Abstract
A goal of the Tox21 program is to transit toxicity testing from traditional in vivo models to in vitro assays that assess how chemicals affect cellular responses and toxicity pathways. A critical contribution of the NIH Chemical Genomics center (NCGC) to the Tox21 program is the implementation of a quantitative high throughput screening (qHTS) approach, using cell- and biochemical-based assays to generate toxicological profiles for thousands of environmental compounds. Here, we evaluated the effect of chemical compounds on mitochondrial membrane potential in HepG2 cells by screening a library of 1,408 compounds provided by the National Toxicology Program (NTP) in a qHTS platform. Compounds were screened over 14 concentrations, and results showed that 91 and 88 compounds disrupted mitochondrial membrane potential after treatment for one or five h, respectively. Seventy-six compounds active at both time points were clustered by structural similarity, producing 11 clusters and 23 singletons. Thirty-eight compounds covering most of the active chemical space were more extensively evaluated. Thirty-six of the 38 compounds were confirmed to disrupt mitochondrial membrane potential using a fluorescence plate reader and 35 were confirmed using a high content imaging approach. Among the 38 compounds, 4 and 6 induced LDH release, a measure of cytotoxicity, at 1 or 5 h, respectively. Compounds were further assessed for mechanism of action (MOA) by measuring changes in oxygen consumption rate, which enabled identification of 20 compounds as uncouplers. This comprehensive approach allows for evaluation of thousands of environmental chemicals for mitochondrial toxicity and identification of possible MOAs.
Keywords: mitochondrial membrane potential assay, mitochondrial toxicity, NTP 1408 compound library, oxygen consumption rate, qHTS, Tox21 collaboration
Introduction
In 2004, the National Toxicology Program (NTP) made public its vision for toxicology testing in the 21st century stating the intention to move toxicology from a mainly observational science at the level of disease-specific, animal-based models to a predictive science focused upon a broad inclusion of target-specific mechanistic approaches.1 To fulfill the vision, the NTP established a High Throughput Screening (HTS) initiative that led in 2008 to the development of the Tox21 collaboration between the NTP, the Environmental Protection Agency (EPA) National Center for Computational Toxicology (NCCT), and the NIH Chemical Genomics Center (NCGC), with the addition of the Food and Drug administration (FDA) in 2010. In 2007, the National Research Council (NRC) published its long-range vision for toxicity testing that emphasized using new tools in molecular toxicology, computational sciences, and information technology to address the challenge of evaluating the thousands of untested chemicals present in the environment,2 stating that traditional toxicity testing methods are inadequate due to the high cost and the vast number of animals that would be needed to characterize these chemicals. Central to both visions is the idea of using chemical profiling strategies to study perturbations of critical cellular responses to xenobiotics using primarily HTS-based in vitro human cell models. The use of cellular targets or “toxicity” pathway perturbations as new, discrete toxicological endpoints will permit the identification of mechanisms of toxicity for each xenobiotic and provide predictive potential. The body of data generated will allow the scientific community to prioritize chemicals for more in-depth, targeted in vitro or in vivo assays that will eventually lead to the development of predictive toxicological models.3
Mitochondria occupy a central role in cellular physiology, making them ideal targets for in vitro toxicity studies. Mitochondria are involved in diverse processes such as energy metabolism,4 calcium homeostasis,5 cell signaling,6 macromolecular synthesis and maturation,7, 8 mitochondrial DNA replication and protein synthesis,9 and cell cycle and apoptosis regulation,10 depending on the tissue. Most of the cellular energy is generated as ATP through oxidative phosphorylation in the mitochondria.11 The protein complexes that transport electrons from reduced intermediates to the final acceptor (oxygen) are embedded in the mitochondrial inner membrane.12 Mitochondria also contain the enzymes involved in the Krebs cycle and fatty acid oxidation.13 Xenobiotic chemicals can perturb a variety of macromolecules in the mitochondria, thereby affecting any one of the several mitochondrial functions; this complexity presents a challenge in identifying unique mechanisms of mitochondrial toxicity, and developing predictive models.14 Numerous examples of molecules that inhibit different respiratory complexes, Krebs cycle or fatty acid metabolism, mtDNA replication and protein synthesis, or dissipate the mitochondrial membrane potential (MMP, Δψm), have been reported, most frequently as off-target effects of drug candidates.15, 16 Chemicals could also generate oxidative stress leading to a redox imbalance, a decrease of the reduced mitochondrial thiol pool and an opening of the mitochondrial permeability transition pore.15–17 Although many environmental toxicants have been studied,18 the work described here constitutes the first large scale study evaluating the mitochondrial toxicological properties of environmental chemicals.
Here, we report on the use of a quantitative high throughput screening (qHTS) approach to evaluate the acute effect of 1,408 chemicals of interest to the NTP on the MMP as a possible first indicator of acute mitochondrial toxicity. These compounds were selected based largely on the availability of toxicity data from standard tests for carcinogenicity, genotoxicity, immunotoxicity, neurotoxicity, and/or reproductive and developmental toxicity among others.19, 20 Based on the results of the primary screening, we selected several compounds for further confirmatory and mechanistic studies including compound-induced cytotoxicity and changes in oxygen consumption. We show that the combination of primary screening with follow-up mechanistic studies is a useful approach for profiling the effect of environmental chemicals on mitochondrial function.
Experimental Procedures
Reagents
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, Chemical Abstracts Service Registry Numbers (CASRN) 370-86-5) and all the compounds listed in Table 1 were purchased from Sigma-Aldrich (St. Louis, MO, USA) with the exception of 2,2'-thiobis(4-chlorophenol), 4,4-thiobis(6-t-butyl-m-cresol), and malachite green oxalate that were purchased from TCI America (Portland, OR, USA) and nitazoxanide that was purchased from Toronto Research Chemicals (North York, ON, Canada). When referring to chemical compounds, CASRN were included either in table 1 or next to the compound name in the text. Black, clear bottom 1536-well plates were purchased from Corning Costar (Acton, MA, USA).
Table 1.
Compound potency results (IC50, µM) for the primary screen at 1 and 5 h and powder compound confirmation at 1 and 5 h of treatment using either a fluorescence plate reader or a high content screen (HCS) imaging approach.
| Compound name | CASRN | Primary screen |
Fluorescence readout |
HCS readout | |||
|---|---|---|---|---|---|---|---|
| 1 h | 5 h | 1 h | 5 h | 1 h | 5 h | ||
| 1,5-Naphthalenediamine | 2243-62-1 | 2.98 | 5.96 | Inactive | Inactive | Inactive | Inactive |
| 1,8-Dihydroxy-4,5-dinitroanthraquinone | 81-55-0 | 4.73 | 3.76 | 1.36 ± 0.09 | 0.97 ± 0.13 | 10.07 ± 1.63 | 10 ± 0 |
| 2,2'-Methylenebis-(4-chlorophenol) | 97-23-4 | 10.59 | 11.88 | 9.29 ± 1.18 | 15.81 ± 0 | 3.98 ± 0 | 5.77 ± 1.85 |
| 2,2'-Thiobis(4-chlorophenol) | 97-24-5 | 8.41 | 7.50 | 9.25 ± 0.63 | 15.81 ± 0 | 4.74 ± 0.39 | 6.35 ± 1.03 |
| 2-Aminoanthraquinone | 117-79-3 | 1.33 | 1.68 | 1.94 ± 0.33 | 2.42 ± 0.16 | 37.62 ± 17.68 | 37.65 ± 3.06 |
| 3,4,5-Trichlorophenol | 609-19-8 | 33.49 | 37.58 | 32.84 ± 2.22 | 30.41 ± 1.98 | 22.02 ± 8.72 | 28.18 ± 0 |
| 3,4-Dichlorophenyl isocyanate | 102-36-3 | 4.22 | 5.96 | 4.3 ± 0.28 | 4.17 ± 0.75 | 2.7 ± 0.65 | 1.82 ± 0.98 |
| 4,4-Thiobis(6-t-butyl-m-cresol) | 96-69-5 | 10.59 | 14.96 | 14.92 ± 3.36 | 15.81 ± 0 | 8.54 ± 2.07 | 11.9 ± 0.97 |
| 4-Cumylphenol | 599-64-4 | 14.96 | 33.49 | 20.72 ± 1.4 | 11.65 ± 0.79 | 17.04 ± 4.12 | 23.75 ± 1.93 |
| 4-Hydroxyphenyl retinamide | 65646-68-6 | 14.96 | 18.83 | 63.24 ± 7.27 | 31.55 ± 0 | 18.26 ± 5.84 | 13.35 ± 6.27 |
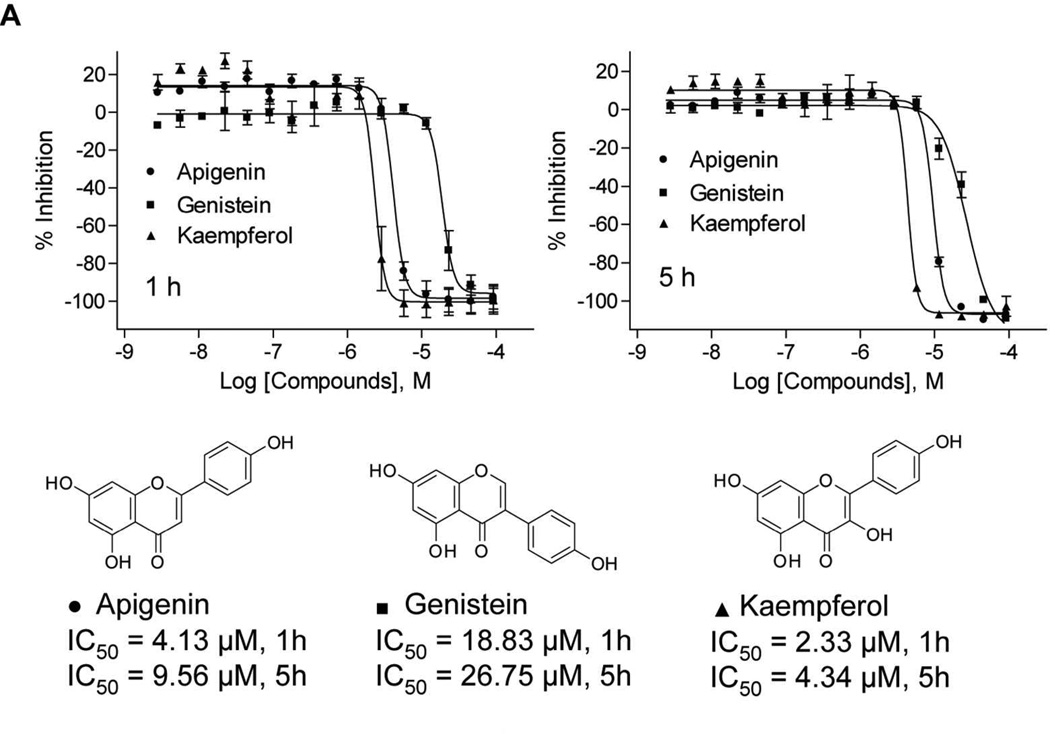
| Apigenin | 520-36-5 | 3.35 | 5.96 | 4.13 ± 0.28 | 9.25 ± 0.63 | 3.57 ± 0.58 | 4.59 ± 1.47 |
| Basic red 9 (p-Rosaniline HCl) | 569-61-9 | 1.33 | 0.60 | 2.21 ± 0.62 | 1.04 ± 0.07 | 26.65 ± 2.17 | 28.18 ± 0 |
| Captan | 133-06-2 | 26.60 | 23.71 | 29.27 ± 1.98 | 32.84 ± 2.22 | 17.49 ± 6.93 | 11.03 ± 4.37 |
| Curcumin | 458-37-7 | 4.22 | 5.96 | 1.75 ± 0.49 | 4.64 ± 0.31 | 11.9 ± 0.97 | 22.54 ± 3.65 |
| Diethylstilbestrol [DES] | 56-53-1 | 33.49 | 33.49 | 68.08 ± 4.44 | 60.68 ± 3.95 | 23.75 ± 1.93 | 25.29 ± 4.1 |
| Digitonin | 11024-24-1 | 26.60 | 26.60 | 31.69 ± 3.64 | 31.55 ± 0 | 64.78 ± 20.73 | 42.24 ± 3.43 |
| Diphenylurea | 102-07-8 | 13.33 | 13.33 | 13.07 ± 0.88 | 10.79 ± 0.7 | 11.22 ± 0 | 11.22 ± 0 |
| Emodin | 518-82-1 | 2.98 | 3.76 | 4.51 ± 0.84 | 7.35 ± 0.5 | 42.24 ± 3.43 | 50.12 ± 0 |
| Formulated fenaminosulf (Dexon) | 140-56-7 | 13.33 | 9.44 | 63.24 ± 7.27 | 56.11 ± 0 | Inactive | 33.53 ± 15.76 |
| Genistein (4',5,7-Trihydroxyisoflavone) | 446-72-0 | 5.96 | 7.50 | 19.28 ± 2.65 | 27.1 ± 1.77 | 18.26 ± 5.84 | 17.49 ± 6.93 |
| Hexachlorophene | 70-30-4 | 2.98 | 3.76 | 10.67 ± 3.14 | 6.07 ± 0.4 | 4.66 ± 3.42 | 2.82 ± 0 |
| Hexamethyl-p-rosaniline chloride (Gentian violet) | 548-62-9 | 0.67 | 1.19 | 0.29 ± 0.02 | 0.61 ± 0.04 | 1.68 ± 0.14 | 2.12 ± 0.17 |
| Kaempferol | 520-18-3 | 5.31 | 8.41 | 2.26 ± 0.47 | 4.3 ± 0.28 | 1.52 ± 0.37 | 2.84 ± 0.46 |
| Kepone | 143-50-0 | 3.35 | 4.22 | 5.64 ± 0.65 | 5.61 ± 0 | 1.83 ± 0.58 | 3.25 ± 1.04 |
| Malachite green oxalate | 2437-29-8 | 0.75 | 0.94 | 1.36 ± 0.09 | 1.85 ± 0.12 | 0.73 ± 0.23 | 1.21 ± 0.29 |
| Nitazoxanide | 55981-09-4 | 7.50 | 7.50 | 8.28 ± 1.06 | 7.1 ± 0.82 | 5.39 ± 1.3 | 7.08 ± 0 |
| N-Phenyl-2-naphthylamine | 135-88-6 | 37.58 | 42.16 | 56.11 ± 0 | 52.04 ± 3.52 | 22.54 ± 3.65 | 33.55 ± 2.73 |
| o,p'-DDD (Mitotane) | 53-19-0 | 33.49 | 37.58 | 32.84 ± 2.22 | 32.84 ± 2.22 | 17.04 ± 4.12 | 14.99 ± 1.22 |
| o-Benzyl-p-chlorophenol | 120-32-1 | 16.79 | 23.71 | 5.41 ± 0.35 | 4.82 ± 0.31 | 5.39 ± 1.3 | 3.81 ± 0.92 |
| p -n -Nonylphenol | 104-40-5 | 10.59 | 16.79 | 58.39 ± 3.95 | 56.11 ± 0 | 21.17 ± 1.72 | 29.9 ± 2.43 |
| p-Aminophenol | 123-30-8 | 7.50 | 7.50 | 22.44 ± 2.58 | 31.55 ± 0 | 35.9 ± 49.35 | 79.43 ± 0 |
| Phenmedipham | 13684-63-4 | 42.16 | 42.16 | 34.12 ± 2.22 | 45.14 ± 8.43 | 28.37 ± 4.6 | 34.89 ± 13.82 |
| Phenyl mercuric acetate | 62-38-4 | 16.79 | 14.96 | 20 ± 2.3 | 23.64 ± 5.33 | 30.3 ± 7.33 | 16.1 ± 12.76 |
| Resveratrol | 501-36-0 | 2.11 | 5.31 | 2.07 ± 0.7 | 8.25 ± 0.56 | 0.95 ± 0.08 | 1.91 ± 0.46 |
| Riboflavin | 83-88-5 | 6.68 | 7.50 | 22.44 ± 2.58 | 15.24 ± 0.99 | 20.27 ± 16.06 | 31.1 ± 12.32 |
| Silibinin | 22888-70-6 | 18.83 | 26.60 | Inactive | 65.52 ± 4.44 | Inactive | Inactive |
| tetra-N-Octylammonium bromide | 14866-33-2 | 3.76 | 2.66 | 1.6 ± 0.3 | 1.86 ± 0.34 | 2.3 ± 0.74 | 1.91 ± 0.46 |
| Zearalenone | 17924-92-4 | 37.58 | 42.16 | 42.31 ± 11.96 | 30.41 ± 1.98 | 22.02 ± 8.72 | 32.46 ± 10.39 |
Each value of potency [half maximal inhibitory concentration (IC50), µM] from MMP assays including HCS is the mean ± standard deviation from 4 independent experiments (except for the primary screen).
The NTP 1,408 compound library
This study was performed using an NTP library collection of 1,408 compounds.19, 20 Of the 1,408 compounds in the NTP collection, 1,340 were unique, 62 were present in duplicate, and 2 were present in triplicate. Briefly, this collection of 1,408 compounds included 1,206 compounds that had been tested by the NTP in one or more in vitro and/or in vivo assays (including those for bacterial mutagenicity (68%), chronic toxicity/carcinogenicity (23%), reproductive toxicity (3%), developmental toxicity (3%), immunotoxicity (1%)) and 147 reference compounds identified by NTP Interagency Center for the Evaluation of Alternative Toxicological Methods for the development and/or validation of alternative in vitro test methods for dermal corrosion, acute toxicity, and endocrine activity. Functionally, the NTP library of 1,408 compounds includes solvents, fire retardants, preservatives, flavoring agents, plasticizers, therapeutic agents, inorganic and organic pollutants, drinking-water disinfection byproducts, pesticides, and natural products. A complete list of the NTP 1,408 compounds and full chemical descriptions are publicly available at Pubchem.21
Cell culture
Human hepatocellular carcinoma cells (HepG2) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in ATCC complete Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 50 U/mL penicillin, and 50 µg/mL streptomycin (Invitrogen) at 37°C under a humidified atmosphere and 5% CO2. HepG2 cells are a suitable, cost effective, and commonly used hepatocyte model for qHTS. And even though HepG2 cells are a Hepato-carcinoma derived cell line, they are capable of cellular respiration22 and they have been used in many mitochondrial toxicity studies involving liver cells.23, 24 Furthermore the sensitivity of the assay using HepG2 cells was evaluated during the assay optimization and miniaturization using several known mitochondrial poisons and we found the assay sensitive enough for qHTS.25
Quantitative High Throughput Screening (qHTS)
Compound formatting and qHTS were performed as described previously.26, 27 The control plates were plated as follows. A positive control (FCCP) solution in dimethyl sulfoxide (DMSO, CASRN 67-68-5) was dispensed into columns 1, 2 and 3 (Fig 1S). Column 1 contained a sixteen-concentration titration ranging from 2.5 mM to 5.7 µM in duplicates. Columns 2 and 3 contained replicates for the positive control: 2 mM in column 2 (32 replicates) and 1.5 mM and 0.75 mM in column 3 (16 replicates for each concentration). Additionally, DMSO was dispensed into column 4 as a negative control. For testing, 23 nL of the compounds in DMSO solution were transferred via pin tool (Kalypsys, San Diego, CA, USA) from the compound and control plates to the assay plates, resulting in final concentrations of 0.03 nM to 11.5 µM of FCCP and 0.45% DMSO. Test compounds were screened over a 14-point concentration curve ranging from 0.59 nM to 92 µM. To achieve the highest final compound concentration of 92 µM (DMSO concentration 0.90%), 23 nL was transferred twice from the highest concentration of source (compound) plate into the assay plate; control plates with DMSO transferred twice were also included for comparison. The final concentration of DMSO in the assay was 0.45% or 0.90% in wells that were dosed once or twice, respectively.
MMP assay
MMP was measured using the Mitochondrial Membrane Potential Indicator (m-MPI, Codex Biosolutions, Inc., MD, USA).25 This is a fluorescence-based assay that quantifies changes in mitochondrial membrane potential. HepG2 cells were dispensed at 2000 cells/5 µL/well in tissue culture treated 1536-well black wall/clear bottom assay plates (Greiner Bio-One North America, NC, USA) using a Flying Reagent Dispenser (FRD) (Aurora Discovery, CA, USA). After overnight culturing of cells at 37°C to allow for attachment, cells were treated with the compounds at 37°C for 1 and 5 h; after the treatment period, 5 µL of 2X m-MPI reagent (10 µL of Mito-MPS solution + 5 mL of assay buffer) was added into the wells using the FRD and the plates were incubated for an additional 30 min at 37°C. Fluorescence intensities (485 nm excitation/535 nm emission for green fluorescent monomers; 540 nm excitation/590 nm emission for red fluorescent aggregates) were measured using an Envision plate reader (PerkinElmer; Shelton, CT, USA).25 Data were expressed as the ratio of 590 nm/535 nm.
Imaging-based MMP assay
HepG2 cells were dispensed at 2000 cells/5 µL/well in tissue culture treated 1536-well black wall/clear bottom assay plates (Greiner Bio-One North America) using the FRD. After culturing overnight at 37°C, the assay plates were treated with the compounds at 37°C for 1 or 5 h, followed by addition of 5 µL of m-MPI reagent (Codex) with 0.3 µg/mL of Hoechst 33342 (Invitrogen) used to stain DNA. The plates were incubated for 30 min at 37°C. The fluorescence intensities (482 nm excitation/536 nm emission for green fluorescent monomers; 543 nm excitation/593 nm emission for red fluorescent aggregates; 377 nm excitation/447 nm emission for Hoechst 33342) were measured using an ImageXpress Micro Widefield High Content Screening System (Molecular Devices, Sunnyvale, CA, USA). Imaging was processed and analyzed with the MetaXpress® and PowerCore® software (Molecular Devices) using the Multi Wavelength Cell Scoring algorithm. The mean of average fluorescence intensity from each positive cell was calculated per well for both green and red fluorescent colors. Data was expressed as the ratio of 593 nm/536 nm emissions.
LDH (lactate dehydrogenase) release assay
LDH release was measured using the CytoTox-ONE™ Homogenous Membrane Integrity kit (Promega Madison, WI, USA). The HepG2 cells were dispensed at 2,000 cells/5 µL/well in 1,536-well black wall/clear bottom assay plates using an FRD. The cells were incubated overnight at 37°C, followed by the addition of compounds using the pin tool. The assay plates were incubated for 1 or 5 h at 37°C, followed by the addition of 5 µL/well of CytoTox-ONE™ reagent. After a 10 min incubation at room temperature, the fluorescence intensity (560 nm excitation/590 emission) was measured using an Envision plate reader (PerkinElmer).
Oxygen Consumption Studies
Oxygen consumption rates (OCRs) were measured using a Seahorse Biosciences XF96 respirometer (Seahorse Biosciences, North Billerica, MA, USA). The HepG2 cells were plated in XF96 plates at 30,000 cells/well in 100 µL media. After 24 h, OCRs were measured with a 3 min measure, 2 min mix cycle using the Akos algorithm (Seahorse Biosciences) to convert changes in fluorescence to OCR. Basal rates were measured 4 times, followed by injection of compound and an immediate rate measurement that was repeated every 5 m for the next h, after which 1 µM FCCP was injected. Following injection, several OCR measurements were made to assess maximal uncoupled rate.
Data analysis
Primary data analyses were performed as previously described.28 Briefly, raw plate reads for each titration point were first normalized relative to FCCP control (3.5 µM for 1 h and 6.9 µM for 5 h, −100%) and DMSO only wells (basal, 0%), and then corrected by applying a pattern correction algorithm using compound-free control plates (DMSO plates). Concentration-response titration points for each compound were fitted to the Hill equation29 yielding concentrations of half-maximal inhibition (IC50) and maximal response (efficacy) values. Compounds were designated as Class 1–4 according to the type of concentration response curve observed.26, 30 Curve classes are heuristic measures of data confidence, classifying concentration–responses on the basis of efficacy, the number of data points observed above background activity, and the quality of fit. Compounds with ratio (red/green, 590 nm/535 nm) curve classes 1.1, 1.2 or 2.1 and red channel showing inhibition with curve classes different from 4 were considered positive. Compounds with class 4 curves for either the ratio or the red channel were defined as inactive and compounds with other curve classes were considered inconclusive.
The 76 compounds classified as active at both the 1 and 5 h time points were clustered based on structural similarity (Hierarchical (agglomerative) nesting) using the Leadscope software (Leadscope Inc., Columbus, OH, USA), resulting in 11 structural clusters and 23 singletons. Thirty-eight active compounds (at least one per cluster) were selected for follow-up studies based on structural diversity, potency, efficacy, and availability.
For the follow-up OCR studies, 4 measurements of the basal rates were taken as controls and all rates were then normalized to the mean of the 3rd and 4th basal rates for that treatment group. HepG2 cells were treated in triplicate with the 38 compounds at the MMP assay IC50 value, and each compound was tested in 5 or 6 independent experiments. The final concentration of vehicle (DMSO) was 0.1%, the average basal OCR of the vehicle controls were 135 pmol/m/well, and the average maximal uncoupled OCR for the vehicle control treated cells were 210 pmol/m/well. OCR was measured immediately (30 sec) and 1 h after compound treatment and immediately after injection of FCCP. The OCR values were normalized against the basal rate. Given the 12% variance seen in OCR data of known toxicants,31 a change of 20% or higher is considered significant.
Results
Screening of the NTP library for changes in MMP
As part of the Tox21 initiative, we screened the NTP chemical library19, 20 for compounds that decreased the MMP in HepG2 cells using the cationic dye m-MPI.25 Compounds were screened in 14 concentrations from 0.59 nM to 92 µM for 1 and 5 h of treatment; the uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was used as a positive control.31 A concentration titration of FCCP was included as a reference in each plate to examine data quality. The mean IC50 observed for FCCP from all 18 plates, including 4 DMSO control plates, was 0.81 ± 0.20 µM and 1.28 ± 0.32 µM [mean ± SD (standard deviation)] at the 1 and 5 h time points, respectively. The mean signal-to-background ratio from the 18 plates was 23.54 ± 2.01 and 30.39 ± 2.85 and the Z’ factor32 averaged 0.82 ± 0.02 and 0.85 ± 0.02 for 1 and 5 h, respectively. The coefficient of variation (CV) for the DMSO plates was 6.75 ± 0.45% and 5.84 ± 0.29% at 1 and 5 h, respectively.
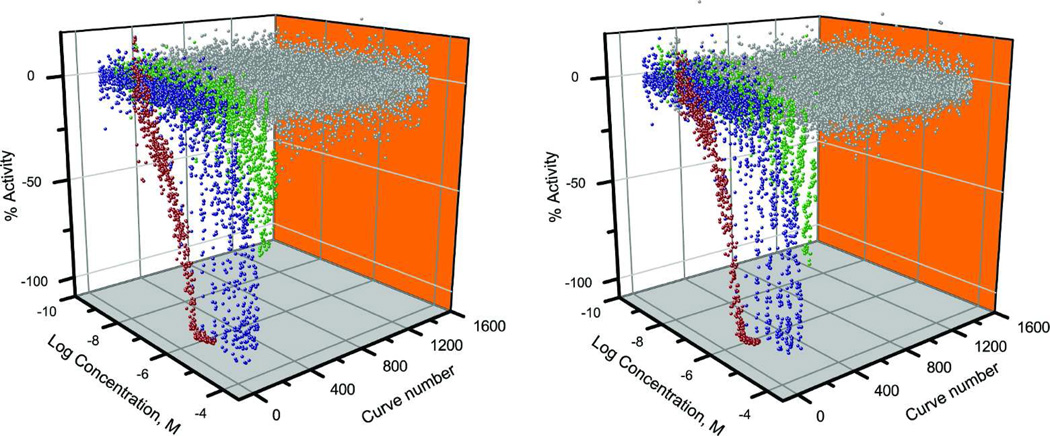
Of the 1340 unique compounds in the NTP library, 190 compounds decreased MMP in a concentration-dependent manner after 1 h of treatment and 151 after 5 h of treatment in HepG2 cells (Fig. 1). Of these compounds, 91 (1 h treatment) and 88 (5 h treatment) compounds with curve classes 1.1, 1.2, or 2.1 (see detail description in method)26 were considered actives. Of the 64 duplicates in the NTP library, 62 pairs (97%) showed the same outcome (either active or inactive) for 1 h, with only parathion (CASRN 56-38-2) and norbixin (CASRN 542-40-5) showing dissimilar responses. Similar results were observed for the 5 h treatment, with 63 (98 %) pairs showing the same results with norbixin being the exception. The two compounds that are present in triplicate were inactive at 1 and 5 h for all replicates. The data generated from the primary screen are available in Pubchem [assay IDs: 651754 (1 h) and 651755 (5 h)].21
Figure 1.
Complete screening dataset. Dose response curves for the FCCP control titrations and 1,408 compounds tested in the primary screening for 1 (left) and 5 (right) h (Red, FCCP positive control; blue, ratio (red/green, 590 nm/535 nm) curve classes 1.1, 1.2 or 2.1; green ratio(red/green) curve classes 1.3, 1.4, 2.2, 2.3, 2.4, and 3, and gray, ratio(red/green) curve class 4).26,30
Seventy-six compounds with curve classes 1.1, 1.2, or 2.1 were active at both 1 and 5 h of treatment. These 76 compounds were clustered by structural similarity resulting in 11 structural clusters and 23 singletons (Supplementary information Table S1). Four of the 5 flavonoids screened — apigenin, kaempferol, genistein (Fig 2), and daidzein (CASRN 486-66-8)— were active at both time points and clustered together (Cluster #4, Supplementary information Table S1). The fifth, flavone (2-phenyl-1,4-benzopyrone, CASRN 525-82-6), was only marginally active at 1 h and inactive at 5 h. Similarly, the three triarylmethane dyes screened — basic red 9 (p-rosaniline HCl), hexamethyl-p-rosaniline chloride, and malachite green oxalate — were positive at both time points and clustered together (Cluster #6, Supplementary information Table S1). Finally 8 of the 11 compounds with an anthraquinone core were positive at both time points and included in the cluster analysis. The exceptions were carminic acid (CASRN 1260-17-9) that was negative for both time points, chrysophanic acid (CASRN 481-74-3) that was inconclusive at both time points, and 2-methyl-1-nitroanthraquinone (CASRN 129-15-7) that was positive only at 5 h. These 8 compounds were grouped into two different clusters depending on the hydroxyl group content (Clusters #3 and #10, Supplementary information Table S1). The inactive compounds from the primary screen were not analyzed further for structure features. The molecular library used here is very diverse structurally and the majority of the library was negative, making the definition of structure features associated with negative results difficult. Furthermore the value of defining structural clusters significantly deficient in active molecules is relative since the activity can be drastically modified by just adding a functional group to the cluster consensus core structure. From the clustering results, we selected 38 active compounds that covered most of the chemical space for confirmatory and further mechanistic studies. Additionally, we selected one inactive close structural analog (wherever available; Supplementary information Table S1) of the active compounds for each cluster, seven total, and subjected them to the confirmatory studies as well, to test for the possibility of false negatives. Powder samples of these compounds were purchased from commercial vendors.
Figure 2.
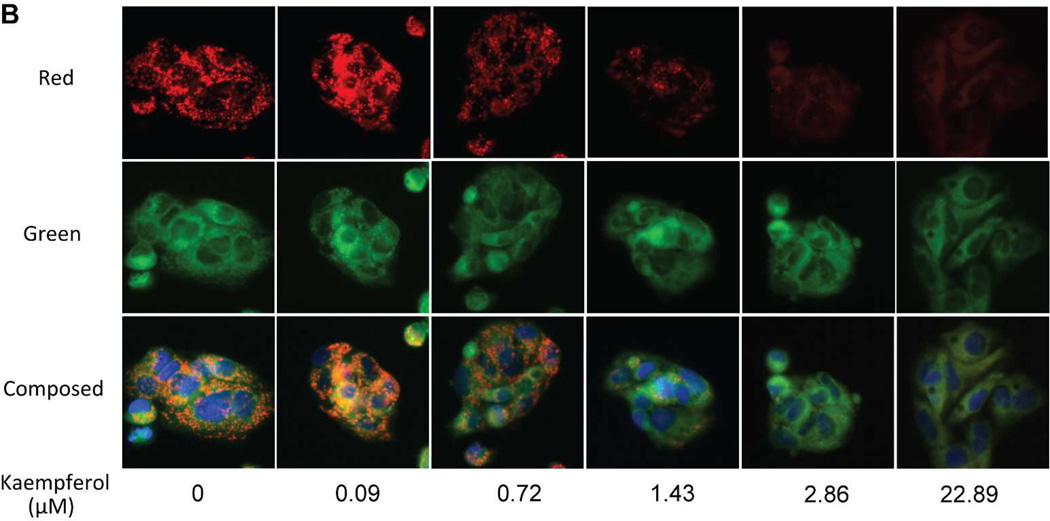
Several flavonoids that decreased mitochondrial membrane potential were identified in the primary screening. A) Changes in mitochondrial membrane potential in intact HepG2 cells generated by three flavonoids identified. (Left, 1 h; right, 5 h). B) A representative image of HepG2 cells stained with Mito-MPS dye in the absence or in the presence of kaempferol. Images acquired in ImageXpress microsystem using a 20× objective. While red fluorescent aggregates are localized in the mitochondria, green fluorescent monomers are mainly in the cytosol. The composed images are the merger of red, blue (nuclei), and green fluorescence.
Confirmatory and mechanistic studies
Of the 38 compounds selected for confirmation and follow-up mechanistic studies, 36 (95%) were confirmed to be positive at 1 h for MMP disruption when tested using 24 concentrations over 6 orders of magnitude (11 pM to 92 µM). Silibinin and 1,5-naphthalenediamine were inactive (Table 1). Thirty-seven compounds (97%) were confirmed as active at the 5 h time point, with 1,5-naphthalenediamine being inactive and silibinin weakly active with low efficacy (38.14%). The inactive analogs were tested at 15 concentrations ranging from 1.2 nM to 92 µM in triplicate and all compounds remained inactive. When the assay was repeated using high content as a readout over a 12-concentration titration (45 nM to 92 µM), 35 of 38 (92%) compounds were confirmed as active at the 1 h time point (silibinin, 1,5-naphthalenediamine, and formulated fenaminosulf were inactive or inconclusive), and 36 (95%) were confirmed for activity at 5 h (silibinin and 1,5-naphthalenediamine were inactive, Table 1).
To rule out cell death as the cause of the observed decrease in membrane potential, we assayed cytotoxicity by measuring LDH release into the media at 1 and 5 h of treatment with the compounds. Four compounds had minimal cytotoxic effect after 1 h of treatment. Only digitonin had similar IC50 values for both a decrease in MMP and an increase in cytotoxicity. Six compounds were cytotoxic after 5 h of treatment, with kaempferol having similar potency for both a decrease in MMP and an increase in cytotoxicity, while phenmedipham was more potent for cytotoxicity than for MMP disruption (Table 2).
Table 2.
Compound potency (IC50, µM) in LDH release assay after 1 or 5 h of treatment.
| Compound | LDH Release | |
|---|---|---|
| 1 h | 5 h | |
| 4,4-Thiobis(6-t-butyl-m-cresol) | Inactive | Inconclusive |
| Digitonin | 45.14±8.43 | 45.14±8.43 |
| Hexamethyl-p-rosaniline chloride (Gentian violet) | Inconclusive | 56.11±0 |
| Kaempferol | Inactive | Inconclusive |
| Kepone | 49.08±12.26 | 58.39±3.95 |
| Malachite green oxalate | Inactive | 62.96±0 |
| Phenmedipham | Inactive | Inconclusive |
| Phenyl mercuric acetate | 68.08±4.44 | 50.01±0 |
| tetra-N-Octylammonium bromide | 35.42±12.64 | 28.12±0 |
Only compounds that were active (curve class 1.1, 1.2, and 2.1) or inconclusive (curve class 1.3, 1.4, 2.2, 2.3, 2.4, and 3) for at least one of the time points are included in the table. Values are the mean ± standard deviation from 3 independent experiments.
To further understand the mechanism of toxicity of the selected compounds, we examined changes in OCR generated after treatment with each compound, using the IC50 concentration determined from the 1 h MMP confirmatory assays. Changes immediately after the addition of the compound and after one hour of treatment were measured (Table 3). Uncoupling agents generally cause an increase in OCR33, 34 and compounds that inhibit the flow of electrons through the respiratory chain have the opposite effect.35 The FCCP-uncoupled OCR (after FCCP addition) was measured at the end of the experiment (Table 3). The FCCP-uncoupled rate approximates maximal mitochondrial capacity and it has been shown that mitochondrial toxicants can decrease this value without an obvious effect on basal rates.34 Twenty compounds seemed to act as uncouplers, increasing the OCR after one hour treatment (Table 3, group 1). Ten of these also demonstrated increased OCR values immediately upon exposure, while the other 10 took 1 h to fully manifest their uncoupling affect, possibly due to slower cellular uptake. Two compounds, gentian violet and malachite green oxalate, caused an immediate uncoupling as evident from increased OCR but by 1 h, the OCR was similar to the initial basal rate (Table 3, group 2). Moreover these two agents significantly suppressed FCCP uncoupled rates showing mitochondrial toxicity. Four compounds - 4-hydroxyphenyl retinamide, curcumin, genistein, and silibinin - had no effect on basal rate at 30 sec or 1 h, but they significantly suppressed the FCCP uncoupled rates, indicating that they are mitochondrial toxicants in a different mechanistic class from the uncouplers (Table 3, Group 3). Another class of toxicants, including basic red 9, diethylstilbestrol (DES), digitonin, phenyl mercuric acetate, and zearalenone, depressed both basal and FCCP uncoupled rates (Table 3, Group 4). Seven of the 38 compounds positive in the 1 h MMP assay showed no effect on the basal or FCCP-uncoupled OCR when tested at their IC50 concentrations (Table 3, Group 5).
Table 3.
Relative percentage oxygen consumption rate (OCR) results generated 30 sec and 1 h after sequential treatment with a concentration of the compound that generated a 50% response in the MMP assay (i.e., the IC50) and FCCP (mean ± standard deviation).
| Group | Compound names | OCR | ||
|---|---|---|---|---|
| 30 Sec | 1 h | FCCP addition |
||
| Control (DMSO) | 108.71 ± 2.92 | 116.54 ± 0.86 | 171.6 ± 1.14 | |
| 1 | 1,8-Dihydroxy-4,5-dinitroanthraquinone | 144.03 ± 7.3 | 150.09 ± 1.27 | 125.05 ± 0.79 |
| 1 | 2,2'-Methylenebis-(4-chlorophenol) | 170.42 ± 33.35 | 168.27 ± 1.85 | 164.18 ± 0.81 |
| 1 | 2,2'-Thiobis(4-chlorophenol) | 273.19 ± 70.36 | 191.27 ± 0.92 | 230.07 ± 2.17 |
| 1 | 3,4,5-Trichlorophenol | 163.21 ± 18.34 | 173.49 ± 1.32 | 202.49 ± 1.47 |
| 1 | 3,4-Dichlorophenyl isocyanate | 102.45 ± 10.76 | 236.03 ± 4.84 | 260.25 ± 2.92 |
| 1 | 4,4-Thiobis(6-t-butyl-m-cresol) | 106.32 ± 4 | 127.15 ± 1.52 | 167.04 ± 1.47 |
| 1 | 4-Cumylphenol | 111.24 ± 4.51 | 121.31 ± 0.33 | 150.32 ± 1.35 |
| 1 | Captan | 110.37 ± 7.97 | 128.41 ± 0.77 | 62.19 ± 0.91 |
| 1 | Diphenylurea | 97.02 ± 4.18 | 143.97 ± 1.85 | 193.8 ± 2.21 |
| 1 | Emodin | 235.59 ± 56.54 | 210.48 ± 1.83 | 247.81 ± 2.96 |
| 1 | Hexachlorophene | 201.72 ± 51.19 | 175.25 ± 1.35 | 171.45 ± 2.44 |
| 1 | Kaempferol | 118.63 ± 4.29 | 138.85 ± 1.28 | 226.35 ± 1.29 |
| 1 | Kepone | 139.53 ± 5.8 | 140.71 ± 0.96 | 188.54 ± 2.11 |
| 1 | Nitazoxanide | 277.15 ± 15.64 | 199.34 ± 2.37 | 127.13 ± 1.38 |
| 1 | N-Phenyl-2-naphthylamine | 103.45 ± 2.35 | 120.42 ± 0.94 | 177.54 ± 1.62 |
| 1 | o-Benzyl-p-chlorophenol | 123.51 ± 12.67 | 126.12 ± 0.47 | 151.46 ± 0.39 |
| 1 | p -n -Nonylphenol | 116.61 ± 6.28 | 131.32 ± 0.61 | 212.58 ± 1.39 |
| 1 | p-Aminophenol | 100.52 ± 5.81 | 158.18 ± 3.31 | 226.06 ± 3.21 |
| 1 | Phenmedipham | 105.47 ± 2.71 | 130.87 ± 1.18 | 173.96 ± 1.01 |
| 1 | tetra-N-Octylammonium bromide | 122.56 ± 40.61 | 215.53 ± 1.14 | 224.07 ± 1.98 |
| 2 | Hexamethyl-p-rosaniline chloride (Gentian violet) | 132.56 ± 3.88 | 105.01 ± 1.65 | 87.45 ± 0.9 |
| 2 | Malachite green oxalate | 163.81 ± 17.5 | 97.14 ± 3.09 | 35.39 ± 0.72 |
| 3 | 4-Hydroxyphenyl retinamide | 96.31 ± 1.42 | 98.83 ± 0.42 | 79.01 ± 2.11 |
| 3 | Curcumin | 86 ± 3.54 | 94.85 ± 0.67 | 111.29 ± 2.82 |
| 3 | Genistein (4',5,7-Trihydroxyisoflavone) | 90.18 ± 3.05 | 105.21 ± 0.19 | 90.29 ± 0.37 |
| 3 | Silibinin | 92.67 ± 0.69 | 95.66 ± 0.46 | 48.37 ± 1.58 |
| 4 | Basic red 9 (p-Rosaniline HCl) | −9.78 ± 16.86 | 35.74 ± 2.42 | 103.83 ± 3.14 |
| 4 | Diethylstilbestrol [DES] | 92.27 ± 8.72 | 75.16 ± 0.85 | 132.65 ± 1.79 |
| 4 | Digitonin | 57.25 ± 6.4 | 54.89 ± 0.48 | 115.17 ± 0.13 |
| 4 | Phenyl mercuric acetate | 67.6 ± 5.55 | 51.54 ± 0.26 | 32.2 ± 1.96 |
| 4 | Zearalenone | 77.68 ± 2.39 | 109.62 ± 0.57 | 120.61 ± 2.48 |
| 5 | 1,5-Naphthalenediamine | 104.68 ± 1.3 | 115.87 ± 0.7 | 193.63 ± 2.33 |
| 5 | 2-Aminoanthraquinone | 97.54 ± 0.63 | 106.44 ± 0.61 | 164.43 ± 3.4 |
| 5 | Apigenin | 99.69 ± 1.13 | 107.51 ± 0.16 | 144.92 ± 0.66 |
| 5 | Formulated fenaminosulf (Dexon) | 102.48 ± 0.9 | 110.45 ± 0.28 | 137.82 ± 0.96 |
| 5 | o,p'-DDD (Mitotane) | 102.84 ± 1.36 | 114.22 ± 0.42 | 200.52 ± 3.25 |
| 5 | Resveratrol | 100.5 ± 0.73 | 110.92 ± 0.36 | 181.11 ± 1.52 |
| 5 | Riboflavin | 103.69 ± 0.37 | 115.15 ± 0.4 | 190.21 ± 1.86 |
The results are normalized to the basal OCR for each compound (basal OCR is arbitrarily set to 100%). Compounds are grouped from 1 to 5 according the overall response. Group 1 includes 20 compounds that caused uncoupling (increased OCR at 1 h). Group 2 compounds caused an immediate uncoupling, but by 1 h the OCR was similar to the initial basal rate. Group 3 compounds had no effect on basal rates at 30 sec or 1 h but they significantly suppressed the FCCP uncoupled rates. Group 4 compounds depressed both basal and FCCP uncoupled rates. Finally, group 5 compounds showed no obvious effect on the basal or FCCP-uncoupled OCR.
Discussion
In the present study, we profiled a diverse collection of chemicals including pesticides, therapeutic agents, preservatives, and flavoring agents among others for their ability to disrupt the MMP (Δψm) in HepG2 cells, as a first step in a systematic study of the mechanisms by which environmental compounds induce mitochondrial toxicity. Compounds that inhibit cellular respiration, the Krebs cycle, or fatty acid oxidation can potentially affect the membrane potential.25 Protonophores can also directly dissipate the membrane potential by facilitating the movement of protons against the electrochemical gradient.25 MMP collapse seems to be a necessary step in the progression of apoptosis via the intrinsic pathway.36 Finally, compounds that directly affect membrane integrity and/or are cytotoxic via other mechanisms will also affect the MMP. Thus, compounds inhibiting a wide range of cellular processes may disrupt the MMP making it a suitable comprehensive first endpoint for screening.
Among the 1340 unique compounds assayed in the primary screen, we identified 91 and 88 active compounds at 1 and 5 h of treatment, respectively, with 76 compounds being active at both time points. Thirty-eight compounds as well as seven inactive analogs were chosen for follow-up studies on the basis of structure–activity relationship (SAR) and biological activity, with over 95% of the 38 active compounds confirmed as active and all inactive analogs confirmed as inactive suggesting low false positive and negative rates of this assay. Of these compounds, only 6 exhibited some level of cytotoxicity (measured by LDH release) at either 1 or 5 h (Table 2) with digitonin being the only compound that showed similar potency for both MMP decrease and LDH release. Digitonin is commonly used in mitochondria isolation since it preferentially binds to membranes containing cholesterol and destabilizes them,37 which could explain both LDH release and, after a prolonged treatment, loss of MMP. All of the other compounds positive for LDH release showed cytotoxicity only at higher concentrations when compared to the concentrations at which they decreased the MMP, indicating either a sequential order of events or just independent events at different concentrations.
To determine if the observed changes in MMP were related to the initiation of apoptotic events we evaluated our previously published HepG2 data for these 38 compounds on the activation of caspase 9 at 5 h and caspase 3/7 at 16 h,38 as well as cytotoxicity at 40 h27 (Supplementary information Table S2). Only one compound (hexamethyl-p-rosaniline chloride [gentian violet]) seems to activate caspase 9 at 5 h of treatment, and at concentrations much higher than the concentration needed to decrease the membrane potential. Therefore, the observed decrease in MMP cannot be explained exclusively by an induction of the intrinsic apoptotic pathway for any of the studied compounds. Moreover, only 5 of these 38 compounds (13%) induced apoptosis after 16 h of treatment and 23 (59%) were cytotoxic for HepG2 cells after 40 h of treatment (Supplementary information Table S2).
To further investigate the possible mechanisms by which these compounds disrupt MMP, changes in the OCR were measured immediately and after 1 h of treatment. Uncouplers are expected to increase oxygen consumption33 while compounds that inhibit the flow of electrons through the respiratory chain decrease the OCR35. Results of these studies allowed us to further classify the 38 mitochondrial toxicants into several major mechanistic groups (Table 3). For example, group 1 consists of 20 compounds that are clearly uncouplers, although some act more slowly than others. This group includes fentichlor [2,2'-thiobis(4-chlorophenol)], trichlorophenol, and captan that have been previously described as uncouplers.39–42 Group two consists of two triarylmethane dyes (gentian violet and malachite green oxalate) that caused an immediate increase on OCR followed by a marked decline at 1 h leading to a decreased FCCP uncoupled rates showing clear signs of mitochondrial toxicity. Compounds in group 3 decreased the maximal capacity without affecting the basal rate, perhaps by suppressing substrate uptake, although other mechanisms cannot be ruled out. Compounds in group 4, possible inhibitors of the electron transport chain (ETC), decreased both the compound treated and uncoupled rates. Finally, 7 compounds in group 5 induced a decrease in MMP but showed no significant effect on OCR at 1 h of treatment and no effect on the uncoupled OCR. Six of these 7 compounds were not cytotoxic at 40 h (the only exception being apigenin), suggesting that cells were able to recover from the change in membrane potential observed after 1 h of treatment. It seems that, at least for a subset of the studied compounds, changes in MMP were more sensitive indicators of acute mitochondrial toxicity than changes in OCR, although this should be evaluated on a case-by-case basis.
Several chemical classes were represented repeatedly within the group of 76 compounds that were positive in the screen at 1 and 5 h of treatment. Three triarylmethane dyes — basic read 9 (p-rosaniline HCl), hexamethyl-p-rosaniline chloride, and malachite green oxalate — were among the most potent compounds, with IC50 values ranging between 0.29 and 2.21 µM. Hexamethyl-p-rosaniline chloride and malachite green oxalate were both cytotoxic at 5 h, and they induced caspase 3/7, while basic red 9 was negative for both endpoints. Moreover, hexamethyl-p-rosaniline chloride and malachite green oxalate produced similar changes in the OCR in HepG2 cells by acting as uncouplers immediately after treatment and eventually decreasing the uncoupled OCR. These two dyes have been shown to accumulate inside the mitochondria, where they inhibit the ETC at complex I leading to mitochondrial swelling and apoptosis.43 Other triarylmethane dyes seem to have different but unclear mechanisms of action that lead to necrosis 43. Compounds that shared a flavone moiety were positive in the primary screen at 1 and 5 h of treatment; these included apigenin, genistein, kaempferol, and daidzein (Figure 2). Three of these compounds were tested in the confirmatory assays and the potency rank was kaempferol>apigenin>genistein at both 1 and 5 h of treatment. Flavonoids have been previously reported to decrease the mitochondrial membrane potential and induce ROS-mediated apoptosis in a variety of cell types, but the specific mechanism is not known.25, 44, 45 The isoflavones genistein and daidzein have been also shown to promote mitochondrial biogenesis after 48h of treatment in renal proximal tubule cells.46 Mitochondrial biogenesis was promoted by activation and/or increase of expression of peroxisome proliferator-activated receptor γ coactivator 1-alpha [(PGC)-1α] and sirtuin (SIRT1).46 Apigenin and kaempferol, as well as other flavonoids, also increase SIRT1 deacetylation activity and could potentially regulate mitochondrial biogenesis.47 In the studies reported here, kaempferol acted as an uncoupler, genistein only affected the uncoupled OCR, and apigenin did not affect the OCR or the FCCP-uncoupled OCR after 1 h of treatment. Thus, flavonoids, although structurally similar, may affect the MMP through a variety of different mechanisms. The eight compounds with an anthraquinone core that were positive for both time points in the screening were grouped into two clusters based on their hydroxyl content (Supplementary information Table S1). Emodin and 1,8-dihydroxy-4,5-dinitroanthraquinone had two or more hydroxy groups and were grouped together. Both were cytotoxic at 40 h and both acted as uncouplers. Alternatively, 2-aminonthraquinone was not cytotoxic at 40 h and did not affect the OCR or the uncoupled OCR in HepG2 cells, suggesting that these two types of anthraquinones have different mechanisms by which they affect the MMP. These results correlate with previous studies that showed that anthraquinones required hydroxyl groups at specific positions to act as uncouplers.48
Evaluating mitochondrial toxicity is a key component in the comprehensive understanding of chemical-induced toxicity.16, 49, 50 Previous efforts to establish high throughput assays included the use of the tetramethylrhodamine methyl ester dye (TMRM) to asses changes in MMP in a 96- or 384-well format, 51 the use of changes in oxygen consumption by live cells 34, 52, the use of differential toxicity when cells are forced to obtain most of their energy via oxidative phosphorylation, 53 and the use of enzymatic assays to study inhibition of cellular respiration 54. Here, we present the first study that screened a large collection of environmental chemicals for their ability to affect mitochondrial function. We used a previously described homogenous assay in a 1536-well plate format to monitor changes in mitochondrial membrane potential25 as a first step to identify compounds that may interfere with mitochondrial physiology. Combining primary qHTS assays with in depth secondary assays constitutes the next steps to elucidate mechanism of compound action. Here, we selected 38 compounds for follow-up confirmatory and mechanistic studies. We examined cytotoxicity and changes in oxygen consumption to further explore mechanisms of action. Overall, we were able to determine discrete mechanisms of action for some of these compounds, demonstrating that a primary screen followed by orthogonal secondary assays can be an effective strategy for evaluation of the toxicological properties of large numbers of environmental chemicals. Nevertheless, the mechanistic hypothesis generated in our study should be further validated using different models (e.g. isolated mitochondria, other cell types) and other endpoints (e.g. metabolic assays, stress pathway assays). The information generated from the in vitro assays would help to prioritize chemicals for in vivo testing using concentrations relevant to human exposures, which would shed light into organ specific toxicity and toxicokinetic and toxicodynamic properties of these compounds. Finally these data can be used to generate computational models to predict the toxic effect of environmental chemicals.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by the Intramural Research Program (Interagency agreement #Y3-ES-7020-01) of Division of the National Toxicology Program, National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health and the NIEHS 2R44ES019378-02 grant (CB).
Abbreviations
- CASRN
Chemical Abstracts Service Registry Number
- CV
coefficient of variation
- DES
diethylstilbestrol
- DMSO
Dimethyl sulfoxide
- o,p’-DDD
1,1-dichloro-2-(o-chlorophenyl)-2-(p-chlorophenyl) ethane
- EPA
Environmental Protection Agency
- FCCP
Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone
- FDA
Food and Drug administration
- IC50
half maximal inhibitory concentration
- h
hour
- LDH
lactate dehydrogenase
- MMP
Mitochondrial Membrane Potential
- MOA
mechanism of action
- NCCT
National Center for Computational Toxicology
- NCGC
NIH Chemical Genomics center
- NTP
National Toxicology Program
- NRC
National Research Council
- OCRs
Oxygen consumption rates
- qHTS
high throughput screening
Footnotes
This article may be the work product of an employee or group of employees of the National Institute of Environmental Health Sciences (NIEHS) and the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), however, the statements, opinions or conclusions contained therein do not necessarily represent the statements, opinions or conclusions of NIEHS, NIH or the United States government.
Supplementary Information
Table S1: A description of the chemical clusters generated by the 76 active compounds in the MMP assay for both the 1 and 5 h time points. The 76 active compounds for 1 and 5 h were clustered based on structural similarity [Hierarchical (agglomerative) nesting] using the Leadscope software (Leadscope Inc., Columbus, OH, USA), resulting in 11 structural clusters and 23 singletons. Table S2: A summary of previously published screening data [compound potency (half maximal activity concentration, µM)] for the 38 compounds. Figure S1: qHTS MMP assay experimental design: a) detailed protocol of the MMP screen. b) A plate map of the control plate used for the screen. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.National Toxicology Program. A National Toxicology Program for the 21st Century. Roadmap to Achieve the NTP Vision. Triangle Park, NC: National Institute of Environmental Health Sciences, Department of Health and Human Services, Research; 2004. http://ntp.niehs.nih.gov/NTP/About_NTP/NTPVision/NTPRoadmap_508.pdf. [Google Scholar]
- 2.National Research Council. Toxicity testing in the 21st century A vision and a strategy. Washington, DC: Committee on Toxicity Testing and Assessment ofEnvironmental Agents, National Research Council, The National Academy press; 2007. http://www.nap.edu/catalog/11970.html. [Google Scholar]
- 3.Krewski D, Acosta D, Jr., Andersen M, Anderson H, Bailar JC, 3rd, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S, Jr., Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L. Toxicity testing in the 21st century: a vision and a strategy. J. Toxicol. Environ. Health B. Crit. Rev. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 5.Vasington FD, Murphy JV. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J. Biol. Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
- 6.Antico Arciuch VG, Alippe Y, Carreras MC, Poderoso JJ. Mitochondrial kinases in cell signaling: Facts and perspectives. Adv. Drug Deliv. Rev. 2009;61:1234–1249. doi: 10.1016/j.addr.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Rossier MF. T channels and steroid biosynthesis: in search of a link with mitochondria. Cell Calcium. 2006;40:155–164. doi: 10.1016/j.ceca.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Lill R, Diekert K, Kaut A, Lange H, Pelzer W, Prohl C, Kispal G. The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. Biol. Chem. 1999;380:1157–1166. doi: 10.1515/BC.1999.147. [DOI] [PubMed] [Google Scholar]
- 9.Clayton DA. Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol. 1992;141:217–232. doi: 10.1016/s0074-7696(08)62067-7. [DOI] [PubMed] [Google Scholar]
- 10.Lemasters JJ, Qian T, He L, Kim JS, Elmore SP, Cascio WE, Brenner DA. Role of mitochondrial inner membrane permeabilization in necrotic cell death, apoptosis, and autophagy. Antioxid. Redox. Signal. 2002;4:769–781. doi: 10.1089/152308602760598918. [DOI] [PubMed] [Google Scholar]
- 11.Chance B, Williams GR. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. Relat. Subj. Biochem. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 12.Gilkerson RW, Selker JML, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. Febs Lett. 2003;546:355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 13.Barnes SJ, Weitzman PDJ. Organization of Citric-Acid Cycle Enzymes into a Multienzyme Cluster. Febs Lett. 1986;201:267–270. doi: 10.1016/0014-5793(86)80621-4. [DOI] [PubMed] [Google Scholar]
- 14.Wallace KB, Starkov AA. Mitochondrial targets of drug toxicity. Annu. Rev. Pharmacol. Toxicol. 2000;40:353–388. doi: 10.1146/annurev.pharmtox.40.1.353. [DOI] [PubMed] [Google Scholar]
- 15.Pereira CV, Moreira AC, Pereira SP, Machado NG, Carvalho FS, Sardao VA, Oliveira PJ. Investigating drug-induced mitochondrial toxicity: a biosensor to increase drug safety? Curr. Drug Saf. 2009;4:34–54. doi: 10.2174/157488609787354440. [DOI] [PubMed] [Google Scholar]
- 16.Wallace KB. Mitochondrial off targets of drug therapy. Trends Pharmacol. Sci. 2008;29:361–366. doi: 10.1016/j.tips.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt CW. Mito-Conundrum Unraveling Environmental Effects on Mitochondria. Environ. Health Persp. 2010;118:A292–A297. [Google Scholar]
- 19.Smith CS, Bucher J, Dearry A, Portier C, Tice R, Witt K, Collins B. Chemical selection for NTP’s high throughput screening initiative. Toxicologist. 2007:247. [Google Scholar]
- 20.Tice RR, Fostel J, Smith CS, Witt K, Freedman JH, Portier CJ, Dearry AD, Bucher JR. The National Toxicology Program high throughput screening initiative: current status and future directions. Toxicologist. 2007:246. [Google Scholar]
- 21.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. PubChem’s BioAssay Database. Nucleic. Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadanaciva S, Rana P, Beeson GC, Chen D, Ferrick DA, Beeson CC, Will Y. Assessment of drug-induced mitochondrial dysfunction via altered cellular respiration and acidification measured in a 96-well platform. J. Bioenerg. Biomembr. 2012;44:421–437. doi: 10.1007/s10863-012-9446-z. [DOI] [PubMed] [Google Scholar]
- 23.Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 24.Desquiret V, Loiseau D, Jacques C, Douay O, Malthiery Y, Ritz P, Roussel D. Dinitrophenol-induced mitochondrial uncoupling in vivo triggers respiratory adaptation in HepG2 cells. Biochim. Biophys. Acta. 2006;1757:21–30. doi: 10.1016/j.bbabio.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Sakamuru S, Li X, Attene-Ramos MS, Huang R, Lu J, Shou L, Shen M, Tice RR, Austin CP, Xia M. Application of a homogenous membrane potential assay to assess mitochondrial function. Physiol. Genomics. 2012;44:495–503. doi: 10.1152/physiolgenomics.00161.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Persp. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia M, Huang R, Sun Y, Semenza GL, Aldred SF, Witt KL, Inglese J, Tice RR, Austin CP. Identification of chemical compounds that induce HIF-1alpha activity. Toxicol Sci. 2009;112:153–163. doi: 10.1093/toxsci/kfp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. (London) 1910;40:4–7. [Google Scholar]
- 30.Southall NT, Jadhav A, Huang R, Nguyen T, Wang Y. Enabling the Large-Scale Analysis of Quantitative High-Throughput Screening Data. In: Seethala R, Zhang L, editors. Handbook of Drug Screening. London: Informa healthcare; 2009. pp. 442–463. [Google Scholar]
- 31.Hopfer U, Lehninger AL, Thompson TE. Protonic conductance across phospholipid bilayer membranes induced by uncoupling agents for oxidative phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 1968;59:484–490. doi: 10.1073/pnas.59.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 33.Lechner V, Siess M, Hoffmann PC. The effect of uncouplers of oxidative phosphorylation on oxygen uptake, ubiquinone redox status and energy-rich phosphate levels of isolated atria. Eur. J. Biochem. 1970;12:117–125. doi: 10.1111/j.1432-1033.1970.tb00828.x. [DOI] [PubMed] [Google Scholar]
- 34.Beeson CC, Beeson GC, Schnellmann RG. A high-throughput respirometric assay for mitochondrial biogenesis and toxicity. Anal. Biochem. 2010;404:75–81. doi: 10.1016/j.ab.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindahl PE, Oberg KE. The effect of rotenone on respiration and its point of attack. Exp. Cell. Res. 1961;23:228–237. doi: 10.1016/0014-4827(61)90033-7. [DOI] [PubMed] [Google Scholar]
- 36.Zamzami N, Marchetti P, Castedo M, Zanin C, Vayssiere JL, Petit PX, Kroemer G. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J. Exp. Med. 1995;181:1661–1672. doi: 10.1084/jem.181.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoppel C, Cooper C. The action of digitonin on rat liver mitochondria. The effects on enzyme content. Biochem. J. 1968;107:367–375. doi: 10.1042/bj1070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho M, Huang R, Leister K, Witt KL, Smith CS, Inglese J, Tice RR, Austin CP, Xia M. Profiling Environmental Chemicals in Apoptosis Pathway Using a High-throughput Format. Toxicologist. 2009;108:59. [Google Scholar]
- 39.McLaughlin SG, Dilger JP. Transport of protons across membranes by weak acids. Physiol. Rev. 1980;60:825–863. doi: 10.1152/physrev.1980.60.3.825. [DOI] [PubMed] [Google Scholar]
- 40.Nelson BD. Action of the fungicides captan and folpet on rat liver mitochondria. Biochem. Pharmacol. 1971;20:737–748. doi: 10.1016/0006-2952(71)90037-2. [DOI] [PubMed] [Google Scholar]
- 41.Bloomfield SF. The effect of the phenolic antibacterial agent fentichlor on energy coupling in Staphylococcus aureus. J. Appl. Bacteriol. 1974;37:117–131. doi: 10.1111/j.1365-2672.1974.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 42.Hugo WB, Bloomfield SF. Studies on the mode of action of the phenolic antibacterial agent fentichlor against Staphylococcus aureus and Escherichia coli. 3. The effect of fentichlor on the metabolic activities of Staphylococcus aureus and Escherichia coli. J. Appl. Bacteriol. 1971;34:579–591. doi: 10.1111/j.1365-2672.1971.tb02320.x. [DOI] [PubMed] [Google Scholar]
- 43.Kowaltowski AJ, Turin J, Indig GL, Vercesi AE. Mitochondrial effects of triarylmethane dyes. J. Bioenerg. Biomembr. 1999;31:581–590. doi: 10.1023/a:1005421112345. [DOI] [PubMed] [Google Scholar]
- 44.George J, Banik NL, Ray SK. Genistein induces receptor and mitochondrial pathways and increases apoptosis during BCL-2 knockdown in human malignant neuroblastoma SK-N-DZ cells. J. Neurosci. Res. 2010;88:877–886. doi: 10.1002/jnr.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang IK, Lin-Shiau SY, Lin JK. Induction of apoptosis by apigenin and related flavonoids through cytochrome c release and activation of caspase-9 and caspase-3 in leukaemia HL-60 cells. Eur. J. Cancer. 1999;35:1517–1525. [PubMed] [Google Scholar]
- 46.Rasbach KA, Schnellmann RG. Isoflavones promote mitochondrial biogenesis. J. Pharmacol. Exp. Ther. 2008;325:536–543. doi: 10.1124/jpet.107.134882. [DOI] [PubMed] [Google Scholar]
- 47.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 48.Betina V, Kuzela S. Uncoupling Effect of Fungal Hydroxyanthraquinones on Mitochondrial Oxidative-Phosphorylation. Chem-Biol. Interact. 1987;62:179–189. doi: 10.1016/0009-2797(87)90089-5. [DOI] [PubMed] [Google Scholar]
- 49.Dykens JA, Will Y. The significance of mitochondrial toxicity testing in drug development. Drug Discov. Today. 2007;12:777–785. doi: 10.1016/j.drudis.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Nadanaciva S, Will Y. Investigating mitochondrial dysfunction to increase drug safety in the pharmaceutical industry. Curr. Drug Targets. 2011;12:774–782. doi: 10.2174/138945011795528985. [DOI] [PubMed] [Google Scholar]
- 51.Huang SG. Development of a high throughput screening assay for mitochondrial membrane potential in living cells. J. Biomol. Screen. 2002;7:383–389. doi: 10.1177/108705710200700411. [DOI] [PubMed] [Google Scholar]
- 52.Schoonen WG, Stevenson JC, Westerink WM, Horbach GJ. Cytotoxic effects of 109 reference compounds on rat H4IIE and human HepG2 hepatocytes. III: Mechanistic assays on oxygen consumption with MitoXpress and NAD(P)H production with Alamar Blue. Toxicol. In Vitro. 2012;26:511–525. doi: 10.1016/j.tiv.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Swiss R, Will Y. Assessment of mitochondrial toxicity in HepG2 cells cultured in high-glucose- or galactose-containing media. Chapter 2, Unit2 20. Curr. Protoc. Toxicol. 2011 doi: 10.1002/0471140856.tx0220s49. [DOI] [PubMed] [Google Scholar]
- 54.Nadanaciva S, Bernal A, Aggeler R, Capaldi R, Will Y. Target identification of drug induced mitochondrial toxicity using immunocapture based OXPHOS activity assays. Toxicol. In Vitro. 2007;21:902–911. doi: 10.1016/j.tiv.2007.01.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.