Abstract
Long thought to be a sterile habitat, the stomach contains a diverse and unique community of bacteria. One particular inhabitant, Helicobacter pylori, colonizes half of the world’s human population and establishes a decades-long infection that can be asymptomatic, pathogenic, or even beneficial for the host. Many host and bacterial factors are known to influence an individual’s risk of gastric disease, but another potentially important determinant has recently come to light: the host microbiota. Although it is unclear to what extent H. pylori infection perturbs the established gastric microbial community, and H. pylori colonization seems generally resistant to disturbances in the host microbiota, it can modulate H. pylori pathogenicity. Interactions between H. pylori and bacteria at non-gastric sites are likely indirect—via programming of the pro-inflammatory vs. regulatory T lymphocytes—which may have a significant impact on human health.
Keywords: Helicobacter pylori, microbiota, microbial community, stomach, disease, rhesus monkey, human, mouse, 16S rDNA
Introduction
The normal human stomach was long thought to be sterile, owing largely to the gastric acid barrier. But after the seminal discovery of Marshall and Warren in 1983,1 it became clear that approximately half the world's human population is chronically colonized with Helicobacter pylori, which causes asymptomatic inflammation (gastritis) in virtually all infected individuals, and peptic ulcer or gastric adenocarcinoma in a few.2 In a sense, H. pylori can be considered a commensal because of its near universal prevalence prior to the antibiotic era (and presently in most developing countries), and because of its extensive co-evolution with humans.3 On the other hand, H. pylori sometimes causes serious disease,2 so pathobiont may be a more appropriate designation.4 Widespread treatment of H. pylori is generally considered impractical, and perhaps harmful, not only because of antibiotic toxicity and off target effects, but also because accumulating evidence suggests that some individuals may actually derive benefit from H. pylori infection. Therefore, it is critical to identify those who are at greatest risk of serious disease. To date the emphasis has been largely on bacterial virulence factors, host genetics, and environmental influences, particularly diet.5-7
But with the recent recognition that the stomach is also colonized by other bacteria, another potential determinant of the outcome of H. pylori infection is the composition or structure of bacterial communities in the stomach, either at the time of exposure or over the course of infection (Fig. 1). This might occur, for example, because the microbial context provides competition or alters the host physiology to render it more or less hospitable to H. pylori. Conversely, introduction of H. pylori to the stomach may disrupt a stable ecosystem and lead to disease indirectly by disrupting beneficial organisms and communities. The gastric microbiota may also affect the host immune response to H. pylori, shifting the balance of Th1, Th17, and regulatory T cell activity that is thought to largely determine the outcome of infection.8
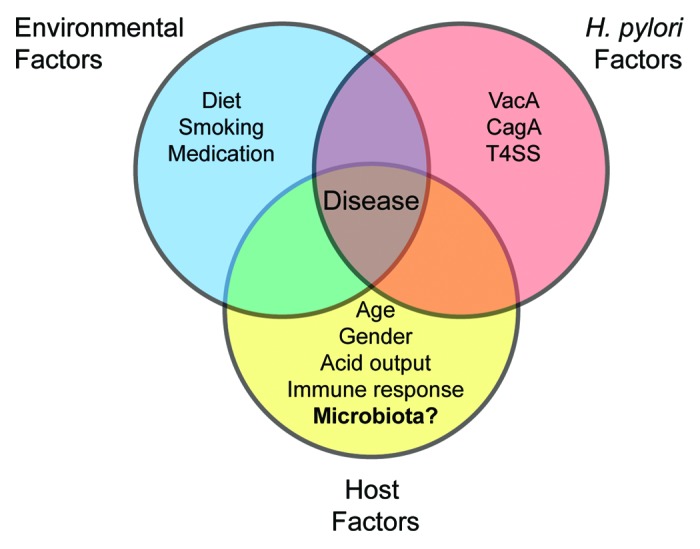
Figure 1. Risk factors for H. pylori-associated disease. Increased risk is associated with environmental factors such as a diet high in salt or low in iron, smoking, and the use of medications such as proton pump inhibitors and nonsteroidal anti-inflammatory drugs. H. pylori factors also play a role, with increased risk attributed to specific alleles of the vacA cytotoxin and the presence of the cag PAI, which encodes a type 4 secretion system (T4SS) and its effector, CagA. Host determinants of disease include the age of acquisition, gender, acid output following infection, an altered immune response due to genetic polymorphisms (IL-1β, IL-10, TNF-α, or IL-1RA), and perhaps the composition or structure of the microbial community.
To begin to probe the interactions between H. pylori and the microbial community of the stomach, we recently characterized the gastric microbiota of specific pathogen (H. pylori) free rhesus monkeys (Macaca mulatta), which serve as a robust and physiological model that closely resembles human H. pylori infection, and compared the results to the microbial communities in the mouth and the gut.9 Deep sequencing of the bacterial 16S rDNA gene identified a community profile of 221 phylotypes that was largely distinct from that of the distal gut and mouth, although there were taxa in common. The gastric community also included a second Helicobacter, the commensal H. suis, which is occasionally seen as a zoonotic infection in humans.10 We then performed longitudinal analyses of the microbiota following inoculation of H. pylori. Although we expected that the community membership and structure would shift in response to H. pylori colonization, we found instead that the gastric microbiota was dominated by H. pylori, leaving the underlying community largely unchanged. The one exception was H. suis, which was in apparent competition with H. pylori, because dynamic increases in levels of the one were accompanied by decreases in the other. Here we put these results in a broader context, and we consider four questions regarding the relationship between H. pylori and the gastric microbial community.
Is There a Truly Distinct, Autochthonous Gastric Microbiota?
There is little doubt that prior to the decline of H. pylori prevalence in the modern era, the native microbiota of the stomach was universally dominated by H. pylori since humans evolved out of Africa more than 60 000 years ago.3 But the question here is whether in the absence of H. pylori, or even in its presence, albeit in lower numbers, there is a distinct, resident gastric microbial community. Answering this question is not easy. DNA sequencing methods are not specific for autochthonous members of the gastric microbiota, but will also identify, for example, naked DNA, environmental bacteria in food or water, oral bacteria, and organisms passing through stomach to take up residence in the gut. These problems are not necessarily unique to analysis of the gastric microbiota. But identification of species that comprise the “normal” microbiota of the stomach is more difficult than in the gut because human gastric contents are not as readily available as feces, especially from asymptomatic hosts. Additionally, the overwhelming numerical dominance of H. pylori in the stomach11,12 means that extremely deep sequencing is required to reliably sample minority species. Additional information could be obtained by sorting bacterial cells on the basis of their activity level in the stomach,13 using, for example, metatranscriptomic analysis to identify genes expressed in the gastric environment, or even culture-based methods, which are increasingly sensitive. Comparison between H. pylori positive and negative individuals may also be difficult because H. pylori is often present in low abundance when detected by DNA methods, even when clinical diagnostic assays suggest that it is absent.11,14
These caveats aside, cultivation-independent methods suggest that the gastric community is in fact distinct, even when H. pylori is not present.11,14-16 This conclusion is supported by both sequence abundance weighted and unweighted phylogeny-based community analyses, irrespective of H. pylori infection status or gastric anatomical site. While communities sampled from oral (dental plaque, tongue, pharynx) and gastric habitats (antrum, corpus) frequently segregate by site, there is some degree of overlap among samples, which is perhaps to be expected given the continual flow of food and saliva from the mouth to the stomach. However, even bacteria that are not autochthonous to the gastric habitat—but are routinely present due to flow from upstream sites—may contribute to the microbial ecology of the stomach and thus deserve consideration as members of this unique assemblage of bacteria.
Does H. pylori Infection Impact the Gastric Microbiota?
In a sense, it is hard to imagine that the profound changes in gastric physiology induced by H. pylori infection would not alter the structure and membership of the gastric microbial community. Infiltration of PMNs, Th1, Th17, and Treg lymphocytes, induction of antimicrobial peptides, chemokines and cytokines, and changes in pH and mucus composition are just a few of the changes that might well be expected to alter the gastric microbiota.
Based on 16S rDNA sequence signatures, the human gastric microbiota is comprised of several hundred operational taxonomic units (OTUs),11,12,15,17 which span 44 bacterial phyla.14 Despite this impressive diversity, the species distribution is highly uneven, with four phyla dominating the human stomach.14 PhyloChip analysis found no difference in phyla richness between H. pylori positive and H. pylori negative stomachs, although there were shifts in the relative abundance of particular taxa.14 In contrast, a clone library-based community analysis estimated higher phylotype diversity and evenness in H. pylori positive samples, once H. pylori sequences were excluded, but did not observe significant changes in community composition.11 These studies were performed on different anatomical sites within the stomach (corpus, largely antrum, respectively), subject number and profiles (10 Amerindians, 2 immigrants, 20 New York citizens), and sensitivity (high, low), which may explain the discrepant conclusions.
The rhesus monkey gastric microbiota was also seemingly indifferent to H. pylori colonization, with the exception of H. suis, a related gastric Helicobacter species that frequently colonizes rhesus macaques and other animals.10 Using six age- and gender-matched rhesus monkeys, we observed no correlation between H. pylori infection status and phylogenetic diversity or community membership at the genus level, in either the antrum or corpus.9 However, there was a striking effect on H. suis levels. While Helicobacter was the most abundant bacterium in post-H. pylori inoculation biopsies, individual biopsies were dominated by only one Helicobacter species, suggesting that H. pylori and H. suis are competitively exclusive (Fig. 2). H. pylori inoculation also knocked down the H. suis 16S rDNA levels in animals that were H. suis PCR-positive prior to inoculation. Both H. pylori and H. suis are ubiquitous among rhesus macaques housed at primate research colonies,10 and although each species predominates in different gastric anatomical sites (antrum corpus, respectively), their distributions clearly overlap. However, they have distinct lifestyles in the rhesus monkey stomach. H. pylori localizes primarily to the mucus layer just above the epithelium and induces inflammation, while H. suis associates with parietal cells and does not provoke histologic inflammation. Consequently, the mechanism of their mutual antagonism is not clear.
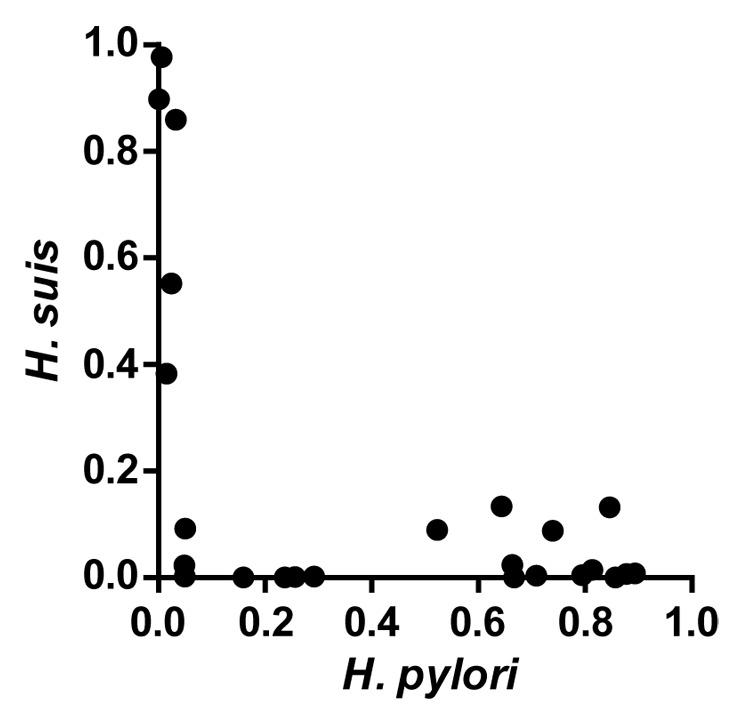
Figure 2.H. pylori and H. suis are competitively exclusive in the gastric mucosa. Biopsies were collected from SPF rhesus monkeys at five different time points following inoculation with H. pylori. DNA extracted from individual biopsies was subjected to pyrosequencing of variable regions 1–3 of the 16S rDNA gene. Shown is the proportion of total sequence reads that mapped to H. pylori or H. suis in individual biopsies. Adapted from Martin et al.9
Rodent models have also been used to assess the impact of H. pylori colonization on the resident gastric microbiota. In wild type mice, culture-independent studies found that neither long nor short-term H. pylori infection had a substantial impact on the gastric microbiota.18,19 Studies in hypergastrinemic INS-GAS mice demonstrated that H. pylori infection caused a reduction in Bacteroidetes and an increase in Firmicutes.20 When these mice were gnotobiotically colonized with members of the altered Schaedler’s flora (ASF), H. pylori infection was accompanied by decreased Bacteroides, and both a decrease (Clostridium) and increase (Lactobacillus) in Firmicutes.21 The Mongolian gerbil is probably the most relevant rodent model for H. pylori-microbiota interactions in the stomach, with a human-like gastric pH of < 2,22,23 and comparable pathology induced by gerbil-adapted H. pylori.24 Although one study suggests that there may be a dynamic interaction between H. pylori and the gastric microbiota in the gerbil model,25 the results are preliminary and based on small numbers of animals.
On balance, current evidence suggests that H. pylori does not have a consistent impact on the underlying structure or composition of the underlying gastric microbiota, though deeper sequencing will be required to confirm that relatively rare taxa are not impacted by H. pylori infection.
Does H. pylori Infection Alter Microbial Communities or Disease at Other Mucosal Sites?
Accumulating data suggest that H. pylori infection can sometimes modulate diseases at mucosal sites outside the stomach. For example, epidemiological studies suggest that while H. pylori infection promotes gastric cancer, it actually protects against esophageal cancer, which is increasing in epidemic proportions in developing countries where H. pylori prevalence is declining.26 H. pylori can also enhance humoral immunity to an oral vaccine of the enteric pathogen Salmonella enterica serovar Typhi in humans,27 and repress S. Typhimurium-induced colitis in mice, through a Treg dependent mechanism.28 The mucosal effects of H. pylori infection may even extend beyond the gastrointestinal tract. For example, H. pylori infection is associated with a decreased risk of asthma,29 particularly in young children.30 These observations have been supported by mechanistic studies demonstrating that H. pylori mitigates allergic airway disease in a murine model of asthma due to the induction of anti-inflammatory regulatory T-cells.31 Observational studies also suggest that H. pylori may protect against active tuberculosis in humans and cynomolgous macaques, which is associated with an increased production of IFNγ and an enhanced Th1 immune response to M. tuberculosis.32 Thus, there is immune “crosstalk” between H. pylori in the stomach and organisms at distant mucosal sites. However, the protection afforded by H. pylori against other mucosal pathogens is not uniform, even among those that also provoke a strong Th1 immune response. For example, H. pylori-infected mice that were subsequently inoculated with influenza A virus had viral titers equivalent to H. pylori-uninfected controls.33
How might H. pylori modulate disease at mucosal surfaces outside the stomach? One possibility is that H. pylori may in fact be resident in these other sites, though it is usually thought to colonize only the stomach and proximal duodenum.2 Perhaps more likely, H. pylori could affect mucosal diseases at distant sites via its effects on immune cells that traffic between the mesenteric lymph nodes and other mucosal sites, a mechanism that has been proposed to explain protection against asthma.31,34 In either case, whether by direct colonization or by immune mediated effects, H. pylori might alter the microbiota at mucosal surfaces outside the stomach. In the rhesus monkey model, the inoculation of H. pylori did not affect the community membership or structure of the oral microbiota.9 However, there are few if any other studies that address this question, and it remains a topic for future investigations.
Does the Gastric Microbiota Change H. pylori Colonization or the Host Response to H. pylori Infection?
The gastric microbiota is another potential factor—in addition to bacterial genetics, host genetics, and environment—that may alter the colonization dynamics of H. pylori and the host response to infection. The critical role of the microbiota in protecting the host from enteric pathogens is increasingly clear; Clostridium difficile colitis in the setting of antibiotics is perhaps the best example, but there are others as well.35-37 Changes in the microbiota can even affect the host response to influenza virus.38 More subtle differences in microbiota have also been linked to host susceptibility to enteric Helicobacter spp. For example, Yang et al. found that H. hepaticus could induce colitis in IL10−/− mice bred at only one of two breeding facilities, and this trait was associated with a distinct intestinal microbiota in uninfected mice.39 A complex gut microbiota has also been associated with enhanced clearance of H. felis, another gastric helicobacter.40
However, studies performed by us and others that have looked for similar vendor or antibiotic effects on H. pylori colonization have met with negative results. A single strain of mouse purchased from different laboratories can have profoundly different microbiota and immune profiles in the lower GI tract,41 yet carry similar levels of H. pylori in the stomach.19 We have made similar observations (Fig. 3). Nevertheless, the composition of the gut microbiota does appear to play a role in the host response to H. pylori infection and severity of disease in mice. For example, antibiotic treatment prior to H. pylori infection led to a reduction in overall gastric inflammation and CD4+ T-cell recruitment compared with untreated mice.19 This phenotype coincided with altered membership in the gastric community, although it is not clear if the differences in pathology were due to changes in the gastric or non-gastric microbial communities. Modulation of H. pylori pathogenesis has also been observed in mice co-infected with non-pylori Helicobacter spp. or with the parasitic roundworm Heligmosomoides polygyrus. These species are all common inhabitants of the murine gut that appear to redirect the T-cell response away from Th1. Co-infection with the H. polygyrus was associated with reduced pathology, possibly through the synergistic increase in immunosuppressive Treg cells,42 and mice co-infected with H. muridarum developed less gastritis and had lower levels of both Th1 and Th17 cytokines.43 Conversely, H. hepaticus co-infection increased gastritis and was associated with a decrease in Th1 and increase in Th17 cytokines.43 These studies add to the mounting evidence that the presence of other species at distant mucosal sites can influence the host response to H. pylori by shifting the balance among Th1, Th17, and Treg lymphocytes.
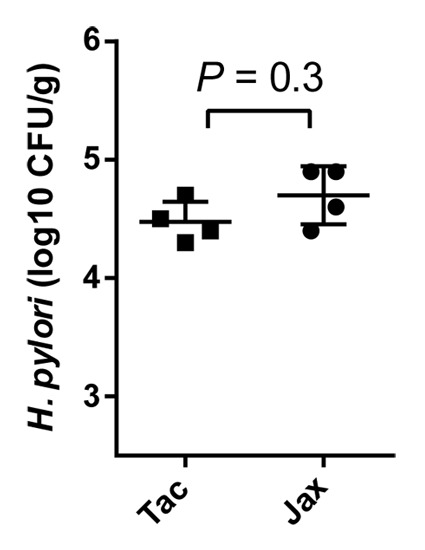
Figure 3.H. pylori colonization is similar in mice from Taconic Farms and from the Jackson Laboratory. C57BL/6 mice from Taconic Farms (Tac) and the Jackson Laboratory (Jax) were inoculated with H. pylori strain J166 and assayed for H. pylori CFU/g gastric tissue five days later. There was no difference in H. pylori colonization level in mice from the two vendors. The Mann-Whitney U test was performed using GraphPad Prism (P > 0.05 is nonsignificant).
Strong support for the effects of host microbiota on the pathology associated with H. pylori infection comes from recent studies of the INS-GAS mouse, a transgenic model of gastric cancer in which gastrin is overexpressed from the insulin promoter.44 Specific pathogen free (SPF) INS-GAS mice, which harbor a complex gut microbiota, progressed more rapidly to gastritis and neoplastic lesions following H. pylori infection than germ free mice colonized with H. pylori alone.20 Strikingly, even colonization with just three members of the altered Schaedler flora (ASF) was sufficient to recapitulate the more aggressive pathology found in conventional INS-GAS mice.21 Since three members of the ASF recapitulate the neoplastic effects of conventional microbiota, it seems unlikely that there is a single or even a few unique species that, together with H. pylori, promote neoplasia. Nevertheless, if the fingerprint of a cancer promoting microbial community could be identified, this might provide another biomarker that could be used to identify H. pylori-infected patients who are at greatest risk of gastric cancer.45
Conclusion
The stomach is not sterile, nor is H. pylori alone. Although there may not be a consistent dynamic between H. pylori colonization and the distinct gastric microbiota, it appears that the constituency of bacterial communities can sometimes promote and other times mitigate H. pylori disease, depending on its composition. If so, this may provide an opportunity for translational application as a biomarker for the risk of serious H. pylori disease, and perhaps even identification of organisms for therapeutic eradication.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
Work in the laboratory of J.V.S. is supported by Public Health Service Grants AI070803, AI081037, CA136647, and AI080788.
References
- 1.Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–5. [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–8. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–76. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wroblewski LE, Peek RM, Jr., Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–39. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, et al. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479–92. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM, Jr., Algood HM, Cover TL. High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis. Infect Immun. 2013;81:2258–67. doi: 10.1128/IAI.01271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Müller A, Solnick JV. Inflammation, immunity, and vaccine development for Helicobacter pylori. Helicobacter. 2011;16(Suppl 1):26–32. doi: 10.1111/j.1523-5378.2011.00877.x. [DOI] [PubMed] [Google Scholar]
- 9.Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA, Solnick JV. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One. 2013;8:e76375. doi: 10.1371/journal.pone.0076375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haesebrouck F, Pasmans F, Flahou B, Chiers K, Baele M, Meyns T, Decostere A, Ducatelle R. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin Microbiol Rev. 2009;22:202–23. doi: 10.1128/CMR.00041-08. [Table of Contents.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–7. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peris-Bondia F, Latorre A, Artacho A, Moya A, D’Auria G. The active human gut microbiota differs from the total microbiota. PLoS One. 2011;6:e22448. doi: 10.1371/journal.pone.0022448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–9. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763–72. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 17.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan MP, Kaparakis M, Galic M, Pedersen J, Pearse M, Wijburg OL, Janssen PH, Strugnell RA. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl Environ Microbiol. 2007;73:1010–3. doi: 10.1128/AEM.01675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolig AS, Cech C, Ahler E, Carter JE, Ottemann KM. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infect Immun. 2013;81:1382–9. doi: 10.1128/IAI.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–20. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teyssen S, Chari ST, Scheid J, Singer MV. Effect of repeated boluses of intravenous omeprazole and primed infusions of ranitidine on 24-hour intragastric pH in healthy human subjects. Dig Dis Sci. 1995;40:247–55. doi: 10.1007/BF02065405. [DOI] [PubMed] [Google Scholar]
- 23.Mollenhauer-Rektorschek M, Hanauer G, Sachs G, Melchers K. Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res Microbiol. 2002;153:659–66. doi: 10.1016/S0923-2508(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 24.Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L, Perez-Perez GI, Hatakeyama M, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:10646–51. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osaki T, Matsuki T, Asahara T, Zaman C, Hanawa T, Yonezawa H, Kurata S, Woo TD, Nomoto K, Kamiya S. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb Pathog. 2012;53:12–8. doi: 10.1016/j.micpath.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 26.de Martel C, Llosa AE, Farr SM, Friedman GD, Vogelman JH, Orentreich N, Corley DA, Parsonnet J. Helicobacter pylori infection and the risk of development of esophageal adenocarcinoma. J Infect Dis. 2005;191:761–7. doi: 10.1086/427659. [DOI] [PubMed] [Google Scholar]
- 27.Muhsen K, Pasetti MF, Reymann MK, Graham DY, Levine MM. Helicobacter pylori Infection Affects Immune Responses Following Vaccination of Typhoid-Naive US Adults With Attenuated Salmonella Typhi Oral Vaccine CVD 908-htrA. J Infect Dis. 2013 doi: 10.1093/infdis/jit625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins PD, Johnson LA, Luther J, Zhang M, Sauder KL, Blanco LP, Kao JY. Prior Helicobacter pylori infection ameliorates Salmonella typhimurium-induced colitis: mucosal crosstalk between stomach and distal intestine. Inflamm Bowel Dis. 2011;17:1398–408. doi: 10.1002/ibd.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167:821–7. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198:553–60. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, Müller A. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology. 2011;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, Hansen LM, Talat N, Hill PC, Hussain R, et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5:e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin ME, Dieter JA, Luo Z, Baumgarth N, Solnick JV. Predicting the outcome of infectious diseases: variability among inbred mice as a new and powerful tool for biomarker discovery. MBio. 2012;3:e00199–12. doi: 10.1128/mBio.00199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, Maxeiner J, Hansson M, Taube C, Quiding-Järbrink M, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122:1082–96. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–45. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–16. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croswell A, Amir E, Teggatz P, Barman M, Salzman NH. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–53. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–9. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz JM, Durham CG, Schoeb TR, Soltau TD, Wolf KJ, Tanner SM, McCracken VJ, Lorenz RG. Helicobacter felis--associated gastric disease in microbiota-restricted mice. J Histochem Cytochem. 2011;59:826–41. doi: 10.1369/0022155411416242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whary MT, Muthupalani S, Ge Z, Feng Y, Lofgren J, Shi HN, Taylor NS, Correa P, Versalovic J, Wang TC, et al. Helminth co-infection in Helicobacter pylori infected INS-GAS mice attenuates gastric premalignant lesions of epithelial dysplasia and glandular atrophy and preserves colonization resistance of the stomach to lower bowel microbiota. Microbes Infect. 2014 doi: 10.1016/j.micinf.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge Z, Feng Y, Muthupalani S, Eurell LL, Taylor NS, Whary MT, Fox JG. Coinfection with Enterohepatic Helicobacter species can ameliorate or promote Helicobacter pylori-induced gastric pathology in C57BL/6 mice. Infect Immun. 2011;79:3861–71. doi: 10.1128/IAI.05357-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang TC, Dangler CA, Chen D, Goldenring JR, Koh T, Raychowdhury R, Coffey RJ, Ito S, Varro A, Dockray GJ, et al. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118:36–47. doi: 10.1016/S0016-5085(00)70412-4. [DOI] [PubMed] [Google Scholar]
- 45.Cooke CL, Torres J, Solnick JV. Biomarkers of Helicobacter pylori-associated gastric cancer. Gut Microbes. 2013;4:532–40. doi: 10.4161/gmic.25720. [DOI] [PMC free article] [PubMed] [Google Scholar]


