Abstract
Membrane proteins have always presented technical challenges for structural studies because of their requirement for a lipid environment. Multiple approaches exist including X-ray crystallography and electron microscopy that can give significant insights into their structure and function. However, nuclear magnetic resonance (NMR) is unique in that it offers the possibility of determining the structures of unmodified membrane proteins in their native environment of phospholipid bilayers under physiological conditions. Furthermore, NMR enables the characterization of the structure and dynamics of backbone and side chain sites of the proteins alone and in complexes with both small molecules and other biopolymers. The learning curve has been steep for the field as most initial studies were performed under non-native environments using modified proteins until ultimately progress in both techniques and instrumentation led to the possibility of examining unmodified membrane proteins in phospholipid bilayers under physiological conditions. This review aims to provide an overview of the development and application of NMR to membrane proteins. It highlights some of the most significant structural milestones that have been reached by NMR spectroscopy of membrane proteins; especially those accomplished with the proteins in phospholipid bilayer environments where they function.
1. Introduction
1.1. Biological membranes
Membranes define the physical boundaries of organelles, cells, unicellular organisms, and some viruses. Under a microscope, cell membranes appear to be continuous round or oval containers, which encase their contents, separating it from the external surroundings while providing a mechanism for selective passage of chemicals and signals between the external and internal environments. It is well known that spherical artificial membranes, also known as liposomes, can form spontaneously from phospholipids in water. Although liposomes appear to be superficially similar to biological membranes, membranes extracted from living organisms consist of approximately 50% protein and 50% lipid by weight. Compartmentalization can be handled by the phospholipids alone, one-third of the proteins expressed from a typical genome are associated with membranes in order to handle the transport and signaling activities. Understanding membrane proteins demands a structural approach to characterize the factors that influence the atomic resolution structures and dynamics of the proteins and their functions within the phospholipid bilayer environment in which they reside. Because of the liquid crystalline nature of the phospholipid bilayer environment, much of this information is available only from nuclear magnetic resonance spectroscopy. This makes studies of membranes among the most meaningful applications of NMR to structural biology.
Structure determination of membrane proteins in general has been hampered by technical difficulties stemming primarily from the preparation of samples suitable for the most widely used methods of structure determination, such as X-ray crystallography and solution NMR spectroscopy. Compared to the more familiar globular proteins, which are generally soluble and crystallizable, membrane proteins are hydrophobic, insoluble in aqueous solution, and difficult to refold into their stable, active conformation. After many years of development, solid-state NMR has matured into an approach fully capable of determining the structures of membrane proteins in their native phospholipid bilayer environment under physiological conditions, and at the present time is the only method with this capability. The initial structures of membrane proteins obtained under near-native conditions are providing a basic understanding of their structures, dynamics, and functions in biological membranes. Along the way, many studies have been performed under a wide variety of sample conditions, the best that could be done at the time, and they have contributed to the development of the spectroscopic methods and have provided background on many issues surrounding the structures and dynamics of these proteins. However, these results have to be interpreted with caution because it is known that non-native environments, such as organic solvents and detergents, can affect the structures and dynamics of membrane proteins.
The characterization of membrane proteins is built on the foundation provided by two of the earliest biophysical chemists, Christian Anfinsen and Charles Tanford. A few of their key ideas are briefly summarized here to provide context for the subsequent applications of NMR spectroscopy to membrane proteins in phospholipid bilayers, which involves the use of many additional layers of technology. Anfinsen (Anfinsen, 1973) noted that “the thermodynamic hypothesis states that the three-dimensional structure of a native protein in its normal physiological milieu (solvent, pH, ionic strength, presence of other components, such as metal ions or prosthetic groups, temperature, and others) is the one in which the Gibbs free energy of the whole system is lowest; that is, that the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment. In terms of natural selection through the design of macromolecules during evolution, this idea emphasized the fact that a protein molecule only makes stable, structural sense when it exists under conditions similar to those for which is was selected – the so-called physiological state.”
Amphipathic molecules, such as phospholipids, form the seamless boundaries of cells and organelles, and the associated proteins control the influx and efflux of metabolites and other biomolecules. This is consistent with the hydrophobic effect, as noted by Tanford (Tanford, 1978), according to whom “the thermodynamics of biological organization does not involve biosynthesis or other chemical transformations, but focuses solely on where molecules prefer to go after they have been synthesized.”
Consistent with these principles, proteoliposomes, protein-containing phospholipid bilayers, are well-characterized and experimentally accessible versions of biological membranes. Although by necessity organic solvents and detergents were used to solubilize membrane proteins for the earliest studies, they violate these tenets, and now that solid-state NMR is providing structures under near-native conditions, many of the structures are found differ in the non-native environments.
1.2. Biogenesis of membranes
Most discussions about the inception of life following the formation of Earth about 4.5 billion years ago are focused on molecules that evolved into the polymers that store information and carry out chemical processes. However, the emergence of membrane-like barriers to separate these molecules from the surrounding environment was a prerequisite for the formation of self-replicating systems. Membrane-like barriers must have been formed by pre-biotic amphiphiles. Notably, single-cell prokaryotes left fossil evidence dating back 3.4 billion years (Wacey et al., 2011). The amphipathic molecules that formed primitive membranes in early living systems served to maintain the proximity of a growing number of interacting functional and information-bearing macromolecules in addition to controlling the influx and efflux of metabolites.
The protein-containing membranes of contemporary organisms did not appear fully formed. Like all other aspects of biology, they are the result of billions of years of evolution and are finely tuned for their specialized biological roles. Consequently, abiogenesis includes amphiphiles for membranes which resulted, along with amino acids, nucleic acids, and carbohydrates and their precursors, from the conditions on the early Earth, as mimicked by the classic Miller-Urey experiment (Miller, 1953).
The pre-biotic amphiphiles that assembled to form a barrier essential for the start of evolution (Lombard et al., 2012) may bear some resemblance to the amphiphilic compounds used for solubilization and crystallization of membrane proteins. Subsequent co-evolution of proteins and phospholipids resulted in contemporary biological membranes with their unique structural and functional characteristics. Our focus in this review is on structure determination of the proteins that are associated with membranes. For the purpose of structure determination, pre-biotic amphiphiles are a starting point in the search for detergents capable of solubilizing membrane proteins, providing a chemical connection between the earliest membranes and those in mammals. In general, sample preparation and spectroscopic studies of bacterial and human membrane proteins are performed using essentially the same experimental protocols.
1.3. Phospholipid Structure
Phospholipids (Cao et al., 2013) and proteins are the two principal macromolecular constituents of biological membranes. Some membranes also contain large amounts of cholesterol. The phospholipids found in biological membranes consist of an amphipathic or charged polar head group attached to two long hydrocarbon chains through a tri-substituted glycerol backbone. A wide range of head groups, chain lengths, number of double bonds, and other chemical characteristics are found in biological membranes. In water, phospholipids have negligible concentrations due to their low solubility, and instead, self-assemble to form bilayers, with the polar head groups exposed to water and the long hydrocarbon chains located in the hydrophobic bilayer interior shielded from the polar, aqueous environment. On the molecular scale, these bilayer sheets are infinitely long in two dimensions, but only two molecules thick in the third dimension, which is sufficient to provide a chemical and physical barrier that is so impermeable that it requires proteins to enable ions, small molecules, biopolymers, and signals to cross. Intra- and inter- molecular motions of the phospholipids add to the complexity of the membrane. In addition to their definitive gel to liquid crystalline phase transitions, different lipids have different amplitudes of motion from the surface to the center of the bilayer (Hubbell & McConnell, 1971).
Both the phospholipids and the proteins undergo fast rotational diffusion about the bilayer normal as well as translational diffusion in the plane of the bilayer and occasionally ‘flip-flop’ motions across the bilayer (Cherry, 1978). These global motions, which are clearly integral to the functions of membranes, cannot be mimicked in proteins solubilized in organic solvents, detergents, or frozen at low temperatures, again emphasizing the importance of studying membrane proteins as part of the membrane assembly under physiological conditions.
The basic understanding of the properties of biological membranes began to emerge in the early 1970s (Singer & Nicholson, 1972). This is in contrast to the dramatic announcements that signaled the birth of molecular biology 15 years – 20 years earlier with the structure of the DNA double helix in 1953 (Watson & Crick, 1953) and that of the protein myoglobin in 1958 (Kendrew et al., 1958). All three of these classes of molecules, DNA, proteins, and lipids, as well as carbohydrates, and their partners and complexes must have evolved in concert, and are inextricably linked. As a result, the underlying theme of this review is how the liquid crystalline environment of phospholipid bilayers affects the properties of membrane proteins. This is the situation even when the membrane is reduced to its minimal chemical composition of a single type of protein-containing phospholipid that self-assembles as described by the fluid mosaic model (Singer & Nicholson, 1972).
1.4. NMR of phospholipid bilayers
NMR played a key role in sorting out the properties of the various lipid assemblies studied in the early 1970s, including their implications for the structures of membrane proteins. In large part this is due to NMR being well suited for the characterization of local and global dynamics. Even when quantitative interpretations may be elusive, there is no mistaking the effects of large-amplitude, rapid motions on NMR spectra because of the dramatic narrowing of resonances that occurs.
The earliest NMR experiment on membranes focused on bilayers of pure phospholipids as well as of mixtures of lecithins. Isotopic labeling was rarely used because of its limited availability as well as the technical limitations of the commercial spectrometers. Thus, at the time, the only viable spectroscopic approach was high-resolution 1H solution NMR, which was performed using instruments and methods developed for liquids. This limitation in spectroscopic technology contributed to their being a fundamental controversy in the field from the very beginning.
The facts were indisputable. Phospholipids in water formed bilayers that yielded very broad 1H resonances (Chan et al., 1971; Chan et al., 1972; Chan et al., 1973; Finer et al., 1972; Keough et al., 1973; Lichtenberg et al., 1975; Veksli et al., 1969). When these same samples were subjected to sonication, forming small, discrete ‘sonicated vesicles’, the spectra contained much narrower 1H resonances. At the time, there were several theoretical interpretations to explain the drastic differences in signal properties from the same molecules in liposomes that appeared to differ only in diameter (Seiter & Chan, 1973). One explanation was that the lipids have basically the same properties in liposomes and vesicles, and the line narrowing resulted from the more rapid isotropic reorientation of vesicles, which could have a diameter as small as 250 Å, compared to the extended two-dimensional arrays in multilamellar liposomes. The other explanation was that the phospholipid environment in small vesicles was perturbed by the effects of sonication and the formation of a highly curved assembly, and that the resulting local disorder led to local motions of the lipid chains and line narrowing of the 1H resonances (Sheetz & Chan, 1972).
Through many studies, the properties of liposomes were characterized and it was understood that the large assemblies of lipids were ill suited for solution NMR experimental methods and theories. The major advances awaited the advent of solid-state NMR spectroscopy, which utilized 2H, 31P, and 13C signals from the phospholipids. The initial studies were from the group of Chapman, which broadly explored the application of NMR to liposomes (Veksli et al., 1969) (Oldfield et al., 1971) (Keough et al., 1973). There were two notable features about these studies. One was the use of a spin S=1 nucleus, deuterium, with its quadrupolar interaction available for analysis. The other was one of the first uses of isotopic labeling in biological NMR to place the deuterium nuclei at informative locations in the lipids. This addressed the limitations of using 1H NMR to study liquid crystalline systems with extended order. In highly anisotropic environments, such as liquid crystals and lipid bilayers, each 2H nucleus gives a doublet in the spectrum. For a bilayer membrane oriented on glass plates with the bilayer normal parallel to the magnetic field, the observed splitting measures the angle between the bond and the bilayer normal (Oldfield et al., 1971). In powder samples, the magnitude of the quadrupolar coupling can also be measured, as well as any reduction from the static value interpreted in terms of dynamics, and this was particularly important in making the transition from the lipid to the protein components of membranes (Blume et al., 1982) (Seelig et al., 1973) (Smith & Oldfield, 1984) (Keniry et al., 1984) (Kinsey et al., 1981a; Kinsey et al., 1981b).
In 1973, Urbina and Waugh described the first application of high resolution solid-state NMR to membranes. They obtained natural abundance 13C NMR spectra of DMPC bilayers above and below the phase transition. Presciently, the spectra showed more signal intensity at the lower temperature where the dipolar couplings are stronger, enabling more efficient cross-polarization from the abundant 1H nuclei to the “dilute” 13C nuclei. The implementation of 13C detection opened up many more sites, as well as the 13C chemical shift anisotropy and heteronuclear 13C-1H dipole-dipole spin-interactions for investigation. Among the first findings about a protein embedded in a membrane bilayer was the dramatic effects of the rotational diffusion, as anticipated in the fluid mosaic model of membrane proteins, by the averaging of the 13C′ powder pattern in labeled bacteriorhodopsin in membrane bilayers (Lewis et al., 1985).
1.5. Protein structure
The precisely folded three-dimensional structure of a protein arranges the chemical functional groups of backbone and side chain sites so that they can carry out the necessary functions with high selectivity and efficiency, for example, catalysis of chemical reactions, transport of ions and organic molecules, binding of signaling molecules, etc. (Petsko & Ringe, 2008). Generally, any variation from the native structure results in an inactive protein with accompanying consequences for biological function. The secondary and tertiary structures of a protein are commonly represented graphically by backbone ribbons (Richardson, 1981). These and other representations of protein structures are typically derived from many individual distances and angles between proximate atoms, as measured in X-ray diffraction or solution NMR experiments. Magic angle spinning (MAS) solid-state NMR provides similar local structural parameters. In contrast, oriented sample (OS) solid-state NMR provides primarily angular constraints measured relative to a single external axis, such as the magnetic field. In all cases, the quality of the structures benefits from the complementary measurements from the various techniques, with the primary issue being that the samples provide comparable membrane environments. Notably, the backbone structure of a protein is equivalently represented by the dihedral angles between the peptide planes in a Ramachandran plot (Ramachandran et al., 1963).
Since protein structure determination started with X-ray diffraction of single crystals (Dickerson et al., 1962; Kendrew et al., 1958; Perutz et al., 1960), and the majority of new structures continue to be determined with this approach, globular proteins are often envisaged as very complex, folded polypeptides, even though they reside and function in aqueous solution where they undergo rapid isotropic reorientation. In addition, large-amplitude side chain motions are superimposed on the backbone structure of the proteins (Gall et al., 1982; Wuthrich & Wagner, 1975). Here we discuss the structure determination of membrane proteins in their native environment of phospholipid bilayers by solid-state NMR spectroscopy (Opella, 2013b; Zhou & Cross, 2013), and the effects of their sequence and environment on their three-dimensional structure (Franks et al., 2012).
Although equally true for all proteins, the combined influence on protein structure from both its sequence of amino acids and its environment is most evident in membrane proteins largely because they are embedded in highly asymmetric, liquid crystalline phospholipids, rather than an isotropic solution or a regular lattice. The nature of the membrane environment used in experimental studies, especially structure determination by NMR, of membrane proteins has been a major factor since the beginning of the field (Bosch et al., 1980; Cross & Opella, 1979; Cross & Opella, 1980; Lauterwein et al., 1979). Environments ranging from mixed organic solvents to protein-containing phospholipid bilayers (proteoliposomes) (Banerjee & Datta, 1983) have been used in structural and other physical and chemical studies of membrane proteins. Because of the difficulty in dealing with these highly hydrophobic proteins, often the goal of the synthetic membrane environment was simply to solubilize the polypeptides so that measurements could be made. Now, with a growing database of membrane protein structures, it is becoming apparent that detergents and organic solvents are generally problematic. The amino acid sequence alone is not sufficient to determine the three-dimensional structure of a protein. Just as globular proteins require an aqueous environment to fold properly, membrane proteins require liquid crystalline phospholipid bilayers. Both classes of proteins require physiological conditions of temperature, pH, concentration, etc. Solid-state NMR is essential for studies of membrane proteins associated with lipid bilayers because they are effectively “immobilized” by their interactions in this large supramolecular membrane structure. A variety of solid-state NMR approaches have been applied to membrane proteins and recent reviews are available on specific examples and aspects of the method (Franks et al., 2012; Judge & Watts, 2011; McDermott, 2009; Murray et al., 2013; Popot, 2010; Tang et al., 2013a; Weingarth & Baldus, 2013) (http://www.drorlist.com/nmr/MPNMR.html) (Cegelski, 2013; Comellas & Rienstra, 2013; Goncalves et al., 2013; Hong et al., 2012; Judge & Watts, 2011; Knight et al., 2013; Mueller & Dunn, 2013; Murray et al., 2013; Opella, 2013a; Opella, 2013b; Parthasarathy et al., 2013; Saito et al., 2010; Sengupta et al., 2013; Shi & Ladizhansky, 2012; Ullrich & Glaubitz, 2013; Ulrich, 2005; Weingarth & Baldus, 2013; Yan et al., 2013).
Proteoliposomes are nearly ideal samples for structure determination of membrane proteins, since the proteins are embedded in chemically defined, fully hydrated, liquid crystalline phospholipid bilayers. This is illustrated in the iconic representation by Singer and Nicholson of the “fluid mosaic” model of a biological membrane (Singer & Nicholson, 1972). It is important that the forces exerted by the phospholipids on the polypeptides in proteoliposomes are likely to be very similar to those encountered in a biological membrane. Significantly, the proteins in membranes are highly constrained in most directions while undergoing two types of large-amplitude, rapid, global motions. These are fast rotational diffusion about the bilayer normal and lateral diffusion in the plane of the bilayer.
Most of the previous characterizations of membrane protein dynamics have focused on local backbone motions and those of the sidechains, many of which have important implications for the functions of the proteins (Gall et al., 1982; Herzfeld et al., 1987; Keniry et al., 1984; Kinsey et al., 1981a; Leo et al., 1987; Wuthrich & Wagner, 1975). Here, we focus on the influence of global rotational diffusion of the membrane proteins about the bilayer normal, not only to understand its effects on the protein structures, dynamics, and functions, but also in order to exploit this fundamental property of membrane proteins to expand the repertoire of solid-state NMR spectroscopy for protein structure determination (Das et al., 2012; Lewis et al., 1985; Park et al., 2011b).
1.6. Global motions of membrane proteins
In aqueous solution, globular, soluble proteins undergo rapid isotropic reorientation; the rate or frequency of the motion is generally described in terms of the rotational correlation time, which is typically only somewhat slower than the timescale of a liquid, in the range of 1 nsec – 20 nsec. The diameter of the protein and viscosity of the solution determine the rotational correlation time. In cases where the protein structure is non-spherical, the reorientation is found to be anisotropic. The overall rotational motions of soluble, globular proteins are well characterized by NMR and other biophysical experiments (Bauer et al., 1975) and in most cases, it is readily possible to distinguish any local backbone motions from the overall reorientation of the protein based on their differences in frequencies, amplitudes, and directions, which generally result in a subset of narrower, more intense, resonances superimposed on the background of broad resonances associated with the folded, structured bulk of the protein (Bogusky et al., 1987).
In contrast, the extremely asymmetric chemical and physical environment of phospholipid bilayers drastically affects the global motions of membrane proteins (Edidin, 1974). There are three major classes of motions of the protein in the bilayer to consider, two of which the NMR experiment is insensitive to. First, there are “flip-flop” motions where the molecule goes from one side of the bilayer to the other. NMR is insensitive to these symmetric inversions, and flip-flop motions have generally been characterized as an infrequent event, too slow to be detected by the vast majority of NMR experiments. The second is lateral diffusion in the plane of the bilayer, which has been thoroughly studied in a number of cases and may be of great importance to biological functions (Devaux & McConnell, 1973; Devaux & McDonnell, 1972; Poo & Cone, 1973; Poo & Cone, 1974). However, NMR is also insensitive to translational motion.
The third type of global motion, rotational diffusion, is crucial for both the functions of the proteins and an approach to the determination of the structures in the bilayer environment. Well-understood physical laws (Durr et al., 2007), two of which are directly pertinent to determining structure from global motions, govern NMR spectroscopy. One is the timescale. The frequency breadth of the spin-interactions, as represented in the spectra as powder patterns, defines a “ruler” for the rate at which motions affect the spectra. Motions that are ‘fast’ compared to the frequency breadth average the powder pattern, and those that are ‘slow’ do not affect the spectra. The second is the angular dependence, which has an equally strong, but entirely different effect on the spectra.
1.7. Effects of motional averaging in solid-state NMR spectroscopy
There are substantial molecular motions in seemingly rigid solids that strongly affect NMR spectra. This is not a generalized narrowing, but rather well-defined motions about a single axis that produced predictable and quantitative changes in the powder patterns that could be directly related to the line shapes observed in the same samples without motions (Gutowsky & Pake, 1950), for example at very low temperatures. Importantly, the motionally averaged powder patterns reflect the precise geometry of the internal motion about a fixed axis, and are highly relevant to the situations encountered with proteins used as samples for 1H/13C/15N triple-resonance experiments. This is the key concept for deriving structural information from rotationally averaged powder patterns, which we refer to as rotationally aligned solid-state NMR. In the case of membrane proteins, we consider the vast majority of backbone N-H and C-H bonds to be rigid on the time scales of the dipole-dipole and chemical shift interactions, thus it is the motion from the global rotational diffusion of the protein in the liquid crystalline bilayer that averages the powder pattern as determined by the angle between the principal axis of the coupled pair of nuclei (Gutowsky & Pake, 1950) or the chemical shift tensor (Mehring et al., 1971) of a site and the bilayer normal (McLaughlin et al., 1975).
With the increasing interest in the analysis of the functions of membrane proteins in terms of their structures, there is a heightened concern about the influence of the surrounding environment when it is provided by detergents, non-natural lipids, organic solvents, crystal packing, etc. (Durr et al., 2013; Judge & Watts, 2011; Kruger-Koplin et al., 2004; Page et al., 2006; Poget & Girvin, 2007; Sanders & Oxenoid, 2000; Vinogradova et al., 1998). Nonetheless, considerable progress has been made in describing the structures of membrane proteins, primarily by focusing on exceptional cases, for example bacteriorhodopsin, which as a bacterial photo transducer is an extraordinarily stable protein. The early applications of electron microscopy of tilted, unstained samples to identify the seven trans-membrane helices of bacteriorhodopsin in its native purple membrane environment were crucial first steps in understanding the architecture of membrane proteins (Henderson & Unwin, 1975). As shown by a variety of experimental measurements, bacteriorhodopsin protein molecules are constrained in their highly ordered bilayer environment containing diphytanyl lipids. However, when the protein is purified and reconstituted into bilayers consisting primarily of phosphatidyl choline lipids, it undergoes rapid rotational diffusion about the bilayer normal at temperatures above that of the gel to liquid crystalline phase transition temperature of the lipids (Lewis et al., 1985). In a spectrum obtained at 3°C, well below the DMPC phase transition temperature, the chemical shift powder pattern from the carbonyl carbons has the full width and asymmetry expected in a rigid lattice. In contrast, the spectrum of the same sample obtained at 30°C, above the phase transition temperature, is much narrower and axially symmetric as the result of the motional narrowing from the rotational diffusion about the bilayer normal. This shows that the majority of labeled carbonyl groups are aligned in the protein with the C-O bond almost parallel to the bilayer normal, which is consistent with the arrangement of helices in the static structures. Similar results are obtained for other highly helical membrane proteins because the helices are aligned approximately parallel to the bilayer normal. Indeed, the 13C′ resonance line shape in an unoriented, stationary sample is often used as a ‘control’ experiment to ensure that the protein is undergoing rapid rotational diffusion about the bilayer normal under the experimental conditions for the structure determination.
Taken together, these early structural and dynamic studies of bacteriorhodopsin demonstrated that fast rotational diffusion of membrane proteins about the bilayer normal depends on lipid composition and temperature. Typically, the rate of rotational diffusion about the bilayer normal can be switched between frequencies that are “slow” or “fast” on the relevant NMR timescales defined by the frequency breadth of the static powder patterns by changing the temperature between values that are below or above, respectively, the gel to liquid crystalline phase transition temperature of the phospholipids. These experiments serve to map out the basic parameters of lipid and protein motions in artificial and biological membrane bilayers. Of particular importance to structure determination is the finding that under most conditions even quite large membrane proteins undergo fast rotational diffusion about the bilayer normal that is highly relevant to NMR spectroscopy (Park et al., 2011b).
1.8. Equivalence of rotational diffusion and sample alignment
Early studies indicated that the membrane was in a ‘fluid’ state, a term that was often used synonymously with liquid crystalline state (McConnell & Hubbell, 1971; McConnell et al.; Wien et al., 1972). Certainly some of the magnetic resonance experiments on synthetic and biological bilayers gave results compatible with large amplitude effectively isotropic motions because of the extensive line narrowing. However, as the research moved towards unperturbed bilayers, the asymmetric nature of phospholipid bilayers dominated the effects. As mentioned above, early critical experiments utilized rhodopsin, especially those of Cone (Cone, 1972), which measured the relaxation due to rotation about an axis normal to the plane of the disc membrane.
The rotational diffusion of membrane proteins merges oriented sample solid-state NMR and magic angle spinning solid-state NMR experimental methods enabling the complementary use of aligned and unoriented samples, and increasing the potential for synergy between isotropic and anisotropic nuclear spin interaction contributions to protein structure determination. This is feasible because the same rotationally averaged powder patterns observed in stationary unoriented samples can be measured in MAS solid-state NMR spectra of unoriented samples through the application of ‘slow spinning’ (Herzfeld & Berger, 1980; Wylie et al., 2007) or recoupling methods (Chan & Tycko, 2003; Wylie et al., 2006). Here we use the spectral parameters associated with the 15N and 13C chemical shifts and 1H-15N and 1H-13C dipolar couplings in specifically labeled backbone and side chain sites to demonstrate the equivalence of mechanical and magnetic uniaxial alignment of protein-containing bilayers and “rotational alignment” of the membrane proteins resulting from their rapid rotational diffusion about the bilayer normal in unoriented samples when the temperature is higher than that of gel to liquid crystalline transition for the phospholipids. Because the resonance frequencies are determined by the orientations of functional groups (chemical shift) or chemical bonds (dipole-dipole coupling) relative to a single axis defined by the direction of the magnetic field or by rotational diffusion about the bilayer normal, the protein structure is mapped onto the resulting spectra by the anisotropy of the spin interactions.
These measurements are well suited for membrane proteins that undergo rapid rotational diffusion in phospholipid bilayers. Also, these measurements are particularly forgiving with respect to experimental errors and uncertainties regarding the principal values and orientations of the tensors in the molecular frame because site-to-site errors do not accumulate. Each measurement is made independently relative to a common external axis, in this case the bilayer normal. In complementary approaches, the axis can be in the direction of the magnetic field, orthogonal to the field, or relative to the mechanical alignment on glass plates (Park et al., 2010a).
Uniaxial sample alignment, where the direction of the molecular alignment is parallel to that of the magnetic field, is the next most general type of sample after a single crystal. In this case, the sample has one direction of orientation and the other two directions are random or undefined. What is most notable is that while these samples are capable of yielding very high resolution spectra when they are aligned parallel to the field, the line shapes are two-dimensional powder patterns when they are tilted away from parallel (Opella & Waugh, 1977). This is seen most dramatically when the axis of alignment is perpendicular to the direction of the field. However, the situation is greatly simplified when the molecule of interest undergoes fast rotational diffusion about a single axis, in which case single line spectra are observed in all directions of alignment (Park et al., 2005).
Membrane proteins in phosphopholipid bilayers can be aligned mechanically between glass plates (Bechinger et al., 1991; Murray et al., 2013). As with uniaxially aligned polymers, when the axis of alignment is parallel to the field single line resonances are observed regardless of whether any molecular motion is present. Magnetically aligned samples of membrane proteins were originally sought as direct replacements for mechanically aligned samples (De Angelis et al., 2004) and once it was discovered that the lanthanide ions added to the lipids could “flip” the alignment so that the bilayer normals were parallel to the field (Prosser et al., 1996), then that was the case. However, the presence of detergents to form the ends of the bilayer discs and lanthanides is of concern for their possible effects on the protein structures of interest.
2. Examples of Membrane Proteins
A variety of solid-state NMR approaches to study of membrane proteins have been reviewed. The field is fortunate in having an up-to-date web site where the structures of membrane proteins determined by solid-state NMR (http://www.drorlist.com/nmr/SPNMR.html) are summarized. The structures of proteins obtained by heterologous expression and reconstituted in phospholipids are providing the foundation for the future of the field, since they incorporate the principal advantages of NMR. Taken from the DrorList in January 2014, the structures of domains and full-length membrane proteins determined by oriented sample solid-state NMR of stationary, aligned samples include the M2 channel-lining segment of the acetylcholine receptor (Opella et al., 1999), the Influenza M2 channel (Sharma et al., 2010), the membrane-bound form of the coat protein from filamentous bacteriophage fd (Marassi & Opella, 2003), the transmembrane channel-forming domain of Vpu from HIV-1 (Park et al., 2003), and the truncated form of the mercury transport membrane protein MerFt (De Angelis et al., 2006). By combining OS solid-state NMR data with solution NMR data obtained on the same proteins in micelles, there are additional examples of membrane proteins which have had their structures determined, including stannin (Buck-Koehntop et al., 2005), phospholamban (Traaseth et al., 2009; Verardi et al., 2011), the membrane-bound form of the coat protein from filamentous bacteriophage Pf1 (Park et al., 2010b), and cytochrome b5 (Ahuja et al., 2013). Of greatest relevance to the topic of deriving structure from motion in this article are the membrane protein structures determined by rotationally aligned solid-state NMR, which include the truncated form of the mercury transport membrane protein MerFt (Das et al., 2012), the full-length mercury transport membrane protein MerF (Lu et al., 2013), the viral membrane protein p7 from the human hepatitis C virus (unpublished results), and the 350-residue chemokine receptor CXCR1 (Park et al., 2012).
2.1 Filamentous bacteriophage coat proteins
Studies of filamentous bacteriophages have provided many insights into fundamental biological processes. They have played early and important roles in structural biology, in large part because the major coat protein is both a membrane bound protein and a structural protein of a DNA-protein complex at different stages of the lifecycle. Moreover, the high efficiency of their self-propagation in bacteria provides the capability of preparing milligram quantities for NMR experimental studies. Indeed, the major coat protein of both class I (fd, M13) and class II (Pf1) filamentous bacteriophages has been used in many aspects of the development and application of NMR to both membrane proteins and structural proteins of biological supramolecular assemblies.
Each bacteriophage particle is a long, thin filament, which consists of several thousand copies of the major coat protein wrapped around its single stranded DNA genome that is extended lengthwise within the particle. The particle is about 90% by weight major coat protein, therefore the vast majority of the signals in NMR spectra of uniformly labeled bacteriophage particles come from the structural form of the coat protein. Additionally, at high concentrations the intact bacteriophage particles have liquid crystalline properties and align in the presence of a strong magnetic field. Since the coat protein subunits are packed symmetrically with respect to the longitudinal axis of the phage particles, each site in the coat protein gives rise to a single resonance. The particles have masses of several million daltons, which immobilizes the coat proteins and DNA on the requisite NMR timescales, even though the samples are clear, pourable solutions. As a result, the structural form of the protein satisfies all of the sample requirements of oriented sample solid-state NMR, and with about fifty residues has proven to be an invaluable model protein for the development of the methodology and ultimately for structure determination of structural (Zeri et al., 2003) and membrane proteins (Marassi & Opella, 2003) in supramolecular complexes (Opella et al., 2008). An important benefit of using oriented sample solid-state NMR is that the structural studies can be carried out directly on intact, infectious phage particles, ensuring that the information is biologically relevant.
The major coat protein of M13 bacteriophage, which is widely used in molecular biology laboratory procedures, differs from that of fd in only a single amino acid, and they can be thought of as mutants of each other. Early structural and dynamics studies of the membrane bound forms of the coat proteins were performed in detergent micelles, and included experiments that utilized 1H NMR, 13C NMR, and 19F NMR (Henry & Sykes, 1990) (Cross & Opella, 1985; McDonnell et al., 1993; Opella et al., 1980). The first high-resolution structure of the membrane-bound form of the fd bacteriophage phage coat protein was determined in SDS detergent micelles by solution NMR (Almeida & Opella, 1997) and subsequently, the membrane-bound form of the coat protein in POPC/POPG lipid bilayers (Marassi & Opella, 2003) was determined. The structure of the membrane bound form of the fd coat protein is shown in Figure 1.
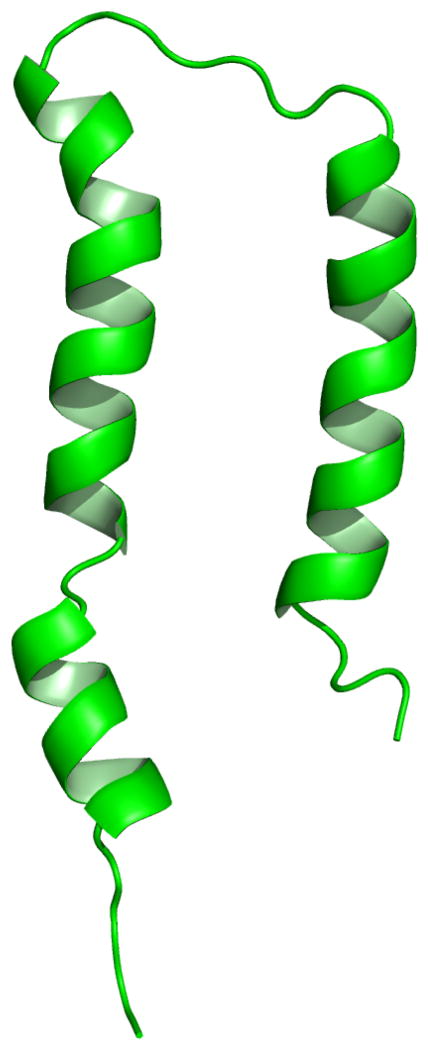
Figure 1.
Three views of the structure of the membrane-bound form of fd coat protein in POPC/POPG phospholipid bilayers. The amphipathic in-plane helix is in magenta, the hydrophobic trans-membrane helix is in blue, and the short connecting turn is in yellow. The flexible N- and C- terminal residues are not shown. A. Side view of TM helix, the Trp and Lys sidechains are shown in blue. The dashed gray lines mark the lipid-water boundary. B. Front view and, C. View of in-plane helix looking down from the C-terminus. From Marassi & Opella (2003).
Pf1 infects Pseudomonas aeruginosa and is a class II filamentous bacteriophage. Studies of this protein have also been conducted in detergent micelles by solution NMR and in lipid bilayers by oriented sample solid-state NMR. Using oriented sample solid-state NMR, structures of Pf1 coat proteins have been solved in the intact bacteriophage (Thiriot et al., 2004) (Thiriot et al., 2005) and in lipid bilayers formed by q=3.2 14-O-PC/6-O-PC bicelles (Park et al., 2010b). Complementary studies have been performed using magic angle spinning solid-state NMR on Pf1 bacteriophage (Goldbourt et al., 2007). Similar to fd phage, the Pf1 coat protein in lipid bilayers was found to be composed of a transmembrane helix that has motion restrained by lipids and a water-soluble helix, which undergoes isotropic motion in water (Park et al., 2010b). Notably, the Pf1 assembly mechanism has been illustrated by combining the techniques of solid-state NMR and neutron diffraction (Nambudripad et al., 1991; Shon et al., 1991). During phage assembly, these two helices again elongate to a single long helix with only a few residues in the middle deviating from ideal positions (Thiriot et al., 2004). This is illustrated in Figure 2.
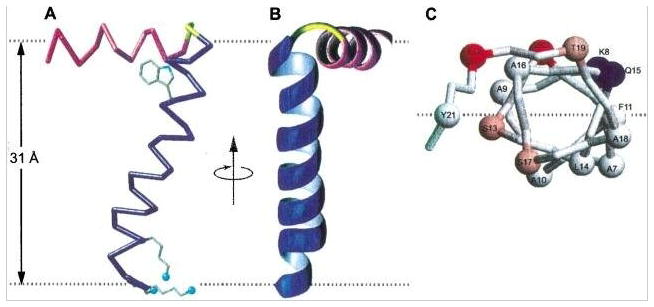
Figure 2.
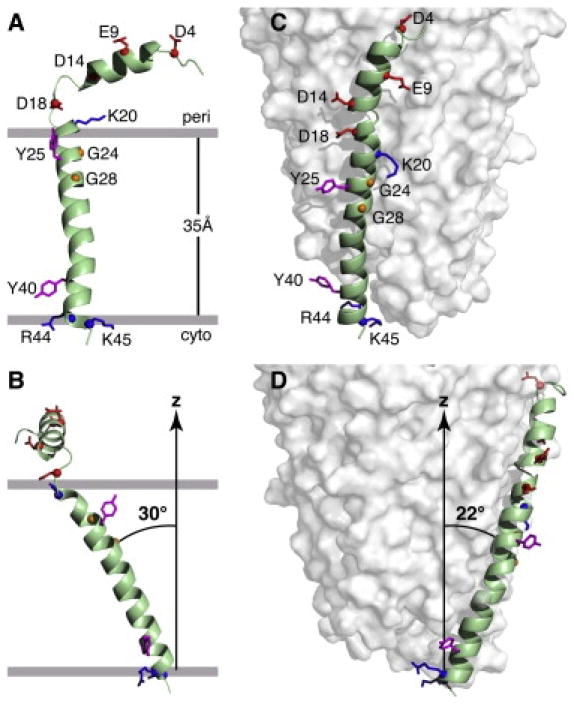
Structures of the Pf1 coat protein. A. and B. The membrane-bound form of the protein. C. and D. The structural form in the intact bacteriophage particle. In the membrane-bound form of the protein, the acidic N-terminus region is exposed to the bacterial periplasmic space (peri) and the basic C-terminal region is exposed to the cytoplasm (cyto). Acidic residues (Asp and Glu) are shown in red, conserved glycine residues in the transmembrane helix are in yellow, basic residues (Arg and Lys) are in blue, and interfacial tyrosines are in pink. Residues R44 and K45 face the cytoplasm in the membrane-bound form and the DNA on the interior of the bacteriophage particles. B. and D. Images obtained by 90o rotation of A and C around the z axis. From Park et al (2010).
M2 segment from nicotinic acetylcholine receptor (nAchR)
The nicotinic acetylcholine receptor (nAchR) resides in the membrane and plays key roles in the transmission of neurological signals. Each monomer of the protein has four transmembrane helices, the second of which, M2, lines the ion pore formed by five of the monomers. Much of the understanding of the nicotinic acetylcholine receptor arises from functional studies and analysis of the ion pore formed by oligomerization of the M2 helical segments. M2 of nAchR was the first mammalian polypeptide domain expressed and labeled in bacteria for structure determination by NMR spectroscopy (Opella et al., 1999). Since this work was performed at an early stage of the development of the field, comparisons were made between the findings in detergent micelles by solution NMR and those in oriented phospholipid bilayers on glass plates by solid-state NMR. In addition, the properties of M2 segments of the nicotinic acetylcholine receptor were compared to those of the N-methyl-D-aspartate (NMDA) receptor. Because of sample preparation difficulties this involved the comparison of synthetic polypeptides to expressed polypeptides, and the spectral results were typical in that much better data could be obtained from the expressed samples than the synthetic samples. This is generally the case and essentially all current studies are performed on heterologously expressed polypeptides in order to avoid the potential problems resulting from incompletely purified synthetic hydrophobic peptides.
A major result of the oriented sample solid-state NMR studies was the determination of the helix tilt angle with respect to the bilayer normal, since this helped to characterize the ion pore in models of the intact receptor (Montal & Opella, 2002; Opella et al., 1999). A pentameric model of the M2 segments showing the central pore was built with the high-resolution NMR structures and the 9Å electron crystallography structure (Unwin, 1995).
Subsequent studies on the nicotinic acetylcholine receptor using solution NMR have been performed; for example, the structures of four-transmembrane-helical subunits have been characterized in organic solvent (Bondarenko et al., 2010) and in lauryldimethylamine-oxide (LDAO) micelles (Bondarenko et al., 2012). However, because of the use of the organic solvent and detergent micelles it is difficult to place these results in the structural context of the intact receptor in phospholipid bilayers.
2.3 Virus protein “u” (Vpu) from HIV-1
Vpu is an 81-residue membrane protein containing a single N-terminal transmembrane helix and a C-terminal cytoplasmic domain containing amphipathic helices. It is found exclusively in HIV-1 (human immunodeficiency virus 1) and has multiple functions, which include facilitating the release of viral particles from infected cells as well as the degradation of CD4 in the host cells (Chen et al., 1993; Gonzalez & Carrasco, 2003; Schubert et al., 1996). Full-length and multiple truncated constructs were created for some of the early studies, enabling their topology and helical orientation to be defined by solid-state NMR (Marassi et al., 1999). Research efforts were devoted to the search for the best candidates for structural characterization, which is exemplified by the expression and functional studies aimed towards defining the minimal folding units (Ma et al., 2002). A well-behaved construct was found to include only the transmembrane helix (residues 2 to 30) and 6 additional residues for optimal expression, purification, and stability (named “Vpu2-30+”). A high-resolution structure of the construct was obtained with oriented sample solid-state NMR (Park et al., 2003). Importantly, the ion-channel activity is retained in the truncated construct. The structural characterization of the transmembrane domain led to many subsequent applications: the structures of Vpu in magnetically aligned bicelles (Park et al., 2006), the effect of the A18H mutation to its conformation (Park & Opella, 2007) (Wang et al., 2013b), and the interaction with the BST-2 protein which is an important step in the molecular mechanism of viral infection (Skasko et al., 2012). MAS solid-state NMR studies by Tycko and co-workers also started with a similar transmembrane helix construct (Sharpe et al., 2005), and the study led to the conclusion that there are mixed oligomerization states of the protein in lipid bilayers (Lu et al., 2010).
The next step in the structural studies was to characterize the full-length protein, which would include the functionally important cytoplasmic domain and be free of structural perturbation from protein truncation. Initial studies (Ma et al., 2002; Marassi et al., 1999) were hindered by the difficulties in obtaining high-resolution spectra, which could arise from the complex behavior of the amphipathic helices in various membrane environments. Solution NMR studies of the truncated cytoplasmic domain were carried out with the protein solubilized in water (Willbold et al., 1997). Willbold and co-workers obtained NOE distance restraints of the cytoplasmic domain in DPC micelles and used paramagnetic relaxation enhancement (PRE) to characterize the protein topology (Wittlich et al., 2009). The resulting structure showed a mixture of well-defined and highly dynamic regions, which reveals the complicated dynamics of the protein. The effort towards characterizing the full-length Vpu in the native-like bilayer environment can be seen in a recent structural study with MAS solid-state NMR of both Vpu and its interaction partner, CD4, in POPC lipid bilayers (Do Hoa et al., 2013).
2.4 Phospholamban
Phospholamban is a small 52-residue membrane protein. It has a C-terminal transmembrane helix with its N-terminal portion exposed to the aqueous environment. Much of the research effort has been devoted to characterizing the structure and dynamics of the N-terminal amphipathic portion of the protein. Phospholamban exists as a homopentamer and dissociates into monomers that bind Ca2+-ATPase (SERCA) in a 1:1 molar ratio.
The NMR studies of phospholamban followed the trend of starting with protein samples in organic solvents (Lamberth et al., 2000; Pollesello & Annila, 2002) and then studying them in detergent micelles and phospholipid bilayers. The structures determined in organic solvents successfully defined the transmembrane helix structure but also displayed features that reflected the non-native environment, for example, that the amphipathic helix is not oriented properly. These features were addressed in subsequent structures of the monomeric (Zamoon et al., 2003) and pentameric states (Oxenoid & Chou, 2005) determined by solution NMR of the polypeptide in DPC micelles. In both cases, part of the N-terminal cytoplasmic region was not well defined due to the local dynamics of the protein. In the monomeric structure, the flexible linker between two cytoplasmic domains made it difficult to determine their relative positions whereas in the pentameric structure, the N-terminal helix was oriented at an unusual angle with respect to the bilayer plane. Besides the technical difficulty of defining helix orientation using solution NMR, these structures are likely to be influenced by the detergent micelle environment and deviate from the structures in the native lipid bilayer (Zhou & Cross, 2013).
Features of both structures were subsequently addressed using solid-state NMR. The monomeric structure was calculated using a hybrid technique, incorporating restraints measured in DPC micelles by solution NMR and in DOPC bilayers by solid-state NMR, followed by molecular dynamics simulations in explicit DOPC bilayers (Traaseth et al., 2009). Here, as well as in later studies (Traaseth & Veglia, 2010), DPC micelles were shown to alter the conformational equilibrium and change the helicity of the cytoplasmic domains, but the transmembrane helix was well conserved in both environments. The pentamer structure determined later also provided well-defined N-terminal domains (Verardi et al., 2011), although it showed a different orientation of the transmembrane helix. Compared to the “bellflower” architecture, the alternative “pinwheel” architecture supports the role of the pentameric state for the regulatory phospholamban protein. Only the “pinwheel” architecture agrees with the spectrum acquired in planar lipid bilayers by solid-state NMR (Verardi et al., 2011), which suggests that the change of transmembrane helix orientation may be associated with exposing the protein to detergents.
In a recent study, the monomeric phospholamban was reconstituted in the native sarcoplasmic reticulum membrane (Gustavsson et al., 2012). The study revealed two structural states, which may explain the contradictory structures observed in detergent micelles and lipid bilayers. The structure of the phosphorylated pentameric form of the protein was also characterized (Vostrikov et al., 2013), and the structure shows the preserved pentamer conformation and an increase in the dynamics of the cytoplasmic region that agrees with previous observations (Oxenoid et al., 2007).
Another direction of research is towards the protein complex structure between the regulatory phospholamban and SERCA. The binding interface between the two proteins was mapped in lipid bilayers and interestingly, the bound form structure of phospholamban was found to be a minor conformation of the protein (Gustavsson et al., 2013).
2.5 Membrane-bound cytochrome b5
Cytochrome P450s are a widely distributed family of membrane proteins that carry out many biological functions, which are modulated by another membrane protein, cytochrome b5. Ramamoorthy and coworkers have used these proteins in the development of new methodologies and to describe their structures and inter-molecular interactions in membrane environments, micelles, and bicelles (Ahuja et al., 2013; Pandey & Ramamoorthy, 2013; Soong et al., 2010; Xu et al., 2010; Yamamoto et al., 2013a; Yamamoto et al., 2013b).
Cytochrome P450 and cytochrome b5 are both membrane anchored proteins with a single trans-membrane helix. Because of their physiological importance, they are both well studied in truncated forms with the trans-membrane helix removed. However, this is a non-physiological state with only partial activities. These two proteins, along with the complex that they form, exemplify the obstacles encountered when trying to work with unmodified membrane proteins that interfere with structure determination by both conventional X-ray crystallography and solution NMR techniques.
The structure of full-length ferric cytochrome b5 was determined using a combination of solution NMR on detergent micelle samples and oriented sample solid-state NMR on magnetically aligned bicelle samples. The protein consists of a globular heme-containing domain, whose structure could be determined by solution NMR methods because it is connected by a 15-residue highly flexible linker to the anchoring trans-membrane helix, whose study required the use of solid-state NMR (Soong et al., 2010; Yamamoto et al., 2013b) because it is immobilized on solution NMR timescales when the protein interacts with micelles. The structure of the full-length protein based on data from these two types of NMR experiments is shown in Figure 6.
Figure 6.
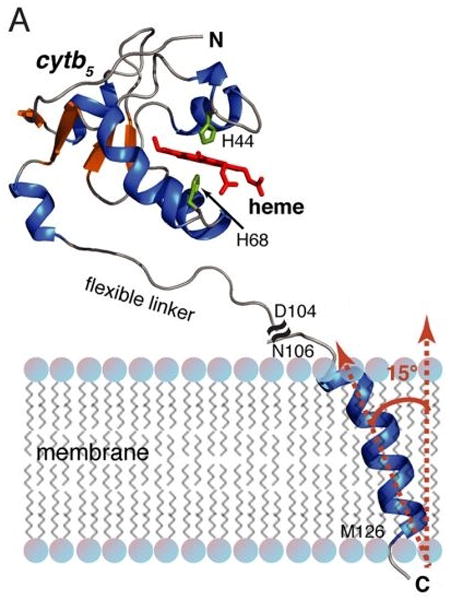
NMR structure of rabbit microsomal cytb5. NMR structure of full-length cytb5 obtained from a combined solution and solid-state NMR approach. The soluble heme domain structure (residues 1–104) of full-length cytb5 was solved in DPC micelles by solution NMR. The transmembrane domain structure (residues 106–126) of full-length cytb5 was determined in aligned DMPC/DHPC bicelles using solid-state NMR spectroscopy. From Ahuja et al (2013).
Using a combination of chemical shift changes observed upon binding, (Pandey & Ramamoorthy, 2013), site-directed mutagenesis, and docking calculations, it was possible to develop a structural model of the interface of the cytochrome b5 – cytochrome P450 complex. In membrane proteins, dynamics are fundamental to the structure and activity of individual polypeptides as well as their interactions with binding partners, and the NMR studies of the cytochrome b5 – cytochrome P450 complex revealed an ensemble of low energy orientations of the two proteins (Ahuja et al., 2013; Yamamoto et al., 2013a).
2.6 M2 channel from Influenza A
The tetramer of the M2 protein from influenza A forms a proton ion channel that promotes the acidification of the virus interior, an important step in the virus life cycle. The protein has been the subject of intensive research for more than a decade by X-ray crystallography, solution NMR, and solid-state NMR. The well-characterized channel blocking drugs amantadine and rimantadine additionally aid in the structural studies of this protein.
As the opening of the M2 channel is triggered at pHs below 6, a process important for the Influenza lifecycle inside the endosome of lung epithelial cells, an important aim of research has been observing the structural and dynamic differences of the M2 channel at neutral and low pH. Structural studies have used X-ray crystallography (Acharya et al., 2010), MAS solid-state NMR (Hu et al., 2010), and oriented sample solid-state NMR (Sharma et al., 2010) to propose that the His37-Trp41 cluster, referred to as the “HxxxW quartet”, guides protons through hydrogen bond formation and breaking (Hu et al., 2006; Sharma et al., 2010). This interpretation was built on computation and structure determination of the M2 channel in lipid bilayers (Sharma et al., 2010). The structure was first calculated as a monomer using the orientational restraints obtained from oriented sample solid-state NMR and a preliminary tetrameric structure was then assembled using PRE restraints and knowledge of the channel architecture. Restrained molecular dynamics simulations were performed on the M2 tetramer embedded in a lipid bilayer of 4:1 DOPC:DOPE, which is the same composition as was used in the oriented sample solid-state NMR sample. The last step was a quantum chemical calculation focusing on the side chains of the His37-Trp41 cluster.
The dimer-of-dimer architecture of the tetrameric M2 channel, which is the core in this proposed conductance mechanism, was further supported in subsequent studies involving a combination of OS solid-state NMR and MAS solid-state NMR on a similar M2 truncated construct and the full-length M2 (Andreas et al., 2012; Can et al., 2012; Miao et al., 2012).
Notably, an important current trend in the research direction is towards the full-length M2 protein, and this is a recurring theme in several other membrane protein studies reviewed here. Early M2 structures have all been done with various constructs of the transmembrane domain containing between 24 and 42 residues, such as (Hu et al., 2007; Sharma et al., 2010; Wang et al., 2001) (Cady et al., 2010; Stouffer et al., 2008) (Schnell & Chou, 2008). Recently, solid-state NMR studies have been carried out on full-length 97-residue M2 proteins reconstituted in DOPC/DOPE lipid bilayers and, more importantly, in native bacterial membrane bilayers (Miao et al., 2012). A second study of the full-length protein has also exploited various lipid bilayer environments, including DMPC and mixtures of POPC, POPG, cholesterol, and sphingomyelin (Liao et al., 2013).
2.7 Mercury transporter MerF
Mercury transporter membrane proteins constitute an essential part of the bacterial mercury detoxification system. These transporter proteins are capable of transporting Hg2+ or organic mercurial compounds into cells and allowing them to be detoxified into elementary mercury by the enzyme mercuric reductase. The structure and topology of mercury transporter proteins from various isolates shows that there are multiple versions of the protein containing two, three, and four transmembrane helices. The two transmembrane helical member of the family is MerF, which has been the target of the most extensive structural studies by NMR.
Studies of MerF began with the analysis of the protein by solution NMR (Howell et al., 2005). Among the isotropic bicelle and detergent micelle conditions, SDS micelles gave the best resolution and were chosen as the solubilizing environment for subsequent studies. A truncated form of the protein was also developed in the study, where the flexible ends of the protein were removed to minimize the complexity and facilitate calculation of the structure primarily from RDC restraints. As was done with previously mentioned proteins, this truncated version of the protein, named MerFt, was subsequently reconstituted into a more native-like lipid bilayer environment, and its structure was determined in magnetically aligned DMPC/DHPC bicelles (De Angelis et al., 2006) and DMPC proteoliposomes (Das et al., 2012). Notably, these two structural determination efforts were also important milestones in the development of solid-state NMR methodologies in that they represent the first multiple helix transmembrane protein determined by oriented sample solid-state NMR as well as the first structure determined using RA solid-state NMR.
The technical challenges of an oriented sample solid-state NMR study (De Angelis et al., 2006) have led to the implementation of new methods including the bicelle technology (De Angelis et al., 2004; De Angelis & Opella, 2007), the new SAMPI4 pulse sequence (Nevzorov & Opella, 2007), and a structural fitting algorithm (Nevzorov & Opella, 2003). These advances were extended to the study of full-length MerF by oriented sample solid-state NMR (Lu & Opella, 2014), where a new round of technology improvement was made including the new MSHOT-Pi4 pulse sequence (Lu et al., 2012), new assignment strategies (Knox et al., 2010; Lu et al., 2011; Nevzorov, 2008), and new structure calculation methods (Marassi et al., 2011; Tian et al., 2012). Also, it was in the structure determination of MerFt (Das et al., 2012) that the general pulse sequences and sample conditions for rotationally aligned solid-state NMR were established (Das et al., 2012; Marassi et al., 2011). In the subsequent extension to the structure determination of full-length MerF (Lu et al., 2013), methods for resonance assignments were further improved.
The full-length MerF structure determined in 14-O-PC lipid bilayers revealed a surprising structural rearrangement in one of the truncated ends, the N-terminal region (Lu et al., 2013). 14-O-PC is the ether linked alternative of the lipid DMPC, which has better stability (De Angelis & Opella, 2007). The first 12 residues that had previously shown a large degree of dynamics in SDS micelles turned out to be on the same time scale of dynamics as the transmembrane helices. In fact, these truncated residues form a continuing helix together with the subsequent 13 residues, which strongly suggests that the SDS detergent micelle environment completely alters the conformation of this domain of the protein. This is a classic example showing that the replacement of the native lipid bilayer environment by detergents can modify the structure and dynamics of membrane proteins.
2.8 p7 from Hepatitis C virus
p7 is one of the ten proteins encoded in the RNA genome of the human hepatitis C virus. Since p7 forms a small conductive channel in the membrane, it is categorized as a viroporin protein; however, p7 is involved in multiple protein-protein interactions in addition to forming the channel.
Structural studies of p7 were initially carried out in organic solvent, a recurring theme from previous sections. The first structure of p7 was obtained for a construct containing only the second transmembrane helix solubilized in a TFE/water mixture (Montserret et al., 2010) and the structure of full-length p7 was determined in methanol (Foster et al., 2013). More recently, structures of p7 from two different genotypes were determined in DPC micelles (OuYang et al., 2013) and in DHPC micelles (Cook et al., 2013). Interestingly, the structure of p7 in DPC micelles was determined to be a hexamer with an unusual architecture where part of each p7 protein crosses over to interact with two p7 proteins that are not its neighbors. In contrast, the structure in DHPC micelles is monomeric and shows the topology typical of two transmembrane helical proteins. However, a recent analysis on the hexamer p7 structure shows that the 18 charged side chains of arginine residues may be embedded in the hydrophobic membrane environment (Cross et al., 2013), which is a feature indicating structural distortions as a result of the detergent micelle environment. Notably, these arginine side chains are on the outside of the structure and with the consideration of the spherical shape and limited diameter of detergent micelles, these charged side chains may be exposed to aqueous environment. Additionally, the interaction of helices in the hexameric p7 structure resembles the phenomenon of “domain swapping” seen in the DgkA structure in detergent micelles (Van Horn et al., 2009), which has recently been compared to a crystal structure of the same protein (Li et al., 2013; Zheng & Jia, 2013; Zhou & Cross, 2013). Lastly, the packing of the hexamer structure has revealed several cavities embedded in the hydrophobic lipid environment that expose some of the hydrophilic backbone or side chain atoms.
In contrast, we have recently determined the three-dimensional structure of p7 in phospholipid bilayers by rotationally aligned solid-state NMR. The resulting structure shown in Figure 9 is of a typical membrane protein with two trans-membrane helices. Three is a break in the N-terminal helix, but it still pass through the bilayer. The C-terminal helix is shorter, and barely brings the carboxyl terminus to the surface of the bilayer. The inter-helical loop adopts a single unique fold on the timescales of the NMR experiments. It is likely that this structure unit interacts with itself to form oligomers, and with other protein to function in infected cells.
Figure 9.
Structure of p7 in phospholipid bilayers.
2.9 DsbB from E. coli
The disulfide bond formation protein B, DsbB, resides in the cytoplasmic membrane of E. coli. It has four transmembrane helices and functions to harvest the oxidizing power of membrane-associated quinones to reoxidize its interaction partner DsbA, which subsequently catalyzes the disulfide bond formation of proteins in the periplasmic space. Homologs of this protein found in several virulent bacteria are potential targets for therapeutics.
Structural studies of DsbB have been performed in detergent and phospholipid environments by X-ray crystallography, solution NMR, and MAS solid-state NMR. The crystal structures include the DsbA-DsbB-ubiquinone complex (Inaba et al., 2006) (Inaba et al., 2009), the charge-transfer intermediate of DsbB, (Malojčić et al., 2008), and the DsbB-Fab complex (Inaba et al., 2009). The detergent DPC has also been used in determining the solution NMR structure (Zhou et al., 2008). Subsequently, Bushweller and co-workers have extended the study of DsbB to nanodiscs, which provide a planar bilayer environment amenable to solution NMR studies (Früh et al., 2010). Rienstra and co-workers have utilized MAS solid-state NMR to study the structure of a DsbB-DsbA complex (Tang et al., 2011a) and features of the DsbB protein in endogenous lipid (Tang et al., 2013b).
The improvement of the structure of the DsbB protein in endogenous lipid (Tang et al., 2013b) over the previous crystal structure or solution NMR structure is of particular interest. Notably, the mutant form (C41S) of DsbB represents a transient intermediate. The NMR structure has defined the loop region between TM3 and TM4, which had been missing in the previous crystal structure due to the intrinsic dynamics of this region of the protein. The new structure (Tang et al., 2013b) was calculated by using dihedral angle restraints and ambiguous distance restraints from solid-state NMR and the X-ray data from the DsbB(Cys41Ser)-Fab complex. MAS solid-state NMR restraints have improved backbone atom precision from 2.36 Å in the crystal structure to 1.35 Å. The improvement is mainly at the loops and turns where the crystal structure is weakly defined. Additionally, the crystal structure was obtained with the Fab antibody fragment attached, while the solid-state NMR structure was obtained with the unmodified protein in native lipid. This comparison of structures is an excellent example of the general trend observed in transitioning from modified to native, full-length protein.
DsbB has also been an important model membrane protein for MAS solid-state NMR methodology development. An important characterization of the temperature effects on the spectral quality of membrane proteins in lipid bilayers, especially in the presence of endogenous lipids, has also been made using a DsbB sample (De Angelis & Opella, 2007; Tang et al., 2011b). Notably, this study aimed to capture the highest-resolution spectra of a membrane protein, which often occurs when the lipids are in the cooler gel phase. Interestingly, the study using DsbB concluded that the best-resolution spectra are acquired slightly below the phase transition temperature. Additionally, three- and four-dimensional experiments for MAS solid-state NMR, newly developed for resonance assignments, have been tested using DsbB (Li et al., 2008). The lipid insertion depth of DsbB has been studied by proton spin diffusion experiments using water and the lipid acyl chain (Tang et al., 2013b), and these experiments have been built upon their earlier applications to the potassium channel KS (Ader et al., 2008). The structural calculation method using joint refinement with solid-state NMR and X-ray crystallography data has also been applied to DsbB studies for the first time (Tang et al., 2011a).
2.10 Sensory Rhodopsin
Sensory rhodopsin II (SRII) is a seven transmembrane helical, retinal containing protein responsible for relaying information to the cell about the intensity and color of light in the environment (Etzkorn et al., 2007; Spudich & Luecke, 2002). Progress towards structure determination of Nastronomonas pharaonis sensory rhodopsin II (pSRII) was initially made using solid state NMR performed on 13C/15N labeled pSRII reconstituted into proteoliposomes (purple membrane lipids) in order to obtain resonance assignments for approximately 40% of the protein (Etzkorn et al., 2007). Soon after, backbone assignments of pSRII in DHPC were reported using solution state NMR methods with more than 98% of the backbone resonances being assigned (Gautier et al., 2008). Ultimately, Nietlispach and co-workers reported the complete structure of pSRII in DHPC micelles using solution state NMR (Gautier et al., 2010). The Ladizhansky group has characterized the Anabaena sensory rhodopsin using solid-state NMR techniques. Their studies have included comparison of the conformation in a lipid environment versus a crystalline environment (Shi et al., 2011), and structure determination (Wang et al., 2013a) which revealed a seven transmembrane helical trimer conformation in the lipid environment as shown in Figure 11.
Figure 11.
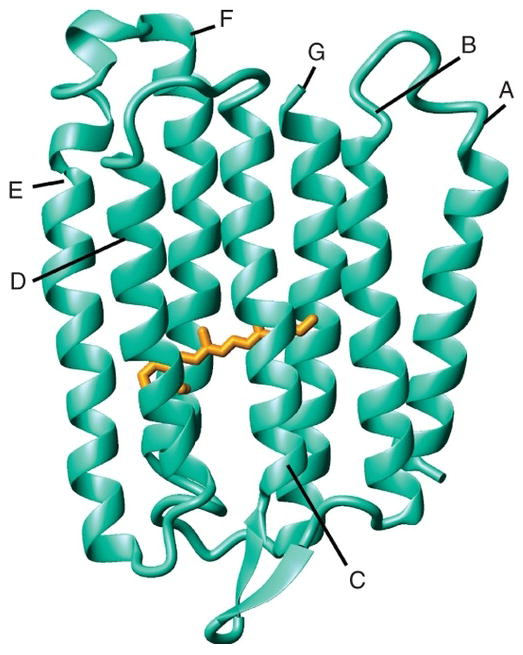
Solid-state NMR structure of Anabaena sensory rhodospsin. Side view of monomeric protein with retinal shown in orange. From Wang et al (2013a).
2.11 CXCR1
G-Protein Coupled Receptors (GPCRs) are a class of integral membrane proteins composed of seven transmembrane (TM) helical domains that are involved in signaling for a number of essential biological processes and functioning as drug receptors. As such, their structural and functional studies are of the essence and, to date, approximately 40% of drugs target GPCRs.
The complete structure of the GPCR CXCR1 has been determined using solid state NMR spectroscopy in phospholipid bilayers without any modification of its sequence (Park et al., 2012). Using rotationally aligned solid-state NMR, it was determined that the structure of CXCR1 has a three dimensional fold characteristic of a GPCR, having seven transmembrane alpha helices connected by three intracellular (ICL1-ICL3) and three extracellular (ELC1-ECL3) loops (Park et al., 2012).
Studies of CXCR1 also involved assessment of the dynamics and assignment of the mobile N- and C- termini using a combination of solution and solid state NMR approaches in q=0.1 DMPC/DHPC isotropic bicelles (Park et al., 2011b) as well as looking at the interactions between the receptor and its ligand, interleukin-8 (IL8) (Park et al., 2011b) which further confirmed previous studies that implied the N-terminal region as being the key site of interaction between ligand and receptor (Park et al., 2011a; Rajarathnam et al., 1995; Skelton et al., 1999).
3. Future Perspectives
The progress in the development of instrumentation, experimental methods, and calculation methods is sufficiently advanced to determine the backbone structures of a number of membrane proteins. Most importantly, initial studies of side chains and of complexes have been reported. This indicates that very high resolution and biologically relevant studies will be feasible in the near future. The ability to determine the structures of unmodified membrane proteins in liquid crystalline phospholipid bilayers under physiological conditions of temperature and pH is an enormous advantage over competitive methods. The field of NMR studies of membrane proteins in their native environment is poised for rapid advances.
Figure 3.
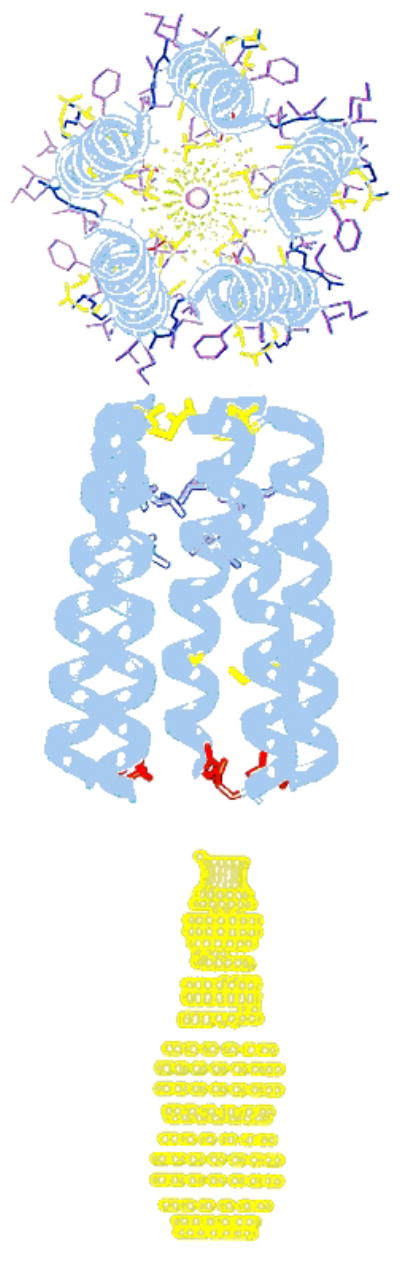
Top and side views of a model of the AChM2 funnel-like pentameric bundle. The channel architecture was calculated using the three-dimensional coordinates of the M2 helix in the lipid bilayer based on restraints from solid-state NMR experiments, and by imposing a symmetric pentameric organization. The top view has the C-terminal synaptic side in front. The wide mouth of the funnel is on the N-terminal, intracellular side of the pore. Middle view shows the side chains from the pore lining residues of Glu1, Ser8, Val15, Leu18, and Gln22. Bottom view contour depicts the pore profile. From Opella et al (1999).
Figure 4.
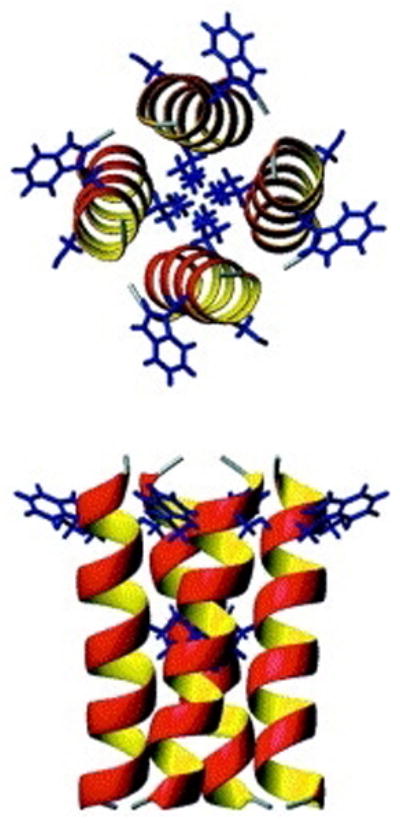
Structural models of the trans-membrane segment of Vpu in a tetramer. The structure was determined by OS solid-state NMR in aligned, stationary phospholipid bilayers. Views of the minimum energy configuration of the tetrameric model; view from above (top) and side view (bottom) with Trp22 protruding out towards lipid head groups. From Park et al, (2003).
Figure 5.
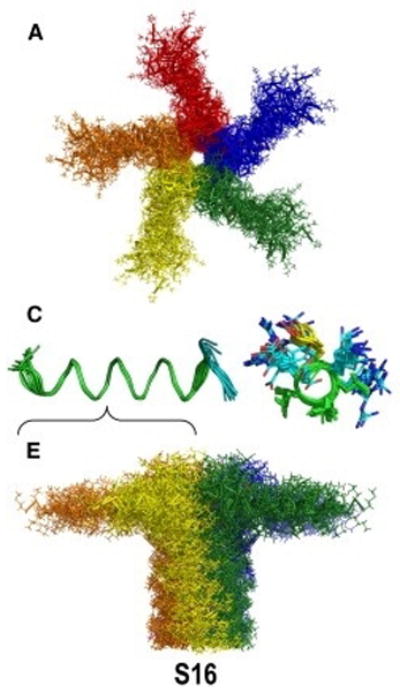
Ensembles of NMR structures of the nonphosphorylated form of phospholamban as pentamer with its amphipathic helix in the plane of the membrane bilayer. Top: View along membrane normal, Middle: view of residues 1-17, and Bottom: view perpendicular to membrane normal. From Vostrikov et al, (2013).
Figure 7.
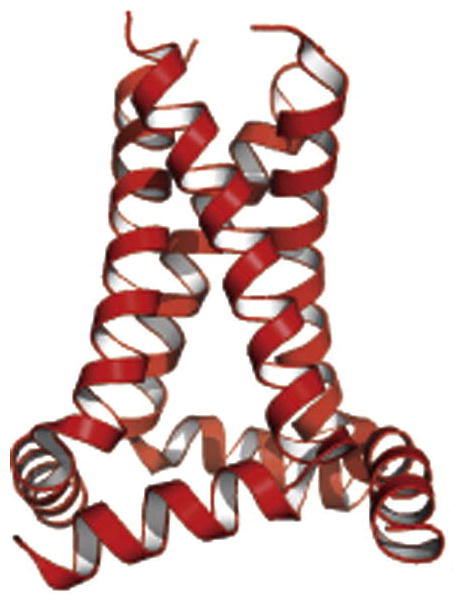
Structure of M2 tetrameter determined by OS solid-state NMR in phospholipid bilayers. Ribbon diagram of the amphipathic and transmembrane helices. From Sharma et al (2010).
Figure 8.
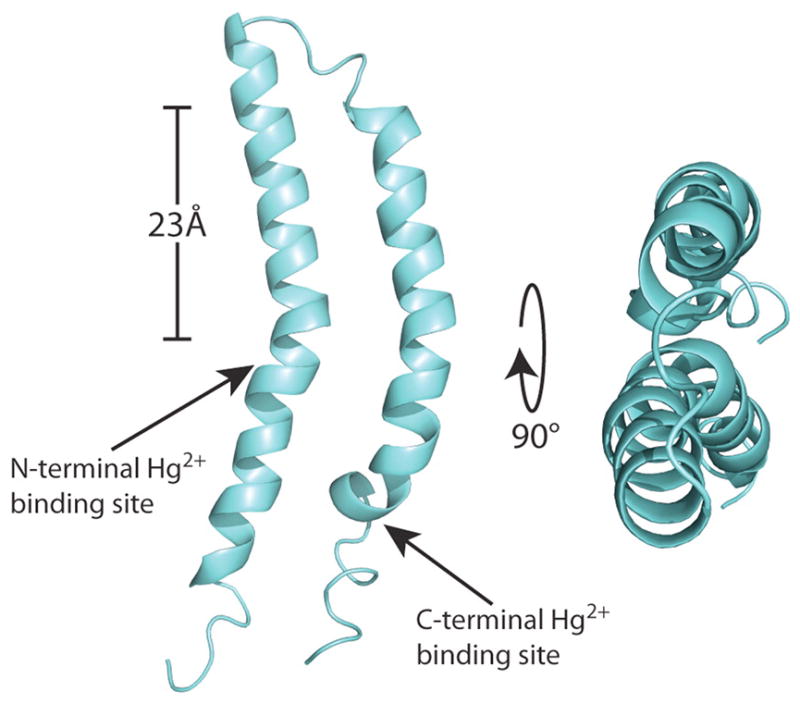
Structure of MerF in phospholipid bilayers determined in 14-O-PC lipid bilayers using solid state NMR. From Lu et al (2013).
Figure 10.
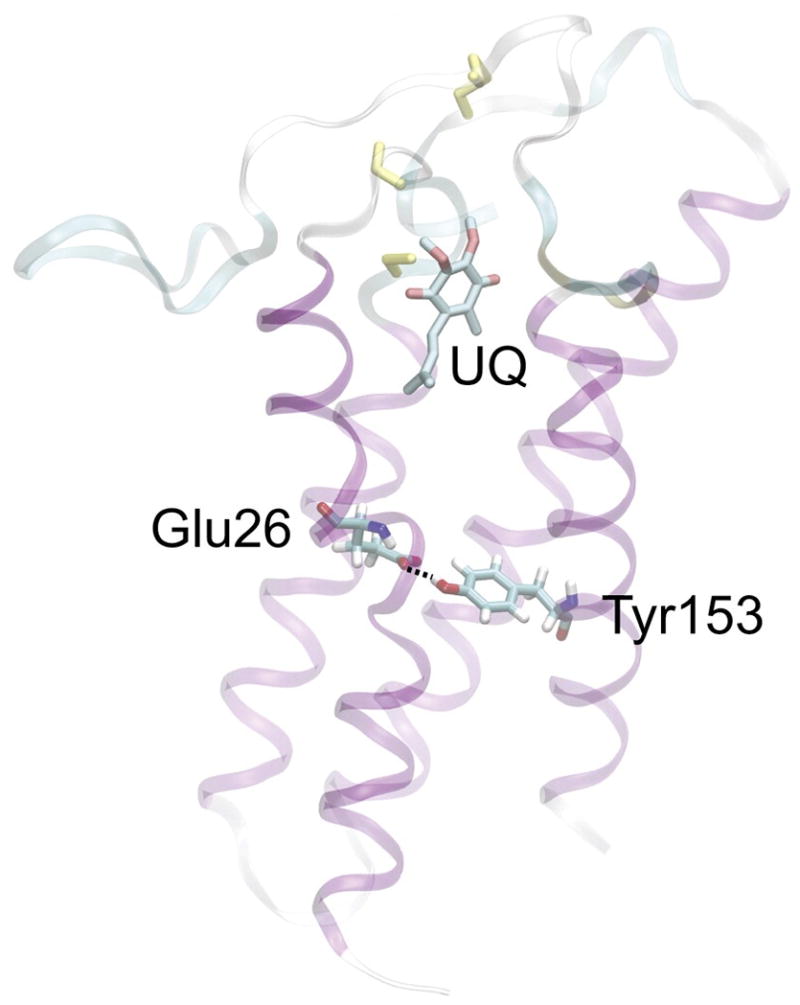
Structure of DsbB determined using a combinatorial approach employing x-ray crystallography, solid state NMR, and MD simulations. The UQ cofactor and bond between Tyr153 and Glu26 are shown. From Tang et al (2013).
Figure 12.
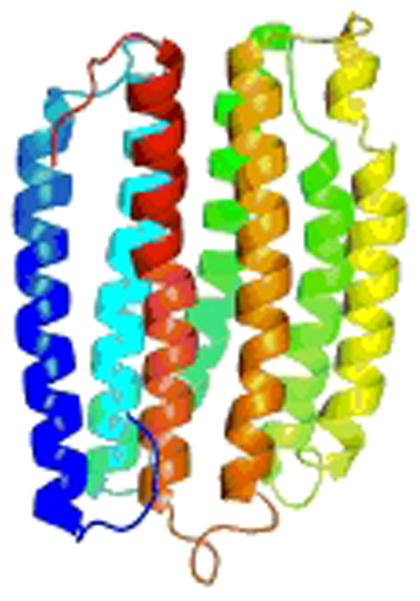
NMR structure of the GPCR CXCR1 in phospholipid bilayers determined using RA solid state NMR. From Park et al (2012).
Acknowledgments
The research described in this review that was performed at the University of California, San Diego was supported by Grants R01GM066978, RO1EB005161, and R01GM099986 from the National Institutes of Health. It utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at the University of California, San Diego supported by P41EB002031.
Abbreviations Used
- 6-O-PC
1,2-di-O-hexyl-sn-glycero-3-phosphocholine
- 14-O-PC
1,2-di-O-tetradecyl-sn-glycero-3-phosphocholine
- C8E8
n-octyltetraoxyethylene
- Cymal-5
5-Cyclohexyl-1-Pentyl-β-D-Maltoside
- CS-Rosetta
Chemical Shift - Rosetta
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPG
1,2-dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphatidylcholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DPC
dodecylphosphatidylcholine
- HMQC
Heteronuclear Multiple Quantum Correlation
- HSQC
Heteronuclear Single Quantum Correlation
- LDAO
n-Dodecyl-N,N-Dimethylamine-N-Oxide
- MAS
Magic angle spinning
- MD
Molecular Dynamics
- MTSL
1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate
- NMR
Nuclear Magnetic Resonance
- NOE
Nuclear Overhauser Effect
- NOESY
Nuclear Overhauser Effect Spectroscopy
- OG
octyl-β-D-glucopyranoside
- PAGE
Polyacrylamide gel electrophoresis
- PDB
Protein data bank
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- PRE
Paramagnetic Relaxation Enhancement
- RA
rotationally aligned
- REDOR
rotational-echo double-resonance
- RDC
Residual dipolar coupling
- RMSD
Root mean square deviation
- SDS
Sodium dodecyl sulfate
- ssNMR
Solid state Nuclear Magnetic Resonance
- TOCSY
TOtal Correlation SpectroscopY
- TRACT
TROSY for Rotational Correlation Times
- TROSY
Transverse relaxation optimized spectroscopy
References
- ACHARYA R, CARNEVALE V, FIORIN G, LEVINE BG, POLISHCHUK AL, BALANNIK V, SAMISH I, LAMB RA, PINTO LH, DEGRADO WF, KLEIN ML. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proceedings of the National Academy of Sciences. 2010;107(34):15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ADER C, SCHNEIDER R, SEIDEL K, ETZKORN M, BECKER S, BALDUS M. Structural Rearrangements of Membrane Proteins Probed by Water-Edited Solid-State NMR Spectroscopy. Journal of the American Chemical Society. 2008;131(1):170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- AHUJA S, JAHR N, IM SC, VIVEKANANDAN S, POPOVYCH N, LE CLAIR SV, HUANG R, SOONG R, XU J, YAMAMOTO K, NANGA RP, BRIDGES A, WASKELL L, RAMAMOORTHY A. A Model of the Membrane-bound Cytochrome b5-Cytochrome P450 Complex from NMR and Mutagenesis Data. Journal of Biological Chemistry. 2013;288(30):22080–22095. doi: 10.1074/jbc.M112.448225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMEIDA FC, OPELLA SJ. fd coat protein structure in membrane environments: structural dynamics of the loop between the hydrophobic trans-membrane helix and the amphipathic in-plane helix. J Mol Biol. 1997;270(3):481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- ANDREAS LB, EDDY MT, CHOU JJ, GRIFFIN RG. MAS NMR of the Drug Resistant S31N M2 Proton Transporter from Influenza A. Journal of the American Chemical Society. 2012 doi: 10.1021/ja3003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANFINSEN CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- BANERJEE R, DATTA A. Proteoliposome as the model for the study of membrane-bound enzymes and transport proteins. Mol Cell Biochem. 1983;50:3–15. doi: 10.1007/BF00225276. [DOI] [PubMed] [Google Scholar]
- BAUER DR, OPELLA SJ, NELSON DJ, PECORA R. Depolarized light scattering and carbon nuclear resonance measurements of the isotropic rotational correlation time of muscle calcium binding protein. J Am Chem Soc. 1975;97:2580–2582. doi: 10.1021/ja00842a067. [DOI] [PubMed] [Google Scholar]
- BECHINGER B, KIM Y, CHIRLIAN LE, GESELL J, NEUMANN JM, MONTAL M, TOMICH J, ZASLOFF M, OPELLA SJ. Orientations of amphipathic helical peptides in membrane bilayers determined by solid-state NMR spectroscopy. J Biomol NMR. 1991;1(2):167–173. doi: 10.1007/BF01877228. [DOI] [PubMed] [Google Scholar]
- BLUME A, WITTEBORT RJ, DAS GUPTA SK, GRIFFIN RG. Phase equilibria, molecular conformation, and dynamics in phosphatidylcholine/phosphatidylethanolamine bilayers. Biochemistry. 1982;21(24):6243–6253. doi: 10.1021/bi00267a032. [DOI] [PubMed] [Google Scholar]
- BOGUSKY MJ, SCHIKSNIS RA, LEO GC, OPELLA SJ. Protein backbone dynamics by solid state and solution 15N NMR spectroscopy. J Magn Reson. 1987;72:186–190. [Google Scholar]
- BONDARENKO V, MOWREY D, TILLMAN T, CUI T, LIU LT, XU Y, TANG P. NMR structures of the transmembrane domains of the α4β2 nAChR. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818(5):1261–1268. doi: 10.1016/j.bbamem.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONDARENKO V, TILLMAN T, XU Y, TANG P. NMR structure of the transmembrane domain of the n-acetylcholine receptor β2 subunit. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2010;1798(8):1608–1614. doi: 10.1016/j.bbamem.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSCH C, BROWN LR, WUTHRICH K. Physicochemical characterization of glucagon-containing lipid micelles. Biochim Biophys Acta. 1980;603:298–312. doi: 10.1016/0005-2736(80)90376-4. [DOI] [PubMed] [Google Scholar]
- BUCK-KOEHNTOP BA, MASCIONI A, BUFFY JJ, VEGLIA G. Structure, Dynamics, and Membrane Topology of Stannin: A Mediator of Neuronal Cell Apoptosis Induced by Trimethyltin Chloride. Journal of Molecular Biology. 2005;354(3):652–665. doi: 10.1016/j.jmb.2005.09.038. [DOI] [PubMed] [Google Scholar]
- CADY SD, SCHMIDT-ROHR K, WANG J, SOTO CS, DEGRADO WF, HONG M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463(7281):689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAN TV, SHARMA M, HUNG I, GOR’KOV PL, BREY WW, CROSS TA. Magic Angle Spinning and Oriented Sample Solid-State NMR Structural Restraints Combine for Influenza A M2 Protein Functional Insights. Journal of the American Chemical Society. 2012;134(22):9022–9025. doi: 10.1021/ja3004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO J, BURKE JE, DENNIS EA. Using hydrogen/deuterium exchange mass spectrometry to define the specific interactions of the phospholipase A2 superfamily with lipid substrates, inhibitors, and membranes. The Journal of biological chemistry. 2013;288(3):1806–1813. doi: 10.1074/jbc.R112.421909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEGELSKI L. REDOR NMR for drug discovery. Bioorg Med Chem Lett. 2013;23:5767–5775. doi: 10.1016/j.bmcl.2013.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAN JCC, TYCKO R. Recoupling of chemical shift anisotropies in solid-state NMR under high-speed magic-angle spinning and in uniformly 13C-labeled systems. J Chem Phys. 2003;118:8378–8389. [Google Scholar]
- CHAN SI, FEIGENSON GW, SEITER CH. Nuclear relaxation studies of lecithin bilayers. Nature. 1971;231(5298):110–112. doi: 10.1038/231110a0. [DOI] [PubMed] [Google Scholar]
- CHAN SI, SEITER CH, FEIGENSON GW. Anisotropic and restricted molecular motion in lecithin bilayers. Biochem Biophys Res Commun. 1972;46:1488–1492. doi: 10.1016/0006-291x(72)90775-9. [DOI] [PubMed] [Google Scholar]
- CHAN SI, SHEETZ MP, SEITER CH, FEIGENSON GW, HSU MC, LAU A, YAU A. Nuclear magnetic resonance studies of the structure of model membrane systems: the effect of surface curvature. Annals of the New York Academy of Sciences. 1973;222:499–522. doi: 10.1111/j.1749-6632.1973.tb15283.x. [DOI] [PubMed] [Google Scholar]
- CHEN MY, MALDARELLI F, KARCZEWSKI MK, WILLEY RL, STREBEL K. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J Virol. 1993;67(7):3877–3884. doi: 10.1128/jvi.67.7.3877-3884.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHERRY RJ. Measurement of protein rotational diffusion in membranes by flash photolysis. Methods Enzymol. 1978;54:47–61. doi: 10.1016/s0076-6879(78)54007-x. [DOI] [PubMed] [Google Scholar]
- COMELLAS G, RIENSTRA CM. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu Rev Biophys. 2013;42:515–536. doi: 10.1146/annurev-biophys-083012-130356. [DOI] [PubMed] [Google Scholar]
- CONE RA. Rotational diffusion of rhodopsin in the visual receptor membrane. Nature New Biology. 1972;236:39–43. doi: 10.1038/newbio236039a0. [DOI] [PubMed] [Google Scholar]
- COOK GA, DAWSON LA, TIAN Y, OPELLA SJ. Three-dimensional structure and interaction studies of Hepatitis C Virus p7 in 1,2-dihexanoyl-sn-glycero-3-phospholcholine by solution nuclear magnetic resonance. Biochemistry. 2013;52(31):5295–5303. doi: 10.1021/bi4006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROSS T, MURRAY D, WATTS A. Helical membrane protein conformations and their environment. European Biophysics Journal. 2013:1–25. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROSS TA, OPELLA SJ. NMR of fd coat protein. J Supramol Struct. 1979;11(2):139–145. doi: 10.1002/jss.400110204. [DOI] [PubMed] [Google Scholar]
- CROSS TA, OPELLA SJ. Structural properties of fd coat protein in sodium dodecyl sulfate micelles. Biochem Biophys Res Commun. 1980;92:478–484. doi: 10.1016/0006-291x(80)90358-7. [DOI] [PubMed] [Google Scholar]
- CROSS TA, OPELLA SJ. Protein structure by solid state nuclear magnetic resonance. Residues 40 to 45 of bacteriophage fd coat protein. Journal of Molecular Biology. 1985;182(3):367–381. doi: 10.1016/0022-2836(85)90197-4. [DOI] [PubMed] [Google Scholar]
- DAS BB, NOTHNAGEL HJ, LU GJ, SON WS, TIAN Y, MARASSI FM, OPELLA SJ. Structure determination of a membrane protein in proteoliposomes. J Am Chem Soc. 2012;134:2047–2056. doi: 10.1021/ja209464f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE ANGELIS AA, HOWELL SC, NEVZOROV AA, OPELLA SJ. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128(37):12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE ANGELIS AA, NEVZOROV AA, PARK SH, HOWELL SC, MRSE AA, OPELLA SJ. High-Resolution NMR Spectroscopy of Membrane Proteins in Aligned Bicelles. Journal of the American Chemical Society. 2004;126(47):15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- DE ANGELIS AA, OPELLA SJ. Bicelle samples for solid-state NMR of membrane proteins. Nature Protocols. 2007;2(10):2332–2338. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- DEVAUX P, MCCONNELL HM. Equality of the rates of lateral diffusion of phosphatidylethanolamine and phosphatidyl choline spin labels in rabbit sarcoplasmic reticulum. Ann NY Acad Sci. 1973;222:489–498. doi: 10.1111/j.1749-6632.1973.tb15282.x. [DOI] [PubMed] [Google Scholar]
- DEVAUX P, MCDONNELL HM. Lateral diffusion in spin-labeled phosphatidylcholine multilayers. J Am Chem Soc. 1972;94:4475–4481. doi: 10.1021/ja00768a600. [DOI] [PubMed] [Google Scholar]
- DICKERSON RE, REDDY J, PINKERTON M, STEINRAUF LK. A 6 angstrom model of triclinic lysozyme. Nature. 1962;196:1178. doi: 10.1038/1961178a0. [DOI] [PubMed] [Google Scholar]
- DO HOA Q, WITTLICH M, GLÜCK JULIAN M, MÖCKEL L, WILLBOLD D, KOENIG BERND W, HEISE H. Full-length Vpu and human CD4(372–433) in phospholipid bilayers as seen by magic angle spinning NMR. Biological Chemistry. 2013;394:1453. doi: 10.1515/hsz-2013-0194. [DOI] [PubMed] [Google Scholar]
- DURR HNU, SOONG R, RAMAMOORTHY A. When detergent meets bilayer: birth and coming of age of lipid bicelles. Prog NMR Spec. 2013;69:1–22. doi: 10.1016/j.pnmrs.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DURR UH, WASKELL L, RAMAMOORTHY A. The cytochromes P450 and b5 and their reductases--promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim Biophys Acta. 2007;1768(12):3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- EDIDIN M. Rotational and translational diffusion in membranes. Annual Review of Biophysics and Bioengineering. 1974;3:179–201. doi: 10.1146/annurev.bb.03.060174.001143. [DOI] [PubMed] [Google Scholar]
- ETZKORN M, MARTELL S, ANDRONESI OC, SEIDEL K, ENGELHARD M, BALDUS M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew Chem Int Ed Engl. 2007;46(3):459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- FINER EG, FLOOK AG, HAUSER H. The nature and origin of the NMR spectrum of unsonicated and sonicated aqueous egg yolk lecithin dispersions. Biochim Biophys Acta. 1972;260(1):59–69. doi: 10.1016/0005-2760(72)90074-4. [DOI] [PubMed] [Google Scholar]
- FOSTER TL, THOMPSON GS, KALVERDA AP, KANKANALA J, BENTHAM M, WETHERILL LF, THOMPSON J, BARKER AM, CLARKE D, NOERENBERG M, PEARSON AR, ROWLANDS DJ, HOMANS SW, HARRIS M, FOSTER R, GRIFFIN S. Structure-guided design affirms inhibitors of hepatitis C virus p7 as a viable class of antivirals targeting virion release. Hepatology. 2013 doi: 10.1002/hep.26685. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKS WT, LINDEN AH, KUNERT B, VAN ROSSUM BJ, OSCHKINAT H. Solid-state magic-angle spinning NMR of membrane proteins and protein-ligand interactions. Eur J Cell Biol. 2012;91(4):340–348. doi: 10.1016/j.ejcb.2011.09.002. [DOI] [PubMed] [Google Scholar]
- FRÜH V, ZHOU Y, CHEN D, LOCH C, AB E, GRINKOVA YN, VERHEIJ H, SLIGAR SG, BUSHWELLER JH, SIEGAL G. Application of Fragment-Based Drug Discovery to Membrane Proteins: Identification of Ligands of the Integral Membrane Enzyme DsbB. Chemistry & Biology. 2010;17(8):881–891. doi: 10.1016/j.chembiol.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALL CM, CROSS TA, DIVERDI JA, OPELLA SJ. Protein dynamics by solid-state NMR: aromatic rings of the coat protein in fd bacteriophage. Proc Natl Acad Sci U S A. 1982;79(1):101–105. doi: 10.1073/pnas.79.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAUTIER A, KIRKPATRICK JP, NIETLISPACH D. Solution-state NMR spectroscopy of a seven-helix transmembrane protein receptor: backbone assignment, secondary structure, and dynamics. Angew Chem Int Ed Engl. 2008;47(38):7297–7300. doi: 10.1002/anie.200802783. [DOI] [PubMed] [Google Scholar]
- GAUTIER A, MOTT HR, BOSTOCK MJ, KIRKPATRICK JP, NIETLISPACH D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nature structural & molecular biology. 2010;17(6):768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDBOURT A, GROSS BJ, DAY LA, MCDERMOTT A. Filamentous phage studied by magic-angle spinning NMR: Resonance assignment and secondary structure of the coat protein in Pf1. J Am Chem Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- GONCALVES JA, EILERS M, SOUTH K, OPEFI CA, LAISSUE P, REEVES PJ, SMITH SO. Magic angle spinning nuclear magnetic resonance spectroscopy of G protein-coupled receptors. Meth Enzymol. 2013;522:365–389. doi: 10.1016/B978-0-12-407865-9.00017-0. [DOI] [PubMed] [Google Scholar]
- GONZALEZ ME, CARRASCO L. Viroporins. FEBS Lett. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- GUSTAVSSON M, TRAASETH NJ, VEGLIA G. Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2012;1818(2):146–153. doi: 10.1016/j.bbamem.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUSTAVSSON M, VERARDI R, MULLEN DG, MOTE KR, TRAASETH NJ, GOPINATH T, VEGLIA G. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1303006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUTOWSKY HS, PAKE GE. Structural investigations by means of nuclear magnetism. II. Hindered rotation in solids. J Chem Phys. 1950;18:162–170. [Google Scholar]
- HENDERSON R, UNWIN PNT. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- HENRY GD, SYKES BD. Structure and dynamics of detergent-solubilized M13 coat protein (an integral membrane protein) determined by 13C and 15N nuclear magnetic resonance spectroscopy. Biochemistry and Cell Biology. 1990;68(1):318–329. doi: 10.1139/o90-044. [DOI] [PubMed] [Google Scholar]
- HERZFELD J, BERGER AE. Sideband intensities in NMR spectra of samples spinning at the magic angle. J Chem Phys. 1980;73:6021–6030. [Google Scholar]
- HERZFELD J, MULLIKEN CM, SIMINOVITCH DJ, GRIFFIN RG. Contrasting molecular dynamics in red and purple membrane fractions of the Halobacterium halobium. Biophysical Journal. 1987;52(5):855–858. doi: 10.1016/S0006-3495(87)83278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG M, ZHANG Y, HU F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWELL SC, MESLEH MF, OPELLA SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44(13):5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- HU F, LUO W, HONG M. Mechanisms of Proton Conduction and Gating in Influenza M2 Proton Channels from Solid-State NMR. Science. 2010;330(6003):505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU J, ASBURY T, ACHUTHAN S, LI C, BERTRAM R, QUINE JR, FU R, CROSS TA. Backbone Structure of the Amantadine-Blocked Trans-Membrane Domain M2 Proton Channel from Influenza A Virus. Biophysical Journal. 2007;92(12):4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU J, FU R, NISHIMURA K, ZHANG L, ZHOU HX, BUSATH DD, VIJAYVERGIYA V, CROSS TA. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proceedings of the National Academy of Sciences. 2006;103(18):6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUBBELL WL, MCCONNELL HM. Molecular motion in spin-labeled phospholipids and membranes. J Am Chem Soc. 1971;93(2):314–326. doi: 10.1021/ja00731a005. [DOI] [PubMed] [Google Scholar]
- INABA K, MURAKAMI S, NAKAGAWA A, II, DA H, KINJO M, ITO K, SUZUKI M. Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB. EMBO Journal. 2009;28(6):779–791. doi: 10.1038/emboj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INABA K, MURAKAMI S, SUZUKI M, NAKAGAWA A, YAMASHITA E, OKADA K, ITO K. Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation. Cell. 2006;127(4):789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- JUDGE PJ, WATTS A. Recent contributions from solid-state NMR to the understanding of membrane protein structure and function. Curr Opin Chem Biol. 2011;15(5):690–695. doi: 10.1016/j.cbpa.2011.07.021. [DOI] [PubMed] [Google Scholar]
- KENDREW JC, BODO G, DINTZIS HM, PARRISH RG, WYCKOFF HW, PHILIPS DC. A three-dimensiona model of the myoglobin molecule obtained by X-ray analysis. Nature. 1958;181:666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- KENIRY MA, GUTOWSKY HS, OLDFIELD E. Surface dynamics of the integral membrane protein bacteriorhodopsin. Nature. 1984;307(5949):383–386. doi: 10.1038/307383a0. [DOI] [PubMed] [Google Scholar]
- KEOUGH KM, OLDFIELD E, CHAPMAN D. Carbon-13 and proton nuclear magnetic resonance of unsonicated model and mitochondrial membranes. Chem Phys Lipids. 1973;10:37–50. doi: 10.1016/0009-3084(73)90039-x. [DOI] [PubMed] [Google Scholar]
- KINSEY RA, KINTANAR A, OLDFIELD E. Dynamics of amino acid side chains in membrane proteins by high field solid state deuterium nuclear magnetic resonance spectroscopy. Phenylalanine, tyrosine, and tryptophan. J Biol Chem. 1981a;256(17):9028–9036. [PubMed] [Google Scholar]
- KINSEY RA, KINTANAR A, TSAI MD, SMITH RL, JANES N, OLDFIELD E. First observation of amino acid side chain dynamics in membrane proteins using high field deuterium nuclear magnetic resonance spectroscopy. The Journal of biological chemistry. 1981b;256(9):4146–4149. [PubMed] [Google Scholar]
- KNIGHT MJ, FELLI IC, PIERATTELLI R, EMSLEY L, PINTACUDA G. Magic angle spinning NMR of paramagnetic proteins. Acc Chem Res. 2013;46:2108–2116. doi: 10.1021/ar300349y. [DOI] [PubMed] [Google Scholar]
- KNOX RW, LU GJ, OPELLA SJ, NEVZOROV AA. A Resonance Assignment Method for Oriented-Sample Solid-State NMR of Proteins. Journal of the American Chemical Society. 2010;132(24):8255–8257. doi: 10.1021/ja102932n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRUGER-KOPLIN RD, SORGEN PL, DRUIEGER-KOPLIN ST, RIVERA-TORRES IO, CAHILL SM, KRULWICH TA, GIRVIN ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:1980–1987. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- LAMBERTH S, SCHMID H, MUENCHBACH M, VORHERR T, KREBS J, CARAFOLI E, GRIESINGER C. NMR Solution Structure of Phospholamban. Helvetica Chimica Acta. 2000;83(9):2141–2152. [Google Scholar]
- LAUTERWEIN J, BOSCH C, BROWN LR, WUTHRICH K. Physicochemical studies of the protein-lipid interactions in melittin-containing micelles. Biochim Biophys Acta. 1979;556(2):244–264. doi: 10.1016/0005-2736(79)90046-4. [DOI] [PubMed] [Google Scholar]
- LEO GC, COLNAGO LA, VALENTINE KG, OPELLA SJ. Dynamics of fd coat protein in lipid bilayers. Biochemistry. 1987;26:854–862. doi: 10.1021/bi00377a029. [DOI] [PubMed] [Google Scholar]
- LEWIS BA, HARBISON GS, HERZFELD J, GRIFFIN RG. NMR structural analysis of a membrane protein: bacteriorhodopsin peptide backbone orientation and motion. Biochemistry. 1985;24(17):4671–4679. doi: 10.1021/bi00338a029. [DOI] [PubMed] [Google Scholar]
- LI D, LYONS JA, PYE VE, VOGELEY L, ARAGAO D, KENYON CP, SHAH STA, DOHERTY C, AHERNE M, CAFFREY M. Crystal structure of the integral membrane diacylglycerol kinase. Nature. 2013;497(7450):521–524. doi: 10.1038/nature12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Y, BERTHOLD DA, GENNIS RB, RIENSTRA CM. Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Science. 2008;17(2):199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAO SY, FRITZSCHING KJ, HONG M. Conformational analysis of the full-length M2 protein of the influenza a virus using solid-state NMR. Protein Science. 2013 doi: 10.1002/pro.2368. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LICHTENBERG D, PETERSEN NO, GIRARDET JL, KAINOSHO M, KROON PA, SEITER CH, FEIGENSON GW, CHAN SI. The interpretation of proton magnetic resonance linewidths for lecithin dispersions. Effect of particle size and chain packing. Biochim Biophys Acta. 1975;382(1):10–21. doi: 10.1016/0005-2736(75)90367-3. [DOI] [PubMed] [Google Scholar]
- LOMBARD J, LOPEZ-GARCIA P, MOREIRA D. The early evolution of lipid membranes and the three domains of life. Nat Rev Microbiol. 2012;10(7):507–515. doi: 10.1038/nrmicro2815. [DOI] [PubMed] [Google Scholar]
- LU GJ, OPELLA SJ. Resonance assignments of a membrane protein in phospholipid bilayers by combining multiple strategies of oriented sample solid-state NMR. J Biomol NMR. 2014 doi: 10.1007/s10858-013-9806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU GJ, PARK SH, OPELLA SJ. Improved 1H amide resonance line narrowing in oriented sample solid-state NMR of membrane proteins in phospholipid bilayers. Journal of Magnetic Resonance. 2012;220:54–61. doi: 10.1016/j.jmr.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU GJ, SON WS, OPELLA SJ. A general assignment method for oriented sample (OS) solid-state NMR of proteins based on the correlation of resonances through heteronuclear dipolar couplings in samples aligned parallel and perpendicular to the magnetic field. Journal of Magnetic Resonance. 2011;209(2):195–206. doi: 10.1016/j.jmr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU GJ, TIAN Y, VORA N, MARASSI FM, OPELLA SJ. The Structure of the Mercury Transporter MerF in Phospholipid Bilayers: A Large Conformational Rearrangement Results From N-terminal Truncation. Journal of the American Chemical Society. 2013;135(25):9299–9302. doi: 10.1021/ja4042115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU JX, SHARPE S, GHIRLANDO R, YAU WM, TYCKO R. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Science. 2010;19(10):1877–1896. doi: 10.1002/pro.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA C, MARASSI F, JONES D, STRAUS S, BOUR S, STREBEL K, SCHUBERT U, OBLATT-MONTAL M, MONTAL M, OPELLA S. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 2002;11(3):546–557. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALOJČIĆ G, OWEN RL, GRIMSHAW JPA, GLOCKSHUBER R. Preparation and structure of the charge-transfer intermediate of the transmembrane redox catalyst DsbB. FEBS Letters. 2008;582(23–24):3301–3307. doi: 10.1016/j.febslet.2008.07.063. [DOI] [PubMed] [Google Scholar]
- MARASSI FM, DAS BB, LU GJ, NOTHNAGEL HJ, PARK SH, SON WS, TIAN Y, OPELLA SJ. Structure determination of membrane proteins in five easy pieces. Methods. 2011;55(4):363–369. doi: 10.1016/j.ymeth.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARASSI FM, MA C, GRATKOWSKI H, STRAUS SK, STREBEL K, OBLATT-MONTAL M, MONTAL M, OPELLA SJ. Correlation of the structural and functional domains in the membrane protein Vpu from HIV-1. Proc Natl Acad Sci USA. 1999;96(25):14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARASSI FM, OPELLA SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12(3):403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCONNELL HM, HUBBELL WL. Molecular motion in spin-labeled phospholipids and membranes. J Am Chem Soc. 1971;93:314–326. doi: 10.1021/ja00731a005. [DOI] [PubMed] [Google Scholar]
- MCCONNELL HM, WRIGHT KL, MCFARLAND BG. The fraction of the lipid in a biological membrane that is in a fluid state: a spin label assay. Biochem Biophys Res Commun. 1972;47(1):273–281. doi: 10.1016/s0006-291x(72)80039-1. [DOI] [PubMed] [Google Scholar]
- MCDERMOTT A. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- MCDONNELL PA, SHON K, KIM Y, OPELLA SJ. fd coat protein structure in membrane environments. J Mol Biol. 1993;233(3):447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- MCLAUGHLIN AC, CULLIS PR, HEMMINGA MA, HOULT DI, RADDA GK, RITCHIE GA, SEELEY PJ, RICHARDS RE. Application of 31P NMR to model and biological membrane systems. FEBS Lett. 1975;57:213–218. doi: 10.1016/0014-5793(75)80719-8. [DOI] [PubMed] [Google Scholar]
- MEHRING M, GRIFFIN RG, WAUGH JS. 19F Shielding tensors from coherently narrowed NMR powder spectra. J Chem Phys. 1971;55:746–755. [Google Scholar]
- MIAO Y, QIN H, FU R, SHARMA M, CAN TV, HUNG I, LUCA S, GOR’KOV PL, BREY WW, CROSS TA. M2 Proton Channel Structural Validation from Full-Length Protein Samples in Synthetic Bilayers and E. coli Membranes. Angewandte Chemie International Edition. 2012;51(33):8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER SL. A production of amino acids under possibe primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- MONTAL M, OPELLA SJ. The structure of the M2 channel-lining segment from the nicotinic acetylcholine receptor. Biochim Biophys Acta. 2002;1565(2):287–293. doi: 10.1016/s0005-2736(02)00575-8. [DOI] [PubMed] [Google Scholar]
- MONTSERRET R, SAINT N, VANBELLE C, SALVAY ASG, SIMORRE JP, EBEL C, SAPAY N, RENISIO JG, BÖCKMANN A, STEINMANN E, PIETSCHMANN T, DUBUISSON J, CHIPOT C, PENIN FS. NMR Structure and Ion Channel Activity of the p7 Protein from Hepatitis C Virus. Journal of Biological Chemistry. 2010;285(41):31446–31461. doi: 10.1074/jbc.M110.122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUELLER LJ, DUNN MF. NMR Crystallography of Enzyme Active Sites: Probing Chemically-Detailed, Three-Dimensional Structure in Tryptophan Synthase. Accounts of Chemical Research. 2013;46:2008–2017. doi: 10.1021/ar3003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURRAY DT, DAS N, CROSS TA. Solid state NMR strategy for characterizing native membrane protein structures. Acc Chem Res. 2013;46(9):2172–2181. doi: 10.1021/ar3003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAMBUDRIPAD R, STARK W, OPELLA SJ, MAKOWSKI L. Membrane-mediated assembly of filamentous bacteriophage Pf1 coat protein. Science. 1991;252(5010):1305–1308. doi: 10.1126/science.1925543. [DOI] [PubMed] [Google Scholar]
- NEVZOROV AA. Mismatched Hartmann-Hahn conditions cause proton-mediated intermolecular magnetization transfer between dilute low-spin nuclei in NMR of static solids. Journal of the American Chemical Society. 2008;130(34):11282–11283. doi: 10.1021/ja804326b. [DOI] [PubMed] [Google Scholar]
- NEVZOROV AA, OPELLA SJ. Structural fitting of PISEMA spectra of aligned proteins. J Magn Reson. 2003;160(1):33–39. doi: 10.1016/s1090-7807(02)00138-6. [DOI] [PubMed] [Google Scholar]
- NEVZOROV AA, OPELLA SJ. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. Journal of Magnetic Resonance. 2007;185(1):59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- OLDFIELD E, CHAPMAN D, DERBYSHIRE W. Deuteron resonance: A novel approach to the study of hydrocarbon chain mobility in membrane systems. FEBS Lett. 1971;16:102–104. doi: 10.1016/0014-5793(71)80343-5. [DOI] [PubMed] [Google Scholar]
- OPELLA S. Structure determination of membrane proteins by nuclear magnetic resonance spectrocopy. Annu Rev Anal Chem. 2013a;6:305–328. doi: 10.1146/annurev-anchem-062012-092631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPELLA SJ. Structure determination of membrane proteins in their native phospholipid bilayer environment by rotationally aligned solid-state NMR spectroscopy. Acc Chem Res. 2013b;46:2145–2153. doi: 10.1021/ar400067z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPELLA SJ, CROSS TA, DIVERDI JA, STURM CF. Nuclear magnetic resonance of the filamentous bacteriophage fd. Biophysical Journal. 1980;32(1):531–548. doi: 10.1016/S0006-3495(80)84988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPELLA SJ, MARASSI FM, GESELL JJ, VALENTE AP, KIM Y, OBLATT-MONTAL M, MONTAL M. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat Struct Biol. 1999;6(4):374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OPELLA SJ, WAUGH JS. Two-dimensional 13C NMR of highly oriented polyethylene. J Chem Phys. 1977;66:4919–4924. [Google Scholar]
- OPELLA SJ, ZERI AC, PARK SH. Structure, dynamics, and assembly of filamentous bacteriophages by nuclear magnetic resonance spectroscopy. Annual Review of Physical Chemistry. 2008;59:635–657. doi: 10.1146/annurev.physchem.58.032806.104640. [DOI] [PubMed] [Google Scholar]
- OUYANG B, XIE S, BERARDI MJ, ZHAO X, DEV J, YU W, SUN B, CHOU JJ. Unusual architecture of the p7 channel from hepatitis C virus. Nature. 2013 doi: 10.1038/nature12283. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OXENOID K, CHOU JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102(31):10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OXENOID K, RICE AJ, CHOU JJ. Comparing the structure and dynamics of phospholamban pentamer in its unphosphorylated and pseudo-phosphorylated states. Protein Science. 2007;16(9):1977–1983. doi: 10.1110/ps.072975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAGE RC, MOORE JD, NGUYEN HB, SHARMA M, CHASE R, GAO FP, MOBLEY CK, SANDERS CR, MA L, SONNICHSEN FD, LEE S, HOWELL SC, OPELLA SJ, CROSS TA. Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J Struct Funct Genomics. 2006;7(1):51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- PANDEY MK, RAMAMOORTHY A. Quantum chemical calculations of amide-15N chemical shift anisotropy tensors for a membrane-bound cytochrome-b5. J Phys Chem B. 2013;117(3):859–867. doi: 10.1021/jp311116p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK S, DAS BB, DEANGELIS AA, SCRIMA M, OPELLA SJ. Mechanically, magnetically, and ‘rotationally aligned’ membrane proteins in phospholipid bilayers give equivalent angular constraints for NMR structure determination. J Phys Chem B. 2010a;114:13995–13003. doi: 10.1021/jp106043w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, CASAGRANDE F, CHO L, ALBRECHT L, OPELLA SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J Mol Biol. 2011a;414(2):194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, CASAGRANDE F, DAS BB, ALBRECHT L, CHU M, OPELLA SJ. Local and global dynamics of the G protein-coupled receptor CXCR1. Biochemistry. 2011b;50(12):2371–2380. doi: 10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, DAS BB, CASAGRANDE F, TIAN Y, NOTHNAGEL HJ, CHU M, KIEFER H, MAIER K, DE ANGELIS AA, MARASSI FM, OPELLA SJ. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491(7426):779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, DE ANGELIS AA, NEVZOROV AA, WU CH, OPELLA SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophysical Journal. 2006;91(8):3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, MARASSI FM, BLACK D, OPELLA SJ. Structure and Dynamics of the Membrane-Bound Form of Pf1 Coat Protein: Implications of Structural Rearrangement for Virus Assembly. Biophysical Journal. 2010b;99(5):1465–1474. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK SH, MRSE AA, NEVZOROV AA, DE ANGELIS AA, OPELLA SJ. Rotational diffusion of membrane proteins in aligned phospholipid bilayers by solid-state NMR spectroscopy. J Magn Reson. 2005;178(1):162–165. doi: 10.1016/j.jmr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- PARK SH, MRSE AA, NEVZOROV AA, MESLEH MF, OBLATT-MONTAL M, MONTAL M, OPELLA SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol. 2003;333(2):409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- PARK SH, OPELLA SJ. Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs. Protein Science. 2007;16(10):2205–2215. doi: 10.1110/ps.073041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARTHASARATHY S, NISHIYAMA Y, ISHII Y. Sensitivity and resolution enhanced solid-state NMR for paramagnetic systems and biomolecules under very fast magic angle spinning. Acc Chem Res. 2013;46:2127–2135. doi: 10.1021/ar4000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERUTZ MF, ROSSMAN MG, CULLIS AF, MUIRHEAD H, GEORG W. Structure of hemoglobin: A three-dimensional Fourier synthesis at 5.5 A resolution, obtaine by X-ray analyss. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- PETSKO GA, RINGE D. Proein structure and function. Oxford University Press; 2008. [Google Scholar]
- POGET SF, GIRVIN ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768(12):3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLLESELLO P, ANNILA A. Structure of the 1–36 N-Terminal Fragment of Human Phospholamban Phosphorylated at Ser-16 and Thr-17. Biophysical Journal. 2002;83(1):484–490. doi: 10.1016/S0006-3495(02)75184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POO MM, CONE RA. Lateral diffusion of rhodopsin in Necturus rods. Exp Eye Res. 1973;17:503–510. doi: 10.1016/0014-4835(73)90079-1. [DOI] [PubMed] [Google Scholar]
- POO MM, CONE RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- POPOT JL. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–777. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- PROSSER RS, HUNT SA, DINATALE JA, VOLD RR. Magnetically Aligned Membrane Model Systems with Positive Order Parameter: Switching the Sign of Szz with Paramagnetic Ions. Journal of the American Chemical Society. 1996;118(1):269–270. [Google Scholar]
- RAJARATHNAM K, CLARK-LEWIS I, SYKES BD. 1H NMR solution structure of an active monomeric interleukin-8. Biochemistry. 1995;34(40):12983–12990. doi: 10.1021/bi00040a008. [DOI] [PubMed] [Google Scholar]
- RAMACHANDRAN GN, RAMAKRISHNAN C, SASISEKHARAN V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- RICHARDSON JS. Anatomy and taxonomy of protein structures. Adv Protein Chemistry. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- SAITO H, ANDO I, RAMAMOORTHY A. Chemical shift tensor - The heart of NMR: Insights into biological aspects of proteins. Prog NMR Spec. 2010;57:181–228. doi: 10.1016/j.pnmrs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANDERS CR, OXENOID K. Customizing model membranes and samples for NMR spectroscopic studies of complex membrane proteins. Biochim Biophys Acta. 2000;1508(1–2):129–145. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- SCHNELL J, CHOU J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451(7178):591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUBERT U, BOUR S, FERRER-MONTIEL AV, MONTAL M, MALDARELL F, STREBEL K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J Virol. 1996;70(2):809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEELIG J, AXEL F, LIMACHER H. Molecular architecture of bilayer membranes. Ann N Y Acad Sci. 1973;222:588–596. doi: 10.1111/j.1749-6632.1973.tb15289.x. [DOI] [PubMed] [Google Scholar]
- SEITER CHA, CHAN SI. Molecular motion in lipid bilayers. A nuclear magnetic resonance line width study. J Am Chem Soc. 1973;95:7541–7553. [Google Scholar]
- SENGUPTA I, NADAUD PS, JARONIEC CP. Protein structure determination with paramagnetic solid-state NMR spectroscopy. Acc Chem Res. 2013;46:2117–2126. doi: 10.1021/ar300360q. [DOI] [PubMed] [Google Scholar]
- SHARMA M, YI M, DONG H, QIN H, PETERSON E, BUSATH DD, ZHOU HX, CROSS TA. Insight into the Mechanism of the Influenza A Proton Channel from a Structure in a Lipid Bilayer. Science. 2010;330(6003):509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARPE S, YAU WM, TYCKO R. Structure and Dynamics of the HIV-1 Vpu Transmembrane Domain Revealed by Solid-State NMR with Magic-Angle Spinning. Biochemistry. 2005;45(3):918–933. doi: 10.1021/bi051766k. [DOI] [PubMed] [Google Scholar]
- SHEETZ MP, CHAN SI. Effect of sonication on the structure of lecithin bilayers. Biochemistry. 1972;11:4573–4581. doi: 10.1021/bi00774a024. [DOI] [PubMed] [Google Scholar]
- SHI L, KAWAMURA I, JUNG KH, BROWN LS, LADIZHANSKY V. Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew Chem Int Ed Engl. 2011;50(6):1302–1305. doi: 10.1002/anie.201004422. [DOI] [PubMed] [Google Scholar]
- SHI L, LADIZHANSKY V. Magic angle spinning solid-state NMR experiments for structural characterization of proteins. Methods Mol Biol. 2012;895:153–165. doi: 10.1007/978-1-61779-927-3_12. [DOI] [PubMed] [Google Scholar]
- SHON KJ, KIM Y, COLNAGO LA, OPELLA SJ. NMR studies of the structure and dynamics of membrane-bound bacteriophage Pf1 coat protein. Science. 1991;252(5010):1303–1305. doi: 10.1126/science.1925542. [DOI] [PubMed] [Google Scholar]
- SINGER SJ, NICHOLSON GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- SKASKO M, WANG Y, TIAN Y, TOKAREV A, MUNGUIA J, RUIZ A, STEPHENS EB, OPELLA SJ, GUATELLI J. HIV-1 Vpu Protein Antagonizes Innate Restriction Factor BST-2 via Lipid-embedded Helix-Helix Interactions. Journal of Biological Chemistry. 2012;287(1):58–67. doi: 10.1074/jbc.M111.296772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SKELTON NJ, QUAN C, REILLY D, LOWMAN H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure. 1999;7(2):157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- SMITH RL, OLDFIELD E. Dynamic structure of membranes by deuterium NMR. Science. 1984;225(4659):280–288. doi: 10.1126/science.6740310. [DOI] [PubMed] [Google Scholar]
- SOONG R, SMITH PE, XU J, YAMAMOTO K, IM SC, WASKELL L, RAMAMOORTHY A. Proton-evolved local-field solid-state NMR studies of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. Journal of the American Chemical Society. 2010;132(16):5779–5788. doi: 10.1021/ja910807e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPUDICH JL, LUECKE H. Sensory rhodopsin II: functional insights from structure. Curr Opin Struct Biol. 2002;12(4):540–546. doi: 10.1016/s0959-440x(02)00359-7. [DOI] [PubMed] [Google Scholar]
- STOUFFER AL, ACHARYA R, SALOM D, LEVINE AS, DI COSTANZO L, SOTO CS, TERESHKO V, NANDA V, STAYROOK S, DEGRADO WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451(7178):596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANFORD C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- TANG M, COMELLAS G, RIENSTRA CM. Advanced solid-state NMR approaches for structure determination of membrane proteins and amyloid fibrils. Acc Chem Res. 2013a;46(9):2080–2088. doi: 10.1021/ar4000168. [DOI] [PubMed] [Google Scholar]
- TANG M, NESBITT AE, SPERLING LJ, BERTHOLD DA, SCHWIETERS CD, GENNIS RB, RIENSTRA CM. Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer. Journal of Molecular Biology. 2013b;425(10):1670–1682. doi: 10.1016/j.jmb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG M, SPERLING L, BERTHOLD D, SCHWIETERS C, NESBITT A, NIEUWKOOP A, GENNIS R, RIENSTRA C. High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data. J Biomol NMR. 2011a;51(3):227–233. doi: 10.1007/s10858-011-9565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG M, SPERLING LJ, BERTHOLD DA, NESBITT AE, GENNIS RB, RIENSTRA CM. Solid-State NMR Study of the Charge-Transfer Complex between Ubiquinone-8 and Disulfide Bond Generating Membrane Protein DsbB. Journal of the American Chemical Society. 2011b;133(12):4359–4366. doi: 10.1021/ja107775w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIRIOT DS, NEVZOROV AA, OPELLA SJ. Structural basis of the temperature transition of Pf1 bacteriophage. Protein Sci. 2005;14(4):1064–1070. doi: 10.1110/ps.041220305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIRIOT DS, NEVZOROV AA, ZAGYANSKIY L, WU CH, OPELLA SJ. Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy. J Mol Biol. 2004;341(3):869–879. doi: 10.1016/j.jmb.2004.06.038. [DOI] [PubMed] [Google Scholar]
- TIAN Y, SCHWIETERS CD, OPELLA SJ, MARASSI FM. AssignFit: A program for simultaneous assignment and structure refinement from solid-state NMR spectra. Journal of Magnetic Resonance. 2012;214:42–50. doi: 10.1016/j.jmr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAASETH NJ, SHI L, VERARDI R, MULLEN DG, BARANY G, VEGLIA G. Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proceedings of the National Academy of Sciences. 2009;106(25):10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAASETH NJ, VEGLIA G. Probing excited states and activation energy for the integral membrane protein phospholamban by NMR CPMG relaxation dispersion experiments. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2010;1798(2):77–81. doi: 10.1016/j.bbamem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ULLRICH SJ, GLAUBITZ C. Perspectives in enzymology of membrane proteins by solid-state NMR. Acc Chem Res. 2013;46(9):2164–2171. doi: 10.1021/ar4000289. [DOI] [PubMed] [Google Scholar]
- ULRICH AS. Solid state 19G NMR methods for studying biomembranes. Prog Nucl Magn Reson Spectrosc. 2005;46:1–21. [Google Scholar]
- UNWIN N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373(6509):37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- VAN HORN WD, KIM HJ, ELLIS CD, HADZISELIMOVIC A, SULISTIJO ES, KARRA MD, TIAN C, SÖNNICHSEN FD, SANDERS CR. Solution Nuclear Magnetic Resonance Structure of Membrane-Integral Diacylglycerol Kinase. Science. 2009;324(5935):1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEKSLI Z, SALSBURY NJ, CHAPMAN D. Physical studies of phospholipids. XII. Nuclear magnetic resonance studies of molecular motion in some pure lecithin-water systems. BBA. 1969;183:434–446. doi: 10.1016/0005-2736(69)90158-8. [DOI] [PubMed] [Google Scholar]
- VERARDI R, SHI L, TRAASETH NJ, WALSH N, VEGLIA G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proceedings of the National Academy of Sciences. 2011;108(22):9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VINOGRADOVA O, SONNICHSEN F, SANDERS CR., 2ND On choosing a detergent for solution NMR studies of membrane proteins. J Biomol NMR. 1998;11(4):381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- VOSTRIKOV VITALY V, MOTE KAUSTUBH R, VERARDI R, VEGLIA G. Structural Dynamics and Topology of Phosphorylated Phospholamban Homopentamer Reveal Its Role in the Regulation of Calcium Transport. Structure. 2013 doi: 10.1016/j.str.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WACEY D, KILBRUN MR, SAUNDERS M, CLIFF J, BRASIER MD. Microfossils of sulphur-metabolizing cells in 3.4-billion-year-old rocks of Western Australia. Nature Geoscience. 2011;4:698–702. [Google Scholar]
- WANG J, KIM S, KOVACS F, CROSS TA. Structure of the transmembrane region of the M2 protein H+ channel. Protein Science. 2001;10(11):2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG S, MUNRO RA, SHI L, KAWAMURA I, OKITSU T, WADA A, KIM SY, JUNG KH, BROWN LS, LADIZHANSKY V. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods. 2013a;10(10):1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- WANG Y, PARK SH, TIAN Y, OPELLA SJ. Impact of histidine residues on the transmembrane helices of viroporins. Molecular Membrane Biology. 2013b;0(0):1–10. doi: 10.3109/09687688.2013.842657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON JD, CRICK FHC. Molecular structure of nucleic acids. Nature. 1953:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- WEINGARTH M, BALDUS M. Solid-State NMR-Based Approaches for Supramolecular Structure Elucidation. Acc Chem Res. 2013;46(9):2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- WIEN RW, MORRISETT JD, MCCONNELLL HM. Spin-label-induced nuclear relaxation. Distances between bound saccharides, histidines-15, and tryptophan-123 on lysozyme in solution. Biochemistry. 1972;11:3707–3716. doi: 10.1021/bi00770a008. [DOI] [PubMed] [Google Scholar]
- WILLBOLD D, HOFFMANN S, RÖSCH P. Secondary Structure and Tertiary Fold of the Human Immunodeficiency Virus Protein U (Vpu) Cytoplasmic Domain in Solution. European Journal of Biochemistry. 1997;245(3):581–588. doi: 10.1111/j.1432-1033.1997.t01-1-00581.x. [DOI] [PubMed] [Google Scholar]
- WITTLICH M, KOENIG BW, STOLDT M, SCHMIDT H, WILLBOLD D. NMR structural characterization of HIV-1 virus protein U cytoplasmic domain in the presence of dodecylphosphatidylcholine micelles. FEBS Journal. 2009;276(22):6560–6575. doi: 10.1111/j.1742-4658.2009.07363.x. [DOI] [PubMed] [Google Scholar]
- WUTHRICH K, WAGNER G. NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett. 1975;50:265–268. doi: 10.1016/0014-5793(75)80504-7. [DOI] [PubMed] [Google Scholar]
- WYLIE B, SPERLING L, FRERICKS H, SHAH G, FRANKS W, RIENSTRA C. Chemical-shift anisotropy measurements of amide and carbonyl resonances in a microcrystalline protein with slow magic-angle spinning NMR spectroscopy. J Am Chem Soc. 2007;129:5318–5319. doi: 10.1021/ja0701199. [DOI] [PubMed] [Google Scholar]
- WYLIE BJ, FRANKS WT, RIENSTRA CM. Determinations of 15N chemical shift anisotropy magnitudes in a uniformly 15N,13C-labeled microcrystalline protein by three-dimensional magic-angle spinning nuclear magnetic resonance spectroscopy. J Phys Chem B. 2006;110(22):10926–10936. doi: 10.1021/jp060507h. [DOI] [PubMed] [Google Scholar]
- XU J, SOONG R, IM SC, WASKELL L, RAMAMOORTHY A. INEPT-based separated local field NMR spectrsocopy: A unique approach to elucidate side-chain dynamics of membrane-associated proteins. J Am Chem Soc. 2010;132:9944–9947. doi: 10.1021/ja103983f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO K, DURR HNU, XU J, IM SC, WASKELL L, RAMAMOORTHY A. Dynamic interaction between membrane bound full-length cytochrome P450 and cytochrome b5 observed by solid-state NMR spectroscopy. Sci Rep. 2013a;3:1–5. doi: 10.1038/srep02538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO K, GILDENBERG M, AHUJA S, IM SC, PEARCY P, WASKELL L, RAMAMOORTHY A. Probing the transmembrane structure and topology of microsomal cytochrome-p450 by solid-state NMR on temperature-resistant bicelles. Sci Rep. 2013b;3:2556. doi: 10.1038/srep02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN S, SUITER CL, HOU G, ZHANG H, POLENOVA T. Probing structure and dynamics of protein assemblies by magic angle spinning NMR spectroscopy. Acc Chem Res. 2013 doi: 10.1021/ar300309s. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAMOON J, MASCIONI A, THOMAS DD, VEGLIA G. NMR Solution Structure and Topological Orientation of Monomeric Phospholamban in Dodecylphosphocholine Micelles. Biophysical Journal. 2003;85(4):2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZERI AC, MESLEH MF, NEVZOROV AA, OPELLA SJ. Structure of the coat protein in fd filamentous bacteriophage particles determined by solid-state NMR spectroscopy. Proc Natl Acad Sci USA. 2003;100(11):6458–6463. doi: 10.1073/pnas.1132059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG J, JIA Z. Structural biology: Tiny enzyme uses context to succeed. Nature. 2013;497(7450):445–446. doi: 10.1038/nature12245. [DOI] [PubMed] [Google Scholar]
- ZHOU HX, CROSS TA. Influences of Membrane Mimetic Environments on Membrane Protein Structures. Annual Review of Biophysics. 2013;42(1):361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU Y, CIERPICKI T, JIMENEZ RHF, LUKASIK SM, ELLENA JF, CAFISO DS, KADOKURA H, BECKWITH J, BUSHWELLER JH. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Molecular Cell. 2008;31(6):896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]