Abstract
In last two decades, the study of epigenetic modification emerged as one of the major areas of cancer treatment targeted by dietary phytochemicals. Recent studies with various types of cancers revealed that the epigenetic modifications are associated with the food source corresponds to dietary phytochemicals. The dietary phytochemicals have been used in Asian countries for thousands of years to cure several diseases including cancer. They have been reported to modulate the several biological processes including histone modification, DNA methylation and non-coding microRNA expression. These events play a vital role in carcinogenesis. Various studies suggest that a number of dietary compounds present in vegetables, spices and other herbal products have epigenetic targets in cancer cells. Dietary phytochemicals have been reported to repair DNA damage by enhancing histone acetylation that helps to restrain cell death, and also alter DNA methylation. These phytochemicals are able to modulate epigenetic modifications and their targets to cure several cancers. Epigenetic aberrations dynamically contribute to cancer pathogenesis. Given the individualized traits of epigenetic biomarkers, the personalized nutrition will help us to prevent various types of cancer. In this review, we will discuss the effect of dietary phytochemicals on genetic and epigenetic modifications and how these modifications help to prevent various types of cancers and improve health outcomes.
Keywords: Epigenetics, Phytochemicals, DNA-methylation, Histone-modification, miRNA, Carcinogenesis
1. Introduction
Dietary intervention experiments and epidemiological studies in humans using laboratory animals have provided evidence to suggest that lifestyle and environmental factors play a critical role in the development of a wide variety of neoplasms. Environmental factors including chemical carcinogens, environmental pollutants, dietary contaminants and physical carcinogens play important role in the etiology of human cancer (Kupchella CE 1986). Additionally, lifestyle factors, such as alcohol consumption, smoking, exposure to sunlight, increased fat consumption and chronic stress can also promote the development and progression of cancer (Stein CJ and Colditz GA 2004). It has further been demonstrated that maternal nutrition imbalance and metabolic disturbances during embryonic development have a persistent effect on the health of the offspring and may be passed down to the next generation (Attig L et al 2010). These studies provide evidence that cancer is a complex disease and manifestation of both genetic and epigenetic modifications (Macaluso M, Paggi MG et al. 2003). Cancer initiation and progression are primarily driven by acquired genetic alterations however microenvironment-mediated epigenetic perturbations play an important role in neoplastic development (Cho HS, Park JH et al. 2007). “Epigenetics” is defined as heritable changes in gene activity and expression that occur without alteration in DNA sequences and are sufficiently powerful to regulate the dynamics of gene expression (Goldberg AD, Allis CD et al. 2007; Stefanska B, Karlic H et al. 2012). Epigenetic modifications are potentially reversible, which makes them attractive and promising avenues for catering cancer preventive and therapeutic strategies. The key processes responsible for epigenetic regulation are DNA methylation, modifications in chromatin [covalent modification of core histones], and post-transcriptional gene regulation by non-coding RNA [micro-RNAs] (Dehan P, Kustermans G et al. 2009; Lim U and Song MA 2012). Additionally, some examples of genetic modifiers that alter epigenetic modifications are discussed in Table 1.
Table 1.
Genetic Factors Which Control Epigenetic Modifications
| S.No | Gene | Epigenetic Modifications | Function | Tumor Type | Alteration | References |
|---|---|---|---|---|---|---|
| 1. | ASXL | Histone Modification | Enhancer of trithorax and polycomb group (EAP) Additional sex combs like 1 | Bohring-Opitz Syndrome, MDS and AML | Mutation | (Hoischen A, van Bon BW et al. 2011; Gelsi-Boyer V, Brecqueville M et al. 2012) |
| 2. | BMI-1 | Histone Modification | PRC1 subunit | Ovarian, mantle cell lymphomas and Merkel cell carcinomas | Overexpression | (Jiang L, Li J et al. 2009; Lukacs RU, Memarzadeh S et al. 2010) |
| 3. | BRD4 | Histone modification | Bromodomain containing 4 | Midline carcinoma, nuclear protein in testis, breast, colon, and AML | Translocation (fusion protein), aberrant expression | (Filippakopoulos P, Qi J et al. 2010; Zuber J, Shi J et al. 2011) |
| 4. | CREBBP | Histone Modification | Histone acetyltransferase | Colorectal, epithelial, gastric and ovarian, lung, esophageal cancer | Mutation, overexpression | (Miremadi A, Oestergaard MZ et al. 2007) |
| 5. | EP300 | Histone Modification | Histone deacetyltransferase | Colorectal, breast, pancreatic cancer | Mutation | (Miremadi A, Oestergaard MZ et al. 2007) |
| 6. | EZH2 | Histone Modification | Histone methyltransferase H3K27 | Colon, pancreas, liver, gastric, uterine tumors, breast, prostate, bladder, melanoma, lymphoma, myeloma, and Ewing’s sarcoma | Mutation, aberrant expression | (Chase A and Cross NC 2011; Tsang DP and Cheng AS 2011) |
| 7. | G9a | Histone Modification | Histone methyltransferase H3K9 | Cervical, uterine, HCC, ovarian, and breast cancer | Aberrant expression | (Varier RA and Timmers HT 2011) |
| 8. | HDAC2 | Histone Modification | Histone deacetyltransferase | Gastric, Colonic, endometrial cancer | Mutation | (Ropero S, Fraga MF et al. 2006) |
| 9. | JARID1B/C | Histone Modification | Histone demethylase H3K4/H3K9 | RCCC, testicular and breast, | Overexpression | (Rotili D 2011) |
| 10. | LSD1 | Histone Modification | Histone demethylase H3K4/H3K9 | Prostate | Mutation | (Rotili D 2011) |
| 11. | MLL1/2/3 | Histone modification | Histone methyltransferase H3K4 | Non-Hodgkin lymphoma, B cell lymphoma, Bladder TCC, ALL and AML, prostate (primary) | Mutation, translocation, aberrant expression | (Morin RD, Mendez-Lago M et al. 2011; Yaoting Gui, Guangwu Guo et al. 2011) |
| 12. | PCAF | Histone Modification | Histone acetyltransferase | Epithelial | Mutation | (Miremadi A, Oestergaard MZ et al. 2007) |
| 13. | PRMT1/5 | Histone Modification | Protein arginine methyltransferase | Breast/gastric | Aberrant expression | (Miremadi A, Oestergaard MZ et al. 2007) |
| 14. | SIRT1, HDAC5/7A | Histone modification | Histone deacetyltransferase | Colorectal, breast, prostate cancer | Mutation, aberrant expression | (Miremadi A, Oestergaard MZ et al. 2007) |
| 15. | UTX (KDM6A) | Histone Modification | Histone demethylase H3K27 | Breast, kidney, lung, pancreas, bladder, esophagus, colon, uterus, brain | Mutation | (Rotili D 2011) |
| 16. | AID | DNA methylation | 5’cytidine deaminase | CML | Aberrant expression | (De Carvalho DD, You JS et al. 2010) |
| 17. | DNMT1 | DNA methylation | DNA methyltransferase | Pancreatic, gastric, breast, colorectal, non-small cell lung cancer | Mutation, Overexpression | (Kanai Y, Ushijima S et al. 2003; Wu SC and Zhang Y 2010) |
| 18. | DNMT3A | DNA methylation | DNA methyltransferase | MDS, AML | Mutation | (Ley TJ, Ding L et al. 2010; Yamashita Y, Yuan J et al. 2010) |
| 19. | DNMT3B | DNA methylation | DNA methyltransferase | ICF syndrome, SNPs in breast and lung adenoma | Mutation | (Wijmenga C, Hansen RS et al. 2000; Shen H, Wang L et al. 2002) |
| 20. | IDH1/2 | DNA methylation | Isocitrate dehydrogenase | Glioma, AML | Mutation | (Figueroa ME, Abdel-Wahab O et al. 2010; Lu C, Ward PS et al. 2012) |
| 21. | MBD1/2 | DNA methylation | Methyl binding protein | Breast and lung cancer | Mutation | (Sansom OJ, Maddison K et al. 2007) |
| 22. | TET1 | DNA methylation | 50methylcytosine hydroxylase | AML | Chromosome translocation | (De Carvalho DD, You JS et al. 2010; Wu SC and Zhang Y 2010) |
| 23. | TET2 | DNA methylation | 50methylcytosine hydroxylase | Myeloid malignancies (AML), MDS, gliomas | Mutation/silenc ing | (Tan AY and Manley JL 2009) |
| 24. | ARID1A (BAF250A) | Chromatin remodeling | BAF subunit | Carcinomas, endometrial carcinomas, ovarian clear cell carcinomas, 30% of endometrioid | Mutation, genomic rearrangement, low expression | (Jones S, Wang TL et al. 2010; Guan B, Mao TL et al. 2011) |
| 25. | ARID2 (BAF200) | Chromatin remodeling | PBAF subunit | Primary pancreatic adenocarcinomas | Mutation | (Li M, Zhao H et al. 2011) |
| 26. | BRD7 | Chromatin remodeling | PBAF subunit | Bladder TCC | Mutation | (Drost J, Mantovani F et al. 2010) |
| 27. | BRG1(SMA CA4) | Chromatin remodeling | ATPase of BAF | Lung, rhabdoid, medulloblastoma | Mutation, low expression | (Wilson BG and Roberts CW 2011) |
| 28. | BRM (SMARCA2) | Chromatin remodeling | ATPase of BAF | Prostate, basal cell carcinoma | Mutation, low expression | (Sun A, Tawfik O et al. 2007; de Zwaan SE and Haass NK 2010) |
| 29. | CHD4/5 | Chromatin remodeling | ATPase of NURD | Gastric and colorectal cancer, ovarian, prostate, neuroblastoma | Mutation | (Bagchi A, Papazoglu C et al. 2007; Kim MS, Chung NG et al. 2011; Wang J, Wu Z et al. 2012) |
| 30. | CHD7 | Chromatin remodeling | ATP-dependent helicase | Gastric and colorectal | Mutation | (Wessels K, Bohnhorst B et al. 2010) |
| 31. | P400/Tip60 | Chromatin remodeling | ATPase of SWR1, acetylase of SWR1 | lymphomas, colon, head-and-neck, breast | Mutation, aberrant expression | (Mattera L, Escaffit F et al. 2009) |
| 32. | PBRM1 (BAF180) | Chromatin remodeling | PBAF subunit | Breast tumor | Mutation | (Varela I, Tarpey P et al. 2011) |
| 33. | SNF5 (SMARCB1, INI1) | Chromatin remodeling | Gastric and colorectal | Kidney malignant rhabdoid tumors, atypical rhabdoid/teratoid tumors (extra-renal), epithelioid sarcomas, small cell hepatoblastomas, extraskeletal myxoid chondrosarcomas, and undifferentiated sarcomas | Mutation, silencing, loss of expression | (Wilson BG and Roberts CW 2011) |
| 34. | SRCAP | Chromatin remodeling | ATPase of SWR1 | Prostate | Aberrant expression | (Balakrishnan A, Bleeker FE et al. 2007) |
2. Histone modification
The histone modifications in chromatin structure play important role in the gene regulations and carcinogenesis (Sawan C and Herceg Z 2010). Chromatin proteins significantly involve in the packaging of eukaryotic DNA into higher order chromatin fibers. Each nucleosome consist of ~146 bp of DNA packed around an octamer of histone proteins, and the octamers mainly consist of double subunits of H2A, H2B, H3 and H4 core histone proteins. Histone proteins are regulators of chromatin dynamics either by providing protein recognition sites by specific modifications or changing chromatic structure by altering electrostatic charge (Mills AA 2010; Suganuma T and Workman JL 2011). Histone modifications are specifically characterized by the genomic regulatory regions, for example inactive promoters which are enriched in trimethylated H3 at lysine 27 (H3K27me3) or trimethylated H3 at lysine 9 (H3K9me3), active promoter regions which are enriched in trimethylated H3 at lysine 4 (H3K4me3) and regulatory enhancers that are enriched in monomethylated H3 at lysine 4 (H3K4me1) and/or acetylated H3 at lysine 27 (H3K27ac) (Hon GC, Hawkins RD et al. 2009; Mills AA 2010; Hawkins RD, Hon GC et al. 2011). The histone proteins coordinate the changes between tightly packed DNA [heterochromatin] and exposed DNA [euchromatin] which are inaccessible to transcription and available for binding to and regulation of transcription factors respectively. These changes occur because of structural characteristics of the nucleosome that are known as ‘histone tails’, which extend from the core octamer. Histone tails are the major sites for posttranslational modifications which consist of Ntermini of the histone proteins. The two opposing group of enzymes (histone deacetylases (HDACs) and histone acetyltransferases (HATs)) involved in chromatin remodeling (Fig. 1). It has been reported that the dietary phytochemicals are involved in chromatin remodeling by acting on the enzymes HDACs and HATs (Hardy TM and Tollefsbol TO 2011). These enzymes are involved in genes deregulations which have been associated with the acetylation of histone proteins by HDACs and HATs. HATs catalyze histone acetylation by neutralizing the positive charge and facilitating the binding of transcription factors to nucleosomal DNA on the ε-amino groups of lysine residues in the N-terminal tails of core histones. On the contrary, HDACs catalyze deacetylation by cleavage of acetyl groups, typically producing a compact chromatin configuration that restricts transcription factor access to DNA and repressing gene expression. HDACs and HATs encompass a large group of enzymes which are classified into several families and control various physiological functions of the cells (Sawan C and Herceg Z 2010; Li Q and Chen H 2012).
Fig. 1. Dietary Inhibitors of Histone Modifications.
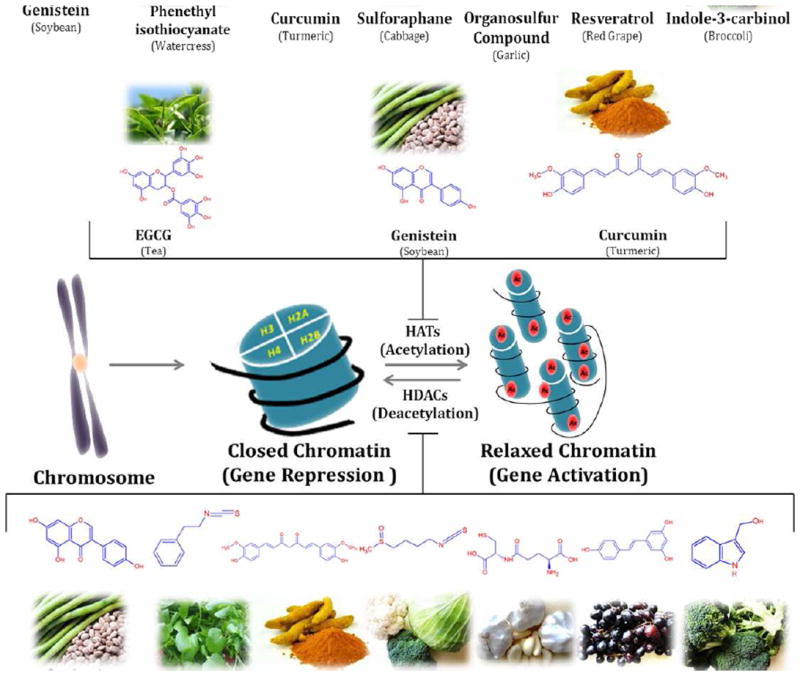
Figure shows the representation of histone modifications (acetylation and deacetylation) by the phytochemicals derived from different food source. Phytochemicals like EGCG, genistein and curcumin play important role in inhibition of histone acetylation by inactivating histone acetyl transferase enzyme. Some other phytochemicals like sulforaphane, curcumin, genistein, phenyl isothiocynate, organosulfur compound, resveratrol and indol-3-carbinol inhibits the deacetylation of relaxed chromatine by inactivating histone deacetylase enzyme. There are several other phytochemicals that alter histone modifications which are not covered in this figure.
3. DNA methylation
DNA methylation is accountable for regulating gene expression and interacting with the nucleosomes that control DNA packaging, and can affect entire domains of DNA (Issa JP and Kantarjian HM 2009). In mammalian cells, DNA methylation occurs within CpG dinucleotides through addition of a methyl group at the 5’ position of the cytosine ring, forming 5-methyl cytosine, in a reaction catalyzed by enzymes known as DNA methyl transferases [DNMTs] (Fig. 2) (Issa JP and Kantarjian HM 2009). There are three principle DNA methyltransferases: DNMT1, DNMT3a and DNMT3b. DNMT1 is the primary maintenance enzyme that preserves existing methylation patterns following DNA replication by adding methyl groups to corresponding daughter strands at the hemi-methylated CpG sites. DNMT3a and DNMT3b are methyltransferases that preferentially target unmethylated CpGs to initiate de novo methylation; they are highly expressed during embryogenesis but minimally expressed in adult tissues. A fourth family member, DNMT-3L, lacks intrinsic methyltransferase activity; however it facilitates methylation of retrotransposons by interaction with DNMT3a and 3b (Denis H, Ndlovu MN et al. 2011). DNA methylation regulates gene expression in normal tissues through genomic imprinting and female Xchromosome inactivation. Contrasting normal tissues, these processes are significantly altered in cancer due to a process known as ‘loss of imprinting’ [LOI]. LOI is the earliest genomic lesion observed in Wilms’ tumors and in stem cell populations of organs and tissues, ultimately leading to additional downstream genetic and epigenetic perturbations (Jelinic P and Shaw P 2007).
Fig. 2. Dietary Inhibitors of DNA Methylations.
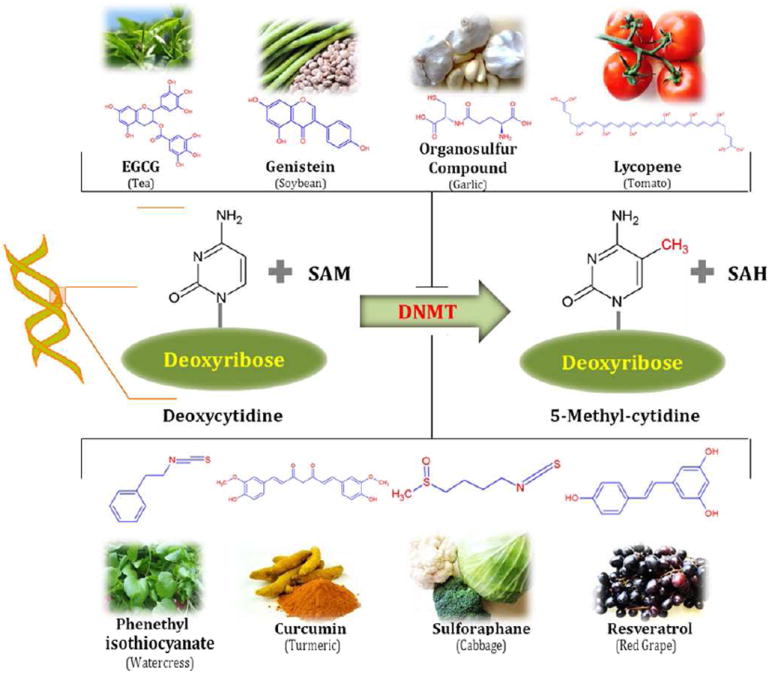
DNA methylation is a biochemical process that is essential for the development of higher organism. Some diatery phytochemicals are reported to inhibit the methylation of cytocine of cytidine. Hypermethylation of cytidine by DNMTs usually results in transcriptional gene silencing and gene inactivation. Several phytochemicals derived from different food source such as: resveratrol from graps and berries, curcumin from turmeric tea phenols from tea leaves, genistein from soybeans, sulforaphane from broccoli, phenethyl isothiocynate from cauliflower, organosulfur compounds from galic, quercetin from citrus fruits and lycopene from tomato act as dietary inhibitors of DNA methyltransferases and also alter gene expression via epigenetic mechanisms.
In addition to regulation by DNA methylation, methylated DNA binding proteins [MBD’s] can bind to methylated cytosine, and sequentially form a complex with histone deacetylase [HDAC] leading to chromatin compaction and gene silencing (Duthie SJ 2011). Up till now, six methyl-CpGbinding proteins, including MECP2, MBD1, MBD2, MBD3, MBD4 and Kaiso, have been identified in mammals. MECP2 binds methylated DNA in vitro and in vivo; it contains a methyl-CpG-binding domain [MBD] at its amino terminus and a transcription repression domain [TRD] in the central domain. MBDs1–4 were cloned on the basis of their sequence homology to MECP2 in the MBD, and all except MBD3 bind preferentially to the methylated CpG islands. MBD1 and MBD2 also function as transcription repressors, whereas MBD4 is a DNA glycosylase and is involved in DNA mismatch repair. Kaiso, although lacking an MBD domain, binds methylated CGCG through its zinc-finger domain. Different methyl-CpG binding proteins may recruit diverse chromatin-remodeling proteins and transcription-regulatory complexes to methylated DNA targets in the genome. Furthermore, it has been demonstrated that nucleosome remodeling complex [NuRD] can methylate DNA by interacting with DNA methylation binding protein MBD2, which directs the NuRD complex to methylate DNA (Lan J, Hua S et al. 2010).
In humans, ~70% of all CpG islands are methylated, primarily in the heterochromatin i.e. tightly packed form regions of the DNA, and these methylated CpG islands are thought to be critical for the control of gene silencing and chromosomal stability. In contrast, euchromatin (relaxed region in the DNA) CpG islands remain locally unmethylated, allowing access to transcription factors and chromatin-associated proteins for the expression of housekeeping genes and other regulatory genes. In cancer cells, global hypomethylation is accompanied by the hypermethylation of localized promoter-associated CpG islands, which are usually unmethylated in normal cells. Global hypomethylation can lead to chromosomal instability, mutations and reactivation of various oncogenes. DNMT1 is responsible for the establishment of the DNA methylation pattern during DNA synthesis, its deficiency in cells may lead to global hypomethylation. Another common alternation observed in cancer cells is DNA hypermethylation of promoter-associated CpG islands of tumor suppressor genes, which could serves as a surrogate for point mutations or deletions to cause transcriptional silencing of these genes (Jones PA 2002; Colacino JA, Arthur AE et al. 2012).
4. Non-coding RNAs
Non-coding RNAs were originally noted to perform enzymatic functions in regulation of gene expression and facilitating RNA splicing. Recently it is recognized that Non-coding RNAs participate in the epigenetic phenomenon of posttranscriptional gene modification. The importance in gene regulation and posttranscriptional gene modification is appreciated after the discovery of miRNAs (microRNAs) and siRNAs (small interfering RNAs), which indicated that ncRNAs are RNAs that are biologically functional, rather than simply being intermediate messengers between DNA and proteins (Parasramka MA, Ho E et al. 2012). They are also known as non-protein coding RNA or microRNA, and are 21-23 nucleotides in length. In the order of 1000 miRNA genes have been predicted in-silico in the human genome, with each miRNA targeting multiple protein coding transcripts. Their significance is demonstrated further by the findings of all transcriptional output in human results from ncRNAs. These ncRNAs are from the exons and introns of non-coding genes as well as from the introns of protein-coding genes, which are synthesized by RNAP II (RNA polymerase II) and RNAP III. Though miRNA are vital to normal cell physiology their misexpression has been linked to carcinogenesis, and miRNA profiles are now being used to classify human cancers (Fig. 3) (Kok TM, Breda SG et al. 2012). The influence of miRNA on the epigenetic machinery and the reciprocal epigenetic regulation of miRNA expression suggest that its deregulation during carcinogenesis has important implications for global regulation of epigenetics and cancer (Garzon R, Calin GA et al. 2009).
Fig. 3. Effect of Dietary Phytochemicals on miRNA.
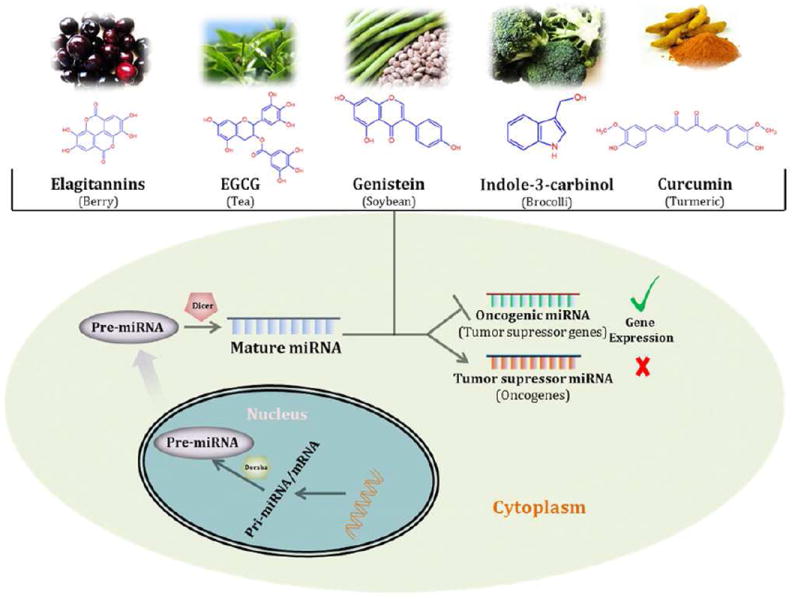
miRNAs are considered to regulates the gene expression. Dietary phytochemicals are reported to inhibit the oncogenic miRNA and promote tumor suppressive miRNA and have impact on the expression level of target RNAs.
5. Dietary agents as epigenetic target
Dietary phytochemicals play an important role in the regulation of pathological progressions and are also involved in normal biological processes. Diseases linked to genetic and epigenetic modifications can be influenced by environmental and dietary factors. In particular, nutritional factors, drugs, chemicals used in pesticides, environmental compounds and inorganic contaminants (i.e., arsenic) can alter the epigenome, and may contribute to the development of abnormalities. Dietary phytochemicals present in fruit, vegetables, beverages and spices have shown to possess potential anticancer properties. There has been considerable interest in the use of naturally occurring phytochemicals for disease prevention including cancer. Previous studies have demonstrated that phytochemicals can work through number of complementary and overlapping mechanisms of action, including induction of detoxification enzymes, antioxidant effects, and inhibition of the formation of nitrosamines, binding/dilution of carcinogens in the digestive tract, alteration of hormone metabolism and modulation of carcinogenic cellular and signaling events (Wang J, Wu Z et al. 2012). However, it was not more than a decade ago, studies demonstrate that phytochemicals could target the activity of various epigenetic factors, such as DNMTs and HDACs and could be useful to prevent and treat various diseases including cancer. Although several dietary agents or nutrients regulate different molecular and epigenetic targets in human cancers, here we summarize the role of some common bioactive dietary phytochemicals and their epigenetic targets in various human cancers. The phytochemicals which we discuss include tea polyphenols, genistein, curcumin, sulforaphane, phenyl isothiocyanate, lycopene, resveratrol, quercetin, indol-3-carbinol, ellagitanin and organosulfur compounds. A brief discussion includes their epigenetic targets in various human cancers leading to their multiple roles in the regulation of cancer prevention and therapy. Additionally, dietary phytochemicals, and their epigenetic targets associated with tumorigenesis are summarized in Table 2.
Table 2.
Epigenetic Regulation by Dietary Phytochemicals
| S.No | Dietary compound | Food source | Epigenetic modification(s) | Epigenetic target(s) | Roles in cancer prevention | References |
|---|---|---|---|---|---|---|
|
| ||||||
| 1. | Resveratrol | Blueberries, mulberries, cranberries, peanuts and grapes | Histone-modifications | TNFα, IL-8, RBP/SIRT1 | Reduce activation of NF kappa B | (Tili E, Michaille JJ et al. 2010) |
| DNA methylation | Unknown/DNMT | |||||
|
| ||||||
| 2. | Curcumin | Turmeric | Histone-modifications | H3 and H4 acetylation, p53, GATA4, GZMB, PRF1, EOMES/HAT, HDAC | Prevention of DNA damage and blocking the inflammatory master molecule NF kappa B | (Mudduluru G, George-William JN et al. 2011) |
| DNA methylation | Unknown/DNMT1 | |||||
| miRNA | SP1, ESR1, PTEN | |||||
|
| ||||||
| 3. | Tea polyphenols | Tea leaves | Histone-modifications | H3 and H4 acetylation, H3K27m3, NF-kB, IL-6, BMI-1, EZH2, SUZ12/HAT, HDAC, HMT | Induce apoptotic cell death and cell cycle arrest in tumor cells | (Tsang WP and Kwok TT 2010) |
| DNA methylation | P16INK4a, RNRβ, MGMT, hMLH1, RECK1, hTERT, WIF-1, RXRα, GSTP1, CDKN2A, RXRβ, CDX2/DNMTI, MBD1, MeCP2 | |||||
| miRNA | Bcl-2, AR | |||||
|
| ||||||
| 4. | Indole-3-carbinol (I3C) and Diindolylmethane (DMI) | Broccoli, cabbage, cauliflower, mustard and radish | Histone-modifications | COX-2/HDAC | Modification of nuclear transcription factors including Sp1, estrogen receptor, NF kappa B and aryl hydrocarbon receptor | (Li Y, VandenBoom TG 2nd et al. 2010) |
| miRNA | ZEB1, EGFR | |||||
|
| ||||||
| 5. | Genistein | Soy beans | Histone-modifications | H3, H4, H2A and H2B acetylation, H3K4me2, H3K9me3, p21, p16, PTEN, p53, FOXA3, BTG3, RARβ, hTERT, CCLD/HAT, HDAC, SIRT1 | Modulation of chromatin configuration and DNA methylation | (Fang MZ, Chen D et al. 2005; King-Batoon A, Leszczynska JM et al. 2008; Majid S, Dar AA et al. 2009) |
| DNA methylation | P16, RARβ2, MGMT, hTERT, BTG3, GSTP1 and EPHB2, HMGNS, CDKN2A/DNMT, MBD1, MBD4, MeCP2 ZEB1, ZBTB10, EGFR | |||||
| miRNA | ZEB1, ZBTB10, EGFR | |||||
|
| ||||||
| 6. | Sulforaphane | Broccoli sprouts, cabbage and kale | Histone-modifications | H3 and H4 acetylation, H3K9ac, H3K9me3, HBD-2, H3K27me3, RARβ, HBD-2, p21, BAX/HDAC | Inhibit the growth of tumor cells and induce apoptosis in cancer cells | (Meeran SM, Patel SN et al. 2010) |
| DNA methylation | Unknown/DNMT1 | |||||
|
| ||||||
| 7. | Phenethyl isothiocynate | Cauliflower, cabbage, cress, bok choy and broccoli | Histone-modifications | H3 and H4 acetylation, p21, GSTP1/HDAC | Induction of cell-cycle arrest | (Wang LG, Beklemisheva A et al. 2007; Izzotti A, Calin GA et al. 2010) |
| DNA methylation | GSTP1/unknown | |||||
|
| ||||||
| 8. | Organosulfur compounds | Garlic | Histone-modifications | H3 and H4 acetylation, p21/HDAC | Inhibition of DNA adduct formation, upregulation of antioxidant defences and DNA repair systems | (Druesne N, Pagniez A et al. 2004) |
| DNA methylation | ||||||
|
| ||||||
| 9. | Quercetin | Citrus fruits and buckwheat | Histone-modifications | IP-10, MIP-2/HAT, SIRT1 | Induction of cell-cycle arrest | (Priyadarsini RV, Vinothini G et al. 2011) |
| DNA methylation | CDKN2A/DNMT | |||||
|
| ||||||
| 10. | Lycopene | Tomato | DNA methylation | GSTP1, RARβ, HIN-1/unknown | Induction of cell-cycle arrest | (King-Batoon A, Leszczynska JM et al. 2008) |
|
| ||||||
| 11. | Ellagitannins | Pomegranate, walnuts and almonds | miRNA | miRNA array | Inhibition of cell proliferation | (Wen XY, Wu SY et al. 2009) |
5.1. Resveratrol
Resveratrol [3, 5, 4’-trihydroxy-trans-stilbene] is a natural poly phenol found in several plants including blueberries, mulberries, cranberries, peanuts and grapes. It is also consumed as a red wine. It has been reported to have anti-cancer, anti-inflammatory and blood-sugar-lowering potential (Savouret JF and Quesne M 2002). It has strong impact on signaling pathways that control cell division, cell growth, apoptosis, angiogenesis and tumor metastasis (Srivastava RK, Unterman TG et al. 2010). Previous studies suggest that in breast cancer MCF7 cells, resveratrol exhibited a weak DNMT inhibitory activity and was unable to reverse the methylation of several tumor suppressor genes (Fig. 1). Effect of resveratrol alone and in combination with adenosine analogues: 2-chloro-2’-deoxyadenosine [2CdA] and 9-beta-d-arabinosyl-2-fluoroadenine [F-ara-A] on methylation and expression of RARbeta2 in MCF-7 breast cancer cell lines was studied. Exposure to resveratrol improved the action of adenosine analogues to inhibit methylation of the promoter of RARb2 gene which correlated with increase expression however resveratrol alone was ineffective (Stefanska B, Rudnicka K et al. 2010).
Previous studies demonstrate that resveratrol targets on the class III HDAC, SIRT1, SIRT2, SIRT3 and p300 (Roy SK, Chen Q et al. 2011). Activated SIRT1 negatively regulates survivin expression through its deacetylase activity. SIRT1 also plays critical role in the aging processes. In breast cancer, human BRCA1 is associated with lower levels of SIRT1 expression. It has been reported that resveratrol can increase the expression of human BRCA1 by altering H3 acetylation, which is an important strategy for targeted therapy for BRCA1-associated breast cancer (Tili E, Michaille JJ et al. 2010). In vivo studies on APC/+ mice demonstrate similar findings that SIRT1- encoded proteins are required for resveratrol-mediated tumor growth inhibition (Wang RH, Zheng Y et al. 2008). In prostate cancer, it has been reported that resveratrol regulates cell survival and/or apoptosis by global modulation of gene expression through deacetylation of FOXO transcription factor (Chen Q, Ganapathy S et al. 2010; Ganapathy S, Chen Q et al. 2010). In vivo study of KrasG12D mice suggested that resveratrol inhibits the expression of transcription factor which are required to maintain pleuripotency and self-renewable capacity of pancreatic CSC cells (Shankar S, Nall D et al. 2011).
In case of human SW480 colon cancer cells, decrease in the levels of several oncogenic miRNAs targeting genes encoding Dicer1, a cytoplasmic RNase III producing mature miRNAs from their immediate precursors and tumor-suppressor factors PDCD4 and PTEN have been shown after treating with the resveratrol. This study on miRNA indicated that resveratrol treatment significantly upregulated the expression of 22 miRNA and downregulated 26 miRNA. Several of the downregulated miRNAs include miR-17, miR-21, miR-25, miR-92a-2, constitutively upregulated in colon cancer. The level of miR-663 was increased after resveratrol treatment, which possess putative tumor-suppressor functions and targets TGF1 transcript. Resveratrol treatment also upregulated components of the TGFβ signaling pathway, including TGFβ receptors type I and type II and downregulated the transcriptional activity of canonical TGFβ key effectors proteins, SMADs. These findings suggest that miR-663 is an intention for resveratrol action which contributes to its anticancer properties (Tili E, Michaille JJ et al. 2010). It has also been shown that resveratrol in combination with tea polyphenols suppress the mouse skin cancer growth via inhibition of activated MAPKs and p53 pathway (George J, Singh M et al. 2011).
5.2. Curcumin
Curcumin, a diferuloylmethane, is a polyphenol that extract from the most popular Indian spices turmeric (Curcuma longa). It is a main component of the spice turmeric and is responsible for the yellow pigmentation of curry. It has been associated with multiple health benefits including cancer prevention. Curcumin has shown ability to modulate many components of intracellular signaling pathways implicated in inflammation, proliferation, invasion, survival, and apoptosis (Teiten MH, Eifes S et al. 2010). In-silico studies of docking between DNMT and curcumin and other related compounds indicate that curcumin has ability to inhibit DNMT1 activity by covalently blocking the catalytic thiol group of C1226 binding site (Medina-Franco JL, López-Vallejo F et al. 2011). Treatment of human leukemia MV4-11 cells with curcumin has shown to cause global hypomethylation, but sequence-specific demethylation at promoter regions of epigenetically silenced genes with curcumin has not been demonstrated (Liu Z, Xie Z et al. 2009).
Curcumin is a potential modulator of histones and modulate the HATs and HDACs enzymes activity (Fig. 1). Results obtained from computational screening algorithms demonstrate that curcumin binds covalently to HATs. Curcumin has been shown to promote proteasome-dependent degradation of p300 and other closely related CBP proteins without affecting HATs such as PCAF or GCN5. This activity effectively blocks histone hyperacetylation in prostate cancer PC3-M cells and peripheral blood lymphocytes induced by the HDAC inhibitor MS-275 (Marcu MG, Jung YJ et al. 2006). Several cell culture studies using various cancer cell types have confirmed that curcumin has potential to inhibit HAT activity of p300/CBP. Inhibition of p300/CBP activity by curcumin suppresses histone acetylation as well as acetylation of non-histone protein like p53 (Balasubramanyam K et al 2004; Ho E, Beaver LM et al. 2011). In another study, exposure of human hepatoma cells to curcumin resulted in a significant decrease in histone acetylation due to inhibition of HAT activity without alterations in HDAC levels (Kang J, Chen J et al. 2005; Rajendran P, Ho E et al. 2011). Recently, it has been shown that curcumin represses the activity of NF-κB and Notch1 in Raji cells by inhibition of HDAC1, HDAC3 and p300/CBP resulting in inhibition of cell proliferation (Chen Y, Shu W et al. 2007). Another study confirmed that curcumin has ability to inhibit the expression of class I HDACs [HDAC1, HDAC3, and HDAC8], and can increase the expression of Ac-histone H4 in Raji cells (Liu HL, Chen Y et al. 2005). HDAC inhibitory activity of curcumin was also investigated using fluorometric assay as well by molecular docking for human HDAC8 enzyme and was found that curcumin is highly potent compared to well-known HDAC inhibitor, sodium butyrate (Bora-Tatar G, Dayangaç-Erden D et al. 2009).
Antitumor activity of curcumin has been linked to its ability to modulate miRNA expression in cancer cells (Fig. 3). Treatment of human pancreatic carcinoma BxPC-3 cells with curcumin resulted in significant change in the expression of 29 miRNAs. Further investigation confirmed curcumin induced upregulation of miRNA-22 and suppresses the expression of its target genes SP1 and ESR1 (Sun M, Estrov Z et al. 2008). In human pancreatic cancer cell lines MIAPaCa-E, MIAPaCa M, and BxPC-3, curcumin and its derivative CFD sensitize these cells to gemcitabine by inhibiting NF-κB, COX-2, and their downstream target molecules, which is in part due to inactivation of miR- 21 and reactivation of miR-200b and miR-200c (Ali S et al 2010). Curcumin promoted apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells by downregulation of miR-186. Down regulation of miR-186 caused an increase in caspase-10 activity (Zhang J, Zhang T et al. 2010). Curcumin has shown to reduce the expression of Bcl-2 in breast cancer MCF-7 cells by upregulating miR-15a and miR-16 expression (Yang J, Cao Y et al. 2010). In another study, curcumin has shown to suppress miR-21 levels in human colon cancer RKO and HCT116 cells, which is overexpressed in several human tumors and promote invasion and metastasis. Curcumin also stabilized the expression of the tumor suppressor Pdcd4 in colorectal cancer (Mudduluru G, George-William JN et al. 2011).
5.3. Tea polyphenols
Tea is the most widely consumed drink in the world followed by water. It is derived from the leaves of tea plant (Camellia sinensis). It has been shown that the polyphenols present in green and black tea may have potential to reduce the risk of several diseases including cancer. The major polyphenols present in green tea are [−]-epicatechin [EC], [−]-epicatechin-3-gallate [ECG], [−]-epigallocatechin [EGC], and [−]-epigallocatechin-3-gallate [EGCG], where EGCG constitutes more than 50% of total catechins present therein. The major polyphenols in black tea are catechins, flavanols, methylxanthines, theaflavins and thearubigens (Siddiqui IA, Adhami VM et al. 2006).
EGCG has been shown to inhibit the DNMT activity and reactivate methylation-silenced genes (Fig. 2). It was reported to reverse the hypermethylation of p16INK4a, RARβ, MGMT, and hMLH1 genes through suppression of DNMT1 activity in human esophageal cancer KYSE 510 cells. EGCG has been shown to bind to the catalytic pocket of DNMT1 and inhibit its enzyme activity (Fang MZ, Wang Y et al. 2003). Besides, EGCG has shown its ability to inhibit dihydrofolate reductase [DHFR] leading to inhibition of DNA and RNA synthesis. Studies have further demonstrated that EGCG-mediated altered DNA methylation could be achieved by enhancing the formation of S-adenosyl-L-homocysteine [SAH], a potent inhibitor of DNMT. SAH is produced from the demethylation of S-adenosyl methionine [SAM] when catechol-O-methyltransferase [COMT] inactivates catechol molecules by introducing methyl group to the catecholamine group, donated by SAM (Lee WJ, Shim JY et al. 2005).
Several studies have confirmed that tea polyphenols can reactivate tumor suppressor genes by promoter demethylation. A large number of studies suggested that the correlation of consumption of EGCG and inhibition of several cancers, such as ovarian, oral, esophageal, breast, gastric, prostate, skin, colorectal, pancreatic, and head and neck cancers (Chuang JC, Yoo CB et al. 2005; Kim JW, Amin AR et al. 2010; Chen PN, Chu SC et al. 2011; Kürbitz C, Heise D et al. 2011; Shanmugam MK, Kannaiyan R et al. 2011; Singh BN, Shankar S et al. 2011; Tu SH, Ku CY et al. 2011; Tang SN, Fu J et al. 2012). Treatment of oral cancer cells with EGCG partially reversed the hypermethylation status of tumor suppressor gene RECK and enhanced the expression of RECK mRNA, which correlated with reduced expression of matrix metalloproteinases: MMP-2 and MMP-9 and suppressed the invasive ability of cancer cells (Kato K, Long NK et al. 2008). Administration of black tea polyphenols [Polyphenon-B] significantly reduced the incidence of DAB-induced hepatomas in male Sprague-Dawley rats, as evidenced by alterations in the expression of MMP-2, MMP-9, and TIMP-2; reversion-inducing cysteine rich protein with Kazal motifs RECK; and suppression of HIF1alpha, VEGF, and VEGFR1 which correlated with HDAC1 levels (Murugan RS, Vinothini G et al. 2009).
EGCG and [-]-epigallocatechin repressed telomerase mRNA in lung, oral cavity, thyroid, and liver cancer cells and telomerase expression may be linked to inhibition of cell growth (Lin SC, Li WC et al. 2006). EGCG also demonstrated anti-neoplastic activity by suppressing the telomerase activity of gastric cancer cells (Ran ZH, Zou J et al. 2005). EGCG can inhibit DNMT activity and reactivate methylation silenced retinoic acid receptor β gene in human colon and prostate cancer cells (Lee WJ, Shim JY et al. 2005). In another study, methylation of CDX2 and other genes involved in gastric carcinogenesis was investigated in relation to the clinico-pathologic and selected lifestyle factors of patients with gastric cancer. An inverse association of CDX2 methylation with the intake of green tea was observed in this study (Yuasa Y, Nagasaki H et al. 2005).
Decreased annexin-I expression is a common event in early-stage bladder cancer development. Comparatively, green tea induced the expression of mRNA and protein levels of the actin binding protein, annexin-I, through demethylation of its promoter and actin remodeling (Xiao GS, Jin YS et al. 2006). EGCG, an efficient inhibitor of human dihydrofolate reductase, altered the p16 methylation pattern [from methylated to unmethylated] after folic acid deprivation resulting in growth inhibition of a human colon carcinoma cell line in a concentration- and time- dependent manner. The same study also demonstrated that through disruption of purine metabolism, EGCG caused adenosine release from the cells, and modulation of different signaling pathways via binding to adenosine-specific receptors (Navarro-Perán E, Cabezas-Herrera J et al. 2007).
EGCG induces apoptosis and inhibits growth in renal cell carcinoma through TFPI-2 mRNA and protein overexpression (Gu B, Ding Q et al. 2009). Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells was also reported (Gao Z, Xu Z et al. 2009). Epigenetic silencing of glutathione-S-transferase pi [GSTP1] by hypermethylation is recognized as being a molecular hallmark of human prostate cancer. Recently, it has been reported that exposure of LNCaP cells to GTP concentrations as low as 1-10 μg/mL up to 7 days caused demethylation in the proximal GSTP1 promoter and regions distal to the transcription factor binding sites. This also caused a concentration- and time- dependent re-expression of GSTP1 and DNMT1 inhibition. GTP exposure also increased mRNA and protein levels of MBD1, MBD4 and MeCP2; HDAC 1-3 whereas levels of acetylated histone H3 [LysH9/18] and H4 decreased. In addition, GTP reduced MBD2 association with accessible Sp1 binding sites causing increased binding and transcriptional activation of the GSTP1 gene. Importantly, GTP treatment did not result in global hypomethylation and promoted maintenance of genomic integrity. Unlike 5-aza-2’deoxycitidine treatment, GTP exposure did not activate prometastatic gene S100P. This study demonstrates the dual potential of tea polyphenols at physiologically attainable non-toxic doses to alter DNA methylation and chromatin modeling, the two global epigenetic mechanisms of gene regulation at physiologically attainable non-toxic doses (Pandey M, Shukla S et al. 2010). Another report showed a significant reduction in the number of newly formed tumors in the Apc [Min/+] mice treated with azoxymethane-treated after they were given a solution of green tea [0.6% W/V] as the only source of beverage for 8 weeks. RXR alpha downregulation was observed as an early event in colorectal carcinogenesis and green tea significantly increased the mRNA and protein levels of RXR alpha. Green tea treatment also significantly decreased CpG methylation in the promoter region of the RXR alpha gene (Volate SR, Muga SJ et al. 2009). Recent reports demonstrated that treatment of breast cancer and promyelocytic leukemia cells with EGCG resulted in a time-dependent decrease in hTERT promoter methylation including E2F-1 binding sites and ablated histone H3Lys9 acetylation which led to increased binding of E2F-1 repressor at the hTERT promoter, and ultimately caused cell death (Berletch JB, Liu C et al. 2008). The Polycomb Group [PcG] proteins are epigenetic repressors of gene expression and their repression is achieved via action of two multi-protein polycomb repressive complex 2 (PRC2) [Eed, Sug12, Ezh1and Ezh2] and PRC1 [Bmi-1]. These complexes increase histone methylation and reduce acetylation that leads to a closed chromatin conformation. Bmi-1 is over-expressed in breast, prostate, colon, pancreatic and non-small cell lung cancers. EGCG treatment caused suppression of two key PcG protein, Bmi-1 and Ezh2 and lead to global reduction in histone H3-K27-trimethylation. This caused reduced expression of key proteins that enhance progression through the cell cycle [cdk1, cdk2, cdk4, cyclin D1, cyclin E, cyclin A, and cyclin B1] and increased expression of proteins that inhibit cell cycle progression [p21 and p27]. EGCG treatment also enhanced apoptosis because of increased caspase 9, 8 and 3 and poly ADP-ribose polymerase [PARP] cleavage, increased Bax, and decreased Bcl-xL expression (Balasubramanian S et al 2010).
Another important epigenetic regulation occurs via modifications of microRNA [miRNA] expression (Fig. 3). Limited studies are available in the literature that explored the influence of tea polyphenols on the expression of miRNAs in various human cancers. One recent report showed that EGCG treatment altered the expression of miRNAs in human hepatocellular carcinoma HepG2 cells. Thirteen miRNAs were upregulated and 48 were downregulated. Among the miRNAs upregulated by EGCG, some target genes include: RAS, Bcl2, E2F, TGFBR2 and c-Kit. Among those miRNAs downregulated by EGCG include the target genes comprised of HOX family proteins, including PTEN, SMAD, MCL1, SLC16A1, TTK, PRPS1, ZNF513, and SNX19 with diversified functions. Further treatment with EGCG down-regulated Bcl-2, an anti-apoptotic protein, and transfection with antimiR- 16 inhibitor suppressed miR-16 expression and counteracted the EGCG effects on Bcl-2 downregulation and induced apoptosis in these cells (Tsang WP and Kwok TT 2010). In another study, treatment with Polyphenon-60 significantly altered the expression of 23 miRNAs which includes downregulation of miR-21 and miR-27. These miRNAs have previously demonstrated to overexpress in MCF-7 breast cancer cells. Furthermore, treatment of hepatocellular carcinoma HepG2 cells with EGCG resulted in the induction of apoptosis by the upregulation of miRNA-16, and downregulation of its target gene Bcl-2, an anti-apoptotic protein. Transfection of cells with antimiR- 16 inhibitor confirmed the role of miR-16 in downregulation of Bcl-2 and induction of apoptosis by EGCG. Recent studies in prostate cancer LNCaP cells demonstrated that EGCG treatment repressed the transcriptional activation of AR. EGCG inhibits AR nuclear translocation and protein expression which correlated with significant down-regulation of androgen regulated miRNA-21 and up-regulation of a tumor suppressor, miRNA-330, in in vivo tumor bearing mice (Fix LN, Shah M et al. 2010). The results obtained for miRNA profiling suggests that EGCG may exert its biologic functions through modulation of miRNA expression. The recent studies of tea polyphenols define and support the photoprotective efficacy of tea polyphenols against UV carcinogenesis. The oral administration of tea polyphenols in drinking water or the topical application of EGCG prevents UVB-induced skin tumor development in mice. This prevention is mediated through the induction of immunoregulatory cytokine interleukin, inhibition of UV-induced immunosuppression through IL-12-dependent DNA repair, IL-12-dependent DNA repair following nucleotide excision repair mechanism, inhibition of angiogenic factors and stimulation of cytotoxic T cells in a tumor microenvironment (Choudhury SR, Balasubramanian S et al. 2011; Katiyar S, Elmets CA et al. 2007). In another study of tea phenols on mice model, it has been shown that tea phenols reduce psoriasiform lesions in the flaky skin mouse by inducing caspase 14 in epidermal keratinocytes followed by MAPK pathways (Hsu S, Dickinson D et al. 2007), reduced skin tumor cell survival by influencing PcG-mediated epigenetic regulatory mechanisms (Balasubramanian S, Adhikary G et al. 2010; Nandakumar V, Vaid M et al. 2011).
5.4. Indole-3-carbinol [I3C] and Diindolylmethane [DIM]
Indole-3-carbinol is a hydrolyzed product of glucosinolate which is mainly derived from the vegetables belongs to brassica genus of crucifery family including broccoli, cabbage, cauliflower, mustard and radish etc. because of the acidic pH of stomach; I3C is converted to many diindolylmethane condensation products. Both I3C and DIM induced apoptosis in cancer cell lines from solid tumors of different organs by modulating various kinases and nuclear receptor mediated signaling (Banerjee S et al 2011). A recent study using various human colon cancer cell lines have shown that DIM selectively induced proteasomal degradation of class I histone deacetylases [HDAC1, HDAC2, HDAC3, and HDAC8] without affecting class II HDAC proteins both in vitro and in vivo. Significant decreases in the levels of HDAC1, HDAC2, and HDAC3 were associated with the promoters of p21 and p27 genes which led to cell cycle arrest and DNA damage in tumor cells (Li Y, Li X et al. 2010).
In another study, DIM treatment of gemcitabine-resistant human pancreatic cancer MiaPaCa-2, Panc-1, and Aspc-1 cells resulted in alteration in miRNA expression. DIM treatment caused upregulation of miR-let-7b, miR- let-7e, miR-200b, and miR-200c. Furthermore, treatment of pancreatic cancer cells with DIM correlated with upregulation of E-cadherin, an epithelial cell marker and downregulation of mesenchymal markers ZEB1 and vimentin (Li Y, VandenBoom TG 2nd et al. 2009). Recent study has shown that DIM treatment influences the invasion capacity of pancreatic cells via a miRNA-regulated mechanism. Treatment of pancreatic cancer cells with DIM caused upregulation of miR-146 which correlated with reduced expression of EGFR, MTA-2 and members of the NF-κB signaling pathway (Li Y, VandenBoom TG 2nd et al. 2010). Another recent study with DIM on estrogen-dependent MCF-7 and estrogen receptor negative p53 mutant MDAMB- 468 human breast cancer cells resulted in upregulation of miR-21 which correlated with downregulation of CDK2, CDK 4 and Cdc25A and cell cycle arrest (Jin Y, Zou X et al. 2010). In vivo studies demonstrate that I3C intake resulted the attenuation of symptoms of cigarette smoke in rats and altered miRNAs involved in p53 functions [miR-34b], TGF-β expression [miR-26a], ERBB2 activation [miR-125a-prec], and angiogenesis [miR-10a] in the lungs (Izzotti A, Calin GA et al. 2010).
5.5. Genistein (Isoflavones)
Genistein is the major isoflavone derived from soy beans (Glycine max). It belongs to the phytoestrogen group. A large number of studies has been reported that genistein can be used as a chemopreventive agent in several types of cancers (Barnes S 1995). Genistein can target various enzymes and pathways which has relevance in cancer (Banerjee S, Li Y et al. 2008). Recent studies demonstrate that genistein is involved in the regulation of gene transcription by modification of epigenetic events including DNA methylation and histone modifications (Figs. 1 and 2). Genistein and other flavonoids of soy are potent modifier of DNA methylation. Genistein, biochanin A and daidzein has shown to cause reversal of DNA hypermethylation and reactivated methylationsilenced genes including p16INK4a, RARβ, and MGMT genes in human esophageal squamous KYSE 510 carcinoma cells; RARβ in human prostate cancer LNCaP and PC-3 cells which correlated with inhibition of DNMT1, 3a and 3b (Fang MZ, Chen D et al. 2005). Studies have shown that low, nontoxic concentrations of genistein partially demethylate promoter of the GSTP1 gene and its expression was restored in human breast cancer MDA-MB-468 cells (King-Batoon A, Leszczynska JM et al. 2008). Genistein treatment has shown to demethylate the promoter region of BTG3, a tumor suppressor gene, downregulated in renal cancer by inhibiting the activity of DNMT and MBD2 in renal cell carcinoma A498, ACHN and HEK-293 cells (Majid S, Dar AA et al. 2009). Treatment with genistein also increased HAT activity and the levels of acetylated histones 3, 4, di and trimethylated H3K 4, and RNA polymerase II at the BTG3 promoter which correlated with the inhibition of prostate cancer cell growth and cell cycle arrest (Majid S, Dar AA et al. 2010). Studies on DNA methylation with genistein have shown inconsistent results. Though studies in cell culture have shown that genistein treatment inhibits DNA methylation by inhibiting DNMT activity in various cancer cells, however in vivo studies have demonstrated opposite findings. For example, a randomized, double-blind trial conducted on 34 healthy premenopausal women conducted to determine the effect of 40 mg or 140 mg of isoflavones [including genistein, daidzein, and glycitein] taken daily through one menstrual cycle on the methylation status of p16, RASSF1A, RARb2, ER, and CCND2 genes which are known to be methylated in breast cancer. The results performed on intraductal specimens showed that RARβ2 and CCND2 methylation was increased after treatment and correlated with serum genistein levels (Qin W, Zhu W et al. 2009).
Genistein has been shown to possess highest histone modifying activity in comparison with other isoflavones. Genistein, daidzein and the daidzein metabolite equol have been reported to exert their effects by elevating histone acetylation through modulating HAT activity and coactivator activity of ER (Hong T, Nakagawa T et al. 2004). Genistein has shown to induce the expression of p21WAF1/CIP1 and p16INK4a tumor suppressor genes in human prostate cancer cells by epigenetic mechanisms involving active chromatin modification including upregulation of the expression of HATs (Majid S, Kikuno N et al. 2008). Furthermore, treatment with genistein caused demethylation and acetylation of histone H3-K9 at the PTEN and the CYLD promoter and acetylation of Histone H3-K9 on p53 and FOXO3a promoter through reduction of SIRT1 activity. Increase expression of these genes reciprocally relate to attenuation of p-AKT and NF-κB binding activity (Kikuno N, Shiina H et al. 2008). In another study, treatment of LNCaP cells with genistein exhibit increased ubiquitination of AR protein which was due to decrease in the chaperone activity and increase acetylation of Hsp90. This study also demonstrated that HDAC6, an Hsp90 deacetylase, was the target of the anti-estrogenic activity of genistein (Basak S, Pookot D et al. 2008).
Soy isoflavones have shown the potential to modulate miRNAs (Fig. 3). In a study using UL- 3A and UL-3B cells established from an ovarian cancer patient treated with genistein, the miRNA profile of untreated and their treated counterpart cells were compared. A total of 53 genes were found to be differentially regulated after genistein treatment. Genistein resulted in the induction of ERα and ERβ mRNA and proteins and reduction in migration and invasion ability of treated cells. The study however did not characterize the involvement of miRNAs in the induction of ERα and ERβ (Parker LP, Taylor DD et al. 2009). Soy isoflavones also suppress the ultraviolet B-induced skin cancer by targeting Cox and MKK4 activity (Lee DE, Lee KW et al. 2011). In another study, treatment with genistein of gemcitabine-resistant human pancreatic cancer cell lines viz. MiaPaCa- 2, Panc-1, and Aspc-1 resulted in downregulation of MiRNA-200, which positively correlated with the mesenchymal markers including ZEB1, slug, and vimentin and reversal of EMT (Li Y, VandenBoom TG 2nd et al. 2009). In prostate cancer cells, genistein treatment caused upregulation of MiRNA-1296 and accumulation of cells in the S phase of the cell cycle along with significant decrease in mRNA and protein levels of mini-chromosome maintenance gene [MCM-2], which is a target of MiRNA-1296 (Majid S, Dar AA et al. 2010). Furthermore, genistein has shown to suppress growth of melanoma C918 cells by inhibition of MiRNA-27a and its target gene ZBTB10 (Sun Q, Cong R et al. 2009).
5.6. Sulforaphane
Sulforaphane [SFN] is a bioactive phytochemical found in broccoli, broccoli sprouts, cabbage and kale. Sulforaphane has ability to alter anti-carcinogenic activity, enhance xenobiotic metabolism, induce cell cycle arrest and apoptosis in various human cancer cells which has relevance in cancer chemoprevention (Clarke JD, Dashwood RH et al. 2008). The effect of sulforaphane on methylation of DNA is not very well understood, whereas downregulation of DNMT1 activity has been demonstrated in human colon cancer CaCo-2 cells (Traka M, Gasper AV et al. 2005). Treatment of breast cancer MCF-7 and MDA-MB-231 cells with SFN resulted in the inhibition of human telomerase reverse transcriptase [hTERT], the catalytic regulatory subunit of telomerase. SFN-mediated decrease in DNMT1 and DNMT3a was observed after treatment and sitespecific CpG demethylation occurred primarily in the first exon of the hTERT gene which facilitated CTCF binding associated with hTERT repression. SFN treatment has shown to increase acetylation of acetyl-H3, acetyl-H3K9 and acetyl-H4; and decrease in the trimethyl-H3K9 and trimethyl-H3K27, respectively. This hyperacetylation enhanced the binding of many hTERT repressor proteins such as MAD1 and CTCF to the hTERT regulatory region resulting in cellular apoptosis. SFN treatment inhibited HDAC activity and modulated histone methylation by increasing the expression of histone demethylase RBP2 (Meeran SM, Patel SN et al. 2010). Treatment of human embryonic kidney, HEK293 and human HCT116 colorectal cancer cells with SFN resulted in inhibition of HDAC activity and increase activity of multiple T-cell factor [TCF]/lymphoid enhancer factor [LEF] binding sites along with increase acetylation of histone and p21 (Myzak MC, Karplus PA et al. 2004). SFN treatment of human prostate epithelial BPH-1, LNCaP and PC-3 cells exhibited inhibition of HDAC activity which was accompanied by increase in acetylated histones and their increased binding on the promoters of p21 and Bax genes. These events correlated with cell cycle arrest and induction of caspase-dependent apoptosis (Myzak MC, Hardin K et al. 2006; Shankar S, Ganapathy S et al. 2008). It has been also reported to induce cell cycle arrest and apoptosis through regulation of FOXO transcription factors (Roy SK, Srivastava RK et al. 2010). In another study, SFN exposure to human breast cancer cell lines namely MDA-MB-231, MDA-MB-468, MCF-7, and T47D resulted in HDAC inhibition and decrease in the protein expression of ER, EGFR, and HER-2 in these cancer cells which correlated with cell growth inhibition and induction of apoptosis. Specifically, SFN treatment did not cause any change in acetylation pattern of histones in this study (Pledgie-Tracy A, Sobolewski MD et al. 2007).
A single oral dose of 10μM SFN in wild-type [C57BL/6J+/+] mice caused significant inhibition in HDAC activity in the colonic mucosa and concomitant transient increase in ac-H3 and ac-H4 levels. In another study using APCMin/+ mice, SFN treatment reduced tumor formation and increased global histone acetylation and increase association of acetylated histone H3 on the promoters of p21 and Bax genes, and increase expression of Bax protein (Myzak MC, Dashwood WM et al. 2006). Consumption of SFN in the diet at an average daily dose of 7.5μM per animal for 21 days resulted in 40% reduced growth in PC-3 tumor xenograft in nude mice. These results correlated with a significant decrease in HDAC activity, increase in global histone acetylation and increase expression of Bax in the tumors and mononuclear blood cells (Myzak MC, Tong P et al. 2007). Furthermore, in a pilot study, 3 human subjects fed with a single dose of 68g broccoli sprouts demonstrated significant inhibition of HDAC activity and induced acetylation of histone H3 and H4 at 3 and 6 h following intake, in their peripheral blood mononuclear cells (Myzak MC, Tong P et al. 2007). It has also been reported that sulforaphane suppresses polycomb group protein (Bmi-1, Ezh2) expression in SCC-13 skin cancer cells and reduces trimethylation of lysine 27 of histone H3 via proteasome-dependent mechanism (Balasubramanian S, Chew YC et al. 2011). The recent studies of effect of sulforaphane on SKH-1 hairless mice has shown that sulforaphane inhibited chemically induced skin carcinogenesis via nuclear factor (erythroid-derived 2)-like 2 (Nrf2) and broccoli sprout extracts containing high SFN protected against UV-induced skin carcinogenesis (Saw CL, Huang MT et al. 2011). Several other studies also support the protective effect of supforaphane over ultra violet induced carcinogenesis (Xu C, Huang MT et al. 2006; Dickinson SE, Melton TF et al. 2009).
5.7. Phenethyl isothiocyanate
Isothiocyanates, such as phenethyl isothiocyanate [PEITC] has shown to inhibit carcinogenic process and as such is a useful chemopreventive agent. The main sources of this phytochemicals are vegetables belong to cruciferi family. PEITC has ability to suppress growth of various cancer cell types and induces apoptosis in cancer cells (Cheung K L and Kong A N 2010). In a recent study, treatment of human prostate cancer LNCaP cells with PEITC resulted in demethylation of GSTP1 gene promoter, inhibited the activity of HDACs, and induced selective histone acetylation and methylation (Wang LG, Beklemisheva A et al. 2007). In another study, treatment of DS19 mouse erythroleukemia cells with allyl isothiocyanate exhibited increase acetylation of histones but had no effect on HDACs (Lea MA, Randolph VM et al. 2001). Recent studies demonstrate that PEITC can modulate miRNAs expression induced by cigarette smoke. Rats were pretreated with PEITC alone or in combination with IC3 for three days, before been exposed to cigarette smoke for 28 days. PEITC strongly counter-regulated the expression of majority of miRNAs downregulated by cigarette smoke. Several of the miRNAs which were modified by PEITC include miR-125b, miR-26a, miR-146-pre, let-7a, let-7c, miR-192, miR-222-pre, miR-99 and miR- 123 designated for TGF-β expression, NF-κB activation, Ras activation, cell proliferation, apoptosis and angiogenesis (Izzotti A, Calin GA et al. 2010).
In another study, the effect of PEITC or the glucocorticoid budesonide treatment either alone or in combination was analyzed on miRNA expression in mouse liver and lungs. Treatment was started after weaning for 2 weeks or directly after birth in combination with exposure to cigarette smoke. PEITC caused modest effect on miRNA expression in the lungs, but in the liver, it significantly downregulated nine and upregulated three miRNAs. Co-treatment group significantly up-regulated 12 and downregulated 11 miRNAs in comparison to the group treated with cigarette smoke only. These differentially expressed miRNAs were shown to be associated with genes regulating stress response, protein repair, cell proliferation, and inflammation (Izzotti A, Larghero P et al. 2010).
5.8. Organosulfur compounds
Allium vegetables, such as garlic (Allium sativum), have been in use in traditional medicine for a long period of time and impart health benefits as hypoglycemic agent, and in improving immunity, cardiovascular health, protection from microbial, radiation and cancer. Their anticancer effects have been attributed to organosulfur compounds [OSCs] released on processing. OSCs are generated upon conversion of alliin to allicin and other alkyl alkane-thiosulfinates by the action of alliinase. These products are highly unstable and decompose to various sulfur compounds such as diallyl sulfide [DAS], diallyl disulfide [DADS] and diallyl trisulfide [DATS]. Regular consumption of Allium vegetables is inversely related to the risk of the development of stomach and colon cancers. DADS has been shown to inhibit growth of cancer cells by causing cell cycle arrest and apoptosis, inhibits angiogenesis, and suppresses metastasis (Ariga T and Seki T 2006). In in vivo models, DADS treatment protected against chemically-induced cancer of various organs and inhibited tumor growth in xenograft models. DADS generates its active metabolite S-allylmercaptocysteine [SAMC] and both are finally metabolized to allyl mercaptan [AM] and other metabolites (Lea MA, Randolph VM et al. 1999).
Studies have shown that DADS and SAMC induces histone acetylation and cell growth inhibition in DS19 mouse erythroleukemia cells and their metabolite AM was found to be a more potent HDAC inhibitor. In silico docking studies predicted their direct binding to the HDAC active site and their HDACs inhibitory potential was confirmed by performing activity assays (Lea MA, Rasheed M et al. 2002). DADS caused increased global acetylation of H3 and H4 histones and increased binding of acetylated histone H3 onto the promoter of p21 gene which correlated with upregulation of p21 and cell cycle arrest and HDAC inhibition. Induction of histone acetylation by Sallylmercaptocysteine was observed in human colon cancer Caco-2 cells and human breast cancer T47D cells, where HDAC activity was inhibited by allyl butyrate (Druesne N, Pagniez A et al. 2004). In another study, treatment of DS19 cells with S-allylmercaptocysteine or allyl isothiocyanate resulted in downregulation of HDACs and HATs. Furthermore, hyperacetylation of histones was induced in a number of cancer cell lines by DADS treatment, causing p21 upregulation, cell-cycle arrest and induction of differentiation and apoptosis. Treatment of colon cancer Caco-2 and HT-29 cells inhibited HDACs and in turn caused acetylation of histones H3 and H4 with increase in the expression of p21/Waf1, resulting in cell cycle arrest (Druesne N, Pagniez A et al. 2004).
5.9. Quercetin
Quercetin is a dietary polyphenol, predominantly present in citrus fruits and buckwheat. It is a multi-potent bioflavonoid with immense potential for the prevention and treatment of cancer (Gibellini L, Pinti M et al. 2011). Quercetin has been shown to activate NAD+ dependent histone deacetylase SIRT1 in yeast. Quercetin has been shown to inhibit the growth of colon cancer RKO cells by reversing the hypermethylation of p16INK4a gene (Tan S, Wang C et al. 2008). Quercetin has been shown to inhibit the expression of TNF-induced interferon-gamma-inducible protein 10 [IP- 10] and macrophage inflammatory protein 2 [MIP-2] which were associated with inhibition of CBP/p300 activity and phosphorylation/acetylation of histone H3 on the promoter region of these genes (Ruiz PA, Braune A et al. 2007). In another study, quercetin induced FasL-mediated apoptosis in human leukemia HL-60 cells by transactivation through activation of c-jun/AP-1 and promotion of histone H3 acetylation (Lee WJ, Chen YR et al. 2011). Recent study demonstrates that administration of quercetin to DMBA-painted hamsters reduced tumor incidence and tumor burden, whereas post-treatment of quercetin resulted in a significant tumor growth delay. Quercetin administration caused cell cycle arrest and apoptosis and blocked invasion and angiogenesis which correlated with the inhibition of HDAC-1 and DNMT1 (Priyadarsini RV, Vinothini G et al. 2011). It has been reported that prostate cancer can be prevented by using quercetin in combination with EGCG (Tang SN, Singh C et al. 2010).
5.10. Lycopene
Lycopene is one of the naturally occurring classes of tetra-terpenoids mainly present in tomato (Solanum lycopersicum) and tomato products. It is a potent antioxidant and has been shown to reduce oxidative DNA damage. Studies with animal cancer models exhibit that lycopene reduced tumor growth in breast, prostate and lungs whereas it was ineffective in preventing colon, kidney and liver cancers (Giovannucci E 1999). In a study using a single dose of 2μM lycopene partially demethylate GSTP1 gene and increased its mRNA expression in MDA-MB-468 breast cancer cell line but RARβ2 gene was not demethylated in either MDA-MB-468 or MCF-7 breast cancer cell lines. Lycopene treatment caused demethylation of the RARβ2 and HIN-1 genes in the non-tumorigenic MCF10A fibrocystic breast cells. Lycopene also acts as a protective agent against ultra violet induced carcinogenesis via inhibition of epidermal ornithine decarboxylase activity, reducing inflammatory responses, maintaining normal cell proliferation, and possibly preventing DNA damage as indicated by blocking the necessitating step of apoptosis (Fazekas Z, Gao D et al. 2003). This data demonstrate that lycopene might have DNA demethylating potential however further investigation is needed to understand the mechanism[s] of demethylation of gene promoter by lycopene (King-Batoon A, Leszczynska JM et al. 2008).
5.11. Ellagitannins
Ellagitannins are phytochemicals present in high concentrations in many fruits and nuts, such as pomegranate, raspberries, walnuts and almonds. These are polyesters of ellagic acid and a sugar moiety and upon hydrolysis release ellagic acid. Ellagitannins exhibit anti-oxidant and radical scavenging, antiviral, antimicrobial, anti-mutagenic, anti-inflammatory, anti-tumor promoting and immunomodulatory properties (Heber D 2008). Ellagitannins elicit their anticancer effects by modulating transcription factors and signaling pathways which inhibit cancer cells proliferation and induces apoptosis. In particular, exposure of liver cancer cells with ellagitannin BJA3121 isolated from a plant Balanophora japonica resulted in cell growth inhibition and alteration in the expression of several miRNAs. BJA3121 treatment resulted in upregulation of miR-let-7e, miR-370, miR-373 and miR-526b and downregulation of let-7a, let-7c, let-7d which correlated with genes involved in cell differentiation and proliferation (Wen XY, Wu SY et al. 2009).
6. Other dietary phytochemicals affecting epigenetic modification(s)
In addition to above mentioned dietary phytochemicals, a number of other natural dietary compounds are under study for their ability to exhibit chemopreventive/therapeutic potential through epigenetic modification(s). Folic acid is a vitamin-B found in many grains, beans, refreshed breakfast cereals, green vegetables and pastas. Folic acid is a key element in the methyl-metabolism pathway. Deficiency of folic acid can modify hepatic DNA methylation patterns and induce liver cancer (Poirier 1994; Pogribny, Ross et al. 2006). Folic acid deficiencies are associated with the development of several different cancers including: brain, lung, breast, cervix, colorectal and ovary (Kim 2007; Yang, Bostick et al. 2009; Duthie 2011). Selenium is a nutrient found in game meat, beef, chicken and Brazil nuts (Lemire, Fillion et al. 2010). It is an essential element with antioxidant, proapoptotic, anticancer and DNA repair properties (Redman, Scott et al. 1998; Combs, Clark et al. 2001; Fischer, Lancia et al. 2006). Selenium is vital for human health; its deficiencies have been linked to several human diseases including cancer (Xiang, Zhao et al. 2008). In addition, several other selenoproteins (i.e., selenium binding protein-1) have been indicated as important in the development of cancers; however, their epigenetic effects have not been clearly defined (Zhuo and Diamond 2009). Several other dietary phytochemicals including apigenin (parsley), biacalein (baikal skullcap), cyanidins (berries), rosmarinic acid (rosemary), silibinin/silymarin (milk thistle seed) and others which have been reported to have either direct or indirect epigenetic targets in cancer prevention and therapy. These compounds are integral part of regular food products and can be integrated in the diet on regular basis leading to reversal in epigenetic modifications.
9. Conclusions and future prospective
The awareness campaign of chemoprevention has led to widespread recognition and use of bioactive phytochemicals around the world. Nutrigenomics refers to the interaction between one’s diet and his/her genes. These interactions can markedly influence digestion, absorption, and the elimination of bioactive food components, as well as influence their site of actions/molecular targets. Nutrigenomics comprises epigenetics, nutrigenetics, and transcriptomics, coupled with other “omic,” such as proteomics and metabolomics, that apparently account for the wide variability in cancer risk among individuals with similar dietary habits. Multiple food components including essential nutrients, phytochemical, zoochemicals, fungochemical, and bacterochemicals have been implicated in cancer risk and tumor behavior, admittedly with mixed results. Such findings suggest that not all individuals respond identically to a diet. The single-nucleotide polymorphism, copy number, epigenetic events, and transcriptomic homeostasis influence the response of food components and ultimately health, including cancer risk.
Based on the studies mentioned in this review, it is clear that these phytochemicals act on the different epigenetic targets leads to the epigenetic modifications. Some of dietary phytochemicals (such as Genistein, Phenethyl isothiocyanate, Curcumin, Sulforaphane, Organosulfur, Compound, Resveratrol and Indole-3-carbinol) act on the inhibition of deacetylation of histone protein, whereas other phytochemicals (such as EGCG, Genistein and Curcumin) act on the inhibition of acetylation of histone protein during epigenetic modifications. Dietary phytochemicals (such as EGCG, Genistein, Organosulfur, Compound, Lycopene, Phenethyl isothiocyanate, Curcumin, Sulforaphane and Resveratrol) inhibit the DNA methylation process by activating DNA methyletranferase enzymatic activity. It has also been reported that some of dietary phytochemicals play an important role in the modulation of overall epigenetic modifications (Histone modifications, DNA methylations and miRNA).
Dietary phytochemicals hold great promise in cancer prevention and in therapy by inducing epigenetic modifications. As the importance of epigenetic modifications in cancer is well recognized, precise contribution of epigenetic mechanisms and cellular targets of epigenetic alterations by dietary phytochemicals in human cancer needs further investigation. Although recent advances in the field of cancer epigenetics has enhanced our understanding of epigenetic changes in normal cellular processes and abnormal events leading to tumorigenesis, however deeper understanding of the global patterns of epigenetic modifications by phytochemicals in cancer will lead to design better strategies to prevent and cure cancer. Moreover, sufficient preclinical and clinical data is required on the epigenetic changes induced by dietary phytochemicals which will to lead to better understanding of the epigenetic targets and pathways altered by these agents to elicit their efficacy in cancer. Additional preclinical and clinical studies are required to analyze the safety profile of doses, route of administration, tissue distribution, bioavailability alone and in combination with other agents in order to obtain maximum beneficial effects. At last, systematic well-designed randomized placebo-controlled trials with adequate power and relevant clinical epigenetic endpoints are needed. Despite these challenges, research on dietary phytochemicals continues to emerge and will offer new epigenetic targets and promising agents with more opportunities for prevention, and perhaps personalized therapy of cancer in the near future. As the concept that “one size fits all” comes to an end and personalized approaches surface, additional research data will be required to identify those who will benefit most from dietary change and any who might be placed at risk because of an adjustment.
Fig. 4. Genetic Mechanism of DNA Methylation and Histone Modification.
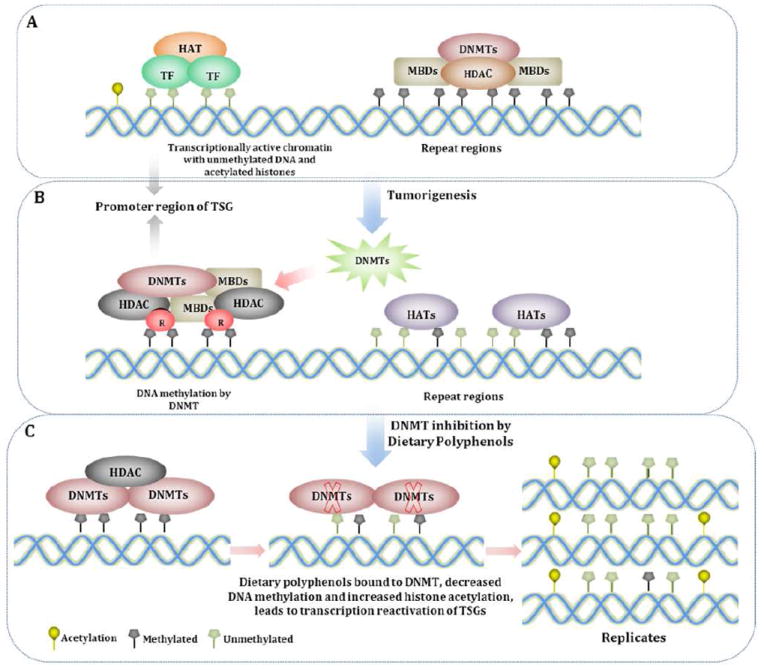
A. Transcriptionally active chromatin with unmethylated DNA and acetylated histones. Genes are unmethylated and packaged with acetylated histone proteins associated with HAT and basal transcription factor machinery. These epigenetic elements constitute an ‘open’ chromatin structure which favors transcription. B. DNA methylation by DNMT. The methylated CpG sites are recognized by the methyl-binding proteins (MBDs), which are associated with repressor (R) and histone deacetyltransferase (HDAC) proteins to remove the acetyl group from the histones, generating a tightly closed chromatin status to shut down gene expression. C. Effects of dietary polyphenols on inhibition of DNMT. DNMT activity is blocked by dietary polyphenols by forming hydrogen bonds with amino acids (Pro, Glu, Cys, Ser, and Arg) in the catalytic pocket of DNMT. Newly synthesized DNA strands are semi-methylated after the first round of DNA replication and become progressively more demethylated after several rounds of replication due to the dilution effect.
Acknowledgments
We thank our lab members for critical reading of the manuscript. This work was supported by the grants from the National Institutes of Health (R01CA125262, RO1CA114469 and RO1CA125262-02S1), Susan G. Komen Breast Cancer Foundation, Department of Defense US Army, and Kansas Bioscience Authority.
Abbreviations
- Akt
v-akt murine thymoma viral oncogene homolog 1
- AP-1
Activator Protein-1
- Bax
BCL2- associated X protein
- Bcl2
B-cell CLL/lymphoma 2
- Bcl-xL
B-cell lymphoma-extra-large
- Bmi-1
B-cell-specific Moloney murine leukemia virus integration site 1
- BRCA1
breast cancer 1, early onset
- CBP
CREB-binding protein
- CCND2
cyclin D2
- Cdc25A
cell division cycle 25 homolog A
- Cdk
cyclin-dependent kinase
- c-Kit
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- COMT
catechol-O-methyltransferase
- COX-2
cyclooxygenase-2
- CYLD
cylindromatosis (turban tumor syndrome)
- DADS
diallyl disulfide
- DAS
diallyl sulfide
- DATS
diallyl trisulfide
- DHFR
dihydrofolate reductase
- DMBA
7,12-dimethylbenz(a)anthracene
- DNMT
DNA methyltransferase
- DNMT-3L
DNA (cytosine-5)-methyltransferase 3-like
- E2F
E2F transcription factor
- EC
[−]- epicatechin
- ECG
[−]-epicatechin-3-gallate
- EGC
[−]-epigallocatechin
- EGCG
[−]-epigallocatechin-3-gallate
- EGFR
epidermal growth factor receptor
- ER
estrogen receptor
- ERBB2
human epidermal growth factor receptor 2
- ERα
estrogen receptor alpha
- FOXO3a
forkhead box protein O3
- GCN5
SAGA complex histone acetyltransferase catalytic subunit Gcn5
- GSTP1
glutathione-S-transferase pi 1
- HATs
histone acetyl transferases
- HDACs
histone deacetylase
- HER-2
human epidermal growth factor receptor 2
- hMLH1
human mutL homolog 1, HOX family proteins- homeobox family proteins
- HSP90
heat shock protein 90
- hTERT
human telomerase reverse transcriptase
- IP-10
TNF-induced interferon-gamma-inducible protein 10
- K
Lysine
- LEF
lymphoid enhancer factor
- LOI
loss of imprinting
- MBD
methylated DNA binding domain proteins
- MCL1
induced myeloid leukemia cell differentiation protein Mcl-1
- MCM-2
minichromosome maintenance gene
- MGMT-O(6)
methylguanine-DNA methyltransferase
- MIP-2
macrophage inflammatory protein 2
- miRNA
microRNA
- MMP
matrix metalloproteinase
- MTA-2
metastasis associated 1 family member 2
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- Notch1
notch homolog 1, translocation-associated (Drosophila)
- NuRD
nucleosome remodeling complex
- OSCs
organosulfur compounds
- p16INK4a
cyclin-dependent kinase 4 inhibitor A
- p21WAF1/CIP1
cyclin-dependent kinase inhibitor 1A
- p53
tumor protein 53
- PARP
Poly ADP-ribose polymerase
- PCAF
K(lysine) acetyltransferase 2B
- PcG
polycomb group proteins
- PDCD4
programmed cell death 4
- PEITC
phenethyl isothiocyanate
- PRMTs
arginine methyltransferases
- PRPS1
phosphoribosyl pyrophosphate synthetase 1
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- R
Arginine
- RAS
rat sarcoma transforming oncogene
- RASSF1A
RAS association domain family 1A
- RECK
reversion-inducing cysteine-rich protein with Kazal motifs repressive complex 3
- RXR alpha
retinoid X receptor, alpha
- SAH
S-adenosyl-L-homocysteine
- SAM
S-adenosyl methionine
- SAMC
Sallylmercaptocysteine
- SIRT1
sirtuin (silent mating type information regulation 2 homolog) 1
- SIRT2
sirtuin (silent mating type information regulation 2 homolog) 2
- SIRT3
sirtuin (silent mating type information regulation 2 homolog) 3
- SLC16A1
solute carrier family 16, member 1
- SNX19
sorting nexin-19
- SP1
transcription Factor Sp1
- TCF
multiple T-cell factor
- TGFBR2
transforming growth factor, beta receptor II
- TGF-β
transforming growth factor, beta
- TIMP-2
tissue inhibitor of metalloproteinase 2
- TTK
phosphotyrosine picked threonine-protein kinase
- VEGF
vascular endothelial cell growth factor
- ZBTB10
zinc finger and BTB domain containing 10
- ZEB1
zinc finger E-box binding homeobox 1
- ZNF513
zinc finger protein 513
Footnotes
CONFLICT OF INTEREST:
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali S, et al. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70(9):3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ariga T, Seki T. Antithrombotic and anticancer effects of garlic-derived sulfur compounds: a review. Biofactors. 2006;26(2):93–103. doi: 10.1002/biof.5520260201. [DOI] [PubMed] [Google Scholar]
- Attig L, et al. P Nutr Soc; Symposium 2: Modern approaches to nutritional research challenges nutritional developmental epigenomics: immediate and long-lasting effects in the Conference on ’Over- and undernutrition: challenges and approaches; 2010. pp. 221–231. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128(3):459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Balakrishnan A, Bleeker FE, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67(8):3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Adhikary G, et al. The Bmi-1 polycomb protein antagonizes the (-)- epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis. 2010;31(3):496–503. doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Balasubramanian S, Chew YC, et al. Sulforaphane suppresses polycomb group protein level via a proteasome-dependent mechanism in skin cancer cells. Mol Pharmacol. 2011;80(5):870–878. doi: 10.1124/mol.111.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, et al. The Bmi-1 polycomb protein antagonizes the (-)-epigallocatechin-3-gallatedependent suppression of skin cancer cell survival. Carcinogenesis. 2010;31(3):496–503. doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Balasubramanyam K, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279(49):51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Li Y, et al. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269(2):226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, et al. Attenuation of multi-targeted proliferation-linked signaling by 3,3’-diindolylmethane (DIM): From bench to clinic. Mutat Res. 2011;728(1-2):47–66. doi: 10.1016/j.mrrev.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes S. Effect of genistein on in vitro and in vivo models of cancer. J Nutr. 1995;125(Suppl 3):777S–783S. doi: 10.1093/jn/125.3_Suppl.777S. [DOI] [PubMed] [Google Scholar]
- Basak S, Pookot D, et al. Genistein down-regulates androgen receptor by modulating HDAC6-Hsp90 chaperone function. Mol Cancer Ther. 2008;7(10):3195–3202. doi: 10.1158/1535-7163.MCT-08-0617. [DOI] [PubMed] [Google Scholar]
- Berletch JB, Liu C, et al. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103(2):509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora-Tatar G, Dayangaç-Erden D, et al. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorg Med Chem Lett. 2009;17(14):5219–5228. doi: 10.1016/j.bmc.2009.05.042. [DOI] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17(9):2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Chen PN, Chu SC, et al. Epigallocatechin-3 gallate inhibits invasion, epithelial-mesenchymal transition, and tumor growth in oral cancer cells. J Agric Food Chem. 2011;59(8):3836–3844. doi: 10.1021/jf1049408. [DOI] [PubMed] [Google Scholar]
- Chen Q, Ganapathy S, et al. Resveratrol induces growth arrest and apoptosis through activation of FOXO transcription factors in prostate cancer cells. PLoS One. 2010;5(12):e15288. doi: 10.1371/journal.pone.0015288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shu W, et al. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007;101(6):427–433. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12(1):87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Park JH, et al. Epigenomics: Novel aspect of genomic regulation. Biochem Mol Biol. 2007;40(2):151–155. doi: 10.5483/bmbrep.2007.40.2.151. [DOI] [PubMed] [Google Scholar]
- Choudhury SR, Balasubramanian S, et al. (-)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis. 2011;32(10):1525–1532. doi: 10.1093/carcin/bgr171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Yoo CB, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2’-deoxycytidine. Mol Cancer Ther. 2005;4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Dashwood RH, et al. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269(2):291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacino JA, Arthur AE, et al. Pretreatment dietary intake is associated with tumor suppressor DNA methylation in head and neck squamous cell carcinomas. Epigenetics. 2012;7(8):1–9. doi: 10.4161/epi.21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, et al. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zwaan SE, Haass NK. Genetics of basal cell carcinoma. Australas J Dermatol. 2010;51:81–92. doi: 10.1111/j.1440-0960.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- Dehan P, Kustermans G, et al. DNA methylation and cancer diagnosis: new methods and applications. Expert Rev Mol Diagn. 2009;9(7):651–657. doi: 10.1586/erm.09.53. [DOI] [PubMed] [Google Scholar]
- Denis H, Ndlovu MN, et al. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO rep. 2011;12(7):647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson SE, Melton TF, et al. Inhibition of activator protein-1 by sulforaphane involves interaction with cysteine in the cFos DNA-binding domain: implications for chemoprevention of UVB-induced skin cancer. Cancer Res. 2009;69(17):7103–7110. doi: 10.1158/0008-5472.CAN-09-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Mantovani F, et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12(4):380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- Druesne N, Pagniez A, et al. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25(7):1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- Duthie SJ. Epigenetic modifications and human pathologies: cancer and CVD. Proc Nutr Soc. 2011;70(1):47–56. doi: 10.1017/S0029665110003952. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Chen D, et al. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11(19 Pt 1):7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–7570. [PubMed] [Google Scholar]
- Fazekas Z, Gao D, et al. Protective effects of lycopene against ultraviolet B-induced photodamage. Nutr Cancer. 2003;47(2):181–187. doi: 10.1207/s15327914nc4702_11. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fix LN, Shah M, et al. MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer Genomics Proteomics. 2010;7(5):261–277. [PubMed] [Google Scholar]
- Ganapathy S, Chen Q, et al. Resveratrol Enhances Antitumor Activity of TRAIL in Prostate Cancer Xenografts through Activation of FOXO Transcription Factor. PLoS One. 2010;5(12):e15627. doi: 10.1371/journal.pone.0015627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Xu Z, et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res. 2009;29(6):2025–2030. [PubMed] [Google Scholar]
- Garzon R, Calin GA, et al. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Brecqueville M, et al. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. doi: 10.1186/1756-8722-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Singh M, et al. Resveratrol and black tea polyphenol combination synergistically suppress mouse skin tumors growth by inhibition of activated MAPKs and p53. PLoS One. 2011;6(8):e23395. doi: 10.1371/journal.pone.0023395. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gibellini L, Pinti M, et al. Quercetin and cancer chemoprevention. Evid Based Complement Alternat Med. 2011:591356. doi: 10.1093/ecam/neq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E. Tomatoes, tomato-based products, lycopene, and cancer: review of the epidemiologic literature. J Natl Cancer Inst. 1999;91(4):317–331. doi: 10.1093/jnci/91.4.317. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, et al. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gu B, Ding Q, et al. EGCG inhibits growth and induces apoptosis in renal cell carcinoma through TFPI- 2 overexpression. Oncol Rep. 2009;21(3):635–640. [PubMed] [Google Scholar]
- Guan B, Mao TL, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35(5):625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3(4):503–518. doi: 10.2217/epi.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21(10):1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber D. Multi-targeted therapy of cancer by ellagitannins. Cancer Lett. 2008;269(2):262–268. doi: 10.1016/j.canlet.2008.03.043. [DOI] [PubMed] [Google Scholar]
- Ho E, Beaver LM, et al. Dietary factors and epigenetic regulation for prostate cancer prevention. Adv Nutr. 2011;2(6):497–510. doi: 10.3945/an.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet. 2011;43(8):729–731. doi: 10.1038/ng.868. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, et al. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18(R2):R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T, Nakagawa T, et al. Isoflavones stimulate estrogen receptor-mediated core histone acetylation. Biochem Biophys Res Commun. 2004;317(1):259–264. doi: 10.1016/j.bbrc.2004.03.041. [DOI] [PubMed] [Google Scholar]
- Hsu S, Dickinson D, et al. Green tea polyphenol induces caspase 14 in epidermal keratinocytes via MAPK pathways and reduces psoriasiform lesions in the flaky skin mouse model. Exp Dermatol. 2007;16(8):678–684. doi: 10.1111/j.1600-0625.2007.00585.x. [DOI] [PubMed] [Google Scholar]
- Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, et al. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila) 2010;3(1):62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Calin GA, et al. Chemoprevention of Cigarette Smoke–Induced Alterations of MicroRNA Expression in Rat. Lungs Cancer Prev Res (Phila) 2010;3(1):62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Larghero P, et al. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31(5):894–901. doi: 10.1093/carcin/bgq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211(3):261–268. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li J, et al. Bmi-1, stem cells and cancer. Acta Biochim Biophys Sin (Shanghai) 2009;41(7):527–534. doi: 10.1093/abbs/gmp040. [DOI] [PubMed] [Google Scholar]
- Jin Y, Zou X, et al. 3,3’-Diindolylmethane negatively regulates Cdc25A and induces a G2/M arrest by modulation of microRNA 21 in human breast cancer cells. Anticancer Drugs. 2010;21(9):814–822. doi: 10.1097/CAD.0b013e32833e53ea. [DOI] [PubMed] [Google Scholar]
- Jones PA. DNA methylation and cancer. Oncogene. 2002;21(35):5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- Jones S, Wang TL, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Ushijima S, et al. Mutation of the DNA methyltransferase (DNMT) 1 gene in human colorectal cancers. Cancer Lett. 2003;192(1):75–82. doi: 10.1016/s0304-3835(02)00689-4. [DOI] [PubMed] [Google Scholar]
- Kang J, Chen J, et al. Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem Pharmacol. 2005;69(8):1205–1213. doi: 10.1016/j.bcp.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Katiyar S, Elmets CA, et al. Green tea and skin cancer: photoimmunology, angiogenesis and DNA repair. J Nutr Biochem. 2007;18(5):287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Kato K, Long NK, et al. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br J Cancer. 2008;99(4):647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno N, Shiina H, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123(3):552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- Kim JW, Amin AR, et al. Chemoprevention of head and neck cancer with green tea polyphenols. Cancer Prev Res. 2010;3(8):900–909. doi: 10.1158/1940-6207.CAPR-09-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chung NG, et al. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58(5):660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- King-Batoon A, Leszczynska JM, et al. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008;49(1):36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- Kok TM, Breda SG, et al. Genomics-based identification of molecular mechanisms behind the cancer preventive action of phytochemicals: potential and challenges. Curr Pharm Biotechnol. 2012;13(1):255–264. doi: 10.2174/138920112798868601. [DOI] [PubMed] [Google Scholar]
- Kupchella CE. Environmental Factors in Cancer Etiology. Semin Oncol Nurs. 1986;2(3):161–169. doi: 10.1016/s0749-2081(86)80004-3. [DOI] [PubMed] [Google Scholar]
- Kurbitz C, Heise D, et al. Epicatechin gallate and catechin gallate are superior to epigallocatechin gallate in growth suppression and anti-inflammatory activities in pancreatic tumor cells. Cancer Sci. 2011;102(4):728–734. doi: 10.1111/j.1349-7006.2011.01870.x. [DOI] [PubMed] [Google Scholar]
- Lan J, Hua S, et al. DNA methyltransferases and methyl-binding proteins of mammals. Acta Biochim Biophys Sin. 2010;42(4):243–252. doi: 10.1093/abbs/gmq015. [DOI] [PubMed] [Google Scholar]
- Lea MA, Randolph VM, et al. Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int J Cancer. 2001;92(6):784–789. doi: 10.1002/ijc.1277. [DOI] [PubMed] [Google Scholar]
- Lea MA, Randolph VM, et al. Increased acetylation of histones induced by diallyl disulfide and structurally related molecules. Int J Oncol. 1999;15(2):347–352. doi: 10.3892/ijo.15.2.347. [DOI] [PubMed] [Google Scholar]
- Lea MA, Rasheed M, et al. Induction of histone acetylation and inhibition of growth of mouse erythroleukemia cells by S-allylmercaptocysteine. Nutr Cancer. 2002;43(1):90–102. doi: 10.1207/S15327914NC431_11. [DOI] [PubMed] [Google Scholar]
- Lee DE, Lee KW, et al. 7,3’,4’-Trihydroxyisoflavone, a metabolite of the soy isoflavone daidzein, suppresses ultraviolet B-induced skin cancer by targeting Cot and MKK4. J Biol Chem. 2011;286(16):14246–14256. doi: 10.1074/jbc.M110.147348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Chen YR, et al. Quercetin induces FasL-related apoptosis, in part, through promotion of histone H3 acetylation in human leukemia HL-60 cells. Oncol Rep. 2011;25(2):583–591. doi: 10.3892/or.2010.1097. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Shim JY, et al. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68(4):1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43(9):828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Chen H. Silencing of Wnt5a during colon cancer metastasis involves histone modifications. Epigenetics. 2012;7(6):551–558. doi: 10.4161/epi.20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, et al. Chemopreventive agent 3,3’-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res. 2010;70(2):646–654. doi: 10.1158/0008-5472.CAN-09-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, 2nd, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, VandenBoom TG, 2nd, et al. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70(4):1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–376. doi: 10.1007/978-1-61779-612-8_23. [DOI] [PubMed] [Google Scholar]
- Lin SC, Li WC, et al. The tea polyphenols EGCG and EGC repress mRNA expression of human telomerase reverse transcriptase (hTERT) in carcinoma cells. Cancer Lett. 2006;236(1):80–88. doi: 10.1016/j.canlet.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Liu HL, Chen Y, et al. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin. 2005;26(5):603–609. doi: 10.1111/j.1745-7254.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xie Z, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, et al. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7(6):682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso M, Paggi MG, et al. Genetic and epigenetic alterations as hallmarks of the intricate road to cancer. Oncogene. 2003;22(42):6472–6478. doi: 10.1038/sj.onc.1206955. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30(4):662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Dar AA, et al. Regulation of minichromosome maintenance gene family by microRNA-1296 and genistein in prostate cancer. Cancer Res. 2010;70(7):2809–2818. doi: 10.1158/0008-5472.CAN-09-4176. [DOI] [PubMed] [Google Scholar]
- Majid S, Dar AA, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2010;116(1):66–76. doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Kikuno N, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68(8):2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- Marcu MG, Jung YJ, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2(2):169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- Mattera L, Escaffit F, et al. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene. 2009;28(12):1506–1517. doi: 10.1038/onc.2008.499. [DOI] [PubMed] [Google Scholar]
- Medina-Franco JL, Lopez-Vallejo F, et al. Natural products as DNA methyltransferase inhibitors: a computer-aided discovery approach. Mol Divers. 2011;15(2):293–304. doi: 10.1007/s11030-010-9262-5. [DOI] [PubMed] [Google Scholar]
- Meeran SM, Patel SN, et al. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5(7):e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miremadi A, Oestergaard MZ, et al. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16:28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudduluru G, George-William JN, et al. Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep. 2011;31(3):185–197. doi: 10.1042/BSR20100065. [DOI] [PubMed] [Google Scholar]
- Murugan RS, Vinothini G, et al. Black tea polyphenols target matrix metalloproteinases, RECK, proangiogenic molecules and histone deacetylase in a rat hepatocarcinogenesis model. Anticancer Res. 2009;29(6):2301–2305. [PubMed] [Google Scholar]
- Myzak MC, Dashwood WM, et al. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006;20(3):506–508. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Hardin K, et al. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27(4):811–819. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Karplus PA, et al. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64(16):5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- Myzak MC, Tong P, et al. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood) 2007;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, et al. (-)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Peran E, Cabezas-Herrera J, et al. Effects of folate cycle disruption by the green tea polyphenol epigallocatechin-3-gallate. Int J Biochem Cell Bio. 2007;39(12):2215–2225. doi: 10.1016/j.biocel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Pandey M, Shukla S, et al. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2010;126(11):2520–2533. doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasramka MA, Ho E, et al. MicroRNAs, diet, and cancer: new mechanistic insights on the epigenetic actions of phytochemicals. Mol Carcinog. 2012;51(3):213–230. doi: 10.1002/mc.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker LP, Taylor DD, et al. Modulation of microRNA associated with ovarian cancer cells by genistein. Eur J Gynaecol Oncol. 2009;30(6):616–621. [PubMed] [Google Scholar]
- Pledgie-Tracy A, Sobolewski MD, et al. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Mol Cancer Ther. 2007;6(3):1013–1021. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- Priyadarsini RV, Vinothini G, et al. The flavonoid quercetin modulates the hallmark capabilities of hamster buccal pouch tumors. Nutr Cancer. 2011;63(2):218–226. doi: 10.1080/01635581.2011.523503. [DOI] [PubMed] [Google Scholar]
- Qin W, Zhu W, et al. Soy isoflavones have an anti-estrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutr Cancer. 2009;61(2):238–244. doi: 10.1080/01635580802404196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran P, Ho E, et al. Dietary phytochemicals, HDAC inhibition, and DNA damage/repair defects in cancer cells. Clin Epigenetics. 2011;3(1):4. doi: 10.1186/1868-7083-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran ZH, Zou J, et al. Experimental study on anti-neoplastic activity of epigallocatechin-3-gallate to digestive tract carcinomas. Chin Med J (Engl) 2005;118(16):1330–1337. [PubMed] [Google Scholar]
- Ropero S, Fraga MF, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38(5):566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- Rotili D. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes & Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Chen Q, et al. Resveratrol inhibits growth of orthotopic pancreatic tumors through activation of FOXO transcription factors. PLoS One. 2011;6(9):e25166. doi: 10.1371/journal.pone.0025166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Srivastava RK, et al. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J Mol Signal. 2010;5:10. doi: 10.1186/1750-2187-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz PA, Braune A, et al. Quercetin inhibits TNF-induced NF-kappaB transcription factor recruitment to proinflammatory gene promoters in murine intestinal epithelial cells. J Nutr. 2007;137(5):1208–1215. doi: 10.1093/jn/137.5.1208. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Maddison K, et al. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Pract Oncol. 2007;4(5):305–315. doi: 10.1038/ncponc0812. [DOI] [PubMed] [Google Scholar]
- Savouret JF, Quesne M. Resveratrol and cancer: a review. Biomed Pharmacother. 2002;56(2):84–87. doi: 10.1016/s0753-3322(01)00158-5. [DOI] [PubMed] [Google Scholar]
- Saw CL, Huang MT, et al. Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Mol Carcinog. 2011;50(6):479–486. doi: 10.1002/mc.20725. [DOI] [PubMed] [Google Scholar]
- Sawan C, Herceg Z. Histone modifications and cancer. Adv Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- Shankar S, Ganapathy S, et al. Sulforaphane enhances the therapeutic potential of TRAIL in prostate cancer orthotopic model through regulation of apoptosis, metastasis, and angiogenesis. Clin Cancer Res. 2008;14(21):6855–6866. doi: 10.1158/1078-0432.CCR-08-0903. [DOI] [PubMed] [Google Scholar]
- Shankar S, Nall D, et al. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelialmesenchymal transition. PLoS One. 2011;6(1):e16530. doi: 10.1371/journal.pone.0016530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam MK, Kannaiyan R, et al. Targeting cell signaling and apoptotic pathways by dietary agents: role in the prevention and treatment of cancer. Nutr Cancer. 2011;63(2):161–173. doi: 10.1080/01635581.2011.523502. [DOI] [PubMed] [Google Scholar]
- Shen H, Wang L, et al. A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002;62(17):4992–4995. [PubMed] [Google Scholar]
- Siddiqui IA, Adhami VM, et al. Beneficial effects of tea and its polyphenols against prostate cancer. Mol Nutr Food Res. 2006;50(2):130–143. doi: 10.1002/mnfr.200500113. [DOI] [PubMed] [Google Scholar]
- Singh BN, Shankar S, et al. Green tea catechin, epigallocatechin-3-gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem Pharmacol. 2011;82(12):1807–1821. doi: 10.1016/j.bcp.2011.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava RK, Unterman TG, et al. FOXO transcription factors and VEGF neutralizing antibody enhance antiangiogenic effects of resveratrol. Mol Cell Biochem. 2010;337(1-2):201–212. doi: 10.1007/s11010-009-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanska B, Karlic H, et al. Epigenetic mechanisms in anti-cancer actions of bioactive food components-the implications in cancer prevention. Br J Pharmacol: Ahead of Print. 2012 doi: 10.1111/j.1476-5381.2012.02002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanska B, Rudnicka K, et al. Hypomethylation and induction of retinoic acid receptor beta 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur J Pharmacol. 2010;638(1-3):47–53. doi: 10.1016/j.ejphar.2010.04.032. [DOI] [PubMed] [Google Scholar]
- Stein CJ, Colditz GA. Modifiable risk factors for cancer. Br J Cancer. 2004;90(2):299–303. doi: 10.1038/sj.bjc.6601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- Sun A, Tawfik O, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67(2):203–213. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- Sun M, Estrov Z, et al. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- Sun Q, Cong R, et al. Genistein inhibits growth of human uveal melanoma cells and affects microRNA- 27a and target gene expression. Oncol Rep. 2009;22(3):563–567. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- Tan AY, Manley JL. The TET Family of Proteins: Functions and Roles in Disease. J Mol Cell Biol. 2009;1(2):82–92. doi: 10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Wang C, et al. Quercetin is able to demethylate the p16INK4a gene promoter. Chemotherapy. 2008;55(1):6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- Tang SN, Fu J, et al. EGCG enhances the therapeutic potential of gemcitabine and CP690550 by inhibiting STAT3 signaling pathway in human pancreatic cancer. PLoS One. 2012;7(2):e31067. doi: 10.1371/journal.pone.0031067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SN, Singh C, et al. The dietary bioflavonoid quercetin synergizes with epigallocathechin gallate (EGCG) to inhibit prostate cancer stem cell characteristics, invasion, migration and epithelialmesenchymal transition. J Mol Signal. 2010;5:14. doi: 10.1186/1750-2187-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teiten MH, Eifes S, et al. Curcumin-The paradigm of a multi-target natural compound with applications in cancer prevention and treatment. Toxins. 2010;2(1):128–162. doi: 10.3390/toxins2010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, et al. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGFβ signaling pathway in SW480 cells. Biochem Pharmacol. 2010;80(12):2057–2065. doi: 10.1016/j.bcp.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Gasper AV, et al. Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr Biochem. 2005;135(8):1865–1872. doi: 10.1093/jn/135.8.1865. [DOI] [PubMed] [Google Scholar]
- Tsang DP, Cheng AS. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. J Gastroenterol Hepatol. 2011;26(1):19–27. doi: 10.1111/j.1440-1746.2010.06447.x. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2010;21(2):140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Tu SH, Ku CY, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits nicotineand estrogeninduced α9-nicotinic acetylcholine receptor upregulation in human breast cancer cells. Mol Nutr Food Res. 2011;55(3):455–466. doi: 10.1002/mnfr.201000254. [DOI] [PubMed] [Google Scholar]
- Varela I, Tarpey P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815(1):75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Volate SR, Muga SJ, et al. Epigenetic modulation of the retinoid X receptor alpha by green tea in the azoxymethane-Apc Min/+ mouse model of intestinal cancer. Mol Carcinog. 2009;48(10):920–933. doi: 10.1002/mc.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu Z, et al. Nutrition, epigenetics, and metabolic syndrome. Antioxid Redox Signal. 2012;17(2):282–301. doi: 10.1089/ars.2011.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LG, Beklemisheva A, et al. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinog. 2007;46(1):24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- Wang RH, Zheng Y, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen XY, Wu SY, et al. Ellagitannin (BJA3121), an anti-proliferative natural polyphenol compound, can regulate the expression of miRNAs in HepG2 cancer cells. Phytother Res. 2009;23(6):778–784. doi: 10.1002/ptr.2616. [DOI] [PubMed] [Google Scholar]
- Wessels K, Bohnhorst B, et al. Novel CHD7 mutations contributing to the mutation spectrum in patients with CHARGE syndrome. Eur J Med Genet. 2010;53(5):280–285. doi: 10.1016/j.ejmg.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hansen RS, et al. Genetic variation in ICF syndrome: evidence for genetic heterogeneity. Hum Mutat. 2000;16(6):509–517. doi: 10.1002/1098-1004(200012)16:6<509::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GS, Jin YS, et al. Annexin-I as a potential target for green tea extract induced actin remodeling. Int J Cancer. 2006;120(1):111–120. doi: 10.1002/ijc.22164. [DOI] [PubMed] [Google Scholar]
- Xu C, Huang MT, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66(16):8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Yang J, Cao Y, et al. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol. 2010;27(4):1114–1118. doi: 10.1007/s12032-009-9344-3. [DOI] [PubMed] [Google Scholar]
- Yaoting Gui, Guangwu Guo, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature Genetics. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa Y, Nagasaki H, et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis. 2005;26(1):193–200. doi: 10.1093/carcin/bgh304. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang T, et al. Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem Biophys Res Commun. 2010;399(1):1–6. doi: 10.1016/j.bbrc.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]


