Most of these genes are repressed partially or completely during the transition to phase 2 (between DN2a and DN2b), although Lyl1 is only silenced in the DN3a stage28, and Erg only during β-selection31, 32.
Bcl11a is crucial for normal lymphoid progenitor proliferative expansion in both fetal liver and bone marrow55. Similarly, although the basic helix–loop–helix (bHLH) factor Lyl1 plays its best-known role in hematopoietic progenitors and B-cells56, 57, deletion of Lyl1 severely impairs development of lymphoid precursors in bone marrow and specifically reduces ETP expansion, progression to DN2, and early T-cell survival56, 58, 59. Lyl1, working in dimers with E2A or HEB, probably helps to maintain expression of the crucial ETP–DN2 growth factor receptor Kit, similar to the function of the related protein SCL (Tal1), and promotes expression of Gfi1, a regulatory factor required for normal generation of ETPs and for proper responses to Notch signaling 58, 60, 61. Erg and Hhex are implicated in early T-cell self-renewal in both normal and malignant contexts62, 63. Thus, despite their transient expression, these factors should be regarded as integral parts of the T-lineage program.
PU.1: linking phase 1 and developmental options
PU.1 is normally associated with differentiation of non-T cells, i.e. dendritic, myeloid, and B-cells64, and its expression correlates very well with the ability of uncommitted T-cell precursors to divert to dendritic cell and myeloid fates when Notch signaling is withdrawn (Fig. 2). However, loss of PU.1 from prethymic or DN1 stages also causes an arrest in T-cell development65. PU.1 is important for expression of Flt3 in immigrants, and it probably sustains the pro-survival actions of Bcl11a in developing thymocytes until they reach commitment (phase 2), as well as regulating other phase 1 regulatory genes66. PU.1 is a highly stable protein and can act as a pioneer factor in some contexts67-70. It binds to most enhancers and promoters active in ETP and DN2a cells in vivo31. Thus, it may in part shape the epigenetic landscape in these hematopoietic progenitors in ways that affect the activities of many other factors. It may also play a more intimate role in controlling the T-cell program, via network interactions with Notch and T-cell commitment genes including Tcf7, Myb and Gata366. PU.1 can dampen the intensity of Notch signaling; this contributes to its ability to restrain the induction of many T-cell identity genes as long as it is expressed66 (A. Champhekar & E.V.R., unpublished results). Because this Notch antagonism is graded and not absolute, PU.1 may itself contribute to the proliferative expansion of T-cell precursor clones by delaying the onset of commitment, as described below.
Notch target genes in early T cell precursors
Notch signaling activates distinct target genes in disparate developmental contexts, and even in DN thymocytes it activates a succession of different target genes at different stages. DN thymocytes are primed for responsiveness to intrathymic Delta ligands by expression of Fringe proteins encoded by Lfng, Mfng and Rfng71. Notch signaling tightly controls the expression of genes encoding the canonical DN2 marker CD25 (Il2ra), the ubiquitination adaptor Deltex1 (Dtx1), pre-TCRα (Ptcra), the ankyrin repeat protein Nrarp, Notch3, and to some extent Notch1 itself, as well as the transcription factors Myc and Hes1, all of which have different patterns of expression in the DN stages39, 72, 73. Notch signaling also strictly controls activation of a distinct promoter for Tcf12 (HEB) that drives the expression of a truncated protein, HEBalt, which appears to be more supportive of cell proliferation than canonical HEB or E2A46, 74.
Hes1 and Nrarp are among the earliest Notch-activated target genes induced in ETPs. Hes1 is a bHLH transcriptional repressor that is needed for early T-cell population expansion, and may also antagonize myeloid cell development75-77. Nrarp is an adaptor protein that can act as a substantial negative feedback regulator of Notch signaling itself78, 79. Whereas Hes1 expression continues from ETP through the DN3a stage, Nrarp is shut off at commitment. Thus in phase 1, Notch signaling is self-limiting and exacerbates the Notch-damping effect of PU.1. Both restraints are relieved, however, when the cells undergo commitment. The later-activated Notch target genes may thus be awaiting an increase in Notch signal strength for their induction as well as other inputs.
Building infrastructure for the Phase 2 network
The regulatory bridge to the phase 2 gene network is built from within the phase 1 network. Notch signaling in ETP cells initiates expression of the crucial regulatory genes Gata3 and Tcf7 (encoding TCF-1), which then transform the developmental status of the cells (Fig. 3a, b). Even in the context of the phase 1 developmental network, these two genes begin to exert T-lineage promoting functions as soon as they are expressed, as knockouts of either gene severely reduce the survival and differentiation of ETP cells as well as their descendants80, 81. They not only begin to antagonize the progenitor-specific, phase 1-restricted factors (Fig. 3b), but also collaborate with the regulatory genes needed for T cell development that were also part of the legacy set, expressed from a prethymic stage (Myb, Runx1/CBFβ, Ikaros, Gfi1, E2A). A particularly important member of this group of inherited factors is the basic helix-loop-helix factor E2A, which is expressed at relatively constant levels throughout T-cell specification, but has its activity substantially modulated by availability of different heterodimerization partners45, 46, 58, 82-86. Despite their stable expression, all these progenitor-inherited factors can also act in developmentally modulated ways, and they become incorporated with TCF-1 and GATA-3 into the network for T-cell lineage commitment.
Activation and function of TCF-1
Tcf7 is activated by Notch directly81, 87, but once activated, its expression is no longer severely affected by Notch signal inhibition29, 66. This suggests that it may become self-sustaining by direct or indirect positive feedback (Fig. 3c). In ETPs, TCF-1 drives expression of a suite of genes needed for survival and expansion81. Some fetal T-cells can develop without it88, 89, but TCF-1 becomes increasingly important in postnatal waves of T-cell precursors90. TCF-1 primarily acts as a crucial positive regulator of T-cell specification, collaborating with Notch to activate T-cell genes through a feed-forward network circuit. Although TCF-1 cannot duplicate all effects of Notch signaling, high amounts of a full-length isoform of TCF-1 can activate many T-cell genes including Gata3, Bcl11b, Il2ra (CD25), Cd3g, Lat, Lck and endogenous Tcf7, even without Notch signals87.
It is still controversial whether TCF-1 acts together with β-catenin in these positive regulatory roles87, 91. Although it can mediate canonical Wnt signaling via the formation of a complex with β-catenin, TCF-1 may operate in a different mode in ETP and DN2a cells. At a later stage TCF-1 can directly or indirectly help to repress Id2, a potent E2A antagonist that can otherwise be induced readily in T-cell precursors89. Certain natural isoforms of TCF-1 also repress or delay activation of the related TCF-1 gene, Lef1, which can otherwise become oncogenic88, 89. Finally, as a highly abundant transcription factor in developing T-cells, TCF-1 may also play an architectural role at cis-regulatory elements across the genome92: its high-mobility group DNA binding domain can bend the DNA to facilitate contacts among other transcription factors93.
In prethymic cells the gene encoding TCF-1 (Tcf7) begins with substantial CpG methylation on some of its putative regulatory elements94, but its promoter and major enhancer become fully active during ETP stage31, and CpG methylation is removed from Tcf7 by the time cells reach DN2 and DN3 stages94. This may reflect the action of Notch/RBPJ(CSL) complexes at the enhancer, in addition to direct and indirect positive feedback signals from TCF-1 and GATA-381, 87. Initially, Notch signals remain important to protect Tcf7 expression against a variety of experimental regulatory perturbations ranging from excessive PU.1 activity66 to excessive GATA-3 activity29. However, in later stages of T-cell development (phase 3 and beyond), Tcf7 can be expressed strongly in a Notch-independent way, in a pattern often correlated with levels of GATA-3.
GATA-3 functions and regulation
GATA-3 has vital dose-dependent roles in early T-cell survival, growth, specification, and commitment, as well as continuing roles in T-cell subset diversification after TCR expression and antigen stimulation95, 96. It plays a key non-redundant role in excluding the B-cell fate97, and probably explains why B-cell potential is lost within the ETP stage24, 98, 99. It is required for full activation of Bcl11b in DN2a-DN2b stage and to prime DN3 cells for β-selection, and may also upregulate Ets1 and repress Spi1 in DN2b stage29, 97, 100(D. D. Scripture-Adams, S. Damle, and E.V.R., unpublished)(Fig. 3c-e). Gata3 expression is activated by a mechanism involving a far downstream enhancer101 and by removal of H3K27me3 repression marks from its promoter102, 103, although the sequence(s) through which it is activated by Notch in ETP cells have not been identified. Part of the regulation of Gata3 could be mediated indirectly, e.g. by TCF-1. Although GATA-3 levels can be maintained when Notch signals are removed, Notch signaling is needed to enable Gata3 to be expressed in the phase 1 context of high PU.1 activity66.
GATA-3 must be tightly regulated at several levels. First, the negative effect of GATA3 overexpression on T-cell development is as severe as loss of this transcription factor29, 104. In precommitment stages, Gata3 expression can be restrained by phase 1 transcription factors PU.1 and Gfi1b66, 105 (Fig. 3a, b). During and after commitment, surprisingly, it is restrained by other T-cell specification regulators including E2A and Bcl11b104(Zhang, J.A., Li, L., and E.V.R., unpublished data)(Fig. 3d, e). Second, GATA-3 is subject to considerable post-transcriptional regulation by signaling pathways that are downstream of many growth factor and activation signals, including Akt pathway regulation of its translation and p38 MAP kinase regulation of its nuclear localization and activity106-108. Thus, GATA-3 activity levels in a cell integrate responses to environmental signals with intrinsic transcriptional programming. Third, the sites that GATA-3 binds across the genome are distinct at different stages of T cell development, even at matched levels of GATA3 expression. Major redistributions of GATA-3 binding are seen between successive DN and DP thymocyte stages, as well as between thymocytes and mature T cells31, 109. This is not because GATA-3 is particularly excluded by repressed (H3K27me3-marked) chromatin sites, but because of shifting preferences among potential open sites. The implication is that GATA-3 “assignments” depend on interactions with changing stage-dependent collaborating factors. Thus, as a member of a regulatory ensemble, this factor may contribute in different ways in each of the three phases from ETP to DP stage.
Phase 2 - commitment
Transition to phase 2
Several cell cycles after TCF-1 and GATA-3 are activated, cells transition to the DN2b stage, when proliferation rates slow, survival becomes strictly Notch signal-dependent, and Kit expression declines28. Bcl11b expression is activated and most phase 1 genes are silenced (Fig. 3c-d). As the cells progress through the DN2b stage there are major shifts in the expression of Ets family transcription factor genes: Ets1 and Ets2 are sharply turned on as Spi1 is turned off48. Lef1 is induced in a highly Notch-dependent way29, 66, adding a partially redundant transcriptional regulation activity to that of its relative Tcf788, 89, 110.
During in vitro differentiation, DN2b cells can continue to proliferate, but in vivo these transformations presage a faster shift to the DN3a stage. The cells become desensitized to IL-7R signaling, in a process dependent on E proteins111. E2A/HEB activity is probably boosted by the silencing of the competitor dimer partner Lyl128, 112. E protein and Notch-dependent gene expression soars (e.g., Rag1, Rag2, Ptcra, Dntt, Cd3e)72, 83, 104, 113-115, and as proliferation slows, RAG-mediated TCR gene recombination intensifies (Fig. 3e).
Bcl11b expression is necessary for many if not all of these changes, and the timing of Bcl11b activation is crucial for lineage commitment116-118. Bcl11b, a six zinc-finger transcription factor with functions including corepressor activity119, is turned on in late DN2a. It may repress Kit directly, thus creating the DN2b phenotype; virtually all DN2b cells express high levels of Bcl11b (H. Y. Kueh and E.V.R., unpublished results). Both the promoter and a far-distant enhancer appear to participate in Bcl11b activation and to contain binding sites for Notch, TCF-1, Runx/CBFβ, and possibly GATA-3120, all of which are positive regulators for Bcl11b expression87, 97, 118, 121. These known positive regulators are expressed before Bcl11b is turned on, but their combined levels may need to exceed a threshold in order to remove initial DNA methylation and H3K27me3 marks from both the promoter and the enhancer of the Bcl11b gene94, 120.
Commitment: end of progenitor status and other options
The commitment transition from DN2a to DN2b marks the end of a process of exclusion of access to different alternative fates (Figure 2). The completion of commitment in DN2b stage terminates access to “innate lymphocyte” (NK or ILC1 and ILC2) development28, 30, while perhaps surprisingly, potential for non-lymphoid pathways to dendritic cell, granulocyte, macrophage and possibly mast-cell development is only extinguished at this point as well (rev. in 26). At least three kinds of molecular mechanisms implement different aspects of alternative lineage exclusion, including the repression of PU.1 (Box 2).
The silencing of PU.1 has an obvious link to commitment, but commitment also silences many other phase 1 factors such as Gfi1b, Hhex, Bcl11a, Mycn, Lmo2, and Mef2c, which have less clear roles in promoting alternative cell fates. Even in some leukemias where these factors are aberrantly expressed (see below), they immortalize pro-T cells without obscuring their T-lineage identity. This implies that T-cell commitment excludes a progenitor-like form of self-renewal as well as specific alternative differentiated fates. Commitment mechanisms that silence phase 1 genes are thus most likely to underlie the cell growth control alteration that intensifies Notch dependence, slows proliferation, and finally brings proliferation under strict TCR dependence28.
The phase 1-specific regulatory genes that are silenced after DN2a show diverse repression patterns. The levels of positive enhancer and promoter marks [H3K4me2, H3K(9,14)Ac] on these genes are reduced as transcription decreases, but H3K27me3 repression marks generally only begin to appear later (if at all), often not until the DP stage31, 103. Bcl11b is required for silencing other phase 1 genes during commitment, including Lyl1, Hhex, Bcl11a, and possibly Spi1 (PU.1), but it is not yet clear whether Bcl11b directly acts on them as a repressor (J. A. Zhang, L. Li and E.V.R., unpublished data)116-118, 122. Thus, a variety of negative regulatory mechanisms ensure that phase 1 proto-oncogenes are silent by the time cells reach the β-selection checkpoint.
DN3a stage: phase 2 beyond commitment
DN3a stage cells are committed, αβ lineage-biased, and primed for TCR gene rearrangement based on a climax of phase 2 regulatory activity. To attain and survive in this stage, cells need Runx1/CBFβ, Notch signals, GATA-3, Gfi1, Bcl11b, and E2A; Myb and Ets1 also play roles in maintaining viability by this stage if not earlier (Fig. 3e). ChIP-seq analysis shows that GATA-3 binding is now redeployed from its phase 1 network targets to a new set of target genes31. Some transcription factors, including Ahr and SpiB, are transiently expressed only during this DN3a stage (Fig. 1b), but their functions at this point are unknown. The maximal expression of the Cd3 gene cluster is regulated by E proteins, GATA-3, and TCF-1; the Rag genes are induced by E proteins and GATA-3; and Hes1, Notch3, and Ptcra are jointly regulated by E proteins and Notch31, 83, 104, 123, 124.
Involvement of E proteins is a hallmark of gene expression in DN3a cells in particular, and E proteins, Notch signaling, and Bcl11b may have a particularly tight collaboration. E proteins are needed to promote and sustain Notch1 expression in DN3a cells, and Notch signaling can be antagonized by E protein antagonist Id266, 125. Bcl11b may thus support the DN3a state by keeping Id2 expression silent117, 118, 122 (Fig. 3d, e). Change in αβ versus γδ T-cell potential between the DN2 and DN3a stages could also be related to their differential needs for Bcl11b and Notch, as well as the strong activation of Id3 or Id2 factors linked to selection of most γδ T-lineages126. The Notch/E protein target gene Ptcra is very poorly activated in Bcl11b-knockout cells. αβ T-cells are accordingly disproportionately affected by Bcl11b deletion127, 128, whereas several γδ T cell lineages, especially fetal lineages and lineages that emerge early from the DN2 stage, are much less affected117, 122, 129, 130. Thus, special features of the DN3a stage may set up the conditions for selection processes specific for the αβ T cell lineage.
Phase 3: Crossing the β-selection checkpoint
Construction of the checkpoint
The proliferation and differentiation checkpoint imposed at DN3a is enforced by E2A and Ikaros84, 131, 132. There are several clues from target gene analyses that suggest why proliferation stops at this point. E2A may slow cell cycling by inducing Cdkn1a123 (coexpressed with Cdkn2d). Other features of the cells suggest that they are progressively starved for growth signals. As they lose support from IL-7R signaling, the cells upregulate a PI3-kinase inhibitor, Pik3ip1, and a growth-suppressive kinase, Dapk1, in response to E2A104, and these may make the cells dependent on ever-stronger viability support signals from Notch and eventually from pre-TCR133. Ikaros, although stably expressed from the earliest stages, also has a vital, dose-dependent role in enforcing the β-selection checkpoint132, 134. The complexes Ikaros forms are pivotal for patterns of nucleosome remodeling activity deployment across the genome135. How exactly Ikaros contributes to cell cycle arrest and TCR gene rearrangement is still unclear, but it acts strongly to suppress Notch response genes rapidly once pre-TCR signals are received136-138, and this is crucial to block leukemogenesis.
β-selection
Signaling through the newly expressed pre-TCR and passage through the β-selection checkpoint disrupt the quiescence of DN3a-stage αβ T-cell precursors. Expression of both Notch target genes and Il7r is shut off as they move to DN3b and beyond (Fig. 1; Fig. 3f)34. Despite the downregulation of these growth-supporting systems, the cells shift into very rapid proliferation, as the cells become highly responsive to chemokine signaling through the receptor Cxcr4139, 140. Proliferation not only expands the potential TCR repertoire pool, it also helps αβ T-cells confirm their lineage commitment by allowing the cells to dilute out the last vestiges of earlier regulatory molecules and to reset epigenetic marks37.
Creating the new DP state, the expression of Ets1, Tcf7 (TCF-1), and Lef1 further increases, whereas that of Ets2 and Tcf12 (canonical HEB) reaches new peaks. Several new transcription factors are turned on during β-selection, including Rorγt, Nfatc3, Pou6f1, and Aiolos (Ikzf3), while some like Egr2 and Id3 are transiently activated in response to the TCR signal during the rapidly proliferating DN4 stages (Fig. 1). Although E protein activity is transiently antagonized by this temporal increase in Id3 expression, E protein dominance is subsequently restored141. RORγt expression in thymocytes is highly specific for DP cells, where it works in a different gene network context than it does in mature Th17 cells142, 143. In DP cells, it works with Myb to promote viability through BclxL induction while suppressing conventional effector responses like proliferation or cytokine production144-146. The roles of TCF-1 and LEF-1 may be qualitatively altered at this point by a stage-specific interaction with the canonical Wnt pathway mediator β-catenin, which is more strongly implicated in effects at β-selection than in earlier stages88, 147-150. This altered regulatory factor ensemble turns on the expression of CD4, CD8, CD2 (in mice), and CD28.
However, arguably the most dramatic feature of the change in the developmental gene network at β-selection is repression. Any residual expression of phase 1 genes vanishes along with DN3-specific regulatory genes like SpiB and Ahr and the Notch-dependent genes. The whole Notch-dependent regulatory framework is dismantled like a redundant assembly scaffold (Fig. 3f). The broad downregulation of multiple Notch targets may result from repression of Lfng, which disables Notch on DP cells from access to its intrathymic ligands71. As cells become DP, H3K27me3 repression marks pile up at the promoters of phase 1 genes and Notch targets such as Hes131, 103. The resultant new regulatory state prepares DP thymocytes for the complex positive and negative selection events that they must undergo.
Although relatively few “new” factors enter the regulatory mix, the β-selection cascade is dangerous because of the way new growth signals impinge on a rapidly altering regulatory state. Cells with inadequate E protein or Ikaros activity cannot control access to this transition or re-establish quiescence correctly, and they become malignantly transformed. Notch target genes must be silenced: inappropriately prolonged Notch activity, in the context of TCR complex activation during β-selection, tilts the cells toward leukemia. Furthermore, as described below, any persistence of phase 1 gene expression through this transition can itself lead to leukemic transformation.
T-cell leukemia
Checkpoint failure and T-cell leukemias
The earliest stages of early T-cell development are replete with potent proto-oncogenes (Fig. 1b; Table 2) inherited from pre-thymic progenitors. Many of these genes encode factors that regulate proliferation, survival and differentiation in the earliest stages of T-cell development in conjunction with the Notch–mediated T-cell specification program, although termination of this progenitor program is imperative for both completing T-cell specification and avoiding leukemogenesis. Overexpression of individual phase 1 transcription factors in mouse hematopoietic stem and progenitor cells commonly lead to self-renewal and transformation, with the specific leukemia type dependent upon the cell in which it is first overexpressed (Table 2). In addition, recent data show that if early thymic cells are not displaced from their intrathymic niches by new immigrants, they retain self-renewal potential coupled with a threat of leukemic transformation 15-17.
Repression of legacy proto-oncogenes occurs at several developmental points (Figure 1). While a subset of these genes terminates expression within the ETP stage, others are expressed through to the commitment checkpoint, after which time most of these phase 1-specific genes are repressed. By the time cells transit through the β-selection checkpoint, remnants of the stem/progenitor program are fully and permanently extinguished. Epigenetic mechanisms likely play important roles in establishing and maintaining repression of the progenitor gene programs, although the process needs clarification and likely varies between stages and specific genes. For example, progenitor genes that continue to be expressed until commitment are generally not strongly marked by H3K27me3 until after β-selection, even though most are repressed in the DN2 stage31. These results hint that proliferation may be needed for establishment of repressive chromatin.
T-ALLs in humans are a heterogeneous group of aggressive T-cell malignancies that are thought to arise from oncogenic transformation of immature thymocytes151. These leukemias can result from mutation and/or uncontrolled activation of a T-cell proto-oncogene, such as Notch, or from translocation of an exogenous oncogene to the control of T-cell specific promoters or enhancers like those of a TCR locus or BCL11B. Sub-types of human T-ALL, with differing treatment responses and relapse rates, have been identified based on surface phenotypes and genetic signatures, including overexpression of specific transcription factors and surface receptors characteristic of different stages of the T-cell differentiation program152-156. While these genetic signatures provide hints as to the primary and cooperating genetic lesions in the multi-step process that drives malignant transformation, manipulatable mouse models are invaluable for investigating the sequence of genetic lesions that can initiate and sustain T-ALLs. Although some markers of T-cell development differ between humans and mice157, 158, the broader requirements for Notch and cytokines, as well as underlying transcriptional networks are very similar159, making results with mouse models relevant to human T-ALL.
Failure to transit correctly through the β-selection checkpoint due to sustained Notch signaling or loss of E proteins or Ikaros has long been known to result in malignant transformation of early T-cells134, 160-162. Constitutively activated Notch signaling is particularly potent at triggering T-cell leukemia in both humans and mice, with over 50% of human T-ALL cases exhibiting activating mutations in NOTCH1 or its pathway151, 160, 163. However, in both mice and humans distinct “early” subtypes of T-ALL exist, with characteristics of very immature T-cells, and these may arise from a failure to permanently repress legacy phase 1 stem and progenitor genes, which have only recently been recognized to be a major part of early T-cell differentiation156.
High expression of phase 1 genes LYL1 and LMO2 was originally found to distinguish subtypes of human T-ALLs that are associated with higher rates of treatment failure and relapse153. Subsequently, a T-ALL subtype was designated as ETP T-ALL (ETP-ALL), based on overexpression of genes expressed by murine thymic ETP cells, including KIT, GATA2, CEBPA, SPI1 (PU.1), LYL1, and ERG1, 18. ETP-ALLs also express myeloid surface markers and have lower levels of typical T-cell genes. Genome-wide sequencing of human ETP-ALLs showed that they commonly have activating mutations in cytokine receptor genes (FLT3, IL7R) and genes encoding members of RAS signaling pathways, as well as inactivating mutations in some important T-cell genes and specific histone modifying genes, and that they resemble myeloid stem cells and acute myeloid leukemia more than other forms of T-ALL19. Thus, the genetic characteristics of ETP-ALL, including the genes shared with myeloid progenitors, indicate a failure to properly terminate the normal phase 1 regulatory state, despite the onset of the T-cell specification program. As in murine early T cells, the human T-cell development program could be built on a proto-myeloid foundation164.
Specific genes involved in epigenetic silencing are mutated in many ETP-ALL, especially genes EZH2, SUZ12, and EED19, which are part of the Polycomb Repressive Complex 2 (PRC2) that mediates H3K27 trimethylation. This further supports a role for these pathways in extinguishing expression of stem/progenitor phase 1 genes during normal commitment, as implied by repression marking patterns in mice31. Loss-of-function mutations in components of PRC2 are found in other kinds of T-ALLs as well, however, suggesting more diverse stage-specific roles in normal T-cell development and tumor suppression165. Other genes involved in establishing gene repression, such as the DNA methyltransferase DNMT3A, are also mutated in some subsets of T-ALL166, although the role of this gene and other epigenetic factors in early T-cell development and leukemia initiation in mice and humans have yet to be determined.
Three mouse models provide additional insights into the mechanisms of leukemogenesis due to loss of phase 1-specific transcription factor gene repression. First, when overexpressed in mouse DN thymocytes, the ETP-specific gene Lmo2 generates a self-renewing population of Kithi DN thymocytes, and the mice develop T-cell leukemias after a long latency period63, 167. While the frank leukemia probably depends on accumulating additional mutations, the preleukemic self-renewing cells already express other phase 1 genes including Hhex and Lyl1, which play cooperative roles with Lmo2 in self-renewal and leukemogenesis59, 63. Although most T-ALLs also carry activating Notch pathway mutations, it is noteworthy that ETP-ALLs need not do so. T-lineage expression of an Lmo2 transgene induces T-ALL in mice by two distinct pathways: one requires Notch activation but the other, ETP-ALL type, requires only the sustained expression of classic phase 1 genes Lmo2, Lyl1, Hhex, and Mycn155.
Second, early T cells from Cdkn2a-/- mice could be transformed by retroviral transduction with Il7r carrying constitutively activating mutations identified from human ETP-ALLs. The cells generated transplantable leukemias resembling human ETP-ALL168, in which myeloid potential was combined with an early T-cell phenotype. These mutations blocked cell differentiation at an uncommitted DN2a-like stage, and the leukemic cells themselves showed high levels of Lmo2, low to undetectable levels of Bcl11b, and most did not have activating Notch mutations. This mouse model shows that abnormal cytokine receptor signals can prevent or reverse Lmo2 silencing, Bcl11b upregulation and T lineage commitment and that this can efficiently lead to transformation.
Third, Rag-deficient NOD mice also exhibit spontaneous emergence of an aberrant self-renewing thymocyte population from the DN2 stage. These mice also develop T-cell leukemias with high penetrance, which share some gene expression features with human ETP-ALL169, 170. These abnormal thymocytes emerge from the DN2 stage as Kithi cells that fail to repress many of the phase 1-specific genes including Lmo2, Lyl1, Hhex, and Mef2c. These emerging cells also continue some aspects of the T-cell program by activating genes characteristic of committed DN3 cells, like Ptcra, and Spib. However, they bypass the β-selection checkpoint even without TCR gene rearrangements and aberrantly turn on Cd4, Cd8, and Cd5. Thus, this model provides additional evidence that failure to repress the stem/progenitor program at the commitment checkpoint can initiate T-cell transformation. Furthermore, transformation at this checkpoint does not necessarily preclude aberrant activation of some aspects of the T-cell differentiation program.
Taken together, these studies of mouse and human T-ALL suggest that a breakdown of repressive mechanisms normally enforced at the commitment and β-selection checkpoints can lead to specific forms of T-ALL. A more complete understanding of epigenetic and checkpoint control mechanisms in normal T-cell development may shed light on leukemia initiation and subsequent transformational events leading to generation of leukemia stem cells.
Conclusions
The T cell commitment program is not a simple cumulative progression toward endowment of T-cell character, but rather a series of distinct regulatory phases with different, sometimes mutually antagonistic features (Figure 3). An initial phase of proliferative expansion in which differentiation is actively deferred is important to establish an adequate pool of cells, and it is this phase 1 developmental program that is “tuned” to promote varying numbers of cell divisions according to the needs of fetal and postnatal T-cell development. At least some of the stem- and progenitor-cell factors needed to sustain this phase are incompatible with T-lineage commitment and must be repressed. The lasting features of the commitment process include the irreversibility of silencing of phase 1 factors. The commitment process establishes a growth-limited state dominated by different transcription factors, at least one of which, Bcl11b, is only turned on at commitment. Other features (like intensified Notch signaling) are normally temporary, until cells acquire a pre-TCR or γδ TCR, allowing them to exit the commitment process and pass into a third phase. The effective termination of each phase as the cells transition to the next can be viewed as the cells’ vital bulwark against malignant transformation. Failure to terminate the phase 2 program as the cells pass through β-selection, i.e. by continuing Notch activity, has long been recognized to contribute to T-ALL. It is now clear that particularly severe forms of T-ALL are those in which the phase 1–2 transition is porous or too easily re-crossed. Thus, the early lineage commitment events, occurring in only a tiny fraction of cells in the steady-state thymus at any given time, not only offer an illuminating paradigm for developmental fate determination but also take on extreme significance for the fate of the organism as a whole.
Acknowledgments
We apologize to colleagues whose work helped to inspire this review but could not be cited due to space constraints. We thank present and former members of our group whose published and unpublished data, as well as helpful discussion, shaped the ideas presented here. The authors gratefully acknowledge support from NIH grants AI064590 to M.A.Y., AI083514, AI095943, and HD076915 to E.V.R., and the Albert Billings Ruddock Professorship.
Key Terms
- Commitment
Stage when cells give up any intrinsic capability to give rise to more than one kind of descendant. This concept depends on recognizing that precursor cells often begin with intrinsic potential to give rise to a variety of different types of descendants. In embryology and hematopoiesis, the actual choices of fates such precursor cells adopt will be different, depending on signals they receive from the environment. Commitment is the developmental transition within a given pathway during which the cells’ choice of fate becomes intrinsically irreversible, independent of the environment
- Notch signalling
A signalling system comprising highly conserved transmembrane receptors that regulate cell-fate choice in the development of many cell lineages, and so are vital in the regulation of embryonic differentiation and development. Unusually among signaling systems, the cytoplasmic domain of each Notch transmembrane protein can itself become a transcriptional coactivator in the nucleus, because it is proteolytically cleaved from the transmembrane region when Notch interacts with its ligands of the Delta or Jagged family
- T-cell acute lymphoblastic leukemias
Leukemias with an immature T cell phenotype
- Common Lymphoid Progenitor
(CLP). A progenitor cell type that appears to be committed to lymphoid fates as measured by in vivo transfer, and that can give rise to all lymphocyte cell types, including T cells, B cells and natural killer cells
- Lymphoid-primed Multipotent Precursor
(LMPP) A multilineage precursor cell type that can generate myeloid as well as lymphoid descendants in vivo and in vitro, but not erythroid or megakaryocytic cells
- Positive selection
A step in the process of T-cell differentiation in the thymus that selects CD4+CD8+ double-positive T cells for survival and maturation, based on the appropriate degree of interaction between their T-cell receptor and peptide–MHC complexes on thymic epithelial cells. Depending on the class of MHC molecule recognized, thymocytes are positively selected either to a CD4+ or to a CD8+ single-positive cell fate
- Gene regulatory network
A system of relationships among a set of regulatory factor coding genes and the transcription factors that are their products, defined and understood such that the interactions between them explain the stability or change in the developmental properties of a cell type that expresses these genes
- Pioneer Factor
A transcription factor that has the ability to bind to its target site even when the site is initially engaged in nucleosome-packed chromatin, serving as a focal point for recruitment of other transcription factors; crucial for the multi-step process needed to activate some positive regulatory elements in the genome during development
- WNT
A signalling mediator named both for its mutant phenotype in Drosophila melanogaster (Wingless) and for its role as a preferential retrovirus integration site in murine leukaemia virus-induced leukaemias (Int-1). WNT signalling activates the TCF1 and LEF1 family transcription factors through stabilizing their co-activator, β-catenin, and mobilizing it from the cytoplasm to the nucleus
- ChIP-seq analysis
A genome-wide method of mapping the sites where a transcription factor is binding to the DNA in a cell, based on cross-linking proteins to chromatin, immune-precipitating the chromatin with antibodies to the factor of interest, comprehensively sequencing the DNA that is recovered in the immune precipitates, and aligning it to the genome to identify the enriched regions
- Non-obese diabetic (NOD) mice
NOD mice spontaneously develop type 1 diabetes mellitus as a result of autoreactive T-cell-mediated destruction of pancreatic beta-islet cells
Footnotes
The authors declare no competing interests.
References
- 1.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrie HT, Zúñiga-Pflücker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 3.Thompson PK, Zúñiga-Pflücker JC. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–9. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Jeremiah Bell J, Bhandoola A. T-cell lineage determination. Immunol Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–62. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 6.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–77. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu M, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 8.Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–962. doi: 10.1084/jem.20030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belyaev NN, Biro J, Athanasakis D, Fernandez-Reyes D, Potocnik AJ. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. 2012;64:591–604. doi: 10.1007/s00251-012-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 2002;17:749–56. doi: 10.1016/s1074-7613(02)00474-0. This paper transformed the field of early T-cell development by creating a powerful and efficient in vitro system in which the developmental process could be observed and manipulated. [DOI] [PubMed] [Google Scholar]
- 11.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi AW, et al. Identification of Flt3+CD150- myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. Blood. 2011;118:2723–32. doi: 10.1182/blood-2010-09-309989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–16. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudil A, et al. Single-cell analysis of thymocyte differentiation: identification of transcription factor interactions and a major stochastic component in αβ-lineage commitment. PLoS One. 2013;8:e73098. doi: 10.1371/journal.pone.0073098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peaudecerf L, et al. Thymocytes may persist and differentiate without any input from bone marrow progenitors. J Exp Med. 2012;209:1401–8. doi: 10.1084/jem.20120845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins VC, et al. Thymus-autonomous T cell development in the absence of progenitor import. J Exp Med. 2012;209:1409–17. doi: 10.1084/jem.20120846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martins VC, et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature. 2014;509:465–70. doi: 10.1038/nature13317. These three papers overturn the long-standing theory that thymocytes cannot self renew. They show that loss of competition from fresh thymic immigrants leads to extended self renewal and allows transformation of the early stage thymocytes. [DOI] [PubMed] [Google Scholar]
- 18.Coustan-Smith E, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–56. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. These two papers characterize ETP-ALL, a high-risk human T-ALL subtype expressing genes associated with the very earliest stages of normal T cell development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlenner SM, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–9. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 25.Radtke F, Macdonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427–37. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186:6649–55. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda K, et al. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 28.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–93. doi: 10.4049/jimmunol.1000679. These two papers revealed the timing of T-lineage commitment, showing its clear separation from TCR-dependent events and defining its basis in terms of regulatory gene expression changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–55. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong SH, et al. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–36. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–82. doi: 10.1016/j.cell.2012.01.056. This paper presents the first genome-wide analyses of the epigenetic changes and transcriptional dynamics of early T-cell development, based on ChIP-seq and RNA-seq assays across a succession of stages bridging commitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mingueneau M, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–32. doi: 10.1038/ni.2590. This paper reports the transcriptome changes across an extended span of T-cell development from microarray analyses, results generated by the Immunological Genome Project, an invaluable publicly accessible source of gene expression data for all stages of immune cell development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman ES, et al. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 34.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Teague TK, et al. CD28 expression redefines thymocyte development during the pre-T to DP transition. Int Immunol. 2010;22:387–397. doi: 10.1093/intimm/dxq020. [DOI] [PubMed] [Google Scholar]
- 36.Taghon T, et al. Notch signaling is required for proliferation but not for differentiation at a well-defined β-selection checkpoint during human T-cell development. Blood. 2009;113:3254–63. doi: 10.1182/blood-2008-07-168906. [DOI] [PubMed] [Google Scholar]
- 37.Kreslavsky T, et al. β-Selection-induced proliferation is required for αβ T cell differentiation. Immunity. 2012;37:840–53. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zúñiga-Pflücker JC. Stage-specific and differential Notch dependency at the αβ and γδ T lineage bifurcation. Immunity. 2006;25:105–16. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Maillard I, et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–45. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garbe AI, von Boehmer H. TCR and Notch synergize in αβ versus γδ lineage choice. Trends Immunol. 2007;28:124–31. doi: 10.1016/j.it.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Kueh HY, Rothenberg EV. Regulatory gene network circuits underlying T cell development from multipotent progenitors. Wiley Interdiscip Rev Syst Biol Med. 2012;4:79–102. doi: 10.1002/wsbm.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. Int Immunol. 2011;23:661–8. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- 43.Mercer EM, Lin YC, Murre C. Factors and networks that underpin early hematopoiesis. Semin Immunol. 2011;23:317–25. doi: 10.1016/j.smim.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol. 2012;24:132–8. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braunstein M, Anderson MK. HEB in the spotlight: Transcriptional regulation of T-cell specification, commitment, and developmental plasticity. Clin Dev Immunol. 2012;2012:678–705. doi: 10.1155/2012/678705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David-Fung ES, et al. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–67. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabrizifard S, et al. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- 50.Kawazu M, et al. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–5345. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- 51.Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gwin KA, Shapiro MB, Dolence JJ, Huang ZL, Medina KL. Hoxa9 and flt3 signaling synergistically regulate an early checkpoint in lymphopoiesis. J Immunol. 2013;191:745–54. doi: 10.4049/jimmunol.1203294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–98. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riddell J, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–64. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Y, et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209:2467–83. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capron C, et al. The SCL relative LYL-1 is required for fetal and adult hematopoietic stem cell function and B-cell differentiation. Blood. 2006;107:4678–4686. doi: 10.1182/blood-2005-08-3145. [DOI] [PubMed] [Google Scholar]
- 57.Souroullas GP, Salmon JM, Sablitzky F, Curtis DJ, Goodell MA. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 2009;4:180–186. doi: 10.1016/j.stem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zohren F, et al. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–9. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCormack MP, et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood. 2013;122:2093–103. doi: 10.1182/blood-2012-09-458570. [DOI] [PubMed] [Google Scholar]
- 60.Lécuyer E, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 61.Phelan JD, et al. Growth factor independent-1 maintains Notch1-dependent transcriptional programming of lymphoid precursors. PLoS Genet. 2013;9:e1003713. doi: 10.1371/journal.pgen.1003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thoms JAI, et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood. 2011;117:7079–89. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- 63.McCormack MP, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. These authors used fate mapping to demonstrate the role of Lmo2 in inducing self renewal in early T cells leading to leukemia initiation, a prototype for linking natural thymocyte proliferative expansion with oncogenesis. [DOI] [PubMed] [Google Scholar]
- 64.Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010;238:63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 65.Dakic A, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–19. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–92. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostuni R, Natoli G. Lineages, cell types and functional states: a genomic view. Curr Opin Cell Biol. 2013;25:759–64. doi: 10.1016/j.ceb.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 70.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Visan I, et al. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- 72.Georgescu C, et al. A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci U S A. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, et al. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–19. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 75.Obaldia De, E M, et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–84. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wendorff AA, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33:671–84. doi: 10.1016/j.immuni.2010.11.014. These papers demonstrate the critical importance of Hes1 as a critical Notch target gene in the earliest stages of T cell development, through a discrete role in commitment and a role in normal population expansion as well as for Notch-induced T-ALL. [DOI] [PubMed] [Google Scholar]
- 77.Tomita K, et al. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci U S A. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yun TJ, Bevan MJ. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- 80.Hosoya T, et al. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Germar K, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–5. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miyazaki M, et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–42. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones-Mason ME, et al. E protein transcription factors are required for the development of CD4+ lineage T cells. Immunity. 2012;36:348–61. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–70. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–8. doi: 10.1038/nature10279. With ref. 81, this paper demonstrates that TCF-1 is a key mediator of T cell specification, acting downstream of Notch signaling. Here TCF-1 at high levels is shown to drive expression of T cell genes including Gata3 and Bcl11b even in the absence of Notch signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tiemessen MM, et al. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10:e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu S, et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. doi: 10.1016/j.immuni.2012.08.009. These two papers show the complex intrafamily relationships between TCF-1 and LEF-1 in early T-cell development and the importance of certain isoforms of TCF-1 to act as tumor suppressors by repression of Lef1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schilham MW, et al. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 91.Staal FJT, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–94. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dose M, et al. β-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci U S A. 2014;111:391–6. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 94.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. 2010;238:110–25. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.García-Ojeda ME, et al. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–59. doi: 10.1182/blood-2012-06-440065. This paper identifies GATA-3 as the crucial intrinsic regulatory factor in the earliest T-lineage precursor cells that is responsible for the exclusion of access to the B-lineage fate. [DOI] [PubMed] [Google Scholar]
- 98.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 99.Sambandam A, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–70. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 100.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 101.Hosoya-Ohmura S, et al. An NK and T cell enhancer lies 280 kilobase pairs 3′ to the gata3 structural gene. Mol Cell Biol. 2011;31:1894–904. doi: 10.1128/MCB.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 103.Vigano MA, et al. An epigenetic profile of early T-cell development from multipotent progenitors to committed T-cell descendants. Eur J Immunol. 2014;44:1181–93. doi: 10.1002/eji.201344022. [DOI] [PubMed] [Google Scholar]
- 104.Xu W, et al. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–42. doi: 10.1182/blood-2012-08-449447. This paper reveals that GATA-3 activity must be kept under inhibitory restraint by the same basic helix-loop-helix E proteins that also collaborate with it to drive T-cell specification. This is shown to be one important way that E2A promotes successful T-cell commitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu W, Kee BL. Growth factor independent 1B (Gfi1b) is an E2A target gene that modulates Gata3 in T-cell lymphomas. Blood. 2007;109:4406–4414. doi: 10.1182/blood-2006-08-043331. [DOI] [PubMed] [Google Scholar]
- 106.Maneechotesuwan K, et al. Regulation of Th2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. J Immunol. 2007;178:2491–8. doi: 10.4049/jimmunol.178.4.2491. [DOI] [PubMed] [Google Scholar]
- 107.Cook KD, Miller J. TCR-dependent translational control of GATA-3 enhances Th2 differentiation. J Immunol. 2010;185:3209–16. doi: 10.4049/jimmunol.0902544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Frelin C, et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat Immunol. 2013;14:1037–44. doi: 10.1038/ni.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Okamura RM, et al. Redundant regulation of T cell differentiation and TCRβ gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 111.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–26. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhong Y, Jiang L, Hiai H, Toyokuni S, Yamada Y. Overexpression of a transcription factor LYL1 induces T- and B-cell lymphoma in mice. Oncogene. 2007;26:6937–6947. doi: 10.1038/sj.onc.1210494. [DOI] [PubMed] [Google Scholar]
- 113.Welinder E, et al. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A. 2011;108:17402–7. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Takeuchi A, et al. E2A and HEB activate the pre-TCR α promoter during immature T cell development. J Immunol. 2001;167:2157–63. doi: 10.4049/jimmunol.167.4.2157. [DOI] [PubMed] [Google Scholar]
- 116.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 117.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. These three papers define the role of Bcl11b in T-lineage commitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cismasiu VB, et al. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 120.Li L, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–11. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kastner P, et al. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur J Immunol. 2010;40:2143–54. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–81. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Oosterwegel M, et al. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-ε and T cell receptor α enhancers. J Exp Med. 1991;173:1133–1142. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–76. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lauritsen JP, et al. Marked induction of the helix-loop-helix protein Id3 promotes the γδ T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–75. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–9. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 128.Inoue J, et al. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in Bcl11b-/- mice. J Immunol. 2006;176:5871–5879. doi: 10.4049/jimmunol.176.10.5871. [DOI] [PubMed] [Google Scholar]
- 129.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 130.Shibata K, et al. IFN-γ-producing and IL-17-producing γδ T cells differentiate at distinct developmental stages in murine fetal thymus. J Immunol. 2014;192:2210–8. doi: 10.4049/jimmunol.1302145. [DOI] [PubMed] [Google Scholar]
- 131.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–48. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 134.Schjerven H, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. 2013;14:1073–83. doi: 10.1038/ni.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang J, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chari S, Winandy S. Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol. 2008;181:6265–6274. doi: 10.4049/jimmunol.181.9.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kleinmann E, Geimer Le Lay AS, Sellars M, Kastner P, Chan S. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol. 2008;28:7465–75. doi: 10.1128/MCB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Geimer Le, Lay AS, et al. The tumor suppressor Ikaros shapes the repertoire of Notch target genes in T cells. Sci Signal. 2014;7:ra28. doi: 10.1126/scisignal.2004545. [DOI] [PubMed] [Google Scholar]
- 139.Tussiwand R, et al. The preTCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited by IL-7 signaling. Eur J Immunol. 2011;41:3371–80. doi: 10.1002/eji.201141824. [DOI] [PubMed] [Google Scholar]
- 140.Janas ML, et al. Thymic development beyond β-selection requires phosphatidylinositol 3-kinase activation by CXCR4. J Exp Med. 2010;207:247–61. doi: 10.1084/jem.20091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORγt, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 142.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–8. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.He YW, et al. Down-regulation of the orphan nuclear receptor RORγt is essential for T lymphocyte maturation. J Immunol. 2000;164:5668–5674. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- 145.Sun Z, et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 146.Wang R, et al. Transcription factor network regulating CD4+CD8+ thymocyte survival. Crit Rev Immunol. 2011;31:447–58. doi: 10.1615/critrevimmunol.v31.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pongracz JE, Parnell SM, Jones T, Anderson G, Jenkinson EJ. Overexpression of ICAT highlights a role for catenin-mediated canonical Wnt signalling in early T cell development. Eur J Immunol. 2006;36:2376–2383. doi: 10.1002/eji.200535721. [DOI] [PubMed] [Google Scholar]
- 148.Xu M, Sharma A, Wiest DL, Sen JM. Pre-TCR-induced β-catenin facilitates traversal through β-selection. J Immunol. 2009;182:751–8. doi: 10.4049/jimmunol.182.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu M, Sharma A, Hossain MZ, Wiest DL, Sen JM. Sustained expression of pre-TCR induced β-catenin in post-β-selection thymocytes blocks T cell development. J Immunol. 2009;182:759–65. doi: 10.4049/jimmunol.182.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schroeder JH, Bell LS, Janas ML, Turner M. Pharmacological inhibition of glycogen synthase kinase 3 regulates T cell development in vitro. PLoS One. 2013;8:e58501. doi: 10.1371/journal.pone.0058501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–90. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 152.Uckun FM, et al. Clinical features and treatment outcome of childhood T-lineage acute lymphoblastic leukemia according to the apparent maturational stage of T-lineage leukemic blasts: a Children's Cancer Group study. J Clin Oncol. 1997;15:2214–21. doi: 10.1200/JCO.1997.15.6.2214. [DOI] [PubMed] [Google Scholar]
- 153.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 154.Van Vlierberghe P, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smith S, et al. LIM domain only-2 (LMO2) induces T-cell Leukemia by two distinct pathways. PLoS One. 2014;9:e85883. doi: 10.1371/journal.pone.0085883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol. 2013;20:369–73. doi: 10.1097/MOH.0b013e3283623c61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Van de Walle I, et al. An early decrease in Notch activation is required for human TCR-αβ lineage differentiation at the expense of TCR-γδ T cells. Blood. 2009;113:2988–98. doi: 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- 158.Joachims ML, Chain JL, Hooker SW, Knott-Craig CJ, Thompson LF. Human αβ and γδ thymocyte development: TCR gene rearrangements, intracellular TCR β expression, and γδ developmental potential--differences between men and mice. J Immunol. 2006;176:1543–52. doi: 10.4049/jimmunol.176.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 160.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223:262–73. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tremblay CS, Hoang T. Early T cell differentiation lessons from T-cell acute lymphoblastic leukemia. Prog Mol Biol Trans Sci. 2010;92:121–156. doi: 10.1016/S1877-1173(10)92006-1. [DOI] [PubMed] [Google Scholar]
- 162.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 163.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 164.Laurenti E, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–63. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Neumann M, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–52. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 167.Cleveland SM, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells. 2013;31:882–94. doi: 10.1002/stem.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Treanor LM, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med. 2014;211:701–13. doi: 10.1084/jem.20122727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yui MA, et al. Loss of T cell progenitor checkpoint control underlies leukemia initiation in Rag1-deficient nonobese diabetic mice. J Immunol. 2013;190:3276–88. doi: 10.4049/jimmunol.1202970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- 171.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–47. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 172.Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kang J, Volkmann A, Raulet DH. Evidence that γδ versus αβ T cell fate determination is initiated independently of T cell receptor signaling. J Exp Med. 2001;193:689–698. doi: 10.1084/jem.193.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Melichar HJ, et al. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–3. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 175.Feng N, Vegh P, Rothenberg EV, Yui MA. Lineage divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice. J Immunol. 2011;186:826–37. doi: 10.4049/jimmunol.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Van de Walle I, et al. Specific Notch receptor-ligand interactions control human TCR-αβ/γδ development by inducing differential Notch signal strength. J Exp Med. 2013;210:683–97. doi: 10.1084/jem.20121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Haks MC, et al. Attenuation of γδTCR signaling efficiently diverts thymocytes to the αβ lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 178.Hayes SM, Li L, Love PE. TCR signal strength influences αβ/γδ lineage fate. Immunity. 2005;22:583–93. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 179.Narayan K, et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012;13:511–8. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Verykokakis M, et al. Inhibitor of DNA binding 3 limits development of murine Slam-associated Adaptor Protein-dependent “innate” γδ T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kreslavsky T, et al. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–8. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kreslavsky T, Gleimer M, von Boehmer H. αβ versus γδ lineage choice at the first TCR-controlled checkpoint. Curr Opin Immunol. 2010;22:185–92. doi: 10.1016/j.coi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Prinz I, Silva-Santos B, Pennington DJ. Functional development of γδ T cells. Eur J Immunol. 2013;43:1988–94. doi: 10.1002/eji.201343759. [DOI] [PubMed] [Google Scholar]
- 184.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–310. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 185.Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–2624. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–68. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 187.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–44. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 189.Wölfler A, et al. Lineage-instructive function of C/EBPα in multipotent hematopoietic cells and early thymic progenitors. Blood. 2010;116:4116–4125. doi: 10.1182/blood-2010-03-275404. [DOI] [PubMed] [Google Scholar]
- 190.Franco CB, et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU. 1. Proc Natl Acad Sci U S A. 2006;103:11993–8. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huang G, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 192.Zarnegar MA, Chen J, Rothenberg EV. Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–39. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rosenbauer F, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 194.Hoyler T, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–48. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yang Q, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gentek R, et al. Modulation of signal strength switches Notch from an inducer of T cells to an inducer of ILC2. Front Immunol. 2013;4:334. doi: 10.3389/fimmu.2013.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Heng TSP, Painter MW, Consortium TIGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]