Abstract
Transcription factors (TFs) bind in a combinatorial fashion to specify the on-and-off states of genes; the ensemble of these binding events forms a regulatory network, constituting the wiring diagram for a cell. To examine the principles of the human transcriptional regulatory network, we determined the genomic binding information of 119 TFs in 458 ChIP-Seq experiments. We found the combinatorial, co-association of TFs to be highly context specific: distinct combinations of factors bind at specific genomic locations. In particular, there are significant differences in the binding proximal and distal to genes. We organized all the TF binding into a hierarchy and integrated it with other genomic information (e.g. miRNA regulation), forming a dense meta-network. Factors at different levels have different properties: for instance, top-level TFs more strongly influence expression and middle-level ones co-regulate targets to mitigate information-flow bottlenecks. Moreover, these co-regulations give rise to many enriched network motifs -- e.g. noise-buffering feed-forward loops. Finally, more connected network components are under stronger selection and exhibit a greater degree of allele-specific activity (i.e., differential binding to the two parental alleles). The regulatory information obtained in this study will be crucial for interpreting personal genome sequences and understanding basic principles of human biology and disease.
A central goal in biology is to understand how a limited cohort of transcription factors (TFs) is able to choreograph the large diversity of gene-expression patterns in different cell types and conditions. Over the past decade, system-wide analyses of TF binding patterns have been performed in unicellular model organisms, such as E. coli and yeast, and have revealed a great deal of information about the organization of regulatory information1-8. Such studies have provided insights into such features as network hubs1, connectivity correlations9, hierarchical organization10,11, and network motifs12,13. Moreover, more complex networks that integrate disparate forms of genomic and proteomic data, such as protein-protein interactions and phosphorylation, have related gene regulation to other biological processes14-16. However for humans, systems-level analyses have been a challenge due to the size of the TF repertoire and genome, and only specific regulatory sub-networks with a handful of factors have been reported thus far17-19. The large-scale data from the ENCODE project now begins to enable such analyses20. Moreover, with the vast amount of human polymorphism data and genome sequences of many mammals21,22, it is possible to obtain an unprecedented view of how selection relates to networks.
Here, we present an analysis of the genome-wide binding profiles of 119 transcription-related factors, including sequence-specific, general, and chromatin-acting factors. (For simplicity, we abbreviate all of these as TFs, and we use TFSS to denote canonical sequence-specific factors). We first use the TF-binding data to analyze the co-association patterns between different TFs, as well as their differential patterns in promoter-proximal and distal regulatory regions. We then organize the binding patterns into a stratified hierarchy representing the overall systems-level regulatory wiring. To this, we add other forms of network information, including ncRNA regulation (especially miRNAs)23,24, protein-protein interactions25,26, and protein phosphorylation27. We analyzed this “meta-network” for properties that differ based on hierarchical level and connectivity (e.g., hubs vs. non-hubs) and also searched for enriched network motifs. Finally, we surveyed the pattern of sequence variation over the network, examining selective pressure and allelic effects (preferential binding to the maternal or paternal allele).
Several of our key findings include:
* Human TFs co-associate in a combinatorial and context-specific fashion; different combinations of factors bind near different targets, and the binding of one TF often changes the preferred binding partners of others. Moreover, TFs often show different co-association patterns in gene-proximal and distal regions.
* Different parts of the hierarchical TF network exhibit distinct properties. For instance, the middle level has the most information-flow bottlenecks and, offsetting this, tends to have the most regulatory collaboration between TFs. Conversely, higher-level TFs have the greatest connectivity with other networks (e.g., the phosphorylome).
* The occurrence of the feed-forward loops is strongly enriched in the TF network, as are a number of motifs in which two genes co-regulated by a TF are bridged by a protein-protein interaction or regulating miRNA.
* Highly connected network elements (both TFs and targets) are under strong evolutionary selection and exhibit stronger allele-specific activity (this is particularly apparent when multiple TFs are involved). Surprisingly, however, elements with allelic activity are under weaker selection than non-allelic ones.
Overview of Data and Processing
The ENCODE project has generated ChIP-seq datasets for 119 distinct TFs over five main cell lines (SOM/B.1, Tables S1 and S2a). Each dataset contains at least two biological replicates. In addition, for a select set of factors (Fig. S1c), siRNA experiments were performed, where the TF was depleted and expression changes were quantified by RNA-seq (SOM/B.2). Most of the factors (88, 74%) are TFSSs that can be subcategorized based on their DNA-binding domain sequences (Table S2a)28. A small subset (16, 13%) comprises POL2 and general transcriptional machinery; a final subset (15, 13%) consists of chromatin-modifying and remodeling factors.
In order to allow effective integrative analysis of these diverse datasets, we developed a uniform processing pipeline and quality-control measures (SOM/B.1, Figs. S1a,b and S2a, data at www.encodeproject.org). In total, we identified 7,424,765 peaks; 2,948,387 (~40%) were proximal (within +/−2.5 Kbp) to annotated gene transcription start sites (TSSs).
Context-specific TF Co-association
We first examined the genome-wide co-association of all pairs of TFs by analyzing the overlap between peaks of all pairs of factors20. Although many general trends can be identified, this approach does not take into account the context-specificity of TF binding (i.e., the fact that TFs bind together in distinct combinations at different genomic locations, and that the co-binding of one pair of TFs is often affected by the binding of another TF; SOM/C.1). Therefore, we developed a framework focusing on the specific genomic regions bound by a particular TF (the focus-factor) and examined the co-association of all other TFs (partner-factors) within this context (Fig. S2a). For each ~350 bp region in the focus-factor context, we extracted normalized binding signals of overlapping peaks of all TFs, generating a co-binding map. Fig. 1a shows such a map for the GATA1 context. Here, factors that consistently co-associate with each other and a substantial proportion of GATA1 peaks are termed ‘primary partners’ (e.g., group 6 TFs such as GATA2 and TAL1 in Fig. 1a). In addition to these factors, there are also groups of ‘local partners’ that co-associate with each other in the presence of GATA1, but only at specific subsets of GATA1 binding peaks (e.g., JUN in group 7 and MAX in 3; Fig. 1a and S2c-1). These biclusters, typically containing 2 to 5 TFs, can be mutually exclusive or partially overlapping.
Figure 1. TF Co-association.
(a) The co-binding map for the GATA1 focus-factor context in K562 shows the binding intensity of peaks of all TFs in K562 (rows) that overlap each GATA1 peak (columns). The colored rectangles represent 8 key clusters consisting of different combinations of co-associating partner-factors.
(b) The GATA1 context-specific relative importance scores (RI) of all partner-factors (top) and the matrix of co-association scores (CS) between all pairs of TFs (bottom). Primary and local partners of GATA have high RI scores. The co-association score matrix captures the 8 clusters observed in (a).
(c) Different partner-factors are preferentially enriched at gene-distal (positive differential RI) and proximal (negative differential RI) GATA1 peaks.
(d) The aggregate factor importance matrix, obtained by stacking the RI of all partner-factors (columns) from all focus-factor contexts (rows) in K562, shows 9 functionally distinct clusters (C1 to C9) of contexts that can be broadly grouped as distal, proximal, mixed, and repressive. The blue rectangles highlight representative partner-factors with high RI in the clusters. The arrow from (b) to (d) indicates that the GATA1 context-specific RI scores form one row in this matrix.
(e) Co-association variability map of partners (columns) of GATA1 (left panel) and FOS (right panel) over all K562 focus-factor contexts (rows). TAL1 and GATA2 show consistently high CS with GATA1 over most focus-factor contexts, but JUND shows context-specific co-association. FOS shows dramatic changes in CS of partner-factors over different contexts (e.g. FOS-JUND in distal contexts and FOS-SP2 in proximal ones).
(More details in Fig. S2c, S2f-1, S2d, S2l-2.)
To systematically identify all primary and local partners for each focus-factor context, we used a machine-learning approach. We learned non-linear, combinatorial models of each focus-factor's co-binding map relative to randomized control maps (SOM/C.2; Figs. S2a,b). Analysis of multivariate rules in these models, in turn, identified pairs and higher-order clusters of significantly co-associated TFs. Moreover, these co-associations are robust to peak overlap and calling thresholds (SOM/C.4).
The first statistic derived from the models is a relative-importance (RI) score (SOM/C.2.4.2), which gives the overall importance of a TF in the model. It reflects the ‘size’ of the biclusters to which a particular TF belongs, and it is related to the number of co-binding factors and the fraction of peak locations involved. For the GATA1 context (Figs. 1b and S2c-2), primary partners TAL1, GATA2 and POL2, as well as local partners MAX and JUN, have high RI scores. To further manifest the partnering in a particular context, we computed co-association scores between all pairs and higher-order sets of TFs (SOM/C.2.4). These scores measure the impact of the co-dependency implicit in a particular pair on the model as a whole, and they more directly probe the co-occupancy of TFs in the focus-factor context than does the RI score. For the GATA1 context, the co-association scores revealed both expected and novel pairings (e.g., MYCMAX-E2F6 and CCNT2-HMGN3, respectively; Figs. 1b and S2c-2 and SOM/C.3.1.4). Furthermore, GATA1 is usually associated with enhancer activity. However, the co-association score shows that it is connected to both repressive (e.g., NRSF, HDAC2) and activating TFs (e.g., P300). This “two-faced” behavior has been observed previously29; here, it is borne out by expression studies and knockdowns (SOM/C.3.1.4). In particular, after GATA1 knockdown, we found that 94 targets of GATA1 were significantly up-regulated, and only 54 were down-regulated (Fig. S2e-4). Finally, we analyzed the functions of genes that lie near clusters of co-associated TFs, and found that many are enriched for specific biological functions (Fig. S2e-2). For example, one bicluster involving E2F6 (E2F6-GATA1-GATA2-TAL1) was enriched for genes related to myeloid differentiation, while another (E2F6-SP1-SP2-FOS-IRF1) was involved in DNA damage response (SOM/C.3.3). Thus, distinct combinations of factors regulate specific types of genes.
Comparing Co-association Across Contexts
Aggregate RIM & PPM
After establishing the co-binding structure in each TF context, we compared our co-association statistics across contexts. In particular, we combined the RI scores for each TF into a single matrix (RIM, Fig. S2a). Clustering reveals nine functionally distinct classes of TF contexts that fall into four broad groups: proximal, distal, repressive, and mixed (Fig. 1d; S2f-1; SOM/C.3.4.1). Next, combining the co-association scores from all focus-factors across different contexts provides an overall view of all the primary partners of each TF in the form of a primary-partner matrix (PPM, Fig. S2f-4). The RIM reflects the overall similarities in the binding context of focus factors, whereas the PPM highlights the specific factors that tend to co-bind with each other (mutual primary partners). To some degree, one can see the PPM as a subset of the relationships implicit in the RIM. That is, two factors can have similar binding contexts without explicit co-association - e.g., two factors that tend both to bind promoters but near different sets of genes. Overall, the PPM shows well-known sets of co-associated TFs, such as FOS-JUN (the AP1 complex30,31) and CTCF-RAD21-SMC3 (the cohesion complex32,33), as well as many novel co-associations, such as CHD2-ZBTB33, EGR1-ZBTB7A, and ZNF143-CTCF-SIX5 (SOM/C3.6.2). We confirmed one novel co-association (CEBPB-TAL1) using co-immunoprecipitation and mass spectrometry (Table S3a).
Variability Map
The variability map shows the degree of variability in the partners of a given TF over contexts (as determined by the co-association score). For instance, Fig. 1e shows that GATA1 has mostly the same partners in many contexts (e.g., TAL1 and GATA2 are partners over almost all contexts). However, a few partners (e.g., JUND) are present in only some contexts. An extreme example is FOS, which completely changes its partners in different contexts (Fig. 1e, S2l-2 and SOM/C.3.6.1).
Relating Co-association to Cell-type and Genomic Location
Cell-type Differences
We analyzed TF co-association in the 5 main ENCODE cell types (SOM/C.3.4). The GM12878 and K562 cell-lines have the most (31) common TF datasets (SOM/C.3.5). Comparative analysis showed that over 80% of the TF pairs had no significant change in co-association between K562 and GM12878. However, there were a few dramatic examples of cell-line differences. For instance, FOS and JUND co-associate in K562 but not in GM12878 (SOM/3.5.1), despite the fact that most of the other partners of FOS are maintained in both cell lines.
Proximal vs. Distal Differences
Overall, we found distinct partner preferences at proximal and distal sites. These results were robust to the choice of the distance used to define proximal and distal regions (Fig. S2c-3). In particular, for the GATA1 context, we found that RI scores change dramatically between proximal or distal sites (Fig. 1c; S2c-3): typical core promoter TFs (e.g., POL2, E2F6, MAX and ELF1) have a significant proximal promoter bias, while JUND, JUNB, JUN and P300 show preferential co-association with distal sites. Another way of analyzing differences between proximal and distal sites is in the framework of the variability map, in which one can observe the changing partners of a TF in different contexts. For instance, FOS has completely different partners with which it co-associates proximally and distally (Fig. 1e; S2l-2 and SOM/C.3.6.1).
Assembling Pairwise Interactions into Hierarchies
Analysis of co-associations specifies the relationships between the DNA-binding profiles of multiple regulators. To obtain a systems-level perspective, we recast TF associations as a network (Fig. S4a), wherein the nodes are regulators or their targets, and the edges designate regulatory relationships. Here, we focus on the global wiring pattern across all cell types. We expect different subnetworks within this framework to be active to different degrees in different cells.
Using our binding-site list, we identified an initial set of regulatory targets from genes having promoter-proximal binding sites. The resulting raw network consists of 500,542 promoter-associated interactions between TFs and all their putative targets, of which 4,809 are between pairs of TFs (networks at encodenets.gersteinlab.org). We filtered this to identify the most confident interactions using a probabilistic model, giving 26,070 total interactions, with only 338 between TFs34 (SOM/D.1). We validated the performance of the filtering using the siRNA experiments; for each case, the targets identified by our model were more differentially expressed in siRNA-treated cells than were those identified by a simple peak-based method (Figs. S1c-e).
We next computed common connectivity statistics for individual TFs, namely, out-degree (O), in-degree (I), and betweenness, which were then used to identify hubs and information-flow bottlenecks (SOM/K). Of particular interest is the difference between out- and in- degree (O-I), which measures the direction of information flow (Fig. S3a). A positive value suggests that a TF is located “upstream” in the network, whereas a negative value indicates that a TF is “downstream.” We further defined a normalized version of this “hierarchy height” metric, h=(O-I)/(O+I). We found this can be approximated by 3 levels (Fig. S3c), with top-level, “executive” TFs regulating many other factors (h ~ 1), and bottom-level “foreman” TFs more regulated than regulating (h ~ −1). For purposes of visualization, we used a simulated-annealing procedure to optimally and robustly arrange the 119 TFs into 3 discrete levels (with the number of downward-pointing edges maximized) (Fig. 2a, SOM/D.2).
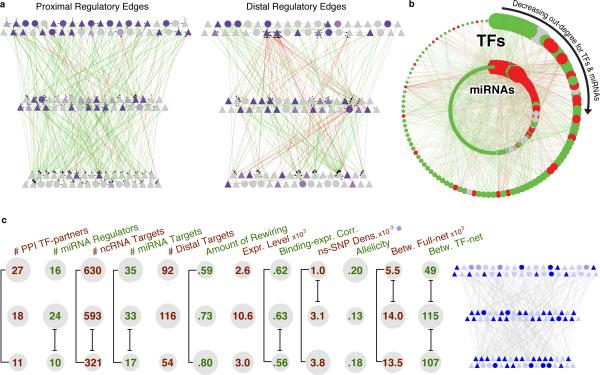
Figure 2. Overall Network.
(a) Close-up of the TF hierarchy. The nodes depict the TFs: TFSSs are triangles, and non-TFSSs are circles. At the left we show the proximal-edge hierarchy with downward pointing edges colored in green, and upward pointing ones colored in red. The nodes are shaded according to their out-degree in the full network (as described in Table 1). The right part shows the TFs placed in the same proximal hierarchy but now with edges corresponding to distal regulation colored green and red, and nodes recolored according to out-degree in the distal network. We see that the distal edges do not follow the proximal-edge hierarchy.
(b) Close-up of TF-miRNA regulation. The outer circle contains the 119 TFs, while the inner circle contains miRNAs. Red edges correspond to miRNAs regulating TFs; green ones, TFs regulating miRNAs. TFs and miRNAs each are arranged by their out-degree, beginning at 12 o'clock and decreasing in order clock-wise. Node sizes are proportional to out-degree. For TFs, the out-degree is as described in Table 1; for miRNAs, it is according to the out-degree in this network. Red nodes are enriched for miRNA-TF edges and green nodes are enriched for TF-miRNA edges. Gray nodes have a balanced number of edges (within ±1).
(c) Average values of various properties (topological, dynamic, expression-related, and selection-related -ordered consistently with Table 1) for each level are shown for the proximal-edge hierarchy. The top, middle, and bottom rows correspond to the top, middle, and bottom of the hierarchy, respectively. The sizing of the grey circles indicate the relative ordering of the values for the three levels. Significantly different values (P<0.05) using the Wilcoxon-rank-sum test are indicated by black brackets. The proximal-edge hierarchy depicted on the right shows non-synonymous SNP density, where the shading corresponds to the density for the associated TF.
(More details in Fig S4.)
Layering on Distal, ncRNA and Protein Interactions
The filtered TF hierarchy consists of the strongest promoter-associated interactions. Building upon this skeleton, we added additional types of connections.
Interactions involving distal regulatory elements (e.g., enhancers) are more difficult to identify than those involving proximal elements. Here, we employed a statistical model 35. This identifies distal sites with potentially many binding TFs using chromatin features. These regions were associated with a gene if their changing pattern of chromatin marks across cell lines correlates with the expression of that gene (SOM/E.1). Overall, the model identified 19258 distal edges (Fig. 2a).
The regulatory interactions between TFs and ncRNAs constitute an additional layer of information to add to the meta-network. We used TF peaks proximal to ncRNAs to identify TF-to-ncRNA regulation. Next, we incorporated miRNA-to-TF regulatory interactions from TargetScan36 (SOM/E.2). Finally, we incorporated physical protein-protein interactions26, as well as predicted phosphorylations (SOM/F.3 and Fig. S7a). Overall, these different interactions form a dense meta-network that was further analyzed for interesting biological properties.
Relating Network Connectivity and Genomic Properties
We next correlated measures for the connectivity and hierarchical position of each TF with a wide variety of genomic and proteomic properties (Fig. 2c, Tables 1 and S4, p-values in the later).
Table 1.
Correlating Properties with Centrality and Hierarchy Height
| Category | Property | Correlation with | ||||
|---|---|---|---|---|---|---|
| Degree centrality^ | Betweenness centrality | |||||
| Full | TF-TF | Full | TF-TF | TF-TF | ||
| Topology | # of TF partners in PPI | 0.28** | 0.27** | 0.25* | 0.33** | 0.08 |
| # of miRNA regulators | 0.24* | 0.33** | −0.02 | 0.00 | 0.29** | |
| # of ncRNA targets | 0.65** | 0.49** | 0.34** | 0.35** | 0.22* | |
| # of miRNA targets | 0.62** | 0.50** | 0.33** | 0.34** | 0.19* | |
| # of distal targets | 0.32** | 0.24* | 0.19* | 0.23* | 0.07 | |
| Dynamics | Amount of rewiring | −0.14 | −0.12 | 0.44* | 0.35 | −0.42* |
| Expression | Expression level | 0.14 | 0.12 | 0.23* | 0.27* | −0.04 |
| Binding-exp. corr. | 0.41** | 0.31** | 0.30** | 0.36** | 0.19* | |
| Selection properties for factors | ns SNP density | −0.19* | −0.27* | −0.01 | −0.03 | −0.22 |
| Allelicity | 0.20 | 0.28* | −0.10 | −0.16 | 0.18 | |
| Selection properties for targets | ns SNP density | −0.05** | ||||
| dN/dS | −0.05** | |||||
Spearman correlation values of various properties (topological, dynamic, expression-related, and selection-related) with centrality measures and hierarchy height (h). Only properties which are significantly correlated with centrality or h are listed (* for P<0.05 and ** for P<0.01). For a full set of properties, p-values, and explanations, see Tables S4 and S6. Degree centrality (note ^) refers to out-degree, except for selection properties on targets, in which case it refers to in-degree. In particular, out-degree in the full TF-target network refers to the “Targets” column in Table S4a, and the same quantity is used throughout Fig. 2.
Correlations with Distal Edges
Distal edges have a different degree distribution than do proximal edges (Figs. 2a and S5). Inspection reveals that many point upward in the TF hierarchy, opposite to most proximal edges. Furthermore, we found many TFs with low in-degree in the proximal network but high in-degree in the distal one, suggesting that they are heavily regulated through enhancers (Fig. S5a). Some of these are well known condition- and tissue-specific regulators (e.g., IRF4 and GATA1)37.
Correlations within the Proximal Network
Upper-level TFs tend to have more targets than lower-level ones, both overall and when considering only other TFs as targets. As measured by betweenness in proximal regulation, middle-level TFs form information-flow bottlenecks (Fig. 2c). Moreover, betweenness in the proximal TF network is correlated with more distal regulation. This tends to increase the information flow through mid-level bottlenecks even more. (See SOM/F.3.6 for clarification on implications.)
Correlation with Protein Interactions and the Phosphorylome
We found that top-level TFs tend to have more partners in the protein-interaction network than do lower-level TFs (Figs. 2c and S4e and Table 1). We further studied how TFs in different levels are regulated by kinases. Though there is no significant difference in terms of the number of kinases regulating TFs at different levels, we found that if the phosphorylome is arranged into a hierarchy using the same approach used for organizing the TF network, kinases at the bottom tend not to phosphorylate TFs, but they tend to be regulated by them (particularly by top-level TFs; Fig. S7).
Correlation with ncRNAs
We found that top- and middle-level TFs have the highest total number of ncRNA targets (Figs. 2c, S6a and Table 1), consistent with our findings for protein-coding targets. We then developed a score indicating the fraction of a TF's total regulation devoted to ncRNAs, relative to protein-coding genes (SOM/E.2); this identified several TFs that preferentially target ncRNAs, such as BDP1 and BRF2 (Figs. S6b,c).
Matching the pattern for ncRNAs in general, most of the TFs involved in miRNA regulation tend to be top- or middle-level TFs (Fig. 2c). Moreover, highly connected TFs tend to regulate more miRNAs and to be more regulated by them (Table 1 and Fig. 2b). However, when we analyze TF-miRNA regulation in detail we find that the TFs most involved in miRNA regulation tend to either largely regulate or be regulated by miRNAs (Fig. 2b, S4d). That is, there are few high-degree TFs with “balanced regulation” (similar numbers of incoming and outgoing edges, relative to a control; Fig. S3m). The same pattern can be seen for miRNAs (Fig. S3l).
Correlation with families and functional categories
Chromatin-related factors are enriched at the top of the hierarchy, while TFSSs are enriched in the middle (Table S5a and SOM/F.1). Also, TFSSs exhibit a greater degree of tissue-specificity and are more highly regulated by miRNAs than are general and chromatin related factors (SOM/F.4), suggesting they may be more finely tuned in their expression. Examining functional enrichment, we found that TFs on the top tend to have more general functions, and TFs on the bottom tend to have more specific functions (Table S5c and SOM/F.1).
Correlation with Network Dynamics
We studied how TFs change their binding patterns among different cell types, principally K562 vs. GM12878. We quantified the amount of “rewiring” as the fraction of unshared targets, normalized by the union of two target sets (SOM/F.3.5). We found that this “rewiring score” is negatively correlated with hierarchy height (Fig. 2c and Table 1). This means that the targets of lower-level TFs tend to change more between cell types, consistent with their role in more specialized processes.
Correlation with Gene Expression
We calculated the average expression levels of TFs across 34 tissues26; highly connected TFs tend to be highly expressed. We further examined the relationship between connectivity and expression by calculating, for each TF, the correlation between its binding signal around its targets and the level of target expression (SOM/F.3.4). This binding-expression correlation is positively correlated with TF connectivity. Moreover, TFs at the top and middle levels exhibit a greater correlation. Thus, more “influential” TFs tend to be better connected and higher in the hierarchy. (This degree of “influence” becomes even clearer when one considers weighting the correlation by the number of TF targets, given that higher-level TFs tend to have more targets.) However, somewhat surprisingly, a model integrating the binding-expression relationships of all the highly connected TFs has about the same predictive power for expression as a model integrating all the less connected ones, indicating that the weak binding-expression relationships of the less influential TFs are collectively quite influential (SOM/F.3.4)38.
Collaboration between Hierarchy Levels
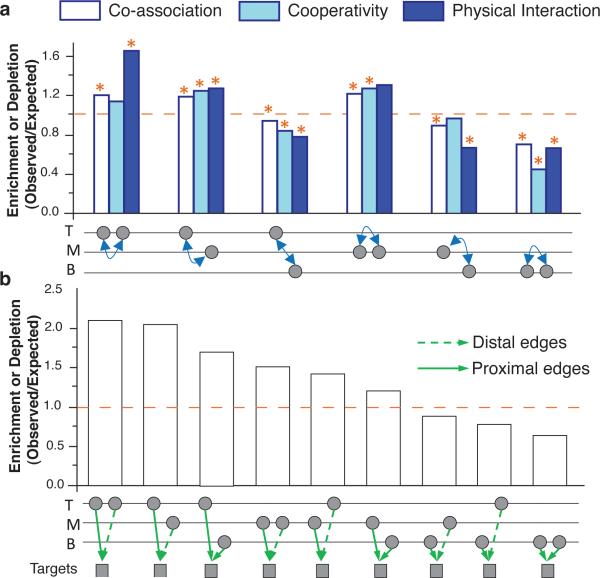
We explored how TFs in the top, middle, and bottom (T, M, and B) levels of the hierarchy collaborate, in terms of both inter- (TM, MB, TB) and intra- (TT, MM, BB) level relationships (Fig. 3a). We examined three kinds of collaboration: co-association (as described earlier), physical interactions , and target-expression cooperativity. We defined two TFs as being cooperative if their shared targets are significantly different in expression from their unshared targets (SOM/G.2). Overall, we find that collaborations involving the middle (and to lesser, extent the top) levels tend to be enriched. In particular, TM and MM TF pairs influence gene expression cooperatively. Next, all co-associations involving top- and middle-level TFs are enriched, whereas those involving the bottom level are depleted. A similar pattern is observed for protein-protein interactions, with TT and TM co-regulation more likely to occur between physically interacting TFs (Fig. 3a and SOM/G.1).
Figure 3. Collaboration between Levels.
(a) Enrichment of collaborating TF pairs from different levels (T,M,B). The TFs are represented by two nodes below each bar graph. The dashed orange line indicates the expected level of collaboration. Significant enrichment above or depletion below that level are marked by asterisks (P<0.05). (More details in SOM/G.1,2.)
(b) Enrichment of proximal and distal co-regulatory pairs in the network hierarchy. Co-regulatory pairs from different levels are shown by the two nodes below each bar.
Finally, we analyzed how proximal and distal sites “collaborate”. We identified pairs of TFs that bind to the promoter and distal regulatory regions of the same target gene (SOM/G.3) and studied their respective locations in the TF hierarchy. We found an asymmetry between proximal and distal regulation, with TFs associated through promoter regulation more likely to reside in upper levels (Fig. 3b).
Enriched Network Motifs
Apart from its global structure, we further studied the network from the perspective of its constituent building blocks - i.e., network motifs, which are small connectivity patterns that carry out canonical functions39. We systematically searched for motifs, first in the promoter-regulation hierarchy and then in the meta-network including distal, miRNA, and protein-protein interactions. Our procedure was to instantiate all possible motifs for broad “template patterns” and then determine which of these were significantly over- or under- represented relative to a random control40 (SOM/H). For instance, starting with all possible “3-TF motifs” in the proximal network (Fig. 4a), we found the most enriched motif to be the well-studied feed-forward loop (FFL)39. In agreement with the observed collaborations within the hierarchy, many FFLs involve the middle level (Fig. S9a). Moreover, by analyzing the expression levels of the constituent genes of the FFLs over many tissues, we found many were positively correlated, highlighting the tight regulation implicit in the motif (Fig. 4a and SOM/H.1). Finally, we found further enriched 3-TF motifs containing an additional regulation on top of that in a FFL. This creates a mutual regulation between a pair of TFs, instantiating a toggle-switch, which has been shown to play an essential role in cell-fate determination41.
Figure 4. Motif Analysis.
Motifs are accompanied by the occurrence frequency, N. Enriched motifs are highlighted in green, and depleted ones, in red. An occurrence frequency with a star means that the corresponding enrichment/depletion is statistically significant (P=1e-5). The motifs are sorted such that those at the ends have more significant p-values. (More details in Fig. S9h.)
(a) Systematic search of 3-TF motifs. The most enriched motif is the FFL. A particular example formed by STAT1, STAT3 and RUNX1 is highlighted. Here, the “+” sign on an edge indicates that the correlation between the gene expression of the source and the target across tissues is positive. Other motifs containing a toggle-switch regulation on top of the basic FFL design are also indicated.
(b) Proximal-Distal-PPI MIMs. Here we searched all motifs involving the co-regulation of two TFs (which could be either proximal or distal) with (or without) a protein-protein interaction between them. We found the motifs containing the protein-protein interaction tended to be enriched.
(c) miRNA-SIMs. This figure shows the 2 enriched motifs resulting from enumerating all motifs in which a miRNA targets two TFs that are connected in various ways. These 2 motifs contain a protein complex of 2 TFs and a cooperative pair of promoter and distal regulatory TFs.
(d) The auto-regulator motif is enriched in the TF-TF network: 28 of all TFs are auto-regulators. Moreover, auto-regulators are more likely to be repressors (-) relative to non-auto regulators, and they tend to have more ncRNAs as their targets.
Next, we analyzed another template: all possible multiple-input modules (MIMs, defined in SOM/K) involving promoter and distal regulation and a protein-protein interaction (proximal-distal-PPI MIMs, Fig. 4b). We found that co-regulating TFs are likely to physically interact, suggesting that they work together as a complex. Moreover, the motif ranking second in enrichment consists of a distal regulatory relationship, a promoter regulatory relationship, and a protein-protein interaction. This is suggestive of a common picture of DNA looping, with an interacting complex of TFs binding to the promoter and enhancer simultaneously.
The connection between co-regulated entities extends to miRNA regulation. We survey all possible instances of a miRNA regulating two TFs (“miRNA-SIM,” Fig. 4c) and find that the miRNAs are more likely regulate a pair of physically interacting TFs. This enrichment suggests that, in order to avoid unwanted cross-talk, a miRNA tends to shut down an entire functional unit (i.e., TF complex) rather than just a single component. Similarly, we found that miRNAs tend to target a pair of TFs binding both proximally and distally (Fig. 4c). This suggests that miRNA represses the expression of both promoter and distal regulators in order to completely shut down a target. Apart from miRNAs, we also studied motifs involving other kinds of ncRNAs. Amongst motifs involving a TF regulating two ncRNAs (“TF-ncRNA-SIM”), there is great enrichment for both ncRNAs to be lincRNAs (SOM/H.2).
Finally, we found the network to be enriched for auto-regulators (28 of 119 TFs), a simple but important motif, which are commonly found in networks exhibiting multi-stability42. Moreover, we found that the auto-regulators tend to be repressors, representing a well-known design principle for maintaining steady state39 (Fig. 4d).
Allelic Behavior in a Network Framework
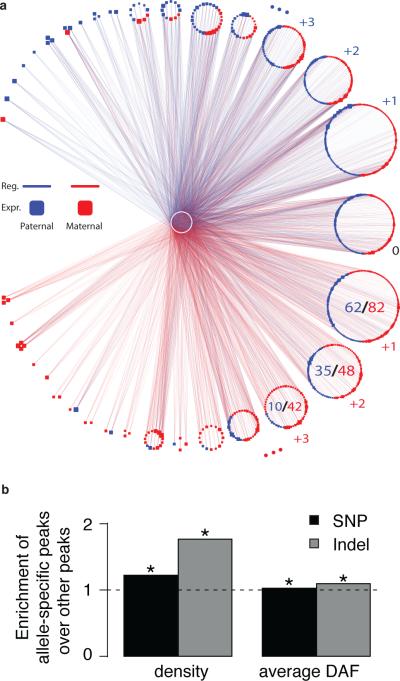
We examined the relationship between sequence variation and TF regulation. In particular, we investigated the coordination between allele-specific binding and expression (ASB and ASE)43,44. We used the sequenced datasets for GM12878, which has a deeply sequenced diploid genome (SOM/I.1). We extended pairwise analysis of allele-specific behavior20 to study higher-order coordination of multiple TFs regulating a common target. We first generated the unfiltered, promoter-regulation network for GM12878 and then identified a sub-network within it with 4,798 TF-target edges showing allele-specific regulation (SOM/I.2). This subnetwork is shown in Fig. 5a, where edges are colored red or blue to represent predominantly maternally or paternally regulated targets; the targets are similarly colored to indicate predominantly maternal or paternal expression. We find that of the 4,798 ASB cases of a single TF regulating its associated target, 57% show coordinated allelic binding and expression. We then find that for the cases in which two TFs regulate a common target, 63% are consistent (i.e., both TFs bind to the same allele that is expressed). For those cases in which triplets of TFs regulate a common target, the consistency increases to 65%. This trend continues, demonstrating that, as one increases the degree of combinatorial regulation, there is a progressively stronger relationship between expressed and regulated alleles.
Figure 5. Allelic Effects.
(a) An “allelic effects network” depicting the increasing coordination between ASB and ASE as the number of TFs regulating a target increases. Central white nodes denote TFs, and peripheral nodes denote targets, which are blue (red) if they are expressed from the paternal (maternal) allele. Blue (red) edges denote ASB to the paternal (maternal) allele. This network represents the strongest differences between the paternal- and maternal-specific regulatory networks. As one goes around the larger circle counterclockwise (clockwise), each of the small circular clusters represents targets with progressively more paternal (maternal) regulation, indicated by the small blue (red) numbers to the side of the clusters. Moreover, within each of the clusters the fraction of predominantly paternally (maternally) expressed targets increases as one goes around the larger circle. As an illustration, this fraction is explicitly indicated by the ratios within three of the larger clusters at bottom right.
(b) Relationship between TF allelicity and selection. The bar height is the ratio of the degree of selection (as measured by SNP density or average DAF) in those TF-binding peaks showing allelic behavior to the degree of selection in all other TF-binding peaks. Asterisks represent significant differences (P<0.05, Wilcoxon-rank-sum test).
(More details in SOM/I.2 and Fig S10b,c.)
The degree of allele-specific behavior of each TF can be quantified by a statistic we call “allelicity”. The allelicity of a TF is defined as the fraction of SNPs that exhibit ASB out of all the SNPs that may potentially exhibit it (SOM/I.3). Thus, qualitatively, allelicity may be thought of as the sensitivity of a TF's binding to maternal-vs-paternal variants. Using our network described here, we find that TFs with higher degrees of allelicity tend to have more target genes, suggesting that less specific TFs tend to vary more in their binding with sequence (Table 1). Finally, and somewhat intriguingly, we find that small insertions and deletions (indels) tend to cause disproportionally more of these allelic events than do SNPs (Table S6g).
Selection in a Network Context
Previous studies have examined the relationship between evolutionary selection and position in the human protein-protein interaction network45. However, the analogous relationship in the regulatory network has not yet been explored.
Selection
To address this, we first analyzed the selective pressure on both TFs and their targets. We predominantly used non-synonymous SNP density from the 1000 Genomes Pilot21 to determine selection amongst modern-day humans (SOM/J). We also verified our results using other measures of selection (i.e. derived allele frequency (DAF) and the pN/pS statistic (SOM/J). For selection over longer time scales, we calculated the ratio of non-synonymous to synonymous substitution in human-chimp ortholog alignments (dN/dS). We find significant negative correlation between the regulatory in-degree of target genes and both their non-synonymous SNP density and dN/dS values (Tables 1 and S6e). Thus, target genes regulated by more TFs are under stronger negative selection. Similarly, we find that there is a significant negative correlation between TF regulatory out-degree and non-synonymous SNP density (Tables 1 and S6d). We observe a consistent result with TF dN/dS values and other measures of selection, although these are not all as statistically significant (Table S6d and SOM/J). This shows that TFs regulating more targets tend to be under stronger negative selection. Moreover, within the TF hierarchy, we find that TFs at the top are under significantly stronger negative selection (Fig. 2c, Tables 1 and S6b).
Consistent with all these results relating connectivity with constraint, we find that genes tolerant of loss-of-function mutations46 are under weaker negative selection and have a significantly lower total degree (I+O) than other genes (SOM/J).
Selection and Allelic Effects
Finally, we attempted to relate selection and allelic effects. We extracted TF binding peaks in promoters and gene bodies showing ASB, and compared the selective pressure in these against a control (binding peaks within the same regions without ASB). We find that TF-binding peaks exhibiting allelic effects have higher SNP densities relative to the control (Fig. 5b). Moreover, binding peaks with no allelic effects show a skew in the DAF spectrum toward rarer SNPs, relative to ASB ones (Fig. 5b and S10c). The same trend holds true for indels and structural variations (Figs. 5b and S10b,c). Interestingly, these results indicate that allelic regulation appears to be under less selective constraint.
Discussion
This study provides the first detailed analysis of how human regulatory information is organized. A number of clear design principles emerge from it. Many of these are shared with model organisms (Table S7), demonstrating that they are general features of TF regulation. First, we find that the connectivity and hierarchical organization of regulatory factors is reflected in many genomic properties. For instance, top-level TFs have their binding more strongly correlated with the expression of their targets, perhaps indicating that they are more “influential”, as reported for model organisms47. Next, the middle-level contains information-flow bottlenecks and much connectivity with miRNA and distal regulation. Targeting these bottlenecks (e.g., by drugs) is likely to most strongly affect the flow of information through regulatory circuits. To some degree, the cell mitigates the effect of bottlenecks by having pairs of middle-level TFs collaborate in regulation. (Co-regulation reduces the degree of “bottleneckness”.) Third, the regulatory network appears to be built from repeated reuse of small, modular motifs. In particular, regulation between levels involves many feed-forward loops, which could be used to filter fluctuations in input stimuli. Again, these properties are shared with model organisms; the network motifs and cooperating middle-level have been observed in yeast48.
In contrast, the differences in proximal and distal regulation appear to be a unique feature of human regulation. This finding is evident in the analysis of both TF co-association and network structure. The proximal-distal differences reflect the much larger intergenic space in humans than model organisms and the commensurately larger amount of distal binding. Finally, analysis of conservation indicates that more highly connected parts of the network are under stronger selection, consistent with results from model organisms. However, one unique finding for humans is the “allelic” effects. More highly connected TFs are more likely to exhibit allele-specific binding. Interestingly, we found that the actual allele-specific binding sites tend to be under less selection. Unraveling this interplay between selection and regulatory networks will be crucial to interpreting variants in the many personal genome sequences expected in the future.
Methods Summary
Detailed methods associated with each section of the paper are in a similarly titled section of the Supplementary Online Material (SOM); see SOM Table of Contents and overview (SOM/A). In particular, an overview of our data processing pipeline is in SOM/B.
Supplementary Material
Acknowledgements
We thank the ENCODE Project (NIH/NHGRI) for funding. We thank Peter Bickel and Ben Brown at Berkeley for helpful conversations. Funding has also been provided by the NIH Predoctoral Training Program in Biophysics (Declan Clarke; T32 GM008283-24) and the Sackler institute (Gili Zilberman-Schapira). Manoj Hariharan wishes to thank Gouri Nair for designing database operations.
Footnotes
Author Contributions
Work on the paper was divided between data production and analysis. The analysts were A Abyzov, R Alexander, P Alves, S Batzoglou, N Bhardwaj, D Chen, C Cheng, D Clarke, Y Fu, M Hariharan, A Harmanci, E Khurana, A Kundaje, J Leng, R Min, X Mu, J Rozowsky, A Sidow, J Wang, K Yan, K Yip, and G Zilberman-Schapira. The data producers were N Addleman, A Boyle, P Cayting, A Charos, Y Cheng, C Eastman, G Euskirchen, P Farnham, S Frietze, J Gertz, F Grubert, P Jain, M Kasowski, P Lacroute, S Landt, J Lian, H Monahan, R Myers, H O'Geen, Z Ouyang, E Partridge, D Patacsil, F Pauli, D Raha, L Ramirez, T Reddy, B Reed, M Shi, T Slifer, S Weissman, L Wu, and X Yang. Larger efforts in analysis and data production are ascribed to the joint first authors. Author contributions to specific exhibits and files are shown in SOM/N and SOM/O. Overall project management was carried out by the two corresponding authors (M Gerstein and M Snyder).
Data sets described here can be obtained from the ENCODE project website at encodeproject.org and from encodenets.gersteinlab.org. More detail on data availability is in SOM/B and SOM/N. The authors declare competing financial interests: M Snyder is the founder of Personalis, and serves on the Scientific Advisory Boards of Personalis, and Genapsys. He is also a consultant for Illumina. All other authors declare no competing interests.
References
- 1.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 2.Balazsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu HY, Gerstein M. Genomic analysis of the hierarchical structure of regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14724–14731. doi: 10.1073/pnas.0508637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu ZZ, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nature Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 5.Balaji S, Babu MM, Aravind L. Interplay between network structures, regulatory modes and sensing mechanisms of transcription factors in the transcriptional regulatory network of E. coli. J. Mol. Biol. 2007;372:1108–1122. doi: 10.1016/j.jmb.2007.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jothi R, et al. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol. Syst. Biol. 2009;5:294. doi: 10.1038/msb.2009.52. doi:10.1038/Msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 8.Kim HD, Shay T, O'Shea EK, Regev A. Transcriptional Regulatory Circuits: Predicting Numbers from Alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 10.Ma HW, Buer J, Zeng AP. Hierarchical structure and modules in the Escherichia coli transcriptional regulatory network revealed by a new top-down approach. BMC Bioinformatics. 2004;5:199. doi: 10.1186/1471-2105-5-199. doi:10.1186/1471-2105-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balaji S, Iyer LM, Aravind L, Babu MM. Uncovering a hidden distributed architecture behind scale-free transcriptional regulatory networks. J. Mol. Biol. 2006;360:204–212. doi: 10.1016/j.jmb.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 12.Milo R, et al. Network motifs: Simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 13.Cosentino Lagomarsino M, Jona P, Bassetti B, Isambert H. Hierarchy and feedback in the evolution of the Escherichia coli transcription network. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5516–5520. doi: 10.1073/pnas.0609023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 15.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat. Rev. Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu HY, Xia Y, Trifonov V, Gerstein M. Design principles of molecular networks revealed by global comparisons and composite motifs. Genome Biol. 2006;7:11. doi: 10.1186/gb-2006-7-7-r55. doi:10.1186/gb-2006-7-7-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 18.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-Wide Occupancy of SREBP1 and Its Partners NFY and SP1 Reveals Novel Functional Roles and Combinatorial Regulation of Distinct Classes of Genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ENCODE Project Consortium Initial Analysis of the Encyclopedia of DNA Elements in the Human Genome. Nature. (NCP000 submitted) [Google Scholar]
- 21.Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindblad-Toh K, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barski A, et al. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. doi:10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozsolak F, et al. Chromatin structure analyses identify miRNA promoters. Gene. Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark C, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravasi T, et al. An Atlas of Combinatorial Transcriptional Regulation in Mouse and Man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novershtern N, et al. Densely Interconnected Transcriptional Circuits Control Cell States in Human Hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 29.Kerenyi MA, Orkin SH. Networking erythropoiesis. J. Exp. Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curran T, Franza BR. Fos and Jun - the Ap-1 Connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 31.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 32.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Cheng C, Min R, Gerstein M. TIP: A Probabilistic Method for identifying Transcription Factor Target Genes from ChIP-Seq Binding Profiles. Bioinformatics. 2011;27:3221–3227. doi: 10.1093/bioinformatics/btr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yip KY, et al. Genome-wide analysis of the binding sites of more than 100 transcription factors defines different types of genomic regions with distinct biological properties. Genome Biol. (GBCP033 manuscript in preparation) [Google Scholar]
- 36.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. doi:10.1101/Gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baron MH, Farrington SM. Positive Regulators of the Lineage-Specific Transcription Factor Gata-1 in Differentiating Erythroid-Cells. Mol. Cell. Biol. 1994;14:3108–3114. doi: 10.1128/mcb.14.5.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng C, et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. doi: 10.1101/gr.136838.111. (GRCP032 manuscript in revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alon U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 40.Cheng C, et al. Construction and Analysis of an Integrated Regulatory Network Derived from High-Throughput Sequencing Data. PLoS Comput. Biol. 2011;7:e1002190. doi: 10.1371/journal.pcbi.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang S, Zhou JX. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet. 2011;27:55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Burda Z, Krzywicki A, Martin OC, Zagorski M. Motifs emerge from function in model gene regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17263–17268. doi: 10.1073/pnas.1109435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDaniell R, et al. Heritable Individual-Specific and Allele-Specific Chromatin Signatures in Humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozowsky J, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.54. doi:10.1038/msb.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: Evaluation in terms of structural constraints and cellular context. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20274–20279. doi: 10.1073/pnas.0710183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacArthur DG, et al. A Systematic Survey of Loss-of-Function Variants in Human Protein-Coding Genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhardwaj N, Kim PM, Gerstein MB. Rewiring of Transcriptional Regulatory Networks: Hierarchy, Rather than Connectivity, Better Reflects the Importance of Regulators. Sci. Signal. 2010;3:ra79. doi: 10.1126/scisignal.2001014. [DOI] [PubMed] [Google Scholar]
- 48.Bhardwaj N, Yan K-K, Gerstein MB. Analysis of diverse regulatory networks in a hierarchical context shows consistent tendencies for collaboration in the middle levels. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6841–6846. doi: 10.1073/pnas.0910867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.