Abstract
Temporal patterning of neural progenitors is one of the core mechanisms generating neuronal diversity in the central nervous system. Here, we show that in the tips of the outer proliferation center (tOPC) of the developing Drosophila optic lobes, a unique temporal series of transcription factors not only governs the sequential production of distinct neuronal subtypes, but also controls the mode of progenitor division as well as the selective apoptosis of NotchOFF or NotchON neurons during binary cell fate decisions. Within a single lineage, intermediate precursors initially do not divide and generate only one neuron; subsequently, precursors divide but their NotchON progeny systematically die through Reaper activity whereas later, their NotchOFF progeny die through Hid activity. These mechanisms dictate how the tOPC produces neurons for three different optic ganglia. We conclude that temporal patterning generates neuronal diversity by specifying both the identity and survival/death of each unique neuronal subtype.
INTRODUCTION
A central challenge in developmental neurobiology is to understand how an apparently uniform pool of embryonic progenitors produces the vast diversity of neurons and glial cells found in the adult Central Nervous System (CNS). Studies in vertebrates and insects have revealed that four primary mechanisms generate neural cell diversity. (i) Spatial patterning cues provide a unique lineage identity to each progenitor (Rogulja-Ortmann and Technau, 2008; Technau et al., 2006); (ii) temporal progression of transcription factors in progenitors instructs them to orderly produce different neuronal and glial subtypes (Baumgardt et al., 2009; Brody and Odenwald, 2000; Elliott et al., 2008; Isshiki et al., 2001; Lee et al., 1999; Li et al., 2013; Livesey and Cepko, 2001; Zhu et al., 2006); (iii) binary cell fate decisions mediated by Notch lead to the formation of two post-mitotic cells with different fates during the final mitosis of intermediate precursors (Buescher et al., 1998; Truman et al., 2010), with one of these cells sometimes undergoing apoptosis (Lin et al., 2010; Lundell et al., 2003). (iv) Progenitors can generate intermediate precursors with different proliferation modes (He et al., 2012): Some intermediate precursors divide once to produce two daughter cells (‘type 1’ neuroblasts) (Buescher et al., 1998), some undergo multiple divisions to amplify the lineage (‘type 2’ neuroblasts) (Bello et al., 2008; Boone and Doe, 2008), or some directly differentiate into a neuron without further cell divisions (Karcavich and Doe, 2005; Ulvklo et al., 2012) (which we name ‘type 0’ neuroblasts). Although these mechanisms are well characterized, very little is known about how they interact with each other to specify the unique fate of each neuron. For instance, it has been suggested that Notch integrates spatial signals to determine neuronal survival or apoptosis during binary cell fate decisions (Lin et al., 2010), but these signals and the nature of their interaction with Notch remain enigmatic.
The Drosophila visual system is an excellent model for studying complex neurogenesis. Each optic lobe is composed of four ganglia: the lamina, medulla, lobula and lobula plate (Hofbauer, 1990; Meinertzhagen, 1993). The medulla, which has the largest neuropil, is composed of approximately 40,000 neurons comprising more than 70 subtypes (Fischbach, 1989; Morante and Desplan, 2008). It derives from a single-layered crescent-shaped neuroepithelium in the larval brain called the outer proliferation center (OPC). The OPC is progressively converted into progenitors (called neuroblasts) by a wave of expression of the pro-neural gene lethal of scute that sweeps from the edge toward the center of the crescent over time (Egger et al., 2007; Egger et al., 2010; Ngo et al., 2010; Yasugi et al., 2010)(Fig. S1A). We and others have recently shown that neuronal diversity in the medulla is generated by a combination of spatial patterning, temporal patterning and binary cell fate decisions (Li et al., 2013; Suzuki et al., 2013). Indeed, the expression of four genes, Vsx1, optix, decapentaplegic (dpp) and wingless (wg) divides the OPC into four spatial regions along the antero-posterior axis (Kaphingst and Kunes, 1994)(Erclik et al. in preparation). Each of these spatially distinct regions produces different neuronal subtypes. In addition, neuroblasts deriving from the main OPC (which includes the regions defined by Vsx1, Optix and Dpp) all sequentially express five temporal factors - Homothorax (Hth), Eyeless (Ey), Sloppy-paired (Slp), Dichaete (D) and Tailless (Tll)- as they age (Fig. S1A,B). These factors and their overlap determine about 12 neuroblast fates that generate distinct neuronal subtypes. Finally, these neuroblasts produce intermediate precursors called ganglion mother cells (GMCs) that divide asymmetrically to generate two distinct neurons, one NotchOFF, and one NotchON neuron expressing Apterous (Ap) (Fig. S1B). As a result, the larval medulla cortex is composed of several layers, each composed of NotchON Ap-positive neurons intermingled with NotchOFF neurons.
Here, we focus on the development of the tips of the OPC defined by wingless expression (tOPC, Fig. 1A,B and S1A), which provides a unique opportunity for studying how temporal patterning of progenitors interplays with Notch to control neuronal survival. We show that tOPC progenitors undergo complex neurogenesis involving two dramatic transitions, one in the mode of intermediate precursor division and the other in systematic apoptosis of one of their neuronal progeny during Notch-mediated binary cell fate decisions. We provide evidence that, in addition to specifying distinct neuronal subtypes over time, temporal patterning of tOPC progenitors also controls the transition in apoptosis by specifying the systematic death of NotchON neurons in a first phase and of NotchOFF neurons in a second phase, through the regulation of Reaper (Rpr) and Head involution defective (Hid), respectively. This complex neurogenesis dictates how the tOPC produces neurons for three different neuropils of the adult optic lobes. We conclude that temporal patterning of progenitors generates neuronal diversity by controlling multiple aspects of neurogenesis including neuronal identity and Notch-mediated cell survival.
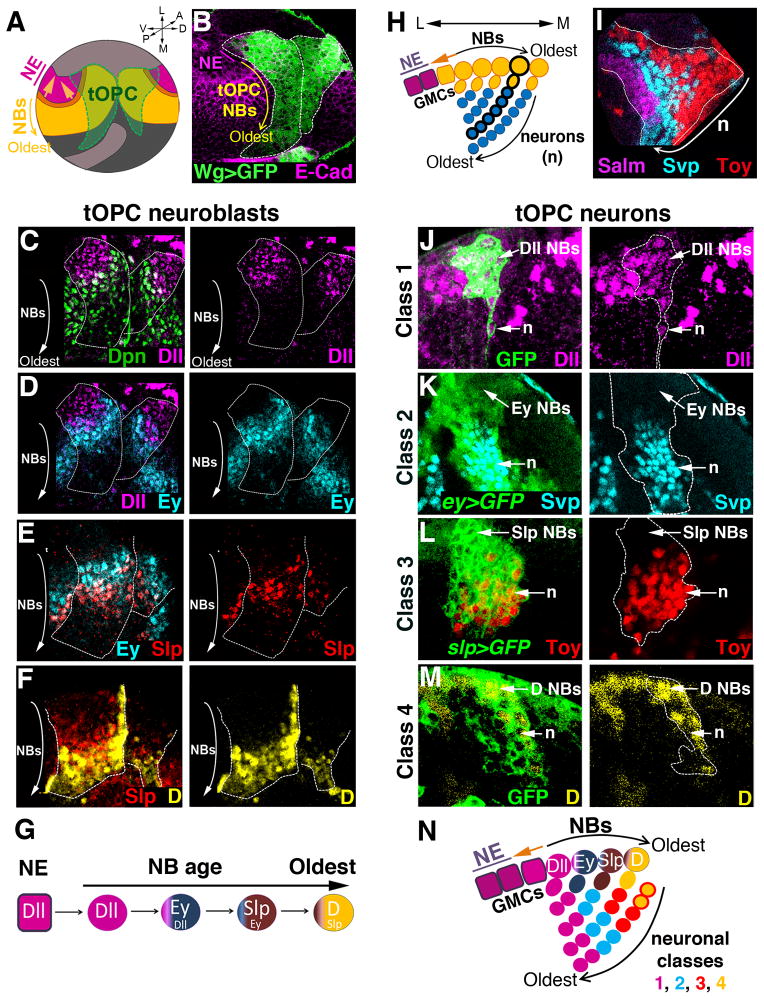
Figure 1. The tOPC temporal series sequentially produces four neuronal classes.
Unless specified, all pictures are posterior views of third instar larvae (L3). A, Model of a larval optic lobe showing that a wave of neurogenesis (orange) converts OPC neuroepithelial cells (NE, purple) into neuroblasts (NBs, yellow). The tOPC is defined by wg-gal4 expression (green). A, anterior; P, posterior; V, ventral; D, dorsal; L, lateral; M, medial. B, The tOPC region (wg>GFP, green, dashed lines). E-Cad stains the neuroepithelium (purple). C–G, tOPC neuroblasts sequentially express Dll (magenta in C, D), Ey (cyan in D, E), Slp (red in E, F) and D (yellow in F). White dashed lines show the tOPC. H, Model of a cross-section showing neuroblasts (yellow circles), GMCs (yellow) and neurons (cyan). A single neuroblast clone is shown with black thick outlines. I, Cross-section view showing the clustered organization of three of the four medulla neuronal classes: Class 1 (Salm, magenta), Class 2 (Svp, cyan) and Class 3 (Toy, red). J, Cross-section view. Early neuroblast clone (GFP, dashed lines) showing that Dll (magenta) is transmitted from neuroblasts (NBs) to neurons (n). K, Perduring GFP driven by ey-gal4 (dashed lines) is expressed in almost all Svp neurons (cyan). L, GFP driven by slp-gal4 (dashed lines) is expressed in all Toy neurons (red). M, Cross section view. Late neuroblast clone (dashed lines) showing that D expression (yellow) is transmitted to the neurons. N, Schematic model of tOPC neurogenesis. Only one neuroblast/GMC is shown for each stage. See also Figure S1.
RESULTS
tOPC neuroblasts undergo temporal patterning
In order to understand how medulla neuronal diversity is generated, we conducted an antibody screen to identify transcription factors expressed in the developing optic lobes (Li et al., 2013). The medulla derives from the larval OPC, where the progressive conversion of neuroepithelium into neuroblasts allows us to visualize in one snapshot neuroblasts at different temporal stages. Oldest neuroblasts are found at the medial edge of the OPC near the neuroepithelium while newly specified neuroblasts are found more laterally (Fig. 1A,B).
This screen led us to identify four transcriptions factors, Distalless (Dll), Eyeless (Ey), Sloppy-paired (Slp) and Dichaete (D), expressed in consecutive stripes in tOPC neuroblasts of increasing ages (Fig. 1C–F), with Dll expressed in the youngest tOPC neuroblasts (Fig. 1C,D) and D expressed in the oldest neuroblasts (Fig. 1F). Neighboring stripes partially overlap suggesting a gradual replacement of one transcription factor by the next as neuroblasts age. Therefore, tOPC neuroblasts express a unique series of transcription factors, Dll, Ey, Slp and D (Fig. 1G) which is different from the main OPC where neuroblasts sequentially express Hth, Ey, Slp, D and Tll (Fig. S1A,B). Next, we examined the progeny of tOPC neuroblasts. tOPC neuroblasts divide asymmetrically multiple times to self-renew and produce a GMC which, in turn, generates neurons. The progeny of each tOPC neuroblast therefore forms a chain, with newly specified neurons occupying the most superficial layer and the oldest neurons the deepest layer (Fig. 1H). Despite screening over 200 antibodies, we found only four different classes of larval tOPC neurons based on their expression of transcription factors. More importantly, in contrast to the main OPC where different neuronal classes are intermingled, neuronal classes are organized as homogeneous clusters in the tOPC (Fig. 1I), suggesting that this region has a distinct mode of neurogenesis.
To decipher the mode of tOPC neurogenesis, we first determined which temporal windows produce each of the four neuronal clusters, which are localized in different layers that correlate with their birth order. Class 1 neurons are localized in the deepest layer and co-express Dll, Spalt major (Salm), Runt and D (the latter only in the ventral tOPC) (Fig. S1D–F). Young neuroblast clones in which the neuroblasts are at the Dll+ stage include Dll+ GMCs and neurons (Fig. 1J), indicating that Class 1 Salm/Runt neurons are produced during the Dll time window. Class 2 neurons are localized in the layer above Class 1 neurons and express Seven-up (Svp). Although Ey expression is not transmitted to these neurons, GFP driven by ey-gal4 is expressed in almost all of them due to perdurance of Gal4 and GFP (Fig. 1K and S1G). This suggests that Class 2 Svp neurons are produced during the Ey time window. Class 3 neurons are localized above Class 2 neurons and express Twin-of-eyeless (Toy). Slp is not transmitted to these neurons but we identified a slp-Gal4 line expressed in almost all Slp-expressing neuroblasts (Fig. S1H). This Gal4 line drives GFP expression in the majority of Toy neurons (also due to perdurance, Fig. 1L), suggesting that Class 3 Toy neurons are produced during the Slp time window. Finally, Class 4 neurons are localized in the most superficial layer near the neuroblasts and co-express Toy and D (Fig. S1I). Late neuroblast clones in which the neuroblasts are at the D+ stage include D+ GMCs and neurons, indicating that Class 4 Toy/D neurons are produced at the end of tOPC lineages, during the D time window (Fig. 1M).
In summary, four temporal windows in neuroblasts sequentially produce four classes of neurons in the larval tOPC (Fig. 1N): Initially, Dll is expressed and produces Class 1 Salm/Runt neurons (magenta). These neuroblasts then switch to Ey expression and produce Class 2 Svp neurons (cyan) followed by expression of Slp to produce Class 3 Toy neurons (red) and finally D expression to produce Class 4 Toy/D neurons (yellow).
Three groups of tOPC neurons based on their Notch status
The clustered organization of tOPC neurons could be due to systematic apoptosis of one of the neuronal progeny of ‘type 1’ neuroblasts. Alternatively, it could result from a ‘type 0’ mode of division of tOPC neuroblasts (Fig. S1C). To test this, we first determined the Notch status of the four classes of tOPC neurons. We monitored the expression of the bHLH-O protein Hairy/enhancer-of-split like a Y (Hey), which has been used as a Notch sensor in the Drosophila CNS (Monastirioti et al., 2010). Hey expression is lost when Notch signaling is abolished (suppressor of hairless, su(H) mutant clones, Fig. S2A) and is expanded when Notch signaling is constitutively active (over-expression of the Notch intracellular domain, NotchICD, Fig. S2B,C). Therefore, Hey is also a good sensor of Notch activity in optic lobe neurons.
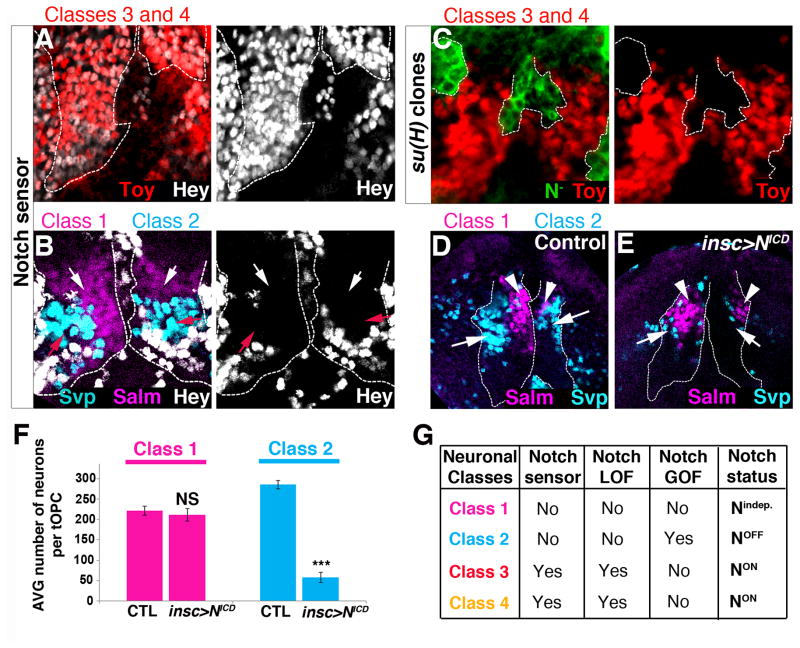
In the tOPC, Hey marks Class 3 Toy and Class 4 Toy/D neurons, which must therefore be NotchON (Fig. 2A). The absence of Hey in Class 1 Salm/Runt and Class 2 Svp neurons suggests that they are NotchOFF (Fig. 2B). Consistent with this, Class 1 Salm/Runt and Class 2 Svp neurons are not affected in su(H) mutant clones (Fig. S2D,E), whereas Class 3 Toy and Class 4 Toy/D neurons are completely lost (Fig. 2C). Conversely, ectopic activation of Notch signaling in the tOPC decreases the number of Class 2 Svp neurons (Fig. 2D,E&F: control 282 ± 16; insc-gal4>NotchICD 54 ± 12; P<0.001) but does not affect Class 3 Toy and Class 4 Toy/D neurons (Fig. S2F). Surprisingly, although Class 1 Salm/Runt neurons appear to be NotchOFF, they are not affected by high levels of Notch signaling, suggesting that these neurons acquire their fate in a Notch-independent manner (Fig. 2D,E&F: control 211±10; insc-gal4>NotchICD 221 ±10; P=0.1). Thus, based on their Notch status, the tOPC neurons can be divided into 3 groups (Fig. 2G): Notchindependent neurons (Class 1), NotchOFF neurons (Class 2) and NotchON neurons (Classes 3 and 4).
Figure 2. Notch status of tOPC neurons.
Posterior views of L3 optic lobes. A, B, Hey (white) is expressed by Class 3 Toy and Class 4 Toy/D neurons (A) but not by Class 1 Salm/Runt (magenta, white arrows) and Class 2 Svp (cyan, red arrows) neurons (B). Dashed lines indicate the tOPC. C, Class 3 Toy and Class 4 Toy/D neurons (red) are lost in su(H) mutant clones (GFP, dashed lines). D, Control brains showing Class 1 Salm/Runt (magenta, arrowheads) and Class 2 Svp (cyan, arrows) neurons. E, Ectopic Notch signaling (insc>NICD) leads to the loss of Class 2 Svp neurons (cyan, arrows) but has no effect on Class 1 Salm/Runt neurons (magenta, arrowheads). Dashed lines indicate the tOPC. F, Average number of tOPC neurons in control and Notch gain-of-function (N=7 brains/genotype). Magenta=Class 1 Salm/Runt neurons, Cyan=Class 2 Svp neurons. All data represent mean ± s.d. NS, not significant. *** P<0.001. G, Summary. See also Figure S2.
Origin of the clustered organization of tOPC neurons
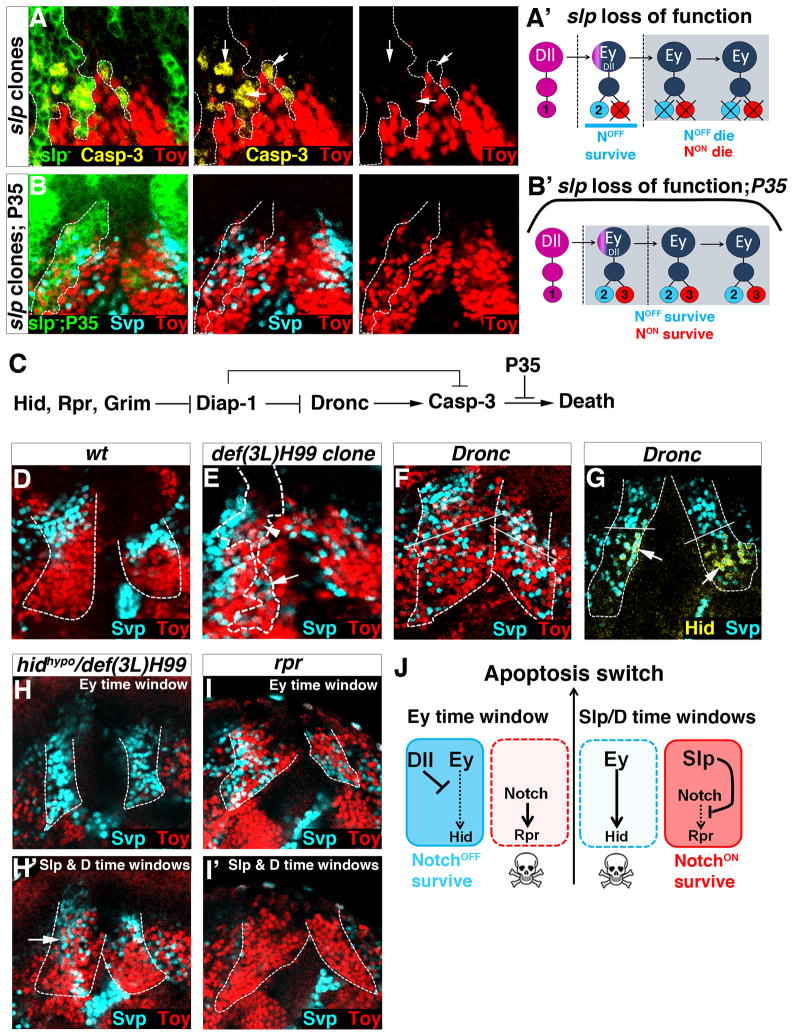
The presence of both NotchOFF and NotchON neurons suggests that most tOPC neuroblasts are ‘type 1’ neuroblasts and that one of the progeny of their GMCs undergoes apoptotic cell death. To test this, we stained tOPC neurons with the cell death marker cleaved Caspase-3.
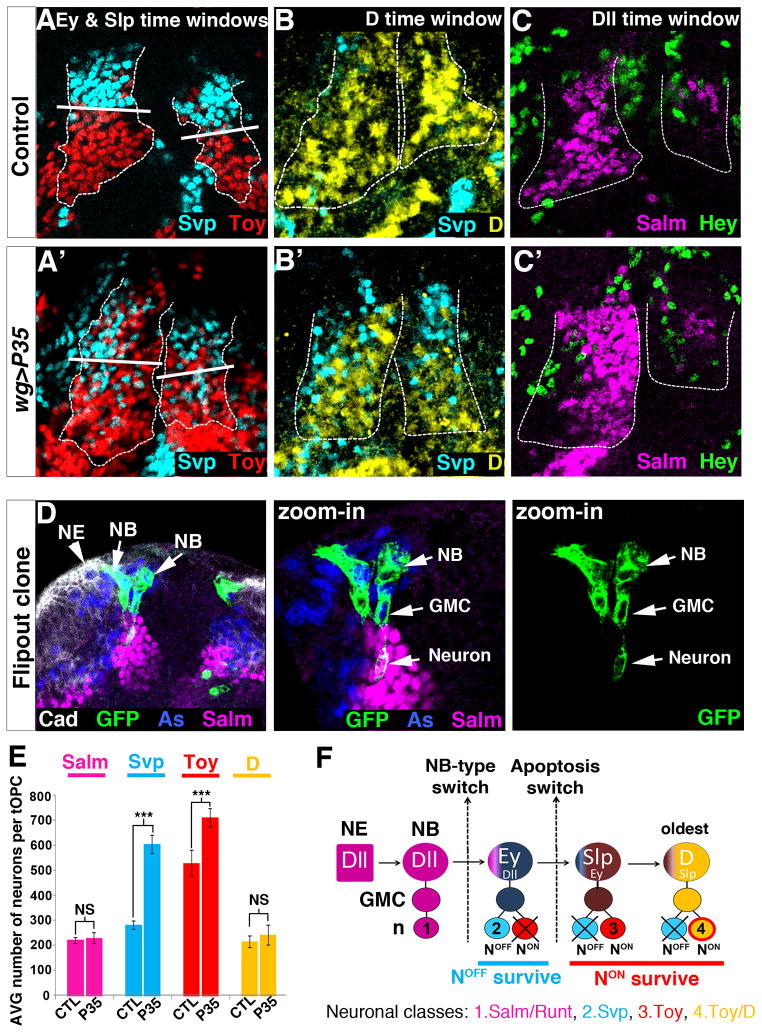
Cleaved Caspase-3 is expressed in a few neurons of the Class 2 cluster that forms during the Ey time window. Indeed, although this cluster is almost exclusively composed of NotchOFF Svp neurons, we detected close to the neuroblast layer, a small number of NotchON neurons that all express cleaved Caspase-3 as well as Toy (white arrows, Fig. S3A). This suggests that Ey neuroblasts are ‘type 1’ neuroblasts which, at each round of division, produce one NotchOFF Svp neuron that survives and one NotchON Toy neuron that undergoes apoptosis. Consistent with this, forcing the survival of NotchON Toy neurons via P35 expression increased their number (Fig. 3E: Control 529±51; wg>P35 711±36; P<3.10−4) and creates an intermingling between these neurons (red) and the NotchOFF Class 2 Svp neurons (cyan, compare top parts of Fig. 3A and A′; Fig. S3B,C).
Figure 3. Origin of the clustered organization of tOPC neurons.
Posterior views of L3 optic lobes. A–C, P35 expression in tOPC neurons. Dashed lines show the tOPC. A, B Control brains showing the NotchOFF Class 2 Svp (cyan in A), NotchON Class 3 Toy (red in A) and NotchON Class 4 Toy/D clusters (yellow in B). In P35-expressing brains, NotchOFF Svp neurons are completely intermingled with NotchON Toy (A′) and NotchON Toy/D (B′) neurons in the Ey, Slp and D time windows. White bars in A and A′ delimitate the border between the Ey and the Slp time windows. C, Control brain stained with Hey (green) and Salm (magenta). C′ P35 expression has no effect on the Notchindep Salm/Runt cluster. D, Single neuroblast clones showing that newly specified neuroblasts generate GMCs (Asense, blue) that directly differentiate into one Class 1 neuron (Salm, magenta). Neuroepithelium (NE, arrowhead) is stained with DE-Cad (white). Asterisk indicates a newly formed neuroblast that has not produced any neuron yet. E, Average number of Salm (magenta), Svp (cyan), Toy (red) and D (yellow) neurons in control and wg>P35 (N=6 brains/genotype). All data represent mean ± s.d. NS, not significant. *** P<0.001. F, tOPC neurogenesis mode. See also Figure S3.
Similarly, we found evidence that cell death is implicated in the formation of Class 3 and Class 4 clusters. In these two NotchON clusters, we detected a small number of NotchOFF neurons expressing cleaved Caspase-3 and Svp (yellow arrows, Fig. S3A). Forcing the survival of these neurons via P35 expression strongly increased their number (Fig. 3E: Control 282 ±16; wg>p35 603 ±36; P<5.10−6.). Moreover, P35 expression creates an intermingling between these NotchOFF Svp neurons (cyan) and the NotchON Class 3 Toy neurons (red) in the Slp time window (compare bottom parts of Fig. 3A and A′). These NotchOFF Svp neurons are also intermingled with NotchON Class 4 Toy/D neurons (yellow) (compare Fig. 3B and B′) and, since there is no cell migration during tOPC neurogenesis, they must be produced locally during the D time window. Thus, during the Slp and D time windows, an inversion in the apoptotic pattern causes NotchOFF Svp-expressing neurons to die, leading to the formation of the NotchON Class 3 Toy and Class 4 Toy/D clusters.
We next examined the mode of division of Dll neuroblasts. These neuroblasts produce Class 1 Salm/Runt neurons independently of Notch and show no indication of apoptosis since no new neurons appear in the presence of P35 (Fig. 3C,C′&E: control 221±12; wg>p35 230±22; P=0.12). This opened the possibility that Dll neuroblasts undergo a ‘type 0’ mode of division and produce GMCs that do not divide but instead directly differentiate into a neuron. To test this, we generated single neuroblast clones and analyzed the progeny of newly specified Dll-expressing neuroblasts (Fig. 3D). These clones always contain one GMC (stained with Asense) and only one Class 1 Salm/Runt neuron. Thus, during the Dll time-window, tOPC neuroblasts are ‘type 0’ neuroblasts that produce a single neuron at each round of division.
In conclusion, the clustered organization of tOPC neurons results from two phenomena (Fig. 3F): first, tOPC neuroblasts are initially specified as ‘type 0’ neuroblasts (Dll time window) and produce a single class of neurons. Second, they then switch to ‘type 1’ neuroblasts (Ey, Slp and D time windows) but half of their progeny undergo apoptosis, NotchON die first in the Ey time-window while NotchOFF die later, in the Slp and D time windows. This leads to the production, within the same lineage, of four hemilineages composed of neuronal Classes 1, 2, 3 and 4, respectively.
Cross regulations between tOPC temporal factors
We next examined whether, like in the main OPC, cross-regulation among Dll, Ey, Slp and D contributes to the transition from one factor to the next. The main OPC and tOPC temporal series begin with different factors, Hth and Dll respectively, raising the possibility that Dll represses Hth in the tOPC. However, this is not the case since Hth is not expressed in tOPC dll mutant clones (Fig. S4A). In addition, ey, slp and D expression is not affected in dll clones (not shown), indicating that Dll is not required to turn on subsequent factors of the temporal cascade. In ey loss of function (mutants and RNAi, see M&M), dll expression remains restricted to the youngest neuroblasts (Fig. S4B), whereas slp and D are lost (compare Fig. S4C and C′). Therefore, Ey is required to turn on the next factor in the series, slp, but is not required to turn off dll. In slp mutant clones, ey expression expands while D is lost (Fig. S4D). Therefore, Slp turns off ey expression and is required to turn on D expression. Finally, in D mutant clones, slp expands, showing that D turns off slp expression (Fig. S4E). Thus, like in the main OPC, Ey, Slp and D are required to turn on the next transcription factor while Slp and D are also required for turning off the preceding transcription factor (Fig. S4F).
tOPC temporal patterning controls Notch-mediated neuronal survival
We then tested the role of Dll, Ey, Slp and D during tOPC neurogenesis. Temporal factors have been shown to specify distinct neuronal identities over time. In addition, in the tOPC, the transition in apoptosis pattern (from NotchON to NotchOFF dying) suggests that this process is also temporally regulated. We therefore tested whether tOPC temporal factors specify both the identity and the survival of the four classes of neurons.
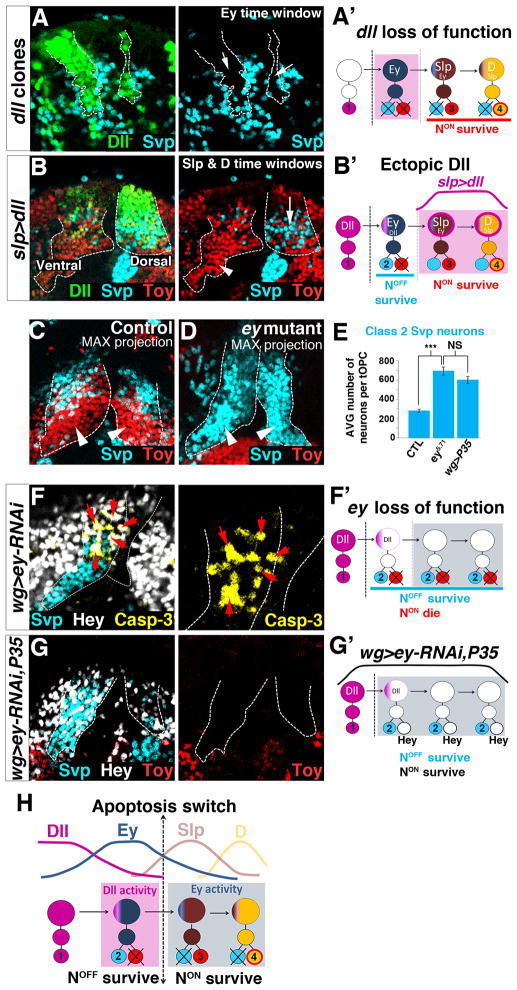
Newly specified neuroblasts express high levels of Dll and produce Class 1 Salm/Runt neurons. However, in dll mutant clones, none of the markers of this neuronal class (Salm, Runt and D) are lost (Fig. S4G and not shown), suggesting that Dll does not specify the identity of Class 1 Salm/Runt neurons. Interestingly, although loss of Dll does not alter Ey expression in neuroblasts, it affects the neurons produced during the Ey time window. Indeed, in dll clones, NotchOFF Class 2 Svp neurons are either partially (~80% of the cases, N=17/22) or completely (~20% of the cases, N=5/22) lost, and they do not appear to be transformed into another subtype (Fig. 4A). This suggests that Dll promotes survival of the NotchOFF neurons produced during the Ey window (Fig. 4A′). To test whether Dll is sufficient to promote the survival of NotchOFF neurons, we ectopically expressed it in the Slp and D later time windows (Fig. 4B,B′) using slp-gal4. If Dll promotes the survival of these neurons, it should rescue the Svp-expressing neurons (the NotchOFF siblings of Class 3 and Class 4 NotchON neurons) that normally die in this part of the lineage. This is indeed the case: although ectopic Dll does not affect the expression of Ey, Slp and D in neuroblasts (not shown), like P35 expression, it creates an intermingling between NotchOFF Svp-expressing neurons and NotchON Class 3 and Class 4 Toy neurons in the dorsal tOPC tip (arrow in Fig. 4B; Svp neurons: control 142±12; slp>dll 204±16; P<10−4), but not in the control ventral tip where slp-gal4 is only weakly expressed (arrowhead in Fig. 4B). These results thus demonstrate that during tOPC neurogenesis, Dll does not specify neuronal identities but instead acts in the Ey temporal window to promote the survival of NotchOFF Class 2 Svp neurons (Fig. 4H).
Figure 4. Dll and Ey control Notch-mediated neuronal survival.
Posterior views of L3 optic lobes. Unless otherwise mentioned, dashed lines show the tOPC. A,A′, dll mutant clones (green, dashed lines). Removing dll affects Class 2 Svp neurons (cyan, arrow). B,B′, Dll over-expression. slp-gal4 is strongly expressed in the dorsal tOPC but weakly expressed in the ventral tOPC that serves as an internal control (B). Expressing Dll (green) in Slp and D neuroblasts rescues the death of Class 2 Svp neurons (cyan, compare arrow with arrowhead on control side). C, Maximal projection of a wt brain showing Class 2 (Svp, cyan) and Class 3 & 4 (Toy, red) neuronal clusters. D, Maximal projection of an ey mutant brain showing the disappearance of NotchON Class 3 and 4 neurons (red) and the expansion of NotchOFF Class 2 Svp neurons (cyan, arrowheads). E, Average number of Class 2 Svp neurons in various genotypes (N=6 brains/genotype). All data represent mean±s.d. NS, not significant. *** P<0.001. F,F′, In wg>ey-RNAi, NotchON Hey-positive neurons (white) all express the death marker cleaved Caspase-3 (yellow, arrows). G,G′, Co-expressing P35 with ey-RNAi rescues the death of NotchON Hey-positive neurons (white) and generates an intermingling between these neurons and NotchOFF Class 2 Svp neurons (cyan). The rescued NotchON neurons do not express Toy (red). H, Timing of Dll and Ey activities. See also Figure S4.
Neuroblasts expressing high levels of Ey produce NotchOFF Class 2 Svp neurons. Removing ey activity does not lead to the loss of these neurons, suggesting that Ey does not specify their identity. However, removing ey activity generates two striking effects. First, it dramatically increases the number of NotchOFF Class 2 Svp neurons (Fig. 4C–E: wt 282±16; wg>ey-RNAi 695±41, P<10−4), indicating that Ey induces apoptosis of NotchOFF neurons. Second, it leads to the complete loss of NotchON Class 3 and 4 Toy-expressing neurons that are normally produced during the later Slp and D time windows (compare Fig. 4C and D). These NotchON Hey-positive neurons are still generated but they all express the apoptosis marker Cleaved Capase-3 and rapidly disappear (Fig. 4F). Co-expression of P35 with ey-RNAi rescues their death and generates an intermingling between these NotchON neurons and NotchOFF Class 2 Svp neurons (Fig. 4G, G′). Thus Ey is required for the survival of NotchON neurons and, in its absence, the switch in the apoptosis pattern no longer happens (Fig. 4F′): NotchOFF neurons survive while NotchON neurons die throughout neurogenesis, resulting in a massive increase of Class 2 Svp neurons. In summary, Ey plays a dual role in the control of neuronal survival: it induces apoptosis of NotchOFF neurons and it promotes survival of NotchON neurons. Interestingly, when we forced the survival of Hey-positive NotchON neurons in ey loss of function (ey RNAi + UAS-P35), we noticed that these neurons no longer express Toy (Fig. 4G), suggesting that Ey is also required for the ‘Toy’ identity.
Removing ey function affects the neurons produced during the later Slp and D time windows (Fig. 4H). Since Ey is required for Slp expression (and thus indirectly for D expression), we next tested whether the ey phenotype could be due to the loss of Slp in neuroblasts. If this hypothesis is correct, removing Slp should produce the same phenotype as ey loss of function, i.e. expansion of NotchOFF Class 2 Svp neurons and loss of NotchON Class 3 Toy neurons. In slp mutant clones, NotchOFF Class 2 Svp neurons do not expand (Fig. S4H), indicating that Slp does not induce apoptosis of NotchOFF neurons. However, NotchON Class 3 neurons (marked by Toy and Hey) express cleaved Caspase-3 and systematically undergo apoptosis (Fig. 5A,A′ and S4I). Expression of P35 in slp clones rescues their death and generates an intermingling between these NotchON Toy neurons and NotchOFF Class 2 Svp neurons (Fig. 5B,B′ and S4J). Thus, Slp promotes survival of NotchON neurons.
Figure 5. tOPC temporal factors and the cell death pathway.
Posterior views of L3 optic lobes. Unless specified, dashed lines show the tOPC. A,A′, In slp mutant clones (dashed lines), NotchON Toy-expressing neurons (red) die as shown by expression of cleaved Caspase-3 (yellow, arrows). B,B′, Expressing P35 in slp clones rescues the death of NotchON Toy neurons (red). C, The cell death pathway. D, NotchOFF Class 2 (Svp, cyan) and NotchON Class 3 & 4 clusters (Toy, red) in a wild-type tOPC. E, Removing hid, rpr and grim (def(3L)H99 clones, dashed lines) rescues the death of NotchON Toy neurons (red) in the Ey time window (arrowhead), and that of NotchOFF Svp neurons (cyan) in the Slp and D time windows (arrow). F, Removing Dronc generates an intermingling between NotchOFF (Svp, cyan) and NotchON neurons (Toy, red) in the Ey, Slp and D time windows. White bars delimitate the border between the Ey and Slp time windows. G, In Dronc mutants, Hid is only expressed in the NotchOFF Svp neurons that normally die in the Slp and D time windows. H, H′, Decreasing Hid activity (hidhypomorph/def(3L)hid99) has no effect on the neurons produced during the Ey time window (H). However, this rescues the death of NotchOFF Svp neurons (cyan) in the Slp and D time windows (H′). This phenotype is more pronounced in the ventral side of the tOPC (arrow). I, I′, Removing rpr rescues the death of NotchON Toy neurons in the Ey time window (red, i) but has no effect on the neurons produced during the Slp and D time windows (I′). J, Interactions between tOPC temporal factors and cell death genes. See also Figure S5.
Ey induces apoptosis of NotchOFF neurons whereas Slp promotes survival of NotchON neurons. We next investigated the potential targets of Ey and Slp. In Drosophila, the cell death pathway is initiated by the expression of the genes reaper (rpr), head involution defective (hid) and grim (Fig. 5C), which by inhibiting DIAP-1 lead to the activation of initiator Caspase-9 (Dronc). Dronc in turn activates Caspase-3, which triggers the apoptotic process (Xu et al., 2009). P35 acts by preventing Casp-3 from killing the cells. Like with P35 over-expression, removing rpr, hid and grim (def(3L)H99 clones, Fig. 5D,E) or Dronc (Fig. 5D,F and S5A,B) blocks apoptosis and generates an intermingling between NotchOFF and NotchON neurons in the Ey, Slp and D time windows. Strikingly, in Dronc mutants where neurons initiate apoptosis but do not complete it, Hid is only expressed in NotchOFF neurons that normally die during the Slp and D time windows (Fig. 5G). This suggests that Hid specifically initiates apoptosis of NotchOFF neurons. Consistent with this, removing Hid activity (Hidhypomorph/def(3L)H99 or wg>hid-RNAi) specifically rescues apoptosis of NotchOFF Svp neurons produced during the Slp and D time windows (Fig. 5H,H′ and S5C,C′). By contrast, removing rpr specifically rescues apoptosis of NotchON neurons produced during the Ey time window (Fig. 5I,I′). Thus, Rpr initiates apoptosis of NotchON neurons whereas Hid initiates apoptosis of NotchOFF neurons. Based on these results, we propose that Ey induces apoptosis of NotchOFF neurons by activating hid whereas Slp promotes survival of NotchON neurons by repressing rpr (Fig. 5J).
The sequential expression of Dll, Ey and Slp governs the switch in apoptosis
Our data suggest that the default fate of NotchON neurons is to undergo apoptosis (through rpr) and that Ey promotes their survival indirectly, by activating Slp expression in neuroblasts. This explains why NotchON neurons die during the Ey time window but survive during the later Slp and D time windows (Fig. 5J). Although Ey also induces apoptosis of NotchOFF neurons, these neurons survive in the Ey time window, suggesting that one or several factors repress Ey activity during the Ey time window. Dll could be one of these factors since it is active in the Ey time window and it is sufficient to promote survival of NotchOFF neurons. To test if we could overcome this repression, we over-expressed Ey in tOPC neuroblasts with insc-gal4. Ectopic Ey has no effect on neurons produced during the Dll, Slp and D time windows but leads to a significant decrease in the number of NotchOFF Class 2 Svp neurons that form during the Ey time-window (control 282±16; UAS-ey 194±17; P<3.10−5). Thus, when Ey levels are very high, its repressors (including Dll) are no longer able to prevent it from inducing apoptosis of NotchOFF neurons.
Altogether, our data demonstrate that Dll, Ey and Slp control neuronal survival and that their sequential expression determines the switch in the pattern of apoptosis. We propose the following model (Fig. 5J): by default, NotchON neurons undergo Rpr-dependent apoptosis. During the Ey time window, Dll and unknown repressors antagonize Ey activity whereas Slp is not expressed yet. As a result, NotchOFF neurons survive whereas NotchON neurons undergo apoptosis. In the Slp time window, Slp promotes survival of NotchON neurons while Ey repressors are no longer expressed/active to prevent Ey-induced apoptosis of NotchOFF neurons (through Hid), therefore causing an inversion in the apoptosis pattern. This inversion lasts until the end of neurogenesis, implying that one or several factors relay Ey and Slp activities. D, which defines the last time window, does not control neuronal survival. Indeed, in D mutant clones, NotchOFF neurons keep dying whereas NotchON Class 4 Toy/D neurons disappear and are replaced by NotchON Class 3 Toy neurons (Fig. S4K). This suggests that D does not relay Ey and Slp activities and instead only specifies neuronal fate. We conclude that temporal patterning of progenitors generates neuronal diversity by controlling both neuronal identity and Notch-mediated survival decisions.
The tOPC produces neurons for three different optic ganglia
In order to understand the consequences of the complex mode of neurogenesis in the tOPC, we finally investigated which types of neurons are produced in this region. The adult optic lobes comprise four neuropils, the lamina, medulla, lobula and lobula plate. The lamina derives from the inner part of the OPC crescent while the main region of the OPC generates the medulla. The Inner Proliferation Center (IPC) and the lobula plug (which derives from the IPC) together generate the lobula and lobula plate (Fig. S6A).
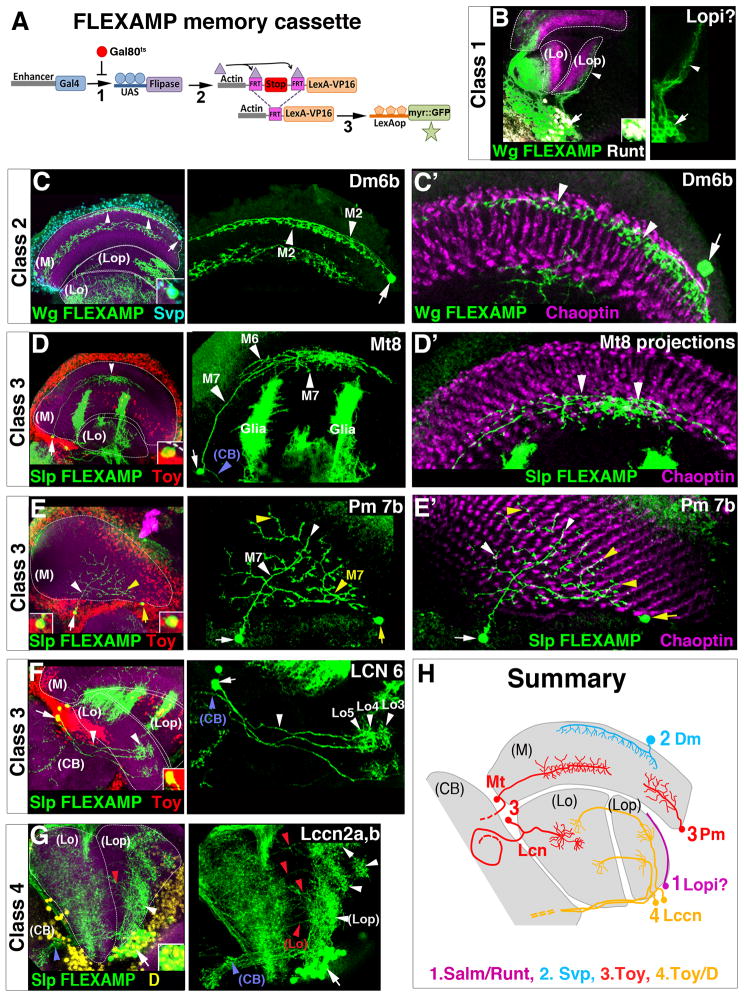
We tested which neurons the tOPC specifically produces. Since wg-gal4 expression is not maintained in adult brains, we generated a new memory cassette tool to determine the adult neuronal subtypes produced by the tOPC that we named FLEXAMP for Flip-out LexA Amplification (Fig. 6A, see M&M). We crossed FLEXAMP to wg-gal4, which is expressed in the entire tOPC and a subset of IPC cells, and with slp-gal4, which is specifically expressed in Slp and D-expressing neuroblasts of the tOPC (due to Gal4 per durance). We generated early clones to label dividing neuroepithelial cells (Fig. S6B–C). Unexpectedly, these clones revealed that the tOPC produces distinct neuronal subtypes for three of the four adult neuropils: Distal medulla neurons (arrow 1), Medulla tangential and Proximal medulla neurons (arrows 2), Lobula Columnar Neurons (arrows 3) and Lobula complex columnar neurons (arrows 4).
Figure 6. Neuronal subtypes produced in the tOPC.
A, The FLEXAMP memory cassette. B–G, FLEXAMP memory clones in mid-pupal (B) and adult brains (C–G); In all panels, dashed lines show the neuropils stained by N-Cad (magenta). B,B′, wg>FLEXAMP clones (green) suggests that Class 1 Salm/Runt neurons (white) are Lpi neurons. C, wg>FLEXAMP clones (green). Class 2 Svp neurons (cyan) are Dm6b neurons projecting in medulla layers M2 and M3. C′, Dm6b projections seem to contact R8 photoreceptors axons (Chaoptin, magenta). D–F, slp->FLEXAMP clones (green) showing the three types of Class 3 Toy neurons (red). D, Mt8 neurons project in M7, M6 (white arrowheads) and the central brain (blue arrowhead). D′, Mt8 projections seem to contact R7 photoreceptors terminations (Chaoptin, magenta). E, Two Pm7b neurons (medium-size projections) identified with white and yellow arrows, projecting into M7 and contacting R7 projections (Chaoptin, magenta in E′). F, LCN6 neurons arborize in lobula layers Lo3, 4 and 5 (white arrowheads) and project towards the central brain (blue arrowhead,). G, slp>FLEXAMP clones (green). Class 4 Toy/D neurons (yellow) are Lccn2a,b neurons that connect the lobula plate (white arrowheads) with the lobula (red arrowheads) and the central brain (blue arrowhead). H, Neuronal subtypes produced by the tOPC (M)=Medulla, (Lo)=Lobula, (Lop)=Lobula plate, (CB)=Central Brain. See also Figure S7.
To determine during which temporal window each of these neuronal subtypes is produced, we stained the nuclei of adult wg> and slp> FLEXAMP clones with antibodies specific to the four larval neuronal classes (Fig. 6). Strikingly, cell bodies of the same neuronal class (i.e. of neurons produced during the same time window) are all localized in the same area of the optic lobes. The identification of these neuronal subtypes is made according to (Fischbach, 1989) and their detailed morphology is described in the figure legends. Because Salm and Runt expression is not maintained in adult tOPC neurons, we could not precisely determine the terminal identity of Class 1 neurons. However, in mid-pupal brains, Salm and Runt are still expressed and we found that cell bodies of Class 1 neurons are localized in the lobula plate and appear to include lobula plate intrinsic neurons (lpi neurons, Fig. 6B). Class 2 Svp neurons migrate from their birthplace in the tOPC and their cell bodies are found in the distal medulla cortex. These neurons resemble previously described Distal-medulla6 neurons (Dm6 neurons, Fig. 6C,C′) but because their arborizations are larger, we named them Dm6b. Cell bodies of Class 3 Toy neurons are all found in the medulla rim, the region between the medulla and the lobula/lobula plate neuropils (Fig. S6A). These neurons can be classified into three groups depending on where they project. The first group is composed of Medulla tangential neurons Mt8 and Mt4 that connect different layers of the medulla with the central brain (Fig. 6D,D′ and S7A). The second group is composed of previously undescribed Proximal medulla neurons (Pm) that project into medulla layer M7 (Fig. 6E,E′). We named these neurons Pm7a, b and c, depending on the length of their arborizations (Fig. S7B–D). The third group is composed of Lobula Columnar Neurons 6 (LCN6), which connect layers of the lobula with the central brain (Fig. 6F and S7D). The presence of three different groups of Class 3 Toy neurons (Mt, Pm and LCN) raises the possibility that unknown temporal genes divide the Slp time window into three smaller windows, each producing one of these groups. Alternatively, this subdivision may be achieved by spatial patterning genes. Finally, cell bodies of Class 4 Toy/D neurons are localized in the lobula plate cortex. These neurons resemble Lobula complex columnar neurons 2 (Lccn2) that connect the lobula and the lobula plate with the central brain. We could not obtain single neuron clones but we could identify at least two subtypes of these neurons projecting into different layers of the lobula plate neuropil (Fig. 6G and S7E). We named them Lcnn2a,b. As in the Slp time window, the fact that we could identify two subtypes of Toy/D neurons suggests the existence of additional temporal and/or spatial patterning genes.
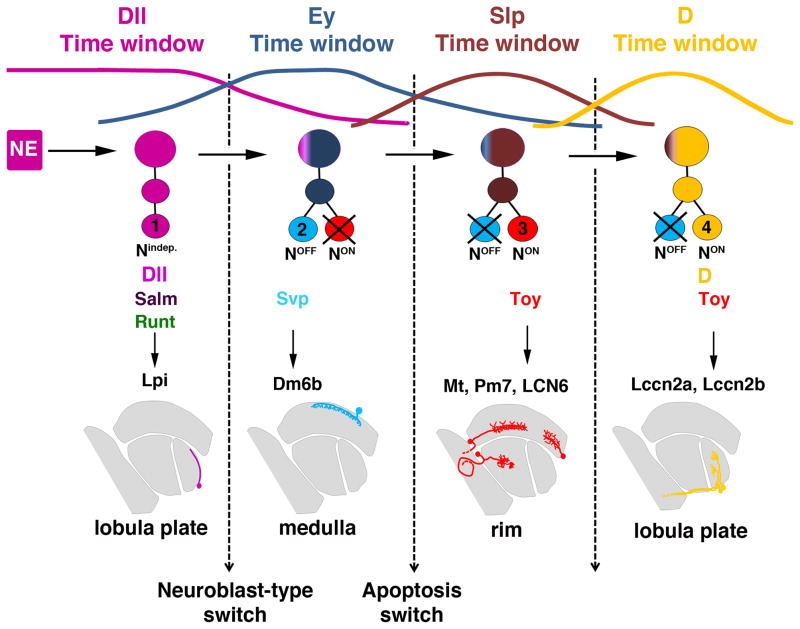
In conclusion, unlike the main OPC, the tOPC region produces several neuronal cell types which innervate three different neuropils of the adult optic lobes, the medulla, lobula and lobula plate (Fig. 6H). Therefore, in addition to leading to the production of distinct neuronal subtypes over time, the four temporal windows defined by Dll, Ey, Slp and D also determine in which area of the optic lobes the neuronal cell bodies are localized (Fig. 7): the Dll time window gives rise to neurons whose cell bodies are in the lobula plate cortex; the Ey time window produces neurons with cell bodies in the medulla cortex; the Slp time window produces neurons with cell bodies in the medulla rim; the D time window produces neurons with cell bodies in the lobula plate cortex.
Figure 7. Summary of tOPC neurogenesis.
The Dll, Ey, Slp and D time windows produce distinct neuronal subtypes, each localized to a specific area of the optic lobes.
DISCUSSION
Although apoptosis is a common feature of neurogenesis in both vertebrates and Drosophila, the mechanisms controlling this process are still poorly understood. For instance, several studies in Drosophila have shown that depending on the context, Notch can either induce neurons to die or allow them to survive during binary cell fate decisions. This is the case in the antennal lobes where Notch induces apoptosis in the antero-dorsal Projecting Neurons lineage (adPN) whereas it promotes survival in the ventral Projecting Neurons lineage (vPN) (Lin et al., 2010). In both of these cases, the entire lineage makes the same decision whether the NotchON or NotchOFF cells survive or die. This suggests that in this system, Notch integrates spatial signals to specify neuronal survival or apoptosis.
Here, we show that during tOPC neurogenesis, neuronal survival is determined by the interplay between Notch and temporal patterning of progenitors. Indeed, within the same lineage, Notch signaling leads to two different fates: it first induces neurons to die whereas later, it allows them to survive. We show that this switch is due to the sequential expression of three highly conserved transcription factors, Dll/Dlx, Ey/Pax-6 and Slp/Fkh, in neural progenitors. These three factors have distinct functions, with Dll promoting survival of NotchOFF neurons, Ey inducing apoptosis of NotchOFF neurons and Slp promoting survival of NotchON neurons. Our data suggest that Ey induces death of NotchOFF neurons by activating the pro-apoptotic factor hid. Thus, Dll probably antagonizes Ey activity by preventing Ey from activating hid. Our data also suggest that Notch signaling induces neuronal death by activating the pro-apoptotic gene rpr. Thus, Slp might promote survival of NotchON neurons by directly repressing rpr expression or by preventing Notch from activating it. In both cases, the interplay between Notch and Slp modifies the default fate of NotchON neurons, allowing them to survive. Further investigations will test these hypotheses and determine how Dll, Ey, Slp and Notch differentially activate/repress hid and rpr.
Although the tOPC and the main OPC have related temporal sequences, their neurogenesis is very different. This difference is in part due to the fact that newly specified tOPC neuroblasts express Dll, which controls neuronal survival, instead of Hth. Why do tOPC neuroblasts express Dll? The tOPC, which is defined by Wg expression in the neuroepithelium, is flanked by a region expressing Dpp (Kaphingst and Kunes, 1994). Previous studies have shown that high levels of Wg and Dpp activate Dll expression in the distal cells of the Drosophila leg disc (Estella et al., 2008). Wg and Dpp could therefore also activate Dll in the neuroepithelium and at the beginning of the temporal series in tOPC progenitors. Another difference between the main OPC and tOPC neurogenesis is that Ey and Slp have completely different functions in these regions. Indeed, unlike in the main OPC, Ey and Slp control the survival of tOPC neurons. This suggests that autonomous and/or non-autonomous signals interact with these temporal factors and modify their function in the tOPC.
Finally, tOPC neuroblasts produce neurons for three different neuropils of the adult visual system, the medulla, the lobula and the lobula plate. This ability could be due to the particular location of this region in the larval optic lobes. Indeed, the tOPC is very close to the two larval structures giving rise to the lobula and lobula plate neuropils: Dll expressing neuroblasts are located next to the lobula plug while D expressing neuroblasts are close to the IPC. Interestingly, Dll and D neuroblasts specifically produce lobula plate neurons. This raises the possibility that these neuroblasts and/or the neurons produced by these neuroblasts receive signals from the lobula plug and the IPC, which instruct them to specifically produce lobula plate neurons. These non-autonomous signals could also modify the function of Ey and Slp in the tOPC.
In summary, this study demonstrates that temporal patterning of progenitors, a well-conserved mechanism from Drosophila to vertebrates, generates neural cell diversity by controlling multiple aspects of neurogenesis including neuronal identity, Notch-mediated cell survival decisions and the mode of intermediate precursor division. In the tOPC temporal series, some factors control two of these aspects (Ey) whereas others have a specialized function (Dll, Slp and D). This suggests that temporal patterning does not consist of a unique series of transcription factors controlling all aspects of neurogenesis, but instead of multiple superimposed series, each with distinct functions.
EXPERIMENTAL PRODECURES
Antibodies and Immunostaining
Standard methods were used for antibody staining (see supplemental methods). The following antibodies were used: rabbit anti Hth (1:500) (R. Mann), guinea pig anti-Dll (1:500) (R. Mann), mouse anti-Ey (1:10) (DSHB), rabbit anti-Slp (1:500) (segmentation antibodies), guinea-pig anti-Runt (1:500) (segmentation antibodies), rabbit anti-D (1:200) (ModENCODE), guinea-pig anti-D (1:50) (J. Nambu), rabbit anti-Salm (1:500) (T. Cook), mouse anti-Svp (1:500) (Y. Hiromi), guinea-pig anti-Toy (1:500) (U. Walldorf), rabbit anti Ey/Toy (1/500) (J. Clements), guinea-pig anti Hey (1:1000) (C. Delidakis), guinea pig anti-Dpn (1:500) (J. Skeath), rabbit anti-Ase (1:100) (Y. Jan), rabbit anti cleaved-Caspase-3 (1:100) (Cell Signaling Technology), guinea pig anti-Hid (D. Ryoo), mouse anti 24B10 (1:20), Rat anti DE-Cadherin (1:20) and Rat anti DN-Cadherin (1:20) (all from DSHB), sheep anti-GFP (1:500, AbD Serotec). Secondary antibodies are from Jackson or Invitrogen.
Genetics and fly strains
The following Gal4 lines were used: wg-gal4 (ND382, K. Basler); eyOK107-gal4 (JB. Connolly); slp-gal4 (R35A08, Janelia Gal4 collection) and insc-gal4 (Li et al., 2013). The following mutant stocks were used: y,w;;; eyJ5.71/Miunc-13MI00468(y+); DroncI24/TM3Sb (D. Ryoo); def(3L)H99,kniP/Tm3Sb (Bloomington #1576); hidA22/TM6c (L. Johnston); y,w,hsFLP;;def(3L)rprXR38/TM6b (L. Johnston). Wild-type and mutant clones were generated by 37°C heat-shocks at early larval stages. Wandering third instar larvae were analyzed. The progeny of over-expression and RNAi experiments (all involving tub-gal80ts) was raised at 18°C, heat-shocked two days at 29°C from early larval stages and dissected right after heat-shock. Genotypes of clonal, RNAi and over-expression experiments are simplified throughout the text and figures. See Table S1 for complete genotypes and crosses.
FLEXAMP memory cassette
FLEXAMP genotype: y,w,UAS-FLP; If/CyO; act>y[+]>LHV2-86Fb,13XlexAop2-myr::GFP/TM6B. The act>y[+]>LHV2-86Fb stop cassette (K. Basler) contains an optimized version of lexVP16 that has a reduced toxicity and cannot be inhibited by Gal80. The lexAop13X-myr::GFP reporter (B. Pfeiffer) is an optimized version of lexAop leading to strong expression levels of myr::GFP without being leaky in the optic lobes. To generate clones, FLEXAMP was crossed to wg-gal4 or slp-gal4 combined with tub-gal80ts. The progeny of these flies was raised at 18°C, incubated several hours at 29°C (to inactivate Gal80) and put back at 18°C until adult stage. The incubation time to obtain an optimal and analyzable number of GFP expressing neurons in adults varied depending on the gal4 line (4 hours with wg-gal4 and 24 hours with slp-gal4).
Quantifications
For each staining, positive cells were counted manually on each z plane using custom-written scripts in Igor Pro (WaveMetrics). Each graph bar represents the average number of neurons for a given genotype. Error bars represent the standard deviation and t-tests are used to determine if the samples are significantly different.
Supplementary Material
HIGHLIGHTS.
Temporal patterning of progenitors controls Notch-mediated cell survival
First, NON neurons systematically die; subsequently, NOFF neurons die
NON neurons die through rpr activity whereas NOFF neurons die through hid activity
The sequential expression of temporal factors governs the switch in apoptosis
Acknowledgments
We thank the fly community and the modENCODE team for gifts of antibodies and fly stocks. We especially thank Don Ryoo for his precious help and advices regarding cell death experiments. We thank the Desplan laboratory members for discussions and support as well as M. Slaidina, L. Christiaen, C. Maurange, N. Vogt, J. Bos, C. Doe, T. Lee and R. Jonhston for critically reading the manuscript. This work was supported by a grant from NIH to C.D. R01 EY017916. Support was also provided by NYU Abu Dhabi grant G-1205C to CD, and fellowships from EMBO (ALTF 680-2009), HFSPO (LT000077/2010-L) and Philippe Foundation to C.B.; The Robert Leet and Clara Guthrie Patterson Trust to X.L.; The Canadian Institutes of Health and Research (CIHR) to T.E.; EMBO (ALTF 249-2009) and The Revson Foundation to M.C; NIH (T32EY022843-02) to B.W.
Footnotes
AUTHORS CONTRIBUTIONS
C.B. designed and performed all experiments. C.B., X.L. and T.E performed the initial antibody screen. X.L. identified the slp-gal4 line. M.C. generated the Igor procedures for quantifications. B.W. contributed to the design of cell death experiments. C.D. contributed to the design of experiments. The manuscript was written by C.B. and C.D. and all authors commented on it.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural development. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental neurobiology. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Developmental biology. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Buescher M, Yeo SL, Udolph G, Zavortink M, Yang X, Tear G, Chia W. Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes & development. 1998;12:1858–1870. doi: 10.1101/gad.12.12.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural development. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Developmental cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KFaD, AP The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell and tissue research. 1989:441–475. [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer ACOJA. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux’s archives of developmental biology. 1990;198:264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Kaphingst K, Kunes S. Pattern formation in the visual centers of the Drosophila brain: wingless acts via decapentaplegic to specify the dorsoventral axis. Cell. 1994;78:437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ. Drosophila neuroblast 7–3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. The Journal of comparative neurology. 2005;481:240–251. doi: 10.1002/cne.20371. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498:456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lai SL, Yu HH, Chihara T, Luo L, Lee T. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nature reviews Neuroscience. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Lee HK, Perez E, Chadwell L. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of Drosophila. Development. 2003;130:4109–4121. doi: 10.1242/dev.00593. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IAH, TE The development of the optic lobes. The Development of Drosophila melanogaster Cold Spring Harbor Laboratory Press. 1993;II:1363–1491. [Google Scholar]
- Monastirioti M, Giagtzoglou N, Koumbanakis KA, Zacharioudaki E, Deligiannaki M, Wech I, Almeida M, Preiss A, Bray S, Delidakis C. Drosophila Hey is a target of Notch in asymmetric divisions during embryonic and larval neurogenesis. Development. 2010;137:191–201. doi: 10.1242/dev.043604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Current biology: CB. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, Olson JM, Banerjee U, Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Developmental biology. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Technau GM. Multiple roles for Hox genes in segment-specific shaping of CNS lineages. Fly. 2008;2:316–319. doi: 10.4161/fly.7464. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Developmental biology. 2013;380:12–24. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Developmental dynamics: an official publication of the American Association of Anatomists. 2006;235:861–869. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvklo C, MacDonald R, Bivik C, Baumgardt M, Karlsson D, Thor S. Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression. Development. 2012;139:678–689. doi: 10.1242/dev.074500. [DOI] [PubMed] [Google Scholar]
- Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly. 2009;3:78–90. doi: 10.4161/fly.3.1.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–3203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.