Abstract
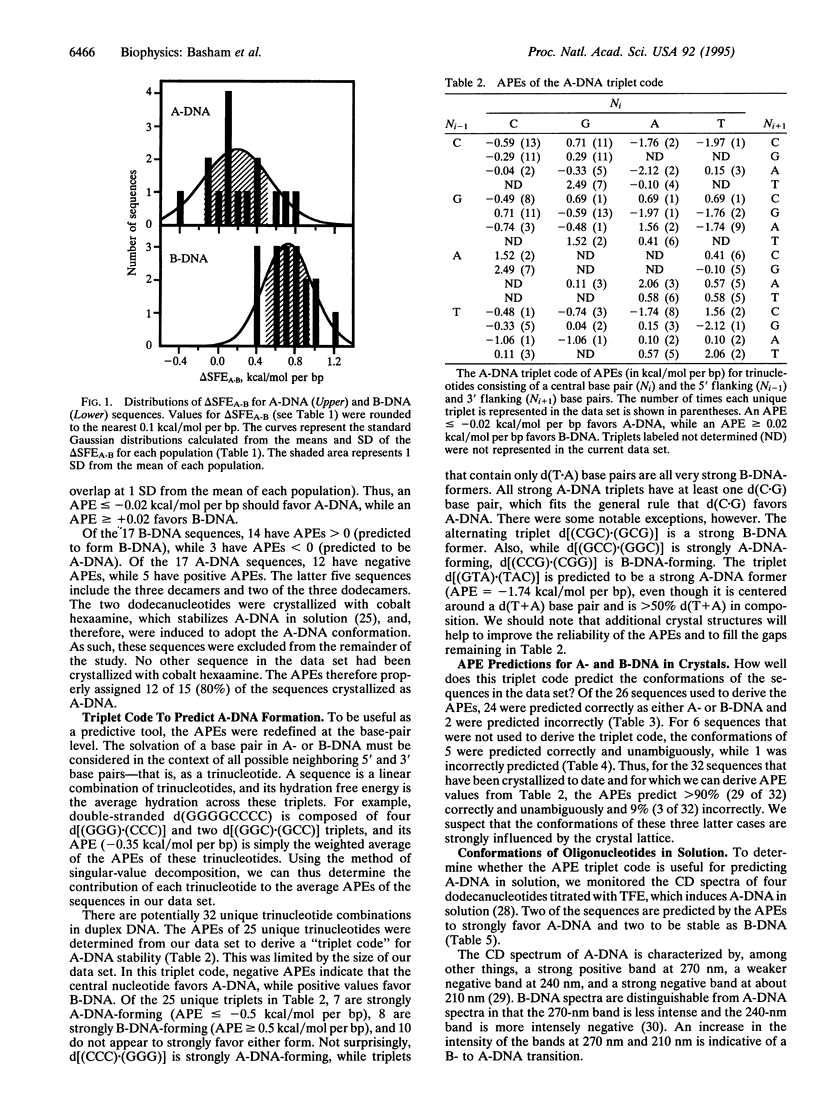
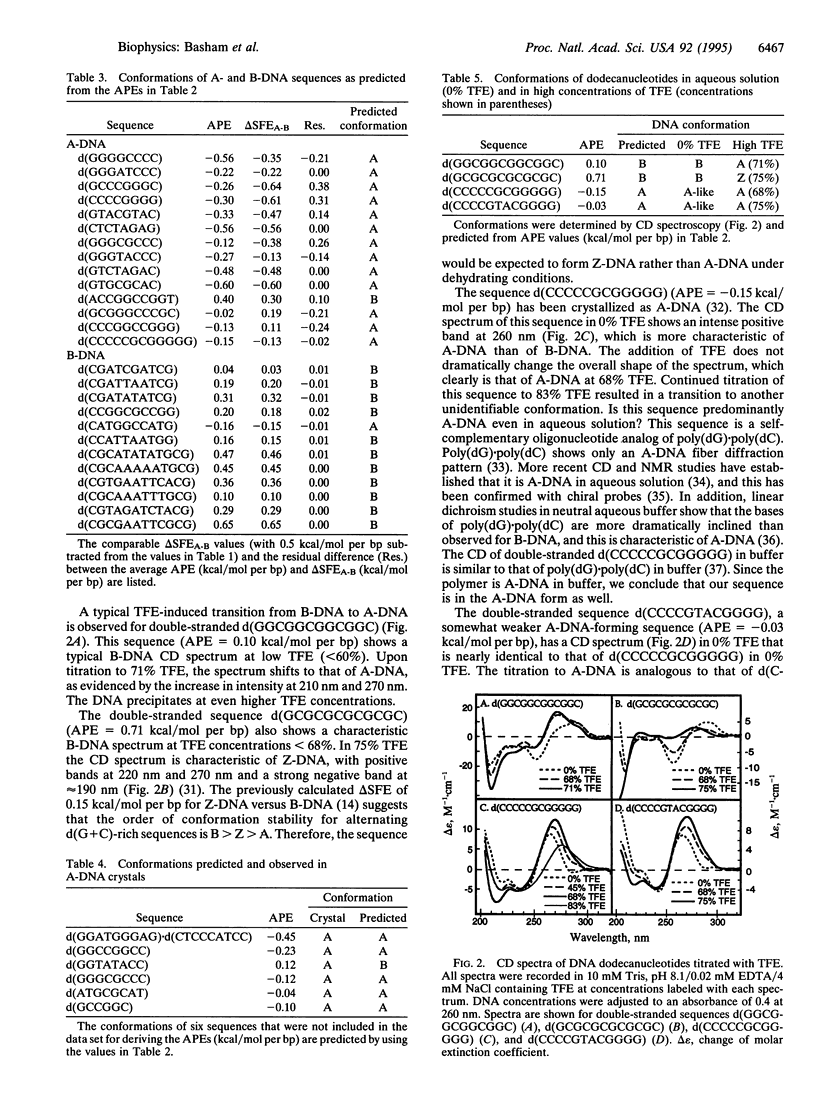
The ability to predict macromolecular conformations from sequence and thermodynamic principles has long been coveted but generally has not been achieved. We show that differences in the hydration of DNA surfaces can be used to distinguish between sequences that form A- and B-DNA. From this, a "triplet code" of A-DNA propensities was derived as energetic rules for predicting A-DNA formation. This code correctly predicted > 90% of A- and B-DNA sequences in crystals and correlates with A-DNA formation in solution. Thus, with our previous studies on Z-DNA, we now have a single method to predict the relative stability of sequences in the three standard DNA duplex conformations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden C. J., Kim S. H. Solvent-accessible surfaces of nucleic acids. J Mol Biol. 1979 Aug 15;132(3):411–434. doi: 10.1016/0022-2836(79)90268-7. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- BRAHMS J., MOMMAERTS W. F. A STUDY OF CONFORMATION OF NUCLEIC ACIDS IN SOLUTION BY MEANS OF CIRCULAR DICHROISM. J Mol Biol. 1964 Oct;10:73–88. doi: 10.1016/s0022-2836(64)80029-2. [DOI] [PubMed] [Google Scholar]
- Barber A. M., Zhurkin V. B., Adhya S. CRP-binding sites: evidence for two structural classes with 6-bp and 8-bp spacers. Gene. 1993 Aug 16;130(1):1–8. doi: 10.1016/0378-1119(93)90339-5. [DOI] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Salisbury S. A., Brown T., Cosstick R., Eckstein F., Kennard O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J Mol Biol. 1986 Dec 20;192(4):891–905. doi: 10.1016/0022-2836(86)90035-5. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. DNA structure from A to Z. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Neidle S. "...the tyranny of the lattice...". Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKLIN R. E., GOSLING R. G. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953 Jul 25;172(4369):156–157. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
- FRANKLIN R. E., GOSLING R. G. Molecular configuration in sodium thymonucleate. Nature. 1953 Apr 25;171(4356):740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- Frederick C. A., Quigley G. J., Teng M. K., Coll M., Van der Marel G. A., Van Boom J. H., Rich A., Wang A. H. Molecular structure of an A-DNA decamer d(ACCGGCCGGT). Eur J Biochem. 1989 May 1;181(2):295–307. doi: 10.1111/j.1432-1033.1989.tb14724.x. [DOI] [PubMed] [Google Scholar]
- Gray D. M. A circular dichroism study of poly dG, poly dC, and poly dG:dC. Biopolymers. 1974;13(10):2087–2102. doi: 10.1002/bip.1974.360131011. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Alings C. Crystallographic study of one turn of G/C-rich B-DNA. J Mol Biol. 1989 Nov 20;210(2):369–381. doi: 10.1016/0022-2836(89)90337-9. [DOI] [PubMed] [Google Scholar]
- Ho P. S., Kagawa T. F., Tseng K. H., Schroth G. P., Zhou G. W. Prediction of a crystallization pathway for Z-DNA hexanucleotides. Science. 1991 Nov 15;254(5034):1003–1006. doi: 10.1126/science.1948069. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Krylov DYu A-DNA in solution as studied by diverse approaches. Methods Enzymol. 1992;211:111–127. doi: 10.1016/0076-6879(92)11008-7. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Kagawa T. F., Howell M. L., Tseng K., Ho P. S. Effects of base substituents on the hydration of B- and Z-DNA: correlations to the B- to Z-DNA transition. Nucleic Acids Res. 1993 Dec 25;21(25):5978–5986. doi: 10.1093/nar/21.25.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T. F., Stoddard D., Zhou G. W., Ho P. S. Quantitative analysis of DNA secondary structure from solvent-accessible surfaces: the B- to Z-DNA transition as a model. Biochemistry. 1989 Aug 8;28(16):6642–6651. doi: 10.1021/bi00442a017. [DOI] [PubMed] [Google Scholar]
- Kang H., Johnson W. C., Jr Infrared linear dichroism reveals that A-, B-, and C-DNAs in films have bases highly inclined from perpendicular to the helix axis. Biochemistry. 1994 Jul 12;33(27):8330–8338. doi: 10.1021/bi00193a021. [DOI] [PubMed] [Google Scholar]
- Mei H. Y., Barton J. K. Tris(tetramethylphenanthroline)ruthenium(II): a chiral probe that cleaves A-DNA conformations. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1339–1343. doi: 10.1073/pnas.85.5.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers B. H., Schroth G. P., Baxter W. W., Ho P. S. Alternating and non-alternating dG-dC hexanucleotides crystallize as canonical A-DNA. J Mol Biol. 1995 Jun 16;249(4):772–784. doi: 10.1006/jmbi.1995.0336. [DOI] [PubMed] [Google Scholar]
- Peticolas W. L., Wang Y., Thomas G. A. Some rules for predicting the base-sequence dependence of DNA conformation. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2579–2583. doi: 10.1073/pnas.85.8.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazance J. H., Johnson W. C., Jr, McIntosh L. P., Jovin T. M. Vacuum UV circular dichroism is diagnostic for the left-handed Z form of poly [d(A-C).d(G-T)] and other polydeoxynucleotides. Nucleic Acids Res. 1987 Sep 25;15(18):7627–7636. doi: 10.1093/nar/15.18.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Gupta G., Sarma R. H. 500-MHz 1H NMR study of poly(dG).poly(dC) in solution using one-dimensional nuclear Overhauser effect. Biochemistry. 1986 Jun 17;25(12):3659–3665. doi: 10.1021/bi00360a028. [DOI] [PubMed] [Google Scholar]
- Schneider B., Cohen D., Berman H. M. Hydration of DNA bases: analysis of crystallographic data. Biopolymers. 1992 Jul;32(7):725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Chou P. J., Ho P. S. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J Biol Chem. 1992 Jun 15;267(17):11846–11855. [PubMed] [Google Scholar]
- Setlow P. DNA in dormant spores of Bacillus species is in an A-like conformation. Mol Microbiol. 1992 Mar;6(5):563–567. doi: 10.1111/j.1365-2958.1992.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Sprecher C. A., Baase W. A., Johnson W. C., Jr Conformation and circular dichroism of DNA. Biopolymers. 1979 Apr;18(4):1009–1019. doi: 10.1002/bip.1979.360180418. [DOI] [PubMed] [Google Scholar]
- Timsit Y., Moras D. Crystallization of DNA. Methods Enzymol. 1992;211:409–429. doi: 10.1016/0076-6879(92)11022-b. [DOI] [PubMed] [Google Scholar]
- Verdaguer N., Aymami J., Fernández-Forner D., Fita I., Coll M., Huynh-Dinh T., Igolen J., Subirana J. A. Molecular structure of a complete turn of A-DNA. J Mol Biol. 1991 Sep 20;221(2):623–635. doi: 10.1016/0022-2836(91)80077-8. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Warne S. E., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the -10 and -35 regions are dependent on spacer DNA sequence. Biochemistry. 1993 Jun 22;32(24):6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- Xu Q., Shoemaker R. K., Braunlin W. H. Induction of B-A transitions of deoxyoligonucleotides by multivalent cations in dilute aqueous solution. Biophys J. 1993 Sep;65(3):1039–1049. doi: 10.1016/S0006-3495(93)81163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]



