Abstract
High extracellular NaCl, such as in the renal medulla, can perturb and even kill cells, but cells mount protective responses that enable them to survive and function. Many high-NaCl-induced perturbations and protective responses are known, but the signaling pathways involved are less clear. Change in protein phosphorylation is a common mode of cell signaling, but there was no unbiased survey of protein phosphorylation in response to high NaCl. We used stable isotopic labeling of amino acids in cell culture coupled to mass spectrometry to identify changes in protein phosphorylation in human embryonic kidney (HEK 293) cells exposed to high NaCl. We reproducibly identify >8,000 unique phosphopeptides in 4 biological replicate samples with a 1% false discovery rate. High NaCl significantly changed phosphorylation of 253 proteins. Western analysis and targeted ion selection mass spectrometry confirm a representative sample of the phosphorylation events. We analyze the affected proteins by functional category to infer how altered protein phosphorylation might signal cellular responses to high NaCl, including alteration of cell cycle, cyto/nucleoskeletal organization, DNA double-strand breaks, transcription, proteostasis, metabolism of mRNA, and cell death.
Keywords: renal medulla, organic osmolytes, stable isotopic labeling of amino acids in cell culture, phosphorylation
interstitial NaCl can vary considerably between tissues and can be very high, particularly in the renal medulla; yet despite such continuous and, at times, variable stress, cells survive and function by means of a number of osmoprotective responses (6). Hypertonicity, such as that caused by high NaCl, shrinks cells, increases intracellular ionic strength, delays the cell cycle, rearranges the cytoskeleton, increases DNA breaks, and alters transcription and translation (6). Protective responses include increase in regulatory volume, increase in heat shock proteins (HSPs), and accumulation of organic osmolytes (6). The signaling involved in the damage and the protective responses is incompletely understood. Phosphorylation of proteins is a common form of cell signaling. Therefore, in the present studies, to further elucidate the signaling pathways involved, we used stable isotopic labeling of amino acids in cell culture (SILAC) (52) coupled to proteomic mass spectrometry [MS; multidimensional liquid chromatography (LC) coupled with tandem MS (LC/LC-MS/MS)] to profile high-NaCl-induced changes in the phosphoproteome of human embryonic kidney (HEK 293) cells. On the basis of these results and known effects of the changes in protein phosphorylation, we infer how changes in phosphorylation of particular proteins might signal damage to cells and cellular protective responses.
METHODS AND MATERIALS
Cell culture in SILAC.
HEK 293 cells (American Type Culture Collection, Manassas, VA) were cultured in 90% Eagle's minimal essential medium (Invitrogen, Carlsbad, CA) plus 10% fetal bovine serum in 5% CO2-95% air at 37°C. Cells were used between passages 38 and 50. Cells were transferred to SILAC medium (Invitrogen, Carlsbad, CA), either [13C6,15N4-arginine,13C6-lysine]DMEM (“heavy”) or [12C6,14N4-arginine,12C6-lysine]DMEM (“light”), at 300 mosmol/kg for six generations. Incorporation of labeled amino acids was measured by LC-tandem MS (LC-MS/MS) analysis of tryptic peptides from heavy amino acid-equilibrated cells. After six generations, 97.5% of cellular proteins had incorporated the heavy amino acids. Osmolality of the heavy medium bathing 80% confluent equilibrated cells was increased to 500 mosmol/kg (NaCl added), while the light medium was exchanged for an identical medium at 300 mosmol/kg. After 1 h, the cells were washed once with PBS of the same osmolality (37°C), harvested in PBS of the same osmolality, and pelleted at 3,000 g for 4 min at 4°C. The experiment was performed three times in this manner. To ensure against bias due to isotope equilibration, a fourth experiment was performed with the same isotopes, but with the labeling reversed, so that medium bathing the light-equilibrated cells was increased to 500 mosmol/kg, while heavy-equilibrated cells remained at 300 mosmol/kg medium.
Sample preparation for MS.
The pelleted cells were lysed in 8 M urea, 50 mM Tris·HCl, and 75 mM NaCl with added protease inhibitor (Roche Diagnostics, Indianapolis, IN) and phosphatase inhibitors I and II (Sigma, St. Louis, MO). Proteins were digested with sequencing-grade trypsin (Promega, Madison, WI) at a ratio of 1:20 at 37°C for 16 h; then the samples were desalted with HLB cartridges (Waters, Milford, MA). The peptide mixture was fractionated by strong cation-exchange chromatography; then phosphopeptides were enriched using immobilized metal ion affinity chromatography columns (Pierce, Rockford, IL) before MS.
MS.
LC-MS/MS was carried out with an Eksigent nanoflow LC system connected to a mass spectrometer (LTQ-Orbitrap XL, Thermo Scientific, Waltham, MA). Data-dependent acquisition mode was enabled, and each survey MS scan was followed by six MS2 scans with dynamic exclusion of 20 s. For targeted ion selection (TIS), peptides were analyzed with an LTQ-Orbitrap Velos (Thermo Scientific), preselecting 50 phosphopeptide targets by specification of their predicted sizes for MS1. The identity of the MS1 peak areas of those phosphopeptides was confirmed by MS2 and quantified as described below.
Protein identification.
MS raw data were searched with the SEQUEST (20) and InsPectT (61) algorithms. Peptides were identified by search against a target-decoy human protein database (downloaded from the National Center for Biotechnology Information website ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/H_sapiens/protein/) with 1% false discovery rate (FDR). Phosphorylation site was assigned using the scoring algorithm phosphate localization score (54). Phosphorylation sites were confirmed using the National Heart, Lung, and Blood Institute in-house programs ProMatch (63) and PhosphoPIC (28).
Phosphopeptide quantification.
Relative phosphopeptide abundance was calculated from the MS1 peak area using in-house software (QUIL) (66) with FDR of 1%. Comparisons were either heavy or light in the SILAC experiments or either 300 or 500 mosmol/kg in the TIS experiments. The weighted means of MS1 peak areas of phosphopeptides were used to calculate relative abundance ratio: heavy-to-light ratio (H/L) = [∑i = 0n(Hi/Li∗√Li∗Hi)]/[∑i = 0n(√Li∗Hi)], where Li is peak area integrated from ion intensity for light peak and Hi is peak area integrated from ion intensity for heavy peak. R and Perl scripts were used for data analysis. Students' t-test was used to determine the significance of changes in abundance of phosphopeptides. We used a threshold of P ≤ 0.05 (t-test) and further selected for log2(500/300 mosmol/kg) less than or equal to −1 or ≥1 to identify important changes. Phosphorylation changes were in satisfactory agreement between the four replicates (Pearson's correlation coefficient = 0.63–0.80, with arginine-to-proline conversion taken into account).
iTRAQ.
For independent quantification of protein abundance, HEK 293 cells were grown in non-SILAC medium at 300 mosmol/kg. Osmolality of the medium bathing 80% confluent cells was increased to 500 mosmol/kg (NaCl added) for 1 h; then proteins were extracted and trypsinized, as described above, and the peptides were labeled differentially with isobaric tags (115 and 117 for 300 and 500 mosmol/kg, respectively) using an iTRAQ 8plex kit (AB Sciex, Framingham, MA). The iTRAQ-labeled peptides were combined and quantified with an Orbitrap Velos mass spectrometer. Results from three biological replicates were analyzed for differences between 300 and 500 mosmol/kg using Proteome Discoverer (Thermo Scientific). Changes were deemed significant if a protein's abundance ratio (500/300) deviated >10% from unity and P < 0.05 in a one-sample t-test. Of the ∼2,500 proteins identified, only 4 met these criteria.
Interpretation.
Gene Ontology functional category enrichment was analyzed by Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/gene2gene.jsp).
Western blotting.
HEK 293 cells at 70% confluence were maintained in 300 or 500 mosmol/kg (NaCl added) medium for 1 h. Whole cell protein was extracted using PhosphoSafe extraction reagent (EMD, La Jolla, CA). Protein (30 μg) was separated on 3–8% gradient Tris-acetate or 4–12% gradient Bis-Tris gel and transferred electrophoretically to nitrocellulose membranes. Membranes were incubated in Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) for 1 h at room temperature. Then membranes were incubated with the following antibodies overnight at 4°C: rabbit anti-phosphorylated (T71) activating transcription factor 2 (ATF-2), rabbit anti-ATF-2, rabbit anti-phosphorylated (S82) HSP27, mouse anti-HSP27, rabbit anti-phosphorylated (S727) STAT1, and mouse anti-STAT1 (Cell Signaling Technology, Danvers, MA); rabbit anti-phosphorylated (S421/S423) histone deacetylase 1 (HDAC1; Millipore); mouse anti-HDAC1, rabbit anti-phosphorylated (S63) c-Jun, rabbit anti-phosphorylated (S73) c-Jun, and mouse anti-c-Jun (Cell Signaling Technology); and rabbit anti-phosphorylated (S232) Ras GTPase-activating protein-binding protein (G3BP), mouse anti-G3BP, rabbit anti-phosphorylated (S964) mediator of DNA damage checkpoint protein 1 (MDC1), and rabbit anti-MDC1 (Abcam, Cambridge, MA). After they were washed with 0.1% Tween 20 in PBS, the membranes were incubated with Alexa Fluor 680-conjugated goat anti-rabbit IgG (Invitrogen, Eugene, OR) or IRDye 800-conjugated goat anti-mouse IgG (Rockland Immunochemicals, Gilbertsville, PA). Blots were visualized and quantified using an Odyssey infrared imager (LI-COR Biosciences).
RESULTS AND DISCUSSION
Phosphoproteome profiling and quantification.
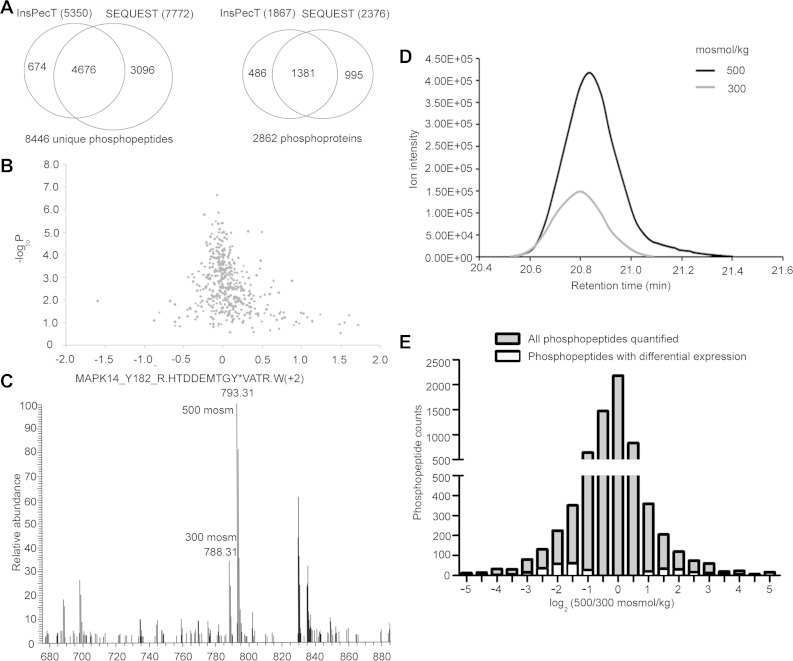
We performed the experiment four times: three times using cells equilibrated with the heavy SILAC label at 300 mosmol/kg and then increased to 500 mosmol/kg for 1 h by addition of NaCl and once with the amino acid labeling reversed. We analyzed the results with SEQUEST (20) and InsPecT (61) algorithms searched against human concatenated real and reversed protein databases with 1% FDR. Combining the results from SEQUEST and InsPecT, we reproducibly identify >8,000 unique phosphopeptides in four biological replicate samples with a 1% FDR (Fig. 1A). The unique phosphopeptides are contained in 2,862 different proteins (Fig. 1A). We assume that abundance of most proteins is not changed by 1 h of high NaCl, so the phosphopeptide changes represent altered phosphorylation at particular sites in proteins, rather than change in amount of the proteins. This assumption is confirmed by our finding that abundance of few proteins (independently measured by iTRAQ) changes significantly after 1 h of high NaCl (Fig. 1B) and none of the phosphopeptides whose abundance changes significantly are contained in proteins whose abundance changes (see Supplemental Table S1 in Supplemental Material for this article, available online at the Journal website). We quantified the effect of high NaCl on phosphorylation by determining the relative areas of the MS1 peaks for each phosphopeptide at 500 vs. 300 mosmol/kg. An example of the peptide containing p38/phosphorylated (Y182) MAPK14 is shown in Fig. 1, C and D. We calculated the means of those ratios, weighted for ion intensity. In all, we are able to quantify 7,362 unique phosphopeptides, containing 8,876 phosphorylation sites; 80% of the peptides are phosphorylated at a single site and 20% are phosphorylated at multiple sites. Abundance of 120 of the phosphopeptides increases significantly (P < 0.05) at 500 mosmol/kg, whereas abundance of 207 phosphopeptides decreases significantly (Fig. 1E; see Supplemental Table S1). The 324 differentially regulated phosphopeptides are contained in 253 different proteins (see Supplemental Table S1).
Fig. 1.
A: number of unique phosphopeptides identified by InsPecT and/or SEQUEST algorithms. B: effect of elevating NaCl for 1 h on abundance of proteins, as measured by iTRAQ. Abundance of only very few proteins changed significantly. C: phosphorylated (Y182) p38/MAPK14 peptide R.HTDDEMTGY*VATR.W(+2). Stable isotopic labeling of amino acids in cell culture (SILAC)-labeled isoforms were identified at 20.78 min. D: high NaCl increased abundance of R.HTDDEMTGY*VATR.W(+2). MS1 peaks are compared. E: phosphopeptides whose abundance was changed by high NaCl. High NaCl significantly increased abundance of 120 and decreased abundance of 207 of the 7,362 unique phosphopeptides that were quantified.
Comparison of quantification by SILAC with quantification by TIS and Western blotting.
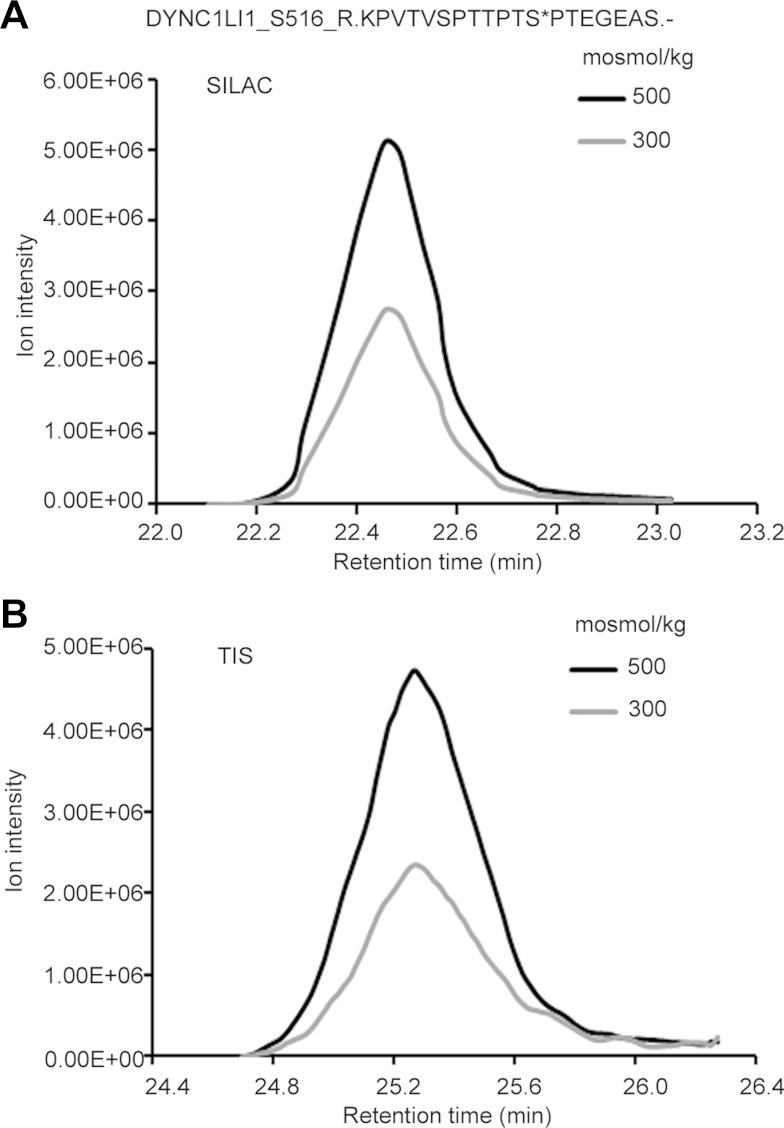
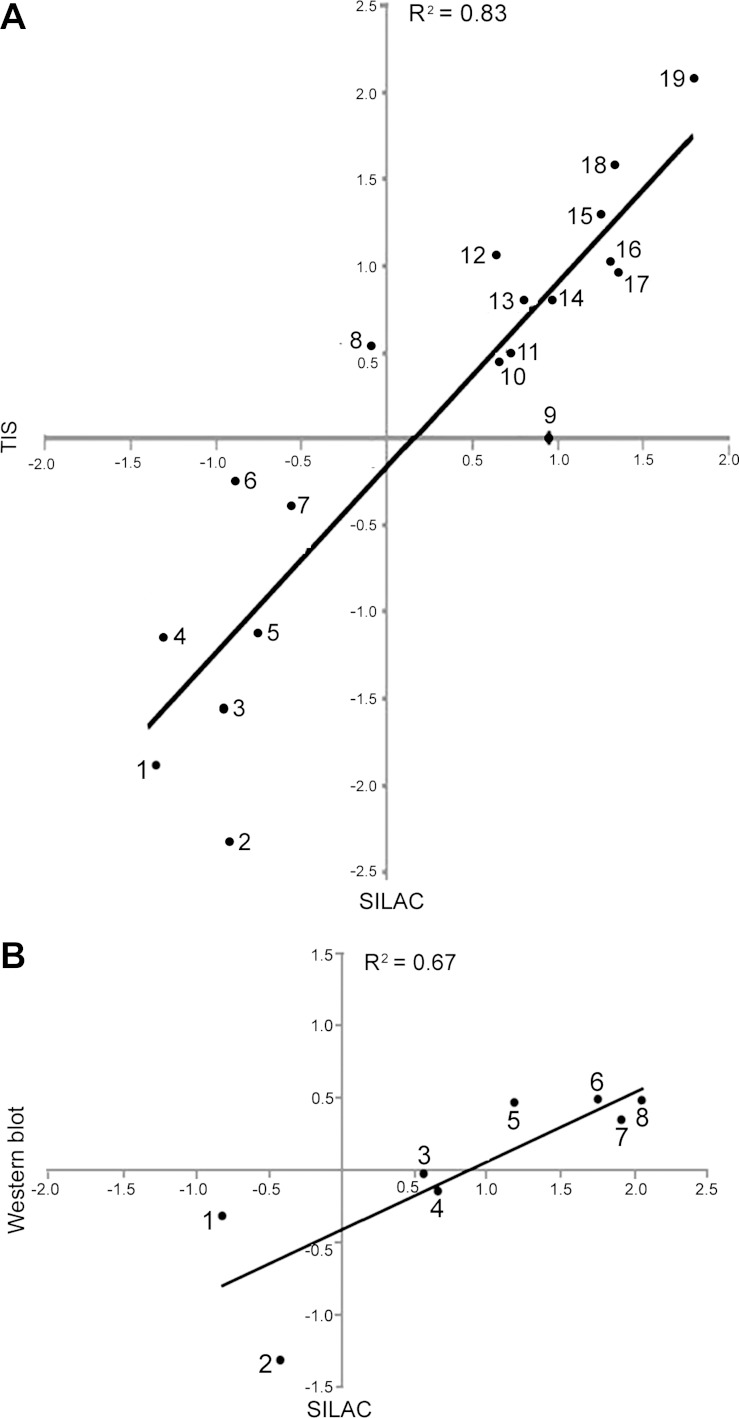
We used TIS, a label-free method, and Western immunoblotting to further test changes in phosphorylation identified by SILAC. Figure 2 shows dynein, cytoplasmic 1, light intermediate chain 1 phosphorylated (S516) at high NaCl, as quantified using SILAC (Fig. 2A) and using TIS (Fig. 2B). In TIS, the MS1 peak area of a selected phosphopeptide quantifies its relative abundance. Using TIS, we were able to quantify the effect of high NaCl on 17 of the differentially changed phosphopeptides indicated by SILAC. The results from TIS correlate highly with those from SILAC (R2 = 0.83; Fig. 3A), which supports the accuracy of both measurements. We also selected eight phosphorylation sites in phosphopeptides whose abundance changed in the SILAC experiment and for which phosphospecific antibodies were available to confirm the differential changes by Western blot analysis. Figure 3B shows that the relative phosphorylation abundance identified in SILAC correlates well (R2 = 0.67) with that shown by Western immunoblotting. However, the magnitude of the changes, as measured by Western blotting, is less than that measured by MS. We previously observed that MS and Western blot analysis give largely equivalent results for changes in protein abundance (44). Our current results extend that conclusion to measurements of protein phosphorylation. The lack of even better agreement may be due to less accuracy of Western blotting (1).
Fig. 2.
High-NaCl-induced change in abundance of the phosphopeptide containing phosphorylated (S516) DYNC1LI1, as measured by SILAC (A) and targeted ion selection (TIS; B). Relative MS1 peak areas are in good agreement between the two methods.
Fig. 3.
Effect of high NaCl on phosphopeptide abundance as determined by TIS vs. SILAC (A) and Western blot with phosphospecific antibodies vs. SILAC (B). Proteins include those whose phosphorylation changed significantly and those whose phosphorylation did not change significantly. Scales are log2(500/300 mosmol/kg). High correlation coefficients between the paired methods support the accuracy of both. Gene symbols-(phosphorylation sites) are as follows: in A, CCNL2(S330) (1), CTTN(T364) (2), GIGYF2(S26) (3), AKT151(S212) (4), TBC1D4(S570) (5), CTNNB1(S552) (6), NEDD4L(S327) (7), GRLF1(S1179) (8), RABEP1 (9), GAPVD1(S929) (10), DTNA(S605) (11), MDC1(S376) (12), MYO9B(S1290) (13), TRIM28(S473) (14), EPS15(S482) (15), DYNC1LI1(S51) (16), MAVS(S222) (17), KIAA1432(S1017) (18), ZFYVE16(S939) (19); in B, G3BP1(S232) (1), MDC1(S964) (2), HDAC1(S421/S423) (3), JUN(S73) (4), STAT1(S727) (5), ATF2(T71) (6), HSPB1/HSP27(S82) (7), JUN(S63) (8).
Role of phosphorylation in signaling known effects of high NaCl.
High NaCl is known to affect cell cycle, cyto/nucleoskeletal organization, DNA double-strand breaks (DSBs), transcription, proteostasis, metabolism of mRNA, and cell death. We used the DAVID bioinformatic tool (http://david.abcc.ncifcrf.gov/) as a basis for identification of proteins whose phosphorylation is altered by high NaCl and whose functions are related to the known effects of high NaCl. This analysis provides insight into the roles of those proteins in response to high NaCl. Where other references are not cited, information about function of specific proteins and of phosphorylation sites within them is from PhosphoSitePlus (30).
High-NaCl-induced cell cycle delay.
Acute elevation of NaCl produces rapid arrest at all phases of the cell cycle (47). We find that high NaCl alters phosphorylation in 20 proteins involved in regulating the cell cycle (Table 1).
Table 1.
Regulation of cell cycle
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| EGFR | 3.46 | T693 | Epidermal growth factor receptor |
| TP53BP2 | 2.90 | S433 | Tumor protein p53-binding protein 2 |
| MAPK14 | 2.87 | Y182 | MAP kinase p38α |
| ATM | 2.67 | S1981 | Ataxia telangiectasia-mutated |
| SIPA1 | 2.13 | S53 | Signal-induced proliferation-associated 1 |
| HCFC1 | 1.57 | S598 | Host cell factor C1 (VP16-accessory protein) |
| 2.42 | S666 | ||
| MDC1 | 1.42 | S1605 | Mediator of DNA-damage checkpoint 1 |
| 1.55 | T1239 | ||
| ANLN | 1.36 | S792 | Anillin, actin-binding protein |
| TTK | 1.21 | S281 | TTK protein kinase |
| DYNC1LI1 | 1.16 | S510, S516 | dynein, cytoplasmic 1, light intermediate chain 1 |
| 1.32 | T515 | ||
| 3.25 | T513, S516 | ||
| RBL1 | −1.11 | S1037, S1041 | Retinoblastoma-like 1 (p107) |
| PML | −1.69 | S518, S527 | Promyelocytic leukemia |
| JUN | −1.86 | S243 | jun proto-oncogene |
| 2.07 | S63 | ||
| 2.13 | T62 | ||
| TPR | −1.87 | S1185 | Translocated promoter region, nuclear basket protein |
| RB1 | −2.16 | S249, T252 | Retinoblastoma 1 |
| TP53BP1 | −2.09 | S557 | Tumor protein p53-binding protein 1 |
| FOXC1 | −2.23 | S235, S241 | Forkhead-related transcription factor 3 |
| CDK13 | −2.33 | S1054, T1058 | Cyclin-dependent kinase 13 |
| DLG1 | −2.61 | S575 | Synapse-associated protein 97 |
| RBL2 | −2.69 | S413, T417 | Retinoblastoma-like 2 (p130) |
Previously, we found that high-NaCl-induced increase in phosphorylation and activation of p38/MAPK14 kinase is responsible for rapid initiation of delay of the G2/M phase (16). We confirm here that high NaCl increases phosphorylated (Y182) MAPK14 (Table 1). We also previously found that high-NaCl-induced activation of p53/tumor protein 53 (TP53) is responsible for delay of the G1/S phase (15). We do not identify changes in phosphorylation of p53 itself in the present experiments, but we do observe altered phosphorylation of related proteins, TP53-binding protein (TB53BP) 2 and TP53BP1 (Table 1). TP53BP2 and TP53BP1 can affect the cell cycle, but their role in high-NaCl-induced cell cycle delay remains to be confirmed.
Retinoblastoma (RB) tumor suppressor proteins (RB1, RBL1, and RBL2) regulate entry into the S phase of the cell cycle after the G1 phase (68). Early in the G1 phase, RB proteins are hypophosphorylated, but they become hyperphosphorylated late in the G1 phase and maintain hyperphosphorylation through the remainder of the cell cycle. Hypophosphorylated RB binds to E2F transcription factors, which prevents transcription of E2F target genes that drive the cell cycle. Hyperphosphorylation of RB disrupts its inhibitory binding to E2Fs. Accordingly, overexpression of RBL1 and RBL2, in which phosphorylation sites have been mutated so that they cannot be phosphorylated, arrests the cell cycle at the G1 phase (22). Phosphorylation inactivates the growth-inhibitory functions of RB. Increased phosphorylation of RB1 at S249 and T252 has been reported to reduce delay of the G1/S phase caused by RB1 (8). Therefore, decreased phosphorylation, such as occurs when NaCl is elevated (Table 1), should increase the delay. We conclude that high-NaCl-induced decrease in phosphorylation of RBL1, RBL2, and RB1 (Table 1) may contribute to high-NaCl-induced G1/S phase delay. Furthermore, rapid high-NaCl/p38-dependent initiation of G2/M phase delay is followed by inhibition of cyclin-dependent kinase 1 (CDK1)/Cdc2 kinase (16). Since CDK1/Cdc2 phosphorylates RB1 at S249 and T252 (43), its inhibition could contribute to termination of G1/S phase delay.
High-NaCl-induced cyto/nucleoskeletal reorganization.
Hypertonicity induces reorganization of the cytoskeleton, including submembranous F-actin assembly, which reinforces the cell structure to withstand the physical challenge of cell shrinkage (14, 29). Hypertonicity also causes nucleoskeletal reorganization involving changes in abundance in the nucleus of several “cytoskeletal” proteins (44). At 1 h after we increased NaCl, changes occurred in phosphorylation of 27 proteins involved in regulating cyto/nucleoskeletal organization (Table 2). The proteins include those involved with GTPase signaling (SIPA1, DOCK7, CDC42EP3, SEPT7, ARHGEF18, and RALGAPA1), actin cyto/nucleoskeleton (ANLN, ZYX, ABLIM1, CTTN, KIF13B, and FARP1), microtubules (MAP2, MAP4, MAP7, MAP1B, CLIP1, and CLASP1), and cell adhesion (CTNNB1, CTNND1, and PKP2) and include a protein kinase (LIMK1), a myosin [myosin heavy chain 9 (MYH9)], a HSP (HSPB1), and adapter proteins (SHKBP1 and DLG1).
Table 2.
Regulation of cytoskeletal and nucleoskeletal organization
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| SIPA1 | 2.13 | S53 | Signal-induced proliferation-associated protein 1 |
| DOCK7 | 1.94 | S30 | Dedicator of cytokinesis 7 |
| HSPB1 | 1.92 | S82 | Heat shock protein β1 |
| MYH9 | 1.69 | S1943 | Myosin-9 |
| ANLN | 1.36 | S792 | Anillin, actin-binding protein |
| CDC42EP3 | 1.18 | S89 | Cdc42 effector protein 3 |
| ZYX | 1.16 | S344 | Zyxin |
| MAP7 | −1.21 | S209 | Microtubule-associated protein 7 |
| CTNNB1 | −1.22 | S552 | Catenin (cadherin-associated protein), β1, 88 kDa |
| SEPT7 | −1.25 | T426 | Cell division cycle 10 isoform 2 |
| ABLIM1 | −1.27 | S655 | Actin-binding LIM protein |
| FARP1 | −1.33 | S427 | FERM, RhoGEF and pleckstrin domain-containing |
| ARHGEF18 | −1.33 | S1103 | Rho-specific guanine nucleotide exchange factor |
| EPB41L3 | −1.59 | S460, T469 | Erythrocyte membrane protein band 4.1-like 3 |
| CTTN | −1.59 | T401 | Cortactin isoform a |
| MAP2 | −1.69 | T306 | Microtubule-associated protein 2 isoform 5 |
| LIMK1 | −1.72 | S310 | LIM domain kinase 1 |
| MAP1B | −1.77 | S1779, S1782, T1788 | Microtubule-associated protein 1B |
| 1.22 | S1785 | ||
| RALGAPA1 | −1.94 | S775 | GTPase-activating Rap/RanGAP domain-like 1 |
| CLIP1 | −2.19 | S204 | Restin isoform a |
| CLASP1 | −2.21 | S1071 | CLIP-associating protein 1 isoform 2 |
| −1.35 | S797 | ||
| SHKBP1 | −2.37 | S587 | SH3-domain kinase-binding protein 1 isoform a |
| DLG1 | −2.61 | S575 | Discs, large homolog 1 isoform 2 |
| PKP2 | −2.62 | S329 | Plakophilin 2 isoform 2a |
| 1.42 | S151 | ||
| CTNND1 | −2.98 | S167 | Catenin, δ1 isoform 3ABC |
| KIF13B | −3.53 | S1644 | Kinesin family member 13B |
| MAP4 | −4.86 | S1073 | Microtubule-associated protein 4 isoform 1 |
| −4.01 | S636 |
Rho family small G proteins are pivotal regulators of actin organization (29). They are highly sensitive to cell volume changes. However, the events between cell volume decrease and Rho protein activation remain enigmatic, at least in part due to the daunting number of upstream regulators of these proteins. Our observation of high-NaCl-induced changes in phosphorylation of proteins involved in GTPase signaling (Table 2) provides clues for identification of additional upstream regulators.
High NaCl decreases phosphorylation of β-catenin/CTNNB1 at S552, catenin D1/CTNND1 at S167, and cortactin/CTTN at T401 (Table 2). β-Catenin links members of the cadherin family of transmembrane cell-cell adhesion receptors to the actin cytoskeleton. AKT (21) and PKA (62) were previously shown to increase phosphorylation of β-catenin/CTNNB1 at S552. AKT (74) and PKA (23) are activated by high NaCl, but their activation should increase phosphorylation of β-catenin/CTNNB1 at S552, not cause the decrease that we found. We speculate that some other (unidentified) kinase or phosphatase may be involved in the decreased phosphorylation of β-catenin/CTNNB1 at S552.
We previously found that high NaCl decreases the abundance of 10 different microtubule proteins in the nucleus within 1 h (44). We now find decreased phosphorylation of six microtubule-associated proteins (MAP2, MAP4, MAP7, MAP1B, CLIP1, and CLASP1) at that time, suggesting that these microtubule-associated proteins might regulate the decrease in nuclear tubulins. High NaCl decreases phosphorylation of MAP7 at S209 (Table 2). The decreased phosphorylation could result from high-NaCl-induced inhibition of CDK1/Cdc2 (16), which phosphorylates MAP7 at S209 (3). These findings reinforce the idea (44) that decrease in nuclear tubulins is associated with rapid high-NaCl-induced cell cycle delay and suggest a previously unknown function of microtubule-associated proteins.
High NaCl increases phosphorylation of MYH9 at S1943 (Table 2), a site whose phosphorylation is known to be involved in cytoskeletal organization, since reduction of phosphorylation of MYH9 at S1943 is associated with its redistribution during ionizing radiation-induced senescence of human mesenchymal stem cells (65). Casein kinase 2 catalyzes phosphorylation of MYH9 at S1943 (65). We do not know, however, whether hypertonicity increases casein kinase 2 activity, which requires further investigation.
High-NaCl-induced DNA damage.
High NaCl increases DNA DSBs in cell culture and in Caenorhabditis elegans (18), renal inner medullas (17), and marine invertebrates (19). The DSBs are not repaired while NaCl remains high but are rapidly repaired when NaCl is lowered. Importantly, high NaCl also decreases efficiency of repair of DNA damage caused by UV radiation, which ordinarily is rapid (17). Some DNA damage response proteins are known to be inhibited by high NaCl. While NaCl remains high, meiotic recombination 11 homolog 1 [Mre11 (MRE11A)] exonuclease is mainly present in the cytoplasm, rather than the nucleus, and histone H2AX (H2AFX) is not phosphorylated, as it normally would be in response to DNA damage (17). If NaCl is subsequently reduced, Mre11 returns to the nucleus and H2AX becomes phosphorylated, accompanying the DNA repair. The changes that we now find in phosphorylation of other DNA damage response proteins (Table 3) can add to our understanding of how high NaCl inhibits DNA repair.
Table 3.
Response to DNA damage stimulus
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| ATM | 2.67 | S1981 | Ataxia telangiectasia mutated protein isoform 1 |
| BAZ1B | 1.87 | S947 | Tyrosine-protein kinase BAZ1B |
| MDC1 | 1.42 | S1605 | Mediator of DNA damage checkpoint 1 |
| 1.55 | T1239 | ||
| SFPQ | 1.11 | S273 | Splicing factor proline/glutamine rich (polypyrimidine tract-binding protein-associated) |
| LIG1 | −1.53 | S76 | DNA ligase I |
| PML | −1.69 | S518, S527 | Promyelocytic leukemia protein isoform 1 |
| TP53BP1 | −2.09 | S557 | Tumor protein p53-binding protein 1 isoform 2 |
| PNKP | −2.26 | T118, T122 | Polynucleotide kinase 3¢-phosphatase |
| CHAF1B | −2.51 | T433 | Chromatin assembly factor 1 subunit B |
Polynucleotide kinase 3′-phosphatase (PNKP) is involved in DSB repair by nonhomologous end-joining. High NaCl reduces phosphorylation of PNKP at T118 and T122 (Table 3). This is opposite to the response in yeast to ionizing radiation, which is a nearly threefold increase in phosphorylation at those sites (2). The possibility that reduced phosphorylation at those sites contributes to inhibition of DNA repair by high NaCl suggests a direction for further investigation.
High NaCl increases phosphorylation of BAZ1B [Williams syndrome transcription factor (WSTF)] at S947 (Table 3). Repair of DSBs induced by genotoxic agents (e.g., ionizing radiation) is initiated by assembly on chromatin of foci that contain H2AX phosphorylated on S139 (“γH2AX”). In the absence of genotoxic stress, BAZ1B phosphorylates histone H2AX at Y142 (70), which prevents formation of γH2AX. This maintains a “standby” mode in which DNA is not repaired unnecessarily. When DSBs are induced by genotoxic agents, WSTF dissociates and is replaced by eyes absent (EYA1/EYA3) phosphatases, which dephosphorylate phosphorylated (Y142) H2AX, facilitating formation of γH2AX (12). In contrast to other genotoxic agents, high-NaCl-induced DSBs do not increase γH2AX (17). The explanation may be that high-NaCl-induced phosphorylation of BAZ1B at S947 maintains its activity and/or that high NaCl inhibits EYA1/EYA2 activity. Both of these possibilities warrant further investigation.
High-NaCl-induced changes in transcription.
High NaCl decreases transcription in general (11) but increases transcription of osmoprotective genes (6, 11). In the present study we find changes in phosphorylation of 44 proteins involved with regulation of transcription (Table 4). Those changes may help us understand why high NaCl specifically increases transcription of the osmoprotective genes. The proteins include transcription factors (FOXK2, YBX1, ZNF446, NFX1, ZNF687, JUN/c-Jun, ATF2, ZMYM2, RFX7, AFF4, CREB5, and STAT1), transcription coactivators/corepressors (MED24, DMAP1, PAWR, DAXX, LRRFIP1, ATF7IP, TLE3, MTA1, HMGA2, HMGA1, SLTM, FOXC1, DNTTIP2, DPF2, PML, HCFC1, GATAD2B, and GTF2F1), regulators of transcription from RNA polymerase III promoter (MAF1 and ARID1A), regulators of transcription from RNA polymerase II promoter (YEATS2, BRWD1, SAFB, ZNF462, and ZNF768), transcription elongation factors (SUPT5H, RDBP, and TCEB3), histone modifiers (RNF20 and SUV39H1), and RNA polymerase II (POLR2A).
Table 4.
Regulation of transcription
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| TCEB3 | 2.91 | S542 | Elongin A |
| CREB5 | 2.69 | T59, T61 | cAMP responsive element-binding protein 5 isoform α |
| ATF7 | 2.64 | T51, T53 | Activating transcription factor 7 isoform 3 |
| JUN | 2.13 | T62 | jun oncogene |
| 2.07 | S63 | ||
| −1.86 | S243 | ||
| ATF7IP | 2.11 | S113, T118 | Activating transcription factor 7-interacting protein |
| PAWR | 1.95 | T230 | PRKC, apoptosis, WT1, regulator |
| ZMYM2 | 1.77 | S305 | Zinc finger protein 198 |
| ATF2 | 1.76 | T69, T71 | Activating transcription factor 2 |
| HCFC1 | 1.57 | S598 | Host cell factor 1 |
| 2.42 | S666 | ||
| STAT1 | 1.24 | S727 | Signal transducer and activator of transcription 1 |
| SUV39H1 | −1.01 | S391 | Suppressor of variegation 3–9 homolog 1 |
| YBX1 | −1.02 | S165 | Nuclease-sensitive element-binding protein 1 |
| BRWD1 | −1.06 | S1475 | Bromodomain and WD repeat domain-containing 1 |
| DMAP1 | −1.13 | T445 | DNA methyltransferase 1-associated protein 1 |
| LRRFIP1 | −1.19 | S88 | Leucine-rich repeat (in FLII)- interacting protein 1 isoform 2 |
| ARID1A | −1.20 | S696, S702 | AT-rich interactive domain 1A isoform a |
| DNTTIP2 | −1.22 | S117 | Deoxynucleotidyltransferase, terminal, interacting protein 2 |
| MED24 | −1.26 | S860 | Mediator complex subunit 24 isoform 2 |
| 2.55 | S849 | ||
| SUPT5H | −1.32 | S666 | Suppressor of Ty 5 homolog isoform a |
| RDBP | −1.34 | S353 | RD RNA-binding protein |
| 2.25 | S115 | ||
| 2.80 | S51 | ||
| 3.10 | S49 | ||
| HMGA1 | −1.37 | S91, S92 | High-mobility group AT-hook 1 isoform b |
| DPF2 | −1.39 | S142 | Zinc finger protein ubi-d4 |
| FOXK2 | −1.40 | S398 | Forkhead box K2 |
| AFF4 | −1.53 | S1043 | ALL1 fused gene from 5q31 |
| DAXX | −1.63 | S683 | Death domain-associated protein isoform b |
| RNF20 | −1.65 | S138 | Ring finger protein 20 |
| PML | −1.69 | S518, S527 | Promyelocytic leukemia protein isoform 1 |
| HMGA2 | −1.69 | S101 | High-mobility group AT-hook 2 isoform a |
| TLE3 | −1.74 | T334 | Transducin-like enhancer protein 3 isoform a |
| SAFB | −1.86 | S604 | Scaffold attachment factor B |
| YEATS2 | −1.86 | S519 | YEATS domain-containing 2 |
| ZNF462 | −1.92 | S688 | Zinc finger protein 462 |
| NFX1 | −1.96 | S50 | Nuclear transcription factor, X-box-binding 1 isoform 2 |
| SLTM | −2.00 | S1002 | Modulator of estrogen-induced transcription isoform a |
| RFX7 | −2.16 | S1178 | Regulatory factor X domain-containing 2 |
| FOXC1 | −2.23 | S235, S241 | Forkhead box C1 |
| ZNF768 | −2.36 | S90, S97 | Zinc finger protein 768 |
| MAF1 | −2.44 | S75 | MAF1 protein |
| ZNF446 | −2.45 | S137 | Zinc finger protein 446 |
| POLR2A | −2.46 | S1878, T1884 | DNA-directed RNA polymerase II polypeptide A |
| GTF2F1 | −2.60 | T389 | General transcription factor IIF subunit 1 |
| ZNF687 | −2.74 | S253 | Zinc finger protein 687 |
| GATAD2B | −2.88 | S333 | GATA zinc finger domain-containing 2B |
| 1.01 | S486 | ||
| MTA1 | −3.21 | T578 | Metastasis-associated protein |
| −1.70 | S576 |
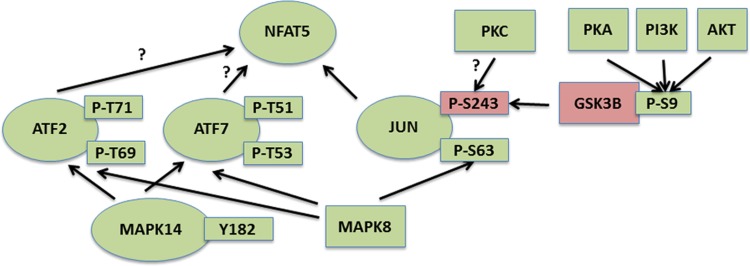
High NaCl activates the transcription factor nuclear factor of activated T cells (NFAT5), which increases transcription of osmoprotective target genes. Promoter regions of those target genes contain not only DNA elements specific for binding NFAT5, but also nearby activator protein 1 (AP-1) sites that bind the AP-1 proteins FOS/c-Fos and JUN/c-Jun (32). FOS/c-Fos and JUN/c-Jun are activated by high NaCl and contribute to increased transcription of NFAT5 target genes (32). Also, inhibition of MAPK14/p38, which is activated by high NaCl, reduces high-NaCl-dependent activation of a transcriptional reporter that contains both the NFAT5-specific DNA element and an AP-1 site, but only if the AP-1 site is intact (32). MAPK14/p38 increases c-Jun abundance by activating the c-jun promoter (46). In the present study we find additional evidence for pathways that activate AP-1 (Fig. 4). High NaCl increases phosphorylation of JUN/c-Jun at T62 and S63 and reduces phosphorylation at S243 (Table 4). We are unaware of any references to the effect on its activity of phosphorylation of JUN/c-Jun at T62. However, increased phosphorylation at S63 (26) and decreased phosphorylation at S243 (5) promote activity of JUN/c-Jun, and identification of the kinases and phosphatases that affect phosphorylation at those sites points to the regulatory pathways that are involved. Considering that MAPK8/JNK activates JUN/c-Jun by phosphorylation at sites including JUN/c-Jun phosphorylated at S63 (26) and high NaCl increases activity of MAPK8/JNK (73), we suggest that the MAPK8/JNK-JUN/c-Jun pathway contributes to high-NaCl-induced activation of NFAT5. Furthermore, reduced phosphorylation of JUN/c-Jun at S243 increases DNA-binding activity of JUN/c-Jun, and, conversely, elevated phosphorylation at that site decreases JUN/c-Jun activity (5). Also, mutation of JUN/c-Jun phosphorylated at S243 to phenylalanine, which cannot be phosphorylated, greatly increases the transactivating ability of JUN/c-Jun. In resting cells, JUN/c-Jun is in a latent, phosphorylated form that is activated by dephosphorylation at S243. GSK3B/GSK-3β phosphorylates JUN/c-Jun at S243 in resting cells, which maintains JUN/c-Jun in an inactive state. High NaCl inhibits GSK3B/GSK-3β by increasing phosphorylation of GSK3B/GSK-3β at S9 (76). Decreased GSK-3β activity, by reducing phosphorylation of JUN/c-Jun at S243, increases JUN/c-Jun activity. Further upstream, the high-NaCl-induced phosphorylation of GSK-3β at S9 depends on PKA, phosphatidylinositol 3-kinase, and AKT (76), which are themselves activated by high NaCl and contribute to activation of NFAT5 (23, 31, 57). PKC may also contribute to high-NaCl-induced activation of JUN/c-Jun. Application of the phorbol ester 12-O-tetradecanoylphorbol-13-acetate, which activates PKC, causes dephosphorylation of c-Jun-S243 (5). High NaCl is reported to activate PKC in most, but not all, studies (reviewed in Ref. 75), and PKCμ contributes to the hypertonicity-induced increase in HSP70 abundance (45). Further studies are necessary to determine whether PKC contributes to high-NaCl activation of JUN/c-Jun and, if so, what isoforms of PKC are involved. High NaCl also increases phosphorylation of two other AP-1 proteins, ATF2 phosphorylated at T69/T71 and ATF7 phosphorylated at T51/T53 (Table 4). ATF2 and ATF7 (25) are highly homologous AP-1 transcription factors that form homodimers or heterodimers with other AP-1 factors, such as c-Fos and c-Jun. ATF2 is activated by phosphorylation at T69 and T71 and ATF7 by phosphorylation at T51 and T53, as we observe in response to high NaCl. MAPK8/JNK and/or MAPK14/p38 phosphorylate those sites (13). However, we do not know what role, if any, ATF2 and ATF7 have in high-NaCl-induced activation of AP-1 sites flanking NFAT5 target genes. Figure 4, which is based on the above-described observations, presents a model of the signaling pathways by which high NaCl activates JUN/c-Jun and other AP-1 factors, including ATF2 and ATF7.
Fig. 4.
Pathways by which high-NaCl-induced changes in phosphorylation of signaling molecules activate activator protein 1 (AP-1), contributing to activation of the osmoprotective transcription factor nuclear factor of activated T cells (NFAT5). Green indicates increase and red indicates decrease in activity or phosphorylation of kinases (rectangles) or transcription factors (ovals).
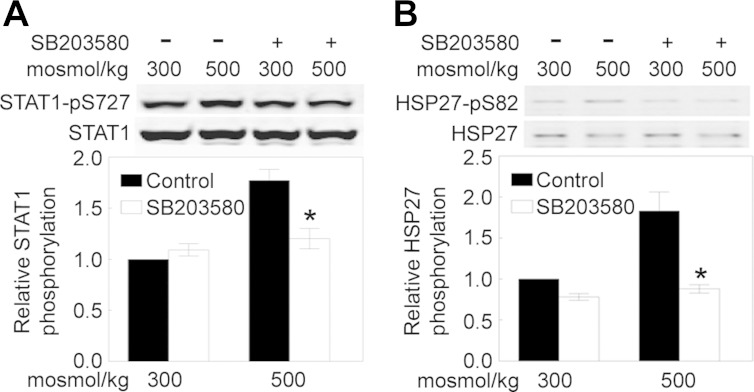
Hypertonicity induced by sorbitol increases activity of STAT1 in COS-7 cells via MAPK14/p38-mediated phosphorylation of STAT1 at T701 (4). We do not find a significant increase in phosphorylation of STAT1 at T701 in HEK 293 cells in response to high-NaCl-induced hypertonicity. However, we do find substantially increased phosphorylation of STAT1 at S727 (Table 4). That may be pertinent, since phosphorylation of STAT1 at S727 increases transactivating activity of STAT1 (69). MAPK14/p38 could be responsible for phosphorylation of STAT1 at S727 in response to high NaCl, since MAPK14/p38 was previously found to phosphorylate STAT1 at S727 in response to interleukin-13 (71). However, we have reservations about that conclusion, since hypertonicity induced by sorbitol only slightly increases phosphorylation of STAT1 at S727 in COS-7 cells, and that phosphorylation is independent of p38 (4). Also, PKC-δ (PRKCD) (64) and phosphatidylinositol 3-kinase/AKT (50) are responsible for phosphorylation of STAT1 at S727 in response to interferon. Therefore, we used the p38 inhibitor SB-203580 to determine whether p38 contributes to high-NaCl-induced increase in phosphorylation of STAT1 at S727 in HEK 293 cells. SB-203580 inhibits high-NaCl-induced phosphorylation of STAT1 at S727 (Fig. 5), indicating that p38 does contribute to high-NaCl-induced phosphorylation of STAT1 at S727 in HEK 293 cells. Nevertheless, additional studies are required to identify the gene targets of the high-NaCl-induced increase in STAT1 activity.
Fig. 5.
p38/MAPK14 activity contributes to high-NaCl-induced increase in phosphorylation of STAT1 and HSP27/HSPB1. HEK 293 cells were preincubated for 30 min with the p38/MAPK14 inhibitor SB-203580 (8 μM) at 300 mosmol/kg; then the medium was changed for 1 h to an identical medium (control) or a medium still containing the inhibitor and with osmolality increased to 500 mosmol/kg by addition of NaCl. Immunoblots were prepared using anti-STAT1 and anti-phosphorylated (S727) STAT1 (A) or anti-HSP27/HSPB1 and anti-phosphorylated (S82) HSP27/HSPB1 antibodies (B). Top: representative Western blots. Bottom: means ± SE; n = 3. *P < 0.05 vs. control.
High NaCl decreases phosphorylation of general transcription factor II F (GTF2F1)/transcription factor II F (TFIIF)/RNA polymerase II-associating protein 74 (RAP74) at T389 (Table 4). GTF2F1/TFIIF/RAP74 is a general transcription factor that binds to RNA polymerase II, helps recruit it to the initiation complex, and promotes transcription elongation. GTF2F1/TFIIF/RAP74 has serine/threonine kinase activity that autophosphorylates it at S385 and T389 (56). The autophosphorylation downregulates RNA polymerase II activity. Therefore, the high-NaCl-induced decrease in phosphorylation of TFIIF at T389 could increase transcriptional activity (56). It is unclear, however, whether transcription of osmoprotective genes would be differentially regulated by the activity of GTF2F1/TFIIF/RAP74.
High-NaCl-induced perturbation of proteostasis.
Proteostasis refers to the biogenesis, folding, trafficking, and degradation of proteins. All phases of proteostasis apparently are affected by hypertonicity. Hypertonicity not only causes dramatic changes in translation in mammalian cells, but evidence has been emerging in C. elegans that high NaCl also induces rapid protein aggregation in vivo and that many of the genes that are essential for survival during hypertonic stress function to prevent accumulation of aggregated proteins (10). Whether hypertonicity induces comparable protein damage in mammalian cells, however, has been less clear.
Hypertonicity, including that produced by high NaCl, rapidly inhibits translation of most proteins, but not translation of osmoprotective proteins. The difference in response apparently depends on whether translation is dependent on the 5′ cap of the mRNA (which is true of most proteins) or is independent of the 5′ cap (which is true of osmoprotective proteins) (55). We find that high NaCl changes phosphorylation of five proteins involved in regulation of translation (Table 5): eukaryotic translation initiation factor (EIF) 4γ 2 (EIF4G2), EIF 4E-binding protein 1 (EIF4EBP1)/PHAS-1, EIF3B, HSPB1, and RPTOR/Raptor.
Table 5.
Perturbation of proteostasis
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| EIF3B | −3.08 | S119 | Eukaryotic translation initiation factor 3, subunit 9η, 116 kDa |
| SQSTM1 | −3.05 | T269 | Sequestosome 1 |
| EIF4EBP1 | −2.57 | S65 | Eukaryotic translation initiation factor 4E-binding protein 1 |
| RPTOR | −1.66 | S859, S863 | Raptor |
| USP47 | −1.65 | S822 | Ubiquitin-specific protease 47 |
| RNF20 | −1.65 | S138 | Ring finger protein 20 |
| UBR1 | −1.60 | T21 | Ubiquitin protein ligase E3 component n-recognin 1 |
| USP42 | −1.55 | S856 | Ubiquitin-specific protease 42 |
| EIF4G2 | −1.31 | T508 | Eukaryotic translation initiation factor 4 γ2 isoform 1 |
| PSMA5 | 1.01 | S56 | Proteasome α5 subunit |
| PSMF1 | 1.02 | S153 | Proteasome inhibitor subunit 1 |
| PSMD1 | 1.12 | T273 | Proteasome 26S non-ATPase subunit 1 |
| HSPB1 | 1.92 | S82 | Heat shock protein β1 |
| DOCK7 | 1.94 | S30 | Dedicator of cytokinesis 7 |
| USP8 | 2.22 | S389 | Ubiquitin-specific peptidase 8 |
The large high-NaCl-induced decrease in phosphorylation of EIF4EBP1 at S65 (Table 5) (48) helps us understand what determines whether hypertonicity decreases or increases translation of particular proteins. Cap-dependent translation (60) is initiated by association of the cap-binding protein eIF4E with eIF4G, which then recruits the ribosomal 43S complex to the 5′ end of mRNA. EIF4EBP1 directly interacts with eIF4E, and that interaction inhibits formation of the eIF4E-eIF4G complex, which represses translation. High-NaCl-induced dephosphorylation of EIF4EBP1 (Table 5) increases its affinity for eIF4E, which inhibits general cap-dependent translation, but not the cap-independent translation of osmoprotective proteins. Interestingly, in C. elegans, the high-NaCl-induced general inhibition of translation may provide the signal for transcription of osmoprotective genes (40).
Genome-wide RNAi screen and in vivo protein aggregation reporters identified degradation of damaged proteins as essential for the hypertonic stress response in C. elegans (10). Hypertonic stress causes loss of cellular water, cell shrinkage, elevated intracellular ionic strength, and macromolecular crowding (6). In vitro studies using simple mixtures of proteins, solutes, and artificial crowding agents have shown that high concentrations of inorganic ions, such as K+, Na+, and Cl−, destabilize protein secondary structure and disrupt enzyme activity and that macromolecular crowding promotes nonnative protein-protein interactions, which can lead to protein aggregate formation. There is little evidence for hypertonicity-induced protein damage similar to that in C. elegans in intact mammalian cells. However, we observe numerous high-NaCl-induced changes in phosphorylation in proteins involved in protein degradation, namely, proteins in the ubiquitin-conjugating system (SQSTM1, USP47, RNF20, UBR1, USP42, and USP8) and proteosome subunits and regulators (PSMA5, PSMF1, and PSMD1) (Table 5).
Phosphorylation of PSMD1 at T273 is increased by hypertonicity produced by sorbitol (41) and NaCl (Table 5). The increased phosphorylation results from hypertonicity-induced activation of MAPK14/p38 (41), and it contributes to an inhibition of proteosomal activity that results in an increase in ubiquitinated proteins (41). We do not know whether altered phosphorylation of the other proteins involved in ubiquitination and proteasomal activity (Table 5) affects their activity, but that seems a likely possibility, pointing to altered proteostasis. Along the same line, HSPB1/HSP27 is a ubiquitin-binding protein proposed to favor the degradation of ubiquitinated proteins (53). Stress-induced activation of MAPK14/p38 promotes phosphorylation of HSPB1/HSP27 at S82 in rabbit muscle (59) and HeLa cells (39). We used the p38 inhibitor SB-203580 to test whether MAPK14/p38 also contributes to high-NaCl-induced increase in phosphorylation of HSPB1/HSP27 at S82 in HEK 293 cells. SB-203580 inhibits high-NaCl-induced phosphorylation of HSPB1/HSP27 at S82 (Fig. 5), confirming that MAPK14/p38 does contribute to high-NaCl-induced phosphorylation of HSPB1/HSP27 at S82 in HEK 293 cells. Phosphomimetic mutation at sites including S82 promotes activity of HSPB1/HSP27 (37). We suggest that increased phosphorylation of HSPB1/HSP27 at S82, enhanced by activation of MAPK14/p38, contributes to changes in proteostasis induced by high NaCl.
High-NaCl-induced effects on metabolism of mRNA.
We were unaware of previous evidence that high NaCl affects mRNA stability or splicing other than increased stability of NFAT5 mRNA (7). However, we now find that high NaCl increases phosphorylation of several proteins involved in metabolism of RNA (Table 6). Known functions of those proteins include mRNA degradation (PARN, SMG9, DCP1A, and DCP1B), pre-mRNA splicing (SRRM2, RBMX, SNRNP70, RBM10, RBM25, SFRS16, KHDRBS1/SAM68, YBX1, SF3B2, SFPQ, ZCCHC8, and HNRNPM), and multiple effects on mRNA processing (PCBP2/hnRNP-E2). High NaCl increases phosphorylation of PCBP2/hnRNP-E2 at S189 (Table 6). Phosphorylation of PCBP2/hnRNP-E2 at S189 contributes to stabilization of PCBP2/hnRNP-E2 protein in murine myeloid cells (9). Phosphorylation at S189 is catalyzed by MAPK1/ERK1/2, which is, in turn, activated by ABL1/c-ABL. High NaCl is known to activate MAPK1/ERK1/2 (34) and ABL1/c-ABL (24), suggesting a pathway by which high NaCl may increase phosphorylation of PCBP2/hnRNP-E2. Other than PCBP2/hnRNP-E2, we do not have information on the effects of site-specific phosphorylation of the proteins involved in metabolism of mRNA. Nevertheless, the results suggest that there may be previously unreported effects of high NaCl on mRNA, including effects on splicing and stability.
Table 6.
Metabolism of mRNA
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| SRRM2 | −2.28 | S1541, S1552 | Splicing coactivator subunit SRm300 |
| 1.35 | S2272 | ||
| 1.71 | S2118 | ||
| 2.08 | T2289 | ||
| PARN2 | −2.08 | S102 | Poly(A)-specific ribonuclease (deadenylation nuclease) isoform 2 |
| SMG9 | −2.04 | S32 | Hypothetical protein LOC56006 |
| RBMX | −1.99 | S328 | RNA-binding motif protein, X-linked |
| SNRNP70 | −1.99 | S410 | U1 small nuclear ribonucleoprotein 70 kDa |
| HNRNPUL1 | −1.83 | S718 | Heterogeneous nuclear ribonucleoprotein U-like 1 isoform a |
| RBM10 | −1.66 | S89 | RNA-binding motif protein 10 isoform 1 |
| DCP1A | −1.62 | S525, T531 | DCP1-decapping enzyme homolog A |
| RBM25 | −1.61 | S677 | RNA-binding motif protein 25 |
| SFRS16 | −1.55 | S547 | Splicing factor, arginine/serine-rich 16 |
| KHDRBS1 | −1.04 | S20 | KH domain-containing, RNA-binding, signal transduction-associated 1 |
| YBX1 | −1.02 | S165 | Nuclease-sensitive element-binding protein 1 |
| SF3B2 | 1.00 | S307, S309 | Splicing factor 3B subunit 2 |
| SFPQ | 1.11 | S273 | Splicing factor proline/glutamine-rich (polypyrimidine tract-binding protein-associated) |
| ZCCHC8 | 1.35 | S658 | Zinc finger CCHC domain-containing protein 8 |
| PCBP2 | 1.62 | S189 | Poly(rC)-binding protein 2 isoform d |
| DCP1B | 1.76 | S275 | Decapping enzyme Dcp1b |
| ADAR | 3.18 | T601 | Adenosine deaminase, RNA-specific isoform a |
| HNRNPM | 4.06 | S579 | Heterogeneous nuclear ribonucleoprotein M isoform b |
High-NaCl-induced effects on cell death.
Acute elevation of NaCl beyond a threshold that depends on the type of cell induces cell death by apoptosis (6). In the present study we raised NaCl to a level that HEK 293 cells survive. Nevertheless, phosphorylation changed in a number of proteins involved in cell death (Table 4), including DPF2/requiem, DNM1L/DRP1, TP53BP2, PML, PAWR, DAXX, CTNNB1, SLTM, SQSTM1, BAG3, SHKBP1, HSPB1, ACIN1, BCL7C and RBM25. Also, JUN/c-Jun (Table 7) is involved in cell death. Some of the specific phosphorylations were previously observed to be proapoptotic, and others were observed to be antiapoptotic. In what follows we analyze the apparently pro- and antiapoptotic changes, since their balance presumably determines cell survival.
Table 7.
Cell death
| Official Gene Symbol | Change, log2(fold) | Phosphorylated Amino Acid Site | Name |
|---|---|---|---|
| BCL7C | −4.23 | S126 | B-cell CLL/lymphoma 7C |
| SQSTM1 | −3.05 | T269 | Sequestosome 1 |
| ACIN1 | −2.43 | S240 | Apoptotic chromatin condensation inducer 1 |
| SHKBP1 | −2.37 | S587 | SH3-domain kinase-binding protein 1 isoform a |
| SLTM | −2.00 | S1002 | Modulator of estrogen-induced transcription isoform a |
| PML | −1.69 | S518, S527 | Promyelocytic leukemia protein isoform 1 |
| DAXX | −1.63 | S683 | Death domain-associated protein isoform b |
| RBM25 | −1.61 | S677 | RNA-binding motif protein 25 |
| DPF2 | −1.39 | S142 | Zinc finger protein ubi-d4 |
| BAG3 | −1.29 | S284, S289 | BCL2-associated athanogene 3 |
| CTNNB1 | −1.22 | S552 | Catenin (cadherin-associated protein), β1, 88 kDa |
| HSPB1 | 1.92 | S82 | Heat shock protein-β1 |
| PAWR | 1.95 | T230 | PRKC, apoptosis, WT1, regulator |
| DNM1L | 2.05 | S616 | Dynamin 1-like protein isoform 1 |
| HTT | 2.47 | S1876 | Huntingtin |
| TP53BP2 | 2.90 | S562 | Tumor protein p53-binding protein, 2 isoform 2 |
High NaCl depolarizes mitochondria and promotes their fission, but the threshold level of NaCl at which that occurs and whether apoptosis occurs at that level of NaCl differ between cell types (6). In the present study we find that high NaCl increases phosphorylation of DNM1L/DRP1 at S616 (Table 7). DNM1L/DRP1 contributes to fission of mitochondria during apoptosis (42). Fission of mitochondria following depolarization depends on DNM1L/DRP1 (33). Phosphorylation of DNM1L/DRP1 at S616, which we find is induced by high NaCl, promotes mitochondrial fission (36). Thus a level of NaCl less than that which causes apoptosis in HEK 293 cells still increases phosphorylation of DNM1L/DRP1 at S616 that is associated with apoptosis.
High NaCl decreases phosphorylation of PML at S518/S527 (Table 7) and JUN/c-Jun at S243 (Table 4). Activation of JUN/c-Jun by PML promotes UV-induced apoptosis (58), and phosphorylation of PML at S518 in response to hypoxia promotes KLHL20-mediated PML destruction (72). If the decreased phosphorylation of JUN/c-Jun and PML that is caused by high NaCl has effects opposite to the previously observed increases, the net result would be antiapoptotic. We do not know whether PML protein is reduced by high NaCl, but, if it is, that would help explain how HEK 293 cells survive high NaCl up to the level in the present study. High NaCl inhibits CDK1 (16), and CDK1 phosphorylates PML at S518 (72). Thus inhibition of CDK1 by high NaCl explains how high NaCl reduces phosphorylation of PML at S518. With regard to JUN/c-Jun, phosphorylation of JUN/c-Jun at S243 primes JUN/c-Jun for phosphorylation of JUN/c-Jun at T239 by GSK3B, and phosphorylation of JUN/c-Jun at S243 creates a high-affinity binding site for FBXW7 E3 ligase. FBXW7 targets JUN/c-Jun for polyubiquitination and proteasomal degradation (67). High NaCl inhibits GSK3B (76). Thus high-NaCl-induced decrease in phosphorylation of JUN/c-Jun at S243 and inhibition of GSK3B could increase JUN/c-Jun protein expression by preventing its FBXW7-mediated degradation. Considering these possibilities, it would be of interest to know the effect of high NaCl on protein expression of JUN/c-Jun and PML.
High NaCl decreases phosphorylation of CTNNB1/-β-catenin at S552 (Table 7). Since phosphorylation of CTNNB1/-β-catenin at S552 increases its transcriptional coregulatory activity (49), decreased phosphorylation could decrease the activity. Decreased activity of CTNNB1/β-catenin could reduce DDK1, which is a transcriptional target of factors regulated by CTNNB1/β-catenin (49). DDK1 is proapoptotic (27). Thus high-NaCl-induced decrease in phosphorylation of CTNNB1/β-catenin at S552 could enhance cell survival by decreasing DDK1. In this respect, it would be of interest to know the effect of high NaCl on protein expression of DDK1.
High NaCl increases phosphorylation of HSPB1/Hsp27 at S82 (Table 7). Hypertonicity-induced phosphorylation of Hsp27 at S82 was previously reported to be catalyzed by activation of MAPK14/p38 in rat brain slices (51) and human epidermal keratinocytes (35). We confirm that MAPK14/p38 is responsible for the high-NaCl-induced phosphorylation of HSPB1/Hsp27 at S82 in HEK 293 cells, finding that inhibition of MAPK14/p38 by SB-203580 prevents the phosphorylation (Fig. 5). Considering that phosphorylation of HSPB1/Hsp27 at S82 protects cells from apoptosis (38), we suggest that that the phosphorylation contributes to survival of HEK 293 cells when osmolality is increased to 500 mosmol/kg by addition of NaCl. In addition to its prosurvival effect, HSPB1/Hsp27 is also involved in cell cycle progression, RNA metabolism, proteostasis, and cytoskeletal organization (38), all of which are affected by high NaCl (see above), so its phosphorylation at S82 could be involved as well in perturbation of those functions by high NaCl.
Perspective.
By combining stable isotope labeling of proteins, multidimensional LC, and high-resolution MS, we performed quantitative global analysis of high-NaCl-induced changes in phosphorylation of proteins. To our knowledge, our work represents the first global analysis of high-NaCl-induced changes in phosphorylation of particular amino acids in proteins. We quantified 7,362 unique phosphopeptides. High salt significantly changed phosphorylation in 324 peptides contained in 253 different proteins. We further analyzed how the specific changes in phosphorylation might be involved in known effects of high NaCl, including altered cell cycle, cyto/nucleoskeletal organization, DNA DSBs, transcription, proteostasis, metabolism of mRNA, and cell death.
Almost all the phosphorylation sites that we found were identified in previous phosphoproteomic screens. Despite so many previous identifications, relatively little has been reported that indicates the kinases and phosphatases involved and the functional consequences of the changes in phosphorylation. From the limited information that is available, we have extrapolated pathways by which high NaCl may alter the specific phosphorylations and the possible functional consequences. A limitation is that most of the studies from which we extrapolate were conducted with cells other than the HEK 293 cells that we used, so we cannot be sure whether certain of the effects depend on cell type. Also, only a few of the previous studies investigated hypertonicity. Despite these limitations, our analysis suggests previously unappreciated possibilities and generates hypotheses for future investigation.
GRANTS
This work was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.W., J.D.F., M.G., and M.B.B. are responsible for conception and design of the research; R.W., Y.I., N.D., K.R., and G.W. performed the experiments; R.W., J.D.F., Y.I., G.W., and M.B.B. analyzed the data; R.W., J.D.F., and M.B.B. interpreted the results of the experiments; R.W. and M.B.B. prepared the figures; R.W. and M.B.B. drafted the manuscript; R.W., J.D.F., Y.I., G.W., M.G., and M.B.B. approved the final version of the manuscript; J.D.F. and M.B.B. edited and revised the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Mark Knepper, Trairaq Pisitkun, and Jason Hoffert for helpful discussion.
REFERENCES
- 1.Aebersold R, Burlingame AL, Bradshaw RA. Western blots vs. selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics 12: 2381–2382, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9: 1314–1323, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci USA 105: 1442–1447, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode JG, Gatsios P, Ludwig S, Rapp UR, Haussinger D, Heinrich PC, Graeve L. The mitogen-activated protein (MAP) kinase p38 and its upstream activator MAP kinase kinase 6 are involved in the activation of signal transducer and activator of transcription by hyperosmolarity. J Biol Chem 274: 30222–30227, 1999. [DOI] [PubMed] [Google Scholar]
- 5.Boyle WJ, Smeal T, Defize LH, Angel P, Woodgett JR, Karin M, Hunter T. Activation of protein kinase C decreases phosphorylation of c-Jun at sites that negatively regulate its DNA-binding activity. Cell 64: 573–584, 1991. [DOI] [PubMed] [Google Scholar]
- 6.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiol Rev 87: 1441–1474, 2007. [DOI] [PubMed] [Google Scholar]
- 7.Cai Q, Ferraris JD, Burg MB. High NaCl increases TonEBP/OREBP mRNA and protein by stabilizing its mRNA. Am J Physiol Renal Physiol 289: F803–F807, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Caldon CE, Lee CS, Sutherland RL, Musgrove EA. Wilms' tumor protein 1: an early target of progestin regulation in T-47D breast cancer cells that modulates proliferation and differentiation. Oncogene 27: 126–138, 2008. [DOI] [PubMed] [Google Scholar]
- 9.Chang JS, Santhanam R, Trotta R, Neviani P, Eiring AM, Briercheck E, Ronchetti M, Roy DC, Calabretta B, Caligiuri MA, Perrotti D. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPα-driven myeloid differentiation. Blood 110: 994–1003, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe KP, Strange K. Genome-wide RNAi screen and in vivo protein aggregation reporters identify degradation of damaged proteins as an essential hypertonic stress response. Am J Physiol Cell Physiol 295: C1488–C1498, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen DM, Wasserman JC, Gullans SR. Immediate early gene and HSP70 expression in hyperosmotic stress in MDCK cells. Am J Physiol Cell Physiol 261: C594–C601, 1991. [DOI] [PubMed] [Google Scholar]
- 12.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458: 591–596, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai S, Laskar S, Pandey BN. Autocrine IL-8 and VEGF mediate epithelial-mesenchymal transition and invasiveness via p38/JNK-ATF-2 signalling in A549 lung cancer cells. Cell Signal 25: 1780–1791, 2013. [DOI] [PubMed] [Google Scholar]
- 14.Di Ciano-Oliveira C, Thirone AC, Szaszi K, Kapus A. Osmotic stress and the cytoskeleton: the R(h)ole of Rho GTPases. Acta Physiol (Oxf) 187: 257–272, 2006. [DOI] [PubMed] [Google Scholar]
- 15.Dmitrieva N, Michea L, Burg MB. p53 tumor suppressor protein protects renal inner medullary cells from hypertonic stress by restricting DNA replication. Am J Physiol Renal Physiol 281: F522–F530, 2001. [DOI] [PubMed] [Google Scholar]
- 16.Dmitrieva NI, Bulavin DV, Fornace AJ, Jr, Burg MB. Rapid activation of G2/M checkpoint after hypertonic stress in renal inner medullary epithelial (IME) cells is protective and requires p38 kinase. Proc Natl Acad Sci USA 99: 184–189, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dmitrieva NI, Cai Q, Burg MB. Cells adapted to high NaCl have many DNA breaks and impaired DNA repair both in cell culture and in vivo. Proc Natl Acad Sci USA 101: 2317–2322, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dmitrieva NI, Celeste A, Nussenzweig A, Burg MB. Ku86 preserves chromatin integrity in cells adapted to high NaCl. Proc Natl Acad Sci USA 102: 10730–10735, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dmitrieva NI, Ferraris JD, Norenburg JL, Burg MB. The saltiness of the sea breaks DNA in marine invertebrates: possible implications for animal evolution. Cell Cycle 5: 1320–1323, 2006. [DOI] [PubMed] [Google Scholar]
- 20.Ducret A, Van O, I, Eng JK, Yates JR, 3rd, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci 7: 706–719, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang D, Hawke D, Zheng Y, Xia Y, Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T, Lu Z. Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J Biol Chem 282: 11221–11229, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas T, Hansen K, Holm K, Lukas J, Bartek J. Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J Biol Chem 277: 26741–26752, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Ferraris JD, Persaud P, Williams CK, Chen Y, Burg MB. cAMP-independent role of PKA in tonicity-induced transactivation of tonicity-responsive enhancer/osmotic response element-binding protein. Proc Natl Acad Sci USA 99: 16800–16805, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallazzini M, Yu MJ, Gunaratne R, Burg MB, Ferraris JD. c-Abl mediates high NaCl-induced phosphorylation and activation of the transcription factor TonEBP/OREBP. FASEB J 24: 4325–4335, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gozdecka M, Breitwieser W. The roles of ATF2 (activating transcription factor 2) in tumorigenesis. Biochem Soc Trans 40: 230–234, 2012. [DOI] [PubMed] [Google Scholar]
- 26.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev 7: 2135–2148, 1993. [DOI] [PubMed] [Google Scholar]
- 27.Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N, Ueno K, Yamamura S, Zaman MS, Khatri G, Chen Y, Saini S, Majid S, Deng G, Ishii N, Dahiya R. Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int J Cancer 128: 1793–1803, 2011. [DOI] [PubMed] [Google Scholar]
- 28.Hoffert JD, Wang G, Pisitkun T, Shen RF, Knepper MA. An automated platform for analysis of phosphoproteomic datasets: application to kidney collecting duct phosphoproteins. J Proteome Res 6: 3501–3508, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev 89: 193–277, 2009. [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 40: D261–D270, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irarrazabal CE, Burg MB, Ward SG, Ferraris JD. Phosphatidylinositol 3-kinase mediates activation of ATM by high NaCl and by ionizing radiation: role in osmoprotective transcriptional regulation. Proc Natl Acad Sci USA 103: 8882–8887, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irarrazabal CE, Williams CK, Ely MA, Birrer MJ, Garcia-Perez A, Burg MB, Ferraris JD. Activator protein-1 contributes to high NaCl-induced increase in tonicity-responsive enhancer/osmotic response element-binding protein transactivating activity. J Biol Chem 283: 2554–2563, 2008. [DOI] [PubMed] [Google Scholar]
- 33.Ishihara N, Jofuku A, Eura Y, Mihara K. Regulation of mitochondrial morphology by membrane potential and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun 301: 891–898, 2003. [DOI] [PubMed] [Google Scholar]
- 34.Itoh T, Yamauchi A, Miyai A, Yokoyama K, Kamada T, Ueda N, Fujiwara Y. Mitogen-activated protein kinase and its activator are regulated by hypertonic stress in Madin-Darby canine kidney cells. J Clin Invest 93: 2387–2392, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonak C, Mildner M, Klosner G, Paulitschke V, Kunstfeld R, Pehamberger H, Tschachler E, Trautinger F. The hsp27kD heat shock protein and p38-MAPK signaling are required for regular epidermal differentiation. J Dermatol Sci 61: 32–37, 2011. [DOI] [PubMed] [Google Scholar]
- 36.Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol 13: 1108–1115, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knapinska AM, Gratacos FM, Krause CD, Hernandez K, Jensen AG, Bradley JJ, Wu X, Pestka S, Brewer G. Chaperone Hsp27 modulates AUF1 proteolysis and AU-rich element-mediated mRNA degradation. Mol Cell Biol 31: 1419–1431, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostenko S, Jensen KL, Moens U. Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell Mol Life Sci 66: 3289–3307, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landry J, Lambert H, Zhou M, Lavoie JN, Hickey E, Weber LA, Anderson CW. Human HSP27 is phosphorylated at serines 78 and 82 by heat shock and mitogen-activated kinases that recognize the same amino acid motif as S6 kinase II. J Biol Chem 267: 794–803, 1992. [PubMed] [Google Scholar]
- 40.Lee EC, Strange K. GCN-2 dependent inhibition of protein synthesis activates osmosensitive gene transcription via WNK and Ste20 kinase signaling. Am J Physiol Cell Physiol 303: C1269–C1277, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SH, Park Y, Yoon SK, Yoon JB. Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. J Biol Chem 285: 41280–41289, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell 15: 5001–5011, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lees JA, Buchkovich KJ, Marshak DR, Anderson CW, Harlow E. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J 10: 4279–4290, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Ferraris JD, Yu D, Singh T, Izumi Y, Wang G, Gucek M, Burg MB. Proteomic analysis of high NaCl-induced changes in abundance of nuclear proteins. Physiol Genomics 44: 1063–1071, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim YS, Lee JS, Huang TQ, Seo JS. Protein kinase Cμ plays an essential role in hypertonicity-induced heat shock protein 70 expression. Exp Mol Med 40: 596–606, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marinissen MJ, Chiariello M, Pallante M, Gutkind JS. A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s, and extracellular signal-regulated kinase 5. Mol Cell Biol 19: 4289–4301, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michea L, Ferguson DR, Peters EM, Andrews PM, Kirby MR, Burg MB. Cell cycle delay and apoptosis are induced by high salt and urea in renal medullary cells. Am J Physiol Renal Physiol 278: F209–F218, 2000. [DOI] [PubMed] [Google Scholar]
- 48.Morley SJ, Naegele S. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J Biol Chem 277: 32855–32859, 2002. [DOI] [PubMed] [Google Scholar]
- 49.Nava P, Koch S, Laukoetter MG, Lee WY, Kolegraff K, Capaldo CT, Beeman N, Addis C, Gerner-Smidt K, Neumaier I, Skerra A, Li L, Parkos CA, Nusrat A. Interferon-γ regulates intestinal epithelial homeostasis through converging β-catenin signaling pathways. Immunity 32: 392–402, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen H, Ramana CV, Bayes J, Stark GR. Roles of phosphatidylinositol 3-kinase in interferon-γ-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression. J Biol Chem 276: 33361–33368, 2001. [DOI] [PubMed] [Google Scholar]
- 51.Niswander JM, Dokas LA. Phosphorylation of HSP27 and synthesis of 14-3-3ε are parallel responses to hyperosmotic stress in the hippocampus. Brain Res 1116: 19–30, 2006. [DOI] [PubMed] [Google Scholar]
- 52.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Parcellier A, Schmitt E, Gurbuxani S, Seigneurin-Berny D, Pance A, Chantome A, Plenchette S, Khochbin S, Solary E, Garrido C. HSP27 is a ubiquitin-binding protein involved in IκBα proteasomal degradation. Mol Cell Biol 23: 5790–5802, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payne SH, Yau M, Smolka MB, Tanner S, Zhou H, Bafna V. Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. J Proteome Res 7: 3373–3381, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rocchi L, Alfieri RR, Petronini PG, Montanaro L, Brigotti M. 5′-Untranslated region of heat shock protein 70 mRNA drives translation under hypertonic conditions. Biochem Biophys Res Commun 431: 321–325, 2013. [DOI] [PubMed] [Google Scholar]
- 56.Rossignol M, Keriel A, Staub A, Egly JM. Kinase activity and phosphorylation of the largest subunit of TFIIF transcription factor. J Biol Chem 274: 22387–22392, 1999. [DOI] [PubMed] [Google Scholar]
- 57.Roth I, Leroy V, Kwon HM, Martin PY, Feraille E, Hasler U. Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-κB activity. Mol Biol Cell 21: 3459–3474, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salomoni P, Bernardi R, Bergmann S, Changou A, Tuttle S, Pandolfi PP. The promyelocytic leukemia protein PML regulates c-Jun function in response to DNA damage. Blood 105: 3686–3690, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett 313: 307–313, 1992. [DOI] [PubMed] [Google Scholar]
- 60.Tait S, Dutta K, Cowburn D, Warwicker J, Doig AJ, McCarthy JE. Local control of a disorder-order transition in 4E-BP1 underpins regulation of translation via eIF4E. Proc Natl Acad Sci USA 107: 17627–17632, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005. [DOI] [PubMed] [Google Scholar]
- 62.Taurin S, Sandbo N, Qin Y, Browning D, Dulin NO. Phosphorylation of β-catenin by cyclic AMP-dependent protein kinase. J Biol Chem 281: 9971–9976, 2006. [DOI] [PubMed] [Google Scholar]
- 63.Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics 40: 167–183, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uddin S, Sassano A, Deb DK, Verma A, Majchrzak B, Rahman A, Malik AB, Fish EN, Platanias LC. Protein kinase Cδ (PKCδ) is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727. J Biol Chem 277: 14408–14416, 2002. [DOI] [PubMed] [Google Scholar]
- 65.Wang D, Jang DJ. Protein kinase CK2 regulates cytoskeletal reorganization during ionizing radiation-induced senescence of human mesenchymal stem cells. Cancer Res 69: 8200–8207, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang G, Wu WW, Pisitkun T, Hoffert JD, Knepper MA, Shen RF. Automated quantification tool for high-throughput proteomics using stable isotope labeling and LC-MSn. Anal Chem 78: 5752–5761, 2006. [DOI] [PubMed] [Google Scholar]
- 67.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8: 25–33, 2005. [DOI] [PubMed] [Google Scholar]
- 68.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 81: 323–330, 1995. [DOI] [PubMed] [Google Scholar]
- 69.Wen Z, Darnell JE., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res 25: 2062–2067, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. WSTF regulates the H2A. X. DNA damage response via a novel tyrosine kinase activity. Nature 457: 57–62, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu B, Bhattacharjee A, Roy B, Xu HM, Anthony D, Frank DA, Feldman GM, Cathcart MK. Interleukin-13 induction of 15-lipoxygenase gene expression requires p38 mitogen-activated protein kinase-mediated serine 727 phosphorylation of Stat1 and Stat3. Mol Cell Biol 23: 3918–3928, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, Tsai CH, Chen HY, Chiang CT, Lai CK, Lu LT, Chen CH, Gu DL, Pu YS, Jou YS, Lu KP, Hsiao PW, Shih HM, Chen RH. A Cullin3-KLHL20 ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 20: 214–228, 2011. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Z, Cohen DM. NaCl but not urea activates p38 and jun kinase in mIMCD3 murine inner medullary cells. Am J Physiol Renal Fluid Electrolyte Physiol 271: F1234–F1238, 1996. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Z, Yang XY, Soltoff SP, Cohen DM. PI3K signaling in the murine kidney inner medullary cell response to urea. Am J Physiol Renal Physiol 278: F155–F164, 2000. [DOI] [PubMed] [Google Scholar]
- 75.Zhao H, Tian W, Cohen DM. Rottlerin inhibits tonicity-dependent expression and action of TonEBP in a PKCδ-independent fashion. Am J Physiol Renal Physiol 282: F710–F717, 2002. [DOI] [PubMed] [Google Scholar]
- 76.Zhou X, Wang H, Burg MB, Ferraris JD. Inhibitory phosphorylation of GSK-3β by AKT, PKA, and PI3K contributes to high NaCl-induced activation of the transcription factor NFAT5 (TonEBP/OREBP). Am J Physiol Renal Physiol 305: F362–F369, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.