Abstract
Context:
Genetics plays a major role in determining an individual's height. Although there are many monogenic disorders that lead to perturbations in growth and result in short stature, there is still no consensus as to the role that genetic diagnostics should play in the evaluation of a child with short stature.
Evidence Acquisition:
A search of PubMed was performed, focusing on the genetic diagnosis of short stature as well as on specific diagnostic subgroups included in this article. Consensus guidelines were reviewed.
Evidence Synthesis:
There are a multitude of rare genetic causes of severe short stature. There is no high-quality evidence to define the optimal approach to the genetic evaluation of short stature. We review genetic etiologies of a number of diagnostic subgroups and propose an algorithm for genetic testing based on these subgroups.
Conclusion:
Advances in genomic technologies are revolutionizing the diagnostic approach to short stature. Endocrinologists must become facile with the use of genetic testing in order to identify the various monogenic disorders that present with short stature.
Short stature is one of the most common reasons for referral to a pediatric endocrinologist. The last two decades have seen tremendous advances in our understanding of the genetic underpinnings of growth. Numerous monogenic causes of growth disorders have been identified. These developments require continuous updating of the endocrinologist's diagnostic approach to children with unexplained short stature.
Consensus guidelines have been assembled focusing on the medical evaluation of children with idiopathic short stature (ISS) (1), those born small for gestational age (SGA) (2), or GH deficiency (GHD) (3). These guidelines rely upon standard physical examination and laboratory parameters that assess organic causes of growth failure, such as renal dysfunction, acid-base disorders, hypothyroidism, celiac disease, or inflammatory disorders. They also propose assessment of the GH axis, either via measurement of GH-dependent factors, such as IGF-1 or IGF binding protein (IGFBP)-3, or through direct measurement of GH levels after a variety of pharmacological stimuli. This standard medical evaluation identifies a pathological cause of short stature in 1–40% (4–10) of individuals, depending on whether the children studied are from a population-based survey or a referral clinic. A recent study found that this standard workup had a diagnostic yield of 1.3% in a population of otherwise healthy children with short stature, mean height of −2.5 SD score (SDS), and a normal growth velocity (10). Ultimately, many patients are characterized with either ISS or isolated GHD, and then a therapeutic decision is made regarding a trial of GH therapy. The differentiation between ISS and idiopathic GHD is often problematic, reflecting the relatively poor discriminatory nature of serum IGF-1 and IGFBP-3 concentrations, as well as the remarkably high false-positive rate observed with GH stimulation tests, especially in the absence of sex-steroid priming (11, 12). Many patients classified with ISS or idiopathic GHD will simply have constitutional delay of growth and puberty, a normal physiological variant that does not require further workup. Genetic testing plays a very small role in the current standard evaluation performed by pediatric endocrinologists, with the exception of assessing females for Turner syndrome and consideration of SHOX (short stature homeobox-containing gene) deficiency or Russell-Silver syndrome (RSS). Recently, some authors have suggested genetic testing algorithms for distinct diagnostic subgroups of patients with short stature (13), but this has not yet permeated routine clinical practice.
In the current article, we will briefly review the current state of knowledge about the genetic underpinnings of height as well as currently available genetic testing modalities. We will then review a number of the prominent classes of growth disorders and the genetic etiologies that should be considered for each class. For this review, a search of PubMed was performed focusing on the genetic diagnosis of short stature using the terms “genetics” and “short stature,” as well as on specific diagnostic subgroups included in this article. References from consensus guidelines and prior comprehensive review articles were reviewed. Appropriate articles were selected for inclusion. We do not attempt to comprehensively review every genetic condition that can lead to short stature, but rather propose a genetic diagnostic framework that encompasses many of these disorders. We strongly believe that genetic evaluation should, and in the near future will, play a much larger role in the evaluation of growth disorders. Pediatric endocrinologists must become facile with modern genetic diagnostics in order to best serve our patients.
Genetic Architecture of Height
Normal variation in adult height is largely due to inherited, genetic factors. Within a population, typically 80% or more of the variation in height is explained by genetic factors (14), although it is clear that environmental factors contribute to differences between populations and to recent increases in average height across generations. The genetic contribution to height is largely attributable to the combined effects of many different genes, meaning that height is typically a polygenic trait (14–16).
Recently, genome-wide association studies have identified hundreds of genetic variants that are common in the population (frequency of approximately 5% or greater) and that each has small effects on height (15). The additive effects of these variants explain about 10% of the variation in adult height, and it is estimated that additional, as yet unidentified common variants will account for another approximately 40% or more of height variation (17). Thus, at least half of the variability in adult height in a population is due to the combined effect of common genetic variants. The source of the remaining genetic contribution to adult height is as yet largely unidentified, but it may arise from variants that are of slightly lower frequency, from much rarer variants, or from genetic interactions among variants.
At the extremes of short stature, by contrast, patients often have mutations in single genes, resulting in large effects on height (for example, achondroplasia; many other causes are discussed below). Thus, some individuals with significant short stature carry variation in a single gene that plays a major role in determining their height. Consistent with a role of rare genetic variation in short stature, the known common variant contributors to height play less of a role in individuals at the short tail of the height distribution (below the 0.5 percentile) (18). Thus, individuals at the low end of the height distribution are short partly due to factors beyond common genetic variation, such as rare variants that could possibly be identifiable by genetic analysis.
It has also been suggested that rare variants of moderate effect could explain many cases of short stature that are less extreme than those seen in many single gene disorders. For example, studies of a family with skeletal dysplasia due to homozygous NPR2 mutations led to a prediction that a small percentage of all individuals with short stature (Z-score <−2) would carry heterozygous mutations in NPR2 that strongly influenced their final height (19). Of note, several studies have also pointed to an excess burden of rare, large (>500 kb) genomic deletions as a contributor to short stature (20). Two recent studies examined the role of rare copy number variants (CNVs) in patients referred for short stature and found pathogenic CNVs in 4–10% (21, 22), indicating that copy number assessment can be an important part of the diagnostic evaluation of patients with short stature. The extent of homozygosity (how much DNA has been inherited identically from both parents) is also a risk factor for short stature, indicating a possible role for rare, recessive mutations (23). In summary, the combined effects of many genes likely explain much of the variation within the normal range of height, but rarer variants in single genes are more likely to play a prominent role as short stature becomes more extreme.
Currently Available Tools for Genetic Diagnostics
Much of the current evaluation for short stature is based on clinical features and laboratory and radiological studies that may be supplemented with an increasing array of genetic tests. These tests evaluate potential etiologies of short statures, including single gene-based tests (where a particular genetic etiology is suspected), tests of panels of genes (for example, to diagnose Noonan syndrome; Ref. 24), and, more recently, exome sequencing to evaluate a comprehensive set of genes across the genome where no specific molecular etiology is strongly suspected. Because most known genetic variants within a given gene that cause short stature are rare and often differ from patient to patient (achondroplasia being a notable exception), most of these tests have sequencing as the major component. However, comparative genomic hybridization can be used for a genome-wide assessment of deletions or duplications that have clinical consequences.
Disorders Presenting with Prenatal Onset Growth Retardation
As part of the evaluation of short stature, it is critical to assess whether growth failure was of prenatal or postnatal onset. Intrauterine growth can be hampered by numerous factors, such as infections, maternal smoking, placental insufficiency, and maternal disease, or it may reflect underlying genetic pathology in the fetus. Most children born SGA have no identifiable etiology at birth. Of those children with idiopathic SGA, approximately 85% will demonstrate catch-up growth to the third percentile for length by 2 years of age (25, 26). The remaining children without catch-up growth require further evaluation, especially the subset with progressive postnatal growth failure.
There are a number of genetic conditions that lead to pre- and postnatal growth retardation. One of the more common causes of SGA with persistent short stature is RSS (27). RSS is characterized by growth retardation of prenatal onset that persists postnatally as proportionate short stature accompanied by a normal head circumference. Other classical features include fifth-finger clinodactyly, body asymmetry often noted as limb-length asymmetry, triangular facies, short arm span, feeding difficulties, and prominent forehead. Children with RSS are at increased risk for developmental delay. RSS is a genetically heterogeneous condition, with the most common etiology being hypomethylation of an imprinting control region on the paternal allele of chromosome 11p15.5 (28). This region controls the methylation of the IGF2 and H19 genes, which are involved in intrauterine growth regulation. The second most common cause of RSS is uniparental disomy of chromosome 7. A recent report identified a single mutation in the paternally imprinted gene CDKN1C in a large family with some characteristics of RSS (29). CDKN1C is a cell cycle regulator also located on chromosome 11p15.5. Missense mutations in a specific domain of this gene cause another condition presenting with SGA, ie, IMAGe syndrome (intrauterine growth restriction, metaphyseal dysplasia, adrenal hypoplasia congenita, and genital anomalies) (30). Another condition that is often confused with RSS is 3-M syndrome (31). Similar to RSS, patients with 3-M syndrome present with SGA, severe postnatal growth retardation, normal head circumference with a prominent forehead, and triangular facies (32). Other characteristic features include a fleshy nose tip, full eyebrows, long philtrum, and prominent mouth and pointed chin, although these facial features become more subtle over time. Intelligence is normal in 3-M syndrome. Radiographic features include gracile long bones, tall vertebral bodies that become foreshortened, small pelvic bones, and a broad thorax with thin ribs. Males with 3-M syndrome have been reported with elevated gonadotropins, small testicular size, and possibly reduced fertility. To date, mutations in three separate genes have been associated with the phenotype of 3-M syndrome: CUL7, OBSL1, and CCDC8. These genes have been shown to interact in the cell, but the exact mechanism leading to short stature is unknown. 3-M syndrome is inherited as a recessive condition and has been found more frequently in consanguineous populations. The prevalence of 3-M syndrome is unknown, but it is likely underdiagnosed (33–35).
In contrast to RSS, 3-M syndrome, and many skeletal dysplasias in which there is sparing of the head size, microcephalic primordial dwarfism constitutes a group of disorders characterized by severe pre- and postnatal growth retardation accompanied by microcephaly (36) (Table 1). These disorders include microcephalic osteodysplastic primordial dwarfism (MOPD) types I and II, Meier-Gorlin syndrome, and the various types of Seckel syndrome. The clinical phenotypes of these disorders range in the severity of growth retardation and microcephaly, as well as in the degree of developmental delay, but there can be significant clinical overlap among syndromes. Seckel syndrome is classically characterized by significant developmental delay, whereas intellect is intact in most cases of MOPD II and Meier-Gorlin syndrome. The genetic bases of many of these disorders have been elucidated recently, and the affected genes have been shown to be necessary for fundamental biological processes, such as DNA replication and damage repair (Table 1).
Table 1.
Microcephalic Primordial Dwarfism Disorders
| Disorder | Clinical Features | Gene | Gene Function |
|---|---|---|---|
| MOPD I (Taybi-Linder syndrome) | Severe brain malformations and microcephaly, short bowed long bones, dry skin, sparse hair, fatal in first few years of life. | RNU4ATAC (86, 87) | Small nuclear RNA that is part of the minor spliceosome and necessary for proper splicing of U12-dependent introns. |
| MOPD II | Preserved intellect, characteristic facial features, small teeth, neurovascular complications (Moyamoya disease), insulin resistance leading to early onset type 2 diabetes. | PCNT (88) | Core centrosomal protein important for mitotic spindle organization. Also interacts with DNA damage response through ATR signaling. |
| Seckel syndrome | Developmental disability ranging in severity, severe microcephaly out of proportion to short stature, characteristic facial features including receding forehead, prominent nose, and micrognathia. | ATR (89), ATRIP (90), CENPJ (91), CEP152 (92), RBBP8 (93), PCNT (94) | Involved in DNA damage response or centrosomal function. |
| Meier-Gorlin syndrome | Small ears, absent or small patellae, preserved intellect (in most individuals), wide range of short stature and microcephaly. | ORC1, ORC4, ORC6, CDT1, CDC6 (95, 96) | Components of the DNA pre-replication complex. |
There are a number of other syndromes associated with intrauterine growth restriction and postnatal short stature, and we refer the reader to the reviews by Wit et al (13) and Seaver and Irons (37) for additional lists of genetic syndromes. Notably, defects in IGF1 and its receptor, IGF1R, can also present with SGA, progressive growth failure, and microcephaly. These will be discussed further below.
Disorders of the GH/IGF-1 Axis
GH plays a critical role in human growth, primarily through its regulation of IGF-1 production. Genetic disorders have been identified throughout the GH/IGF-1 axis, ranging from GHD, either in isolation or as part of multiple pituitary hormone deficiency (MPHD), to primary IGF-1 deficiency, to IGF-1 resistance (38).
Isolated GHD
Although isolated GHD may result from congenital anatomical defects of the pituitary or from pituitary tumors or their treatment, most cases are identified as “idiopathic.” The reported incidence of isolated GHD is 1:4000 to 1:10 000, but given the well-known vagaries involved in the clinical diagnosis of GHD, it is likely that most such cases are children with short stature resulting from a nonpituitary etiology and erroneously diagnosed and labeled as GHD. Of children with true isolated GHD, 3–30% have been reported to be familial; mutations of relevant candidate genes have been identified in 11% of patients with isolated GHD and in 34% of familial cases (39, 40).
Most mutations resulting in isolated GHD involve the genes for GH (GH1) or the GHRH receptor (GHRHR) (Table 2). It is of note that there are no confirmed cases of GHD resulting from mutations of GHRH itself. Heterozygous mutations of HESX1 and SOX3 are rare causes of isolated GHD. Finally, some cases of MPHD (see below) may initially present as isolated GHD and, over time, develop other hormonal deficiencies. A comprehensive review of this topic was recently performed by Alatzoglou et al (41).
Table 2.
Genetic Forms of Isolated GHD
| Type | Gene | Inheritance | Mutations | Phenotype |
|---|---|---|---|---|
| IA | GH1 | AR | Deletions | Severe postnatal growth failure |
| Frameshift | Undetectable serum GH | |||
| Nonsense | Anti-GH antibodies with GH Rx | |||
| IB | GH1, GHRHR | AR | Splice shift | Less severe growth failure than type IA |
| Frameshift | Low, but detectable, serum GH | |||
| Missense | No anti-GH antibodies with GH Rx | |||
| Nonsense | ||||
| II | GH1 | AD | Splice shift | Variability in height deficit |
| Missense | Normal or hypoplastic pituitary | |||
| Splice enhancer | Other pituitary deficits can develop | |||
| Intronic deletions | ||||
| III | BTK, SOX3 | X-linked | Deletions | GHD with agammaglobulinemia; |
| Other genes? | Expansions | GHD with mental retardation | ||
| Other mutations? |
Abbreviations: AR, autosomal recessive; AD, autosomal dominant; Rx, treatment. [Adapted from: K. S. Alatzoglou and M. T. Dattani: Genetic causes and treatment of isolated growth hormone deficiency–an update. Nat Rev Endocrinol. 2010;6:562–576 (97), with permission. © Nature Publishing Group.]
The most severe form of isolated GHD, type IA, is characterized by autosomal recessive transmission and complete absence of detectable serum GH. The most common etiology is homozygosity for GH gene deletions, ranging in size from 6.5–45 kb, but type IA GHD can also result from compound heterozygous frameshift mutations or homozygous nonsense mutations affecting the signal peptide. Most patients develop anti-GH antibodies upon treatment with GH, although this is not a universal finding.
Less severe GHD is observed in type IB GHD, which can result from splice site, frameshift, missense, or nonsense mutations of either GH1 or GHRHR. Serum GH concentrations are typically low, but detectable, and anti-GH antibodies are not observed upon treatment. In contradistinction to types 1A and 1B, type II GHD is transmitted as an autosomal dominant condition. Most of these patients have mutations within the first six nucleotides of intron 3 of GH1, resulting in skipping of exon 3 and production of a 17.5-kDa isoform of GH. This shortened form of GH has a dominant negative effect upon secretion of the full-length GH molecule and may also lead to disordered secretion of other pituitary hormones, such as TSH, LH, and prolactin (PRL). The clinical significance of this observation is not trivial because patients initially diagnosed as isolated GHD may evolve into MPHD. Autosomal dominant transmission of GHD may also be observed with specific missense, splice enhancer, or intronic deletion mutations.
Isolated GHD type III can be seen in association with agammaglobulinemia. Over 600 different mutations of the Bruton agammaglobulinemia tyrosine kinase (BTK) gene have been found to cause agammaglobulinemia, although it remains unclear as to why defects or the absence of this protein cause GHD. Mutations in SOX3, which encodes a transcription factor involved in pituitary development, have been identified in some patients with X-linked GHD (42, 43).
It is of note that recessive and dominant mutations of the gene for the GH secretagogue receptor type I (GHSR), whose natural ligand is ghrelin, have been described in patients with partial, isolated GHD (44). Phenotypes are reportedly variable, however, and mouse models with null deletions of the receptor have not replicated the GHD phenotype.
Multiple pituitary hormone deficiency
The anterior lobe of the pituitary gland, which produces GH, also expresses and secretes five additional hormones: PRL, TSH, FSH, LH, and ACTH. A variety of transcription factors are involved in pituitary ontogeny, differentiation of specific cells within the pituitary, and coordinated expression of specific hormones (Table 3). Defects in these factors lead to variable degrees of MPHD. A full discussion of each syndrome is beyond the scope of this article, but we refer readers to the review article by Pfäffle and Klammt (45) for a more complete discussion.
Table 3.
Established Genetic Defects Resulting in GHD and Potential for MPHD
| Gene | Inheritance | Phenotype |
|---|---|---|
| HESX1 | AR | Septo-optic dysplasia; variable involvement of pituitary hormones |
| PROP1 | AR | GH, PRL, TSH, LH, FSH deficiencies; variable ACTH deficiency |
| POU1F1 (Pit1) | AR, AD | GH, PRL deficiencies; variable degree of TSH deficiency |
| RIEG | AD | Rieger syndrome |
| LHX3 | AR | GH, TSH, LH, FSH, PRL deficiencies |
| LHX4 | AD | GH, TSH, ACTH deficiencies |
| SOX3 | X-linked | GHD, mental retardation |
| GLI2 | AD | Holoprosencephaly, hypopituitarism |
| GLI3 | AD | Pallister-Hall syndrome, hypopituitarism |
| OTX2 | AD | TSH, GH, LH, FSH deficiencies; variable ACTH and PRL deficiency |
| FGF8 | AD | Holoprosencephaly, hypopituitarism |
| FGFR1 | AD | Hypoplasia of pituitary, corpus callosum; ocular defects, hypopituitarism |
| IGSF1 | X-linked | TSH deficiency; variable GH and PRL deficiency; macro-orchidism |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive. [Adapted from P. F. Backeljauw et al: Disorders of growth. In: Sperling MA, ed. Textbook of Pediatric Endocrinology (in press).]
There are several points involved in the characterization of MPHD that should be emphasized:
1) The list shown in Table 3 is constantly expanding, and it is understood that there are probably additional transcription factors involved in pituitary development that will be identified and characterized in coming years. As with isolated GHD, most sporadic cases of MPHD still do not have a recognized gene defect.
2) Many of these factors are not pituitary-specific and, accordingly, are involved in the ontogeny of other organs. The phenotypes characterizing defects of these factors often reflect the variety of organs involved.
3) The time course of pituitary hormone deficiencies can vary greatly from defect to defect and from case to case. In general, TSH and GH deficiencies are seen early, whereas FSH, LH, PRL, and ACTH deficiencies may not be evident until later in life. An early diagnosis of what appears to be isolated GHD or TSH deficiency may, therefore, evolve into a multiple pituitary hormone-deficient state. Molecular evaluation of patients with GHD early in life may help identify patients at risk for evolving pituitary hormone deficiencies. At the very least, such patients require lifelong surveillance, in an effort to avoid late-onset complications and, especially, the development of unrecognized panhypopituitarism.
Primary IGF deficiency (IGFD) and IGF resistance
The earliest identified form of GH insensitivity (GHI) was reported by Laron et al (46) almost 50 years ago and was eventually found to be the consequence of mutations of the gene for the GH receptor (GHR) (47). To date, approximately 300 cases involving defects of GHR have been reported worldwide, with the largest single collection of 99 individuals, one-third of all cases, coming from an inbred population in southern Ecuador (48, 49), all but a few of whom are homozygous for a splicing mutation affecting residue 198 (E198splice; originally reported as E180splice). Multiple mutations and deletions of GHR have now been identified and been shown to result in varying degrees of GHI (50).
As knowledge of GH action expanded, other causes of GHI were identified along the GH-IGF axis. GH action requires binding to a membrane-spanning, homodimeric receptor belonging to the cytokine receptor superfamily. The GHR lacks any intrinsic kinase activity, and consequently, GH action requires recruitment of cytosolic Janus kinase 2 and phosphorylation of several members of the signal transducer and activator of transcription (STAT) family, of which STAT5b appears to be critical for transcriptional regulation of IGF-1 (51, 52). Production of IGF-1 occurs in multiple tissues, and the growth factor may act locally (autocrine/paracrine) or circulate complexed to a family of six high-affinity binding proteins (IGFBP-1 to -6). IGF bound to either IGFBP-3 or IGFBP-5 forms a ternary complex by binding to an acid-labile subunit (ALS), encoded by the gene IGFALS (53); it is in this ternary complex that most IGF-1 and IGF-2 in serum is normally found. The mitogenic and metabolic actions of IGF-1 require binding to a membrane-spanning, homodimeric receptor (IGF1R), resulting in autophosphorylation of the intracellular β-subunit of IGF1R and activation of several intracellular signaling cascades. Molecular defects have been identified at many of the steps described above, with a wide range of clinical and biochemical phenotypes. For detailed descriptions of defects in this pathway, we refer the reader to the review by David et al (38) (Table 4).
Table 4.
Established Genetic Defects Resulting in Primary IGFD or IGF Resistance
| GHR | STAT5B | IGF1 | IGFALS | IGF1R | Bioinactive GH | |
|---|---|---|---|---|---|---|
| Postnatal growth failure | +++ | +++ | +++ | +/− | ++ | ++ |
| IUGR | +/− | +/− | ++ | +/− | ++ | +/− |
| Midfacial hypoplasia | ++ | ++ | − | − | − | +/− |
| Other facial dysmorphism | − | − | ++ | − | +/− | − |
| Microcephaly | − | − | ++ | − | +/− | − |
| Deafness | − | − | +/− | − | − | − |
| Mental retardation | − | − | ++ | − | +/− | − |
| Immunodeficiency | − | ++ | − | − | − | − |
| Serum GH | ↑ | ↑ | ↑ | nl | ↑/nl | ↑ |
| Serum GHBP | ↓/nl/↑ | nl | nl | nl | nl | nl |
| Serum IGF-1 | ↓↓ | ↓↓ | ↑/nl/↓ | ↓↓ | ↑/nl | ↓↓ |
| Serum IGFBP-3 | ↓↓ | ↓↓ | ↑/nl | ↓↓ | ↑/nl | ↓↓ |
| Serum ALS | ↓↓ | ↓↓ | nl | ↓↓↓ | nl | ↓↓ |
| Serum PRL | nl | ↑ | nl | nl | nl | nl |
Abbreviations: IGFALS, IGF-1 acid labile subunit gene; IGF1R, IGF-1 receptor gene; IUGR, intrauterine growth retardation; GHBP, GH binding protein; nl, normal. [Adapted from P. F. Backeljauw et al: Disorders of growth. In: Sperling MA, ed. Textbook of Pediatric Endocrinology (in press).] ++, present; −, absent; +/−, variably present; arrow pointing up, increased; arrow pointing down, decreased (the number of arrows indicates the degree of deviation).
Defects in the GH/IGF-1 axis upstream of IGF1R are characterized by low serum concentrations of IGF-1 and various degrees of growth retardation. Despite this overlap, however, differences in the clinical and biochemical characteristics of these conditions often allow the clinician to focus on specific molecular defects. These variations reflect several key points: 1) production of IGF-1, IGFBP-3, and ALS is largely GH dependent; and 2) IGF-1 production in utero is essentially GH independent. Accordingly, disturbances in GH action, such as molecular defects of GHR or STAT5b, are characterized by normal intrauterine growth but varying degrees of postnatal growth failure, usually manifest in infancy or early childhood. Defects involving IGF1 or IGF1R, on the other hand, display both intrauterine growth failure and poor growth postnatally (54, 55). Because IGF-1 also appears to be involved in central nervous system development in utero, the latter defects are often characterized by some degree of microcephaly, developmental delay, and/or hearing deficiency (54).
All of these molecular defects are transmitted in an autosomal recessive manner. There are, however, some important exceptions to this rule:
1) Virtually all reported cases of IGF1R defects are heterozygotes, presumably reflecting haploinsufficiency. In mouse studies, total knockout of igf1r results in early death, and presumably, this would be true in humans as well. The several reported cases of compound heterozygosity or homozygosity for IGF1R defects in humans tend to have more severe phenotypes than observed in heterozygotes and, presumably, reflect partial defects of IGF1R action.
2) Dominant negative mutations of GHR have been reported, usually involving molecular defects impacting the intracellular domains of the receptor. Phenotypes are somewhat milder than observed in homozygous or compound heterozygous cases.
3) Family studies of patients with mutations of IGF1, IGFALS, or STAT5b suggest that individuals carrying one mutant allele may have mild phenotypic effects, with heights often still within the normal range but significantly lower than family members who are homozygous for wild-type alleles.
A comprehensive database of genetic defects resulting in primary IGFD and IGF resistance can be found at www.growthgenetics.com.
Skeletal Dysplasias
There are over 400 different conditions listed in the most recent nosology of genetic skeletal disorders (56), many of which present with short stature. These conditions are due to molecular defects in hundreds of different genes, with additional genetic causes of skeletal dysplasia being discovered at a rapid pace. A comprehensive review of all of these conditions is beyond the scope of this article, but we will focus on a number of conditions that are most commonly considered in the pediatric endocrine clinic. Proper diagnosis of children with skeletal dysplasias requires close collaboration between the endocrinologist and a geneticist and a radiologist with expertise in skeletal disorders. Clinical identification of a distinct skeletal dysplasia can help direct genetic testing to target a single implicated gene. Recommendations vary as to the extent of radiological assessment that is necessary for a child with short stature, but the general consensus is that a series of skeletal radiographs should be performed in children with disproportionate short stature (57, 58). This series typically includes radiographs of the skull, spine, thorax, pelvis, upper and lower limbs, and hand. It is important to note that whereas some skeletal dysplasias are readily identifiable clinically, clinical findings may be subtle in others, especially in young children.
The SHOX gene, first described in 1997 by Rao et al (59), has been implicated as an important cause of short stature. SHOX is located on the pseudoautosomal region of the X-chromosome and acts as a transcriptional activator. Defects in SHOX lead to a wide phenotypic spectrum, with homozygous loss-of-function mutations in SHOX causing the severe Langer mesomelic dysplasia, whereas heterozygous defects can be found in patients with Leri-Weill dyschondrosteosis (LWD) or ISS. LWD is marked by short stature, mesomelia, and the classic Madelung deformity of the wrist. The disproportionate limb shortening and Madelung deformity often become more evident after puberty and, thus, can be missed in young children. Examination of the child's parents is essential and may reveal a Madelung deformity in an affected parent.
SHOX defects are found in most patients with LWD, but there is significant variability in the reported prevalence of SHOX defects in patients with ISS, with estimates ranging from approximately 2–17% (60–65). This wide range is attributable to variable subject inclusion criteria, as well as the diverse array of methods used for detection of SHOX defects. Most defects are due to whole gene deletions, but sequence variants, intragenic deletions, and deletions and duplications of the downstream enhancer region have also been found in a significant minority of patients (61, 62, 66–68). A comprehensive database listing SHOX variants can be found at http://grenada.lumc.nl/LOVD2/MR/home.php?select_db=SHOX. A number of investigators have created clinical prediction rules to select ISS patients who have a higher probability of having a SHOX defect. These include the extremities-to-trunk ratio (60), the seated height-to-standing height ratio (63), or a more complex scoring system involving multiple anthropometric measurements and dysmorphic features (64). None of these criteria has a high positive predictive value, and clinical utility of these systems is limited by the highly variable clinical presentation of SHOX deficiency.
Although not typically classified as a skeletal dysplasia, Turner syndrome is a relatively common cause of short stature, with some of the short stature likely attributable to SHOX haploinsufficiency. Most experts suggest routine karyotype analysis in all females with unexplained short stature. In a cohort of 972 children referred to a genetics clinic who had heights below −2 SD, 2.8% of males and 9.8% of females had chromosomal anomalies detected by karyotype, with 7.7% of the females having Turner syndrome (69). Males with mosaic 45X/46XY karyotypes with isolated short stature and an otherwise normal male phenotype have been reported (70). The prevalence of this disorder in a well-characterized cohort of children with ISS is not known.
Another condition with a closely overlapping phenotype to SHOX deficiency is hypochondroplasia, due to defects in FGFR3. There are no consensus diagnostic criteria for hypochondroplasia, but the classic phenotype is marked by short stature, mesomelic limb shortening, limitation of elbow extension, brachydactyly, relative macrocephaly, and generalized laxity. The most common radiological features include shortening of the long bones with mild metaphyseal flaring, narrowing of the inferior lumbar interpedicular distances, and shortening of the lumbar pedicles (71–73). The specific Asn540Lys missense variant is found in up to 70% of patients with hypochondroplasia (71), but this prevalence will vary depending on the diagnostic criteria used (72, 73). Hypochondroplasia is likely underdiagnosed, and its prevalence in a clinically ascertained ISS population is unknown.
It is important to note that in addition to the classic presentation of severe recessive skeletal dysplasias, it is possible that heterozygous carriers of damaging variants in a skeletal dysplasia gene may present with apparent ISS (19). A recent report by Vasques et al (74) describes three patients with ISS who carried likely pathogenic heterozygous variants in NPR2, the gene responsible for the severe recessive acromesomelic dysplasia, type Maroteaux. This represented 6% of the patients in their ISS cohort. A second report in a Japanese cohort of 101 patients with short stature identified two heterozygous pathogenic variants in NPR2 for a yield of 2% (75). Additionally, patients with heterozygous defects in ACAN, encoding the cartilage matrix proteoglycan aggrecan, have been reported with short stature, advanced bone age, and premature growth cessation (76). Further work is needed to explore the role that heterozygous variants in NPR2 and other recessive skeletal dysplasia genes play in causing ISS.
Idiopathic Short Stature
ISS represents a clinically heterogeneous group of patients, and thus there are likely a multitude of molecular etiologies for these patients' short stature. In addition to genetic etiologies, patients may present with short stature due to other underlying medical illnesses or environmental influences. We will only focus on patients with a predominantly genetic etiology for their short stature. Many of these patients likely have polygenic short stature and have inherited many common genetic variants of small effect size that, in combination, explain their short stature. However, a subset of these patients is likely to have short stature caused by single genetic variants with large effects, essentially representing undiagnosed monogenic forms of short stature. The genetic etiology may not be clinically apparent because the underlying monogenic cause is quite rare and lacks distinctive features, or the patient may represent a milder phenotypic spectrum of the disorder, without all of the characteristic features. For example, in a recent study using a large-scale, candidate-sequencing approach in patients with undiagnosed short stature, multiple patients with mutations in PTPN11, the gene associated with Noonan syndrome, were identified (77). Noonan syndrome is known to have a very wide phenotypic spectrum (78), and it is unclear how many patients categorized as ISS represent the mild end of this spectrum. Noonan syndrome is typically characterized by short stature, congenital heart defects, variable developmental delay, a broad or webbed neck, cryptorchidism, and distinct facial features. It is caused by defects in a number of genes in the RAS signaling pathway. Candidate gene studies in cohorts of patients with ISS have yielded a low rate of convincingly pathogenic variants in known genes (79), with the possible exception of heterozygous variants in STAT5b in patients with severe short stature and low IGF-1 levels (80). Next-generation sequencing studies of patients with ISS have been undertaken (77, 81), but identifying novel rare genetic causes of short stature is quite challenging and will require large sample sizes. We recently reported the results of exome sequencing in 14 subjects with severe short stature (<−3 SDS), many of whom had additional dysmorphic features (35). Exome sequencing led to the diagnosis of a monogenic disorder in five of the 14 individuals, including two cases of 3-M syndrome.
Why Is Genetic Testing Important for Patients With Short Stature?
Given the current state of knowledge, the purpose of genetic testing in short stature is to identify monogenic causes of short stature due to rare genetic variants with large effects. Identification of polygenic short stature in an individual patient is not currently feasible, nor is it clinically useful. Based on our experience (35), we strongly believe that monogenic causes of short stature are underdiagnosed in the pediatric endocrine clinic. Identification of rare monogenic causes is critical for a variety of reasons. First, identification of a molecular etiology can end the diagnostic workup for the patient and provide the family with an answer as to why their child is not growing normally. Second, the genetic diagnosis may alert the clinician to other medical comorbidities for which the patient is at risk. For example, a male patient with 3-M syndrome will need to be monitored for the development of hypogonadism. Thirdly, determination of a molecular etiology is invaluable for genetic counseling. Finally, the genetic etiology may have implications for therapy. A recent report by Renes et al (82) of two patients with undiagnosed Bloom syndrome provides one such example. The patients presented with SGA without a clear etiology, despite an extensive evaluation. The patients were both treated with GH for multiple years until their clinical presentation evolved and Bloom syndrome was diagnosed. GH was discontinued because it is contraindicated in patients with chromosomal breakage syndromes. Exome sequencing would have readily established this diagnosis.
Who Should Undergo Genetic Testing for Short Stature?
As with all diagnostic testing, the yield of genetic testing will vary significantly based on which patients are selected for testing. Currently, there is no good evidence to support definitive selection criteria for patients who would benefit from genetic testing. Therefore, we propose genetic testing for those individuals for whom the clinician has a high degree of suspicion for underlying genetic alteration at a single locus that is playing a major role in their short stature. Patients suspected of constitutional delay of growth and puberty with a normal predicted adult height should not undergo genetic testing unless other features lead to a strong suspicion of a monogenic disorder. The yield of genetic testing in these patients is likely to be quite low. Factors that may increase the likelihood of a monogenic etiology are shown in Box 1. These factors have not been rigorously validated as predictors yielding a molecular diagnosis, but in our experience, they increase the likelihood of finding a genetic diagnosis. In the absence of one of these features, we suggest consideration of genetic testing in all individuals with heights below −3 SDS, in those below −2.5 SDS with one or more of the additional factors listed above, or in those whose predicted height is more than 2 SD below their midparental target height. In patients who have a single parent with significant short stature, a dominantly inherited rare genetic variant may explain the short stature of both the patient and the parent. If both parents have moderate short stature and the child's short stature is within 2 SD of the midparental target height, it is more likely that the patient has polygenic short stature as opposed to an identifiable monogenic condition. As an admittedly somewhat arbitrary cutoff, we suggest limiting genetic testing in individuals with two short parents to those whose heights are below −3 SDS, unless additional features suggest an underlying genetic etiology.
Box 1.
Factors That Increase the Likelihood for a Monogenic Cause of Short Stature
| Severe GHD |
| MPHD |
| Unequivocal GHI |
| SGA without catch-up growth |
| Additional congenital anomalies or dysmorphic features |
| Evidence of a skeletal dysplasia |
| Associated intellectual disability |
| Microcephaly |
| Height below −3 SD |
A Proposed Diagnostic Testing Algorithm to Identify Major Genetic Causes of Short Stature
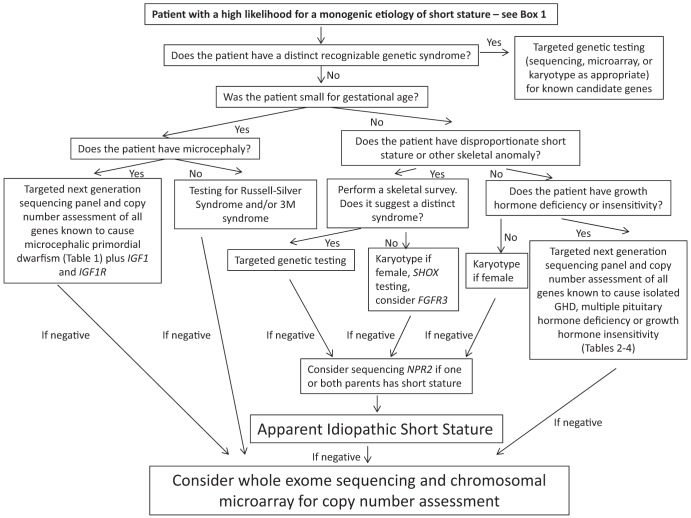
With the advent of next-generation sequencing technologies, it is now possible to rapidly assess a wide array of genes at significantly lower costs. Our diagnostic algorithm (Figure 1) proposes a comprehensive genetic diagnostic approach for a wide array of clinical presentations. The algorithm differentiates patients who were born SGA from those with isolated postnatal short stature. Targeted evaluation of a single gene or panels of genes is recommended for subgroups of patients who fall into the distinct diagnostic categories discussed above. For those patients who do not fit into a distinct subgroup or for whom initial genetic testing is inconclusive, we recommend consideration of genome-wide evaluation through exome sequencing and chromosomal microarray to detect both sequence variants and CNVs. Algorithms have been developed for calling CNVs from exome sequencing data (83, 84), but these have not yet permeated the clinical laboratory. In the future, a single test will likely be available to assess both sequence variants and CNVs on a genome-wide scale. It is important to note that exome sequencing will not identify epigenetic modifications, such as the hypomethylation seen in RSS, and will miss regulatory and intronic variants that may be pathogenic.
Figure 1.
Diagnostic algorithm for the genetic diagnosis of short stature.
A genetic evaluation is not meant to replace a careful medical history and physical examination. Targeted biochemical testing is clearly warranted for select cases, and genetic testing should not replace a reasoned diagnostic approach. Recommendations regarding the biochemical workup of patients with short stature have been addressed by the aforementioned consensus guidelines and are not considered in our algorithm. We acknowledge that some of the testing strategy that we propose is not yet widely available in clinical practice, but we envision that this kind of diagnostic algorithm will become the standard approach for short stature in the coming years. Additionally, this strategy has not yet been validated in clinical trials, and additional research is needed to assess the diagnostic yield of this approach as well as its cost effectiveness. Finally, this strategy will identify many novel genetic variants of unknown clinical significance as well as incidental findings with potential health implications. Recent guidelines have recommended reporting of incidental findings in a subset of genes that may lead to substantial morbidity (85). Rigorous functional studies must be performed to assess the pathogenicity of novel variants. Many variants proposed as pathogenic in the literature are in fact benign rare polymorphisms. Well-curated databases with detailed genotypic and phenotypic information are necessary to assist the clinician in interpreting genetic testing results. One such database that is currently under development is the National Center for Biotechnology Information ClinVar database (www.ncbi.nlm.nih.gov/clinvar/). Before ordering exome sequencing or a chromosomal microarray, a practitioner should become familiar with the benefits and limitations of these techniques, especially with the difficulty in interpreting novel variants of unknown significance as well as the possibility of identifying incidental findings. Often, the optimal care model will include referral to an individual with expertise in these areas. Given the multiple difficulties with interpretation noted above as well as the unknown diagnostic yield in this setting, some individuals prefer that exome sequencing be performed solely in the context of an approved research protocol for the indication of short stature.
Conclusion
Growth is a complex process that is influenced by a multitude of genetic factors both pre- and postnatally. The traditional endocrine evaluation of patients with short stature often does not reveal a definitive etiology. Recent advances in genetic diagnostics coupled with the knowledge gained by those advances are revolutionizing the clinician's ability to obtain a molecular diagnosis for patients with growth disorders. We propose a broad-based genetic approach to the diagnosis of short stature in selected patients who are more likely to have a single, major genetic contributor to their short stature. Additional research is needed to validate this approach and to understand the clinical impact of the numerous genetic variants that are and will continue to be discovered in patients with short stature.
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development at the National Institutes of Health (Grant 1K23HD073351 to A.D.).
Disclosure Summary: J.N.H. received grant support from Pfizer Inc (2011 to present). The remaining authors have no conflicts to report.
Footnotes
- ALS
- acid-labile subunit
- CNV
- copy number variant
- GHD
- GH deficiency
- GHI
- GH insensitivity
- GHR
- GH receptor
- GHRHR
- GHRH receptor
- IGFBP
- IGF binding protein
- IGFD
- IGF deficiency
- ISS
- idiopathic short stature
- LWD
- Leri-Weill dyschondrosteosis
- MOPD
- microcephalic osteodysplastic primordial dwarfism
- MPHD
- multiple pituitary hormone deficiency
- PRL
- prolactin
- RSS
- Russell-Silver syndrome
- SDS
- SD score
- SGA
- small for gestational age
- SHOX
- short stature homeobox
- STAT
- signal transducer and activator of transcription.
References
- 1. Cohen P, Rogol AD, Deal CL, et al. Consensus statement on the diagnosis and treatment of children with idiopathic short stature: a summary of the Growth Hormone Research Society, the Lawson Wilkins Pediatric Endocrine Society, and the European Society for Paediatric Endocrinology Workshop. J Clin Endocrinol Metab. 2008;93:4210–4217 [DOI] [PubMed] [Google Scholar]
- 2. Clayton PE, Cianfarani S, Czernichow P, Johannsson G, Rapaport R, Rogol A. Management of the child born small for gestational age through to adulthood: a consensus statement of the International Societies of Pediatric Endocrinology and the Growth Hormone Research Society. J Clin Endocrinol Metab. 2007;92:804–810 [DOI] [PubMed] [Google Scholar]
- 3. Growth Hormone Research Society. Consensus guidelines for the diagnosis and treatment of growth hormone (GH) deficiency in childhood and adolescence: summary statement of the GH Research Society. GH Research Society. J Clin Endocrinol Metab. 2000;85:3990–3993 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed ML, Allen AD, Sharma A, Macfarlane JA, Dunger DB. Evaluation of a district growth screening programme: the Oxford Growth Study. Arch Dis Child. 1993;69:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grote FK, Oostdijk W, De Muinck Keizer-Schrama SM, et al. The diagnostic work up of growth failure in secondary health care; an evaluation of consensus guidelines. BMC Pediatr. 2008;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voss LD, Mulligan J, Betts PR, Wilkin TJ. Poor growth in school entrants as an index of organic disease: the Wessex growth study. BMJ. 1992;305:1400–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Green AA, MacFarlane JA. Method for the earlier recognition of abnormal stature. Arch Dis Child. 1983;58:535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grimberg A, Kutikov JK, Cucchiara AJ. Sex differences in patients referred for evaluation of poor growth. J Pediatr. 2005;146:212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindsay R, Feldkamp M, Harris D, Robertson J, Rallison M. Utah Growth Study: growth standards and the prevalence of growth hormone deficiency. J Pediatr. 1994;125:29–35 [DOI] [PubMed] [Google Scholar]
- 10. Sisley S, Trujillo MV, Khoury J, Backeljauw P. Low incidence of pathology detection and high cost of screening in the evaluation of asymptomatic short children. J Pediatr. 2013;163:1045–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tauber M, Moulin P, Pienkowski C, Jouret B, Rochiccioli P. Growth hormone (GH) retesting and auxological data in 131 GH-deficient patients after completion of treatment. J Clin Endocrinol Metab. 1997;82:352–356 [DOI] [PubMed] [Google Scholar]
- 12. Marin G, Domené HM, Barnes KM, Blackwell BJ, Cassorla FG, Cutler GB., Jr The effects of estrogen priming and puberty on the growth hormone response to standardized treadmill exercise and arginine-insulin in normal girls and boys. J Clin Endocrinol Metab. 1994;79:537–541 [DOI] [PubMed] [Google Scholar]
- 13. Wit JM, Kiess W, Mullis P. Genetic evaluation of short stature. Best Pract Res Clin Endocrinol Metab. 2011;25:1–17 [DOI] [PubMed] [Google Scholar]
- 14. Hirschhorn JN, Lettre G. Progress in genome-wide association studies of human height. Horm Res. 2009;71(suppl 2):5–13 [DOI] [PubMed] [Google Scholar]
- 15. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hemani G, Yang J, Vinkhuyzen A, et al. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am J Hum Genet. 2013;93:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan Y, Holmen OL, Dauber A, et al. Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals. PLoS Genet. 2011;7:e1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olney RC, Bükülmez H, Bartels CF, et al. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) are associated with short stature. J Clin Endocrinol Metab. 2006;91:1229–1232 [DOI] [PubMed] [Google Scholar]
- 20. Dauber A, Yu Y, Turchin MC, et al. Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet. 2011;89:751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Duyvenvoorde HA, Lui JC, Kant SG, et al. Copy number variants in patients with short stature. Eur J Hum Genet. 2014;22:602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zahnleiter D, Uebe S, Ekici AB, et al. Rare copy number variants are a common cause of short stature. PLoS Genet. 2013;9:e1003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McQuillan R, Eklund N, Pirastu N, et al. Evidence of inbreeding depression on human height. PLoS Genet. 2012;8:e1002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lepri FR, Scavelli R, Digilio MC, et al. Diagnosis of Noonan syndrome and related disorders using target next generation sequencing. BMC Med Genet. 2014;15:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hokken-Koelega AC, De Ridder MA, Lemmen RJ, Den Hartog H, De Muinck Keizer-Schrama SM, Drop SL. Children born small for gestational age: do they catch up? Pediatr Res. 1995;38:267–271 [DOI] [PubMed] [Google Scholar]
- 26. Karlberg J, Albertsson-Wikland K. Growth in full-term small-for-gestational-age infants: from birth to final height. Pediatr Res. 1995;38:733–739 [DOI] [PubMed] [Google Scholar]
- 27. Saal HM. Russell-Silver syndrome. GeneReviews [Internet] In: Pagon RA, Adam MP, Ardinger HH, et al., eds. Seattle, WA: University of Washington, Seattle; http://www.ncbi.nlm.nih.gov/books/NBK1324/ Published November 2, 2002. Updated June 2, 2011 [Google Scholar]
- 28. Netchine I, Rossignol S, Dufourg MN, et al. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab. 2007;92:3148–3154 [DOI] [PubMed] [Google Scholar]
- 29. Brioude F, Oliver-Petit I, Blaise A, et al. CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell Silver syndrome. J Med Genet. 2013;50:823–830 [DOI] [PubMed] [Google Scholar]
- 30. Arboleda VA, Lee H, Parnaik R, et al. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat Genet. 2012;44:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akawi NA, Ali BR, Hamamy H, Al-Hadidy A, Al-Gazali L. Is autosomal recessive Silver-Russell syndrome a separate entity or is it part of the 3-M syndrome spectrum? Am J Med Genet A. 2011;155A:1236–1245 [DOI] [PubMed] [Google Scholar]
- 32. Clayton PE, Hanson D, Magee L, et al. Exploring the spectrum of 3-M syndrome, a primordial short stature disorder of disrupted ubiquitination. Clin Endocrinol (Oxf). 2012;77:335–342 [DOI] [PubMed] [Google Scholar]
- 33. Al-Dosari MS, Al-Shammari M, Shaheen R, et al. 3M syndrome: an easily recognizable yet underdiagnosed cause of proportionate short stature. J Pediatr. 2012;161:139–145.e1 [DOI] [PubMed] [Google Scholar]
- 34. Dauber A, Stoler J, Hechter E, Safer J, Hirschhorn JN. Whole exome sequencing reveals a novel mutation in CUL7 in a patient with an undiagnosed growth disorder. J Pediatr. 2013;162:202–204.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo MH, Shen Y, Walvoord EC, et al. Whole exome sequencing to identify genetic causes of short stature [published online June 20, 2014]. Horm Res Paediatr. doi:10.1159/000360857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seaver LH, Irons M. ACMG practice guideline: genetic evaluation of short stature. Genet Med. 2009;11:465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. David A, Hwa V, Metherell LA, et al. Evidence for a continuum of genetic, phenotypic, and biochemical abnormalities in children with growth hormone insensitivity. Endocr Rev. 2011;32:472–497 [DOI] [PubMed] [Google Scholar]
- 39. Mullis PE. Genetic control of growth. Eur J Endocrinol. 2005;152:11–31 [DOI] [PubMed] [Google Scholar]
- 40. Alatzoglou KS, Turton JP, Kelberman D, et al. Expanding the spectrum of mutations in GH1 and GHRHR: genetic screening in a large cohort of patients with congenital isolated growth hormone deficiency. J Clin Endocrinol Metab. 2009;94:3191–3199 [DOI] [PubMed] [Google Scholar]
- 41. Alatzoglou KS, Webb EA, Le Tissier P, Dattani MT. Isolated growth hormone deficiency (Ghd) in childhood and adolescence: recent advances. Endocr Rev. 2014;35:376–432 [DOI] [PubMed] [Google Scholar]
- 42. Laumonnier F, Ronce N, Hamel BC, et al. Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet. 2002;71:1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burkitt Wright EM, Perveen R, Clayton PE, et al. X-linked isolated growth hormone deficiency: expanding the phenotypic spectrum of SOX3 polyalanine tract expansions. Clin Dysmorphol. 2009;18:218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pantel J, Legendre M, Nivot S, et al. Recessive isolated growth hormone deficiency and mutations in the ghrelin receptor. J Clin Endocrinol Metab. 2009;94:4334–4341 [DOI] [PubMed] [Google Scholar]
- 45. Pfäffle R, Klammt J. Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract Res Clin Endocrinol Metab. 2011;25:43–60 [DOI] [PubMed] [Google Scholar]
- 46. Laron Z, Pertzelan A, Mannheimer S. Genetic pituitary dwarfism with high serum concentration of growth hormone–a new inborn error of metabolism? Isr J Med Sci. 1966;2:152–155 [PubMed] [Google Scholar]
- 47. Amselem S, Duquesnoy P, Attree O, et al. Laron dwarfism and mutations of the growth hormone-receptor gene. N Engl J Med. 1989;321:989–995 [DOI] [PubMed] [Google Scholar]
- 48. Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3:70ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guevara-Aguirre J, Rosenbloom AL, Guevara-Aguirre M, et al. Effects of heterozygosity for the E180 splice mutation causing growth hormone receptor deficiency in Ecuador on IGF-I, IGFBP-3, and stature. Growth Horm IGF Res. 2007;17:261–264 [DOI] [PubMed] [Google Scholar]
- 50. Rosenfeld RG, Rosenbloom AL, Guevara-Aguirre J. Growth hormone (GH) insensitivity due to primary GH receptor deficiency. Endocr Rev. 1994;15:369–390 [DOI] [PubMed] [Google Scholar]
- 51. Kofoed EM, Hwa V, Little B, et al. Growth hormone insensitivity associated with a STAT5b mutation. N Engl J Med. 2003;349:1139–1147 [DOI] [PubMed] [Google Scholar]
- 52. Hwa V, Nadeau K, Wit JM, Rosenfeld RG. STAT5b deficiency: lessons from STAT5b gene mutations. Best Pract Res Clin Endocrinol Metab. 2011;25:61–75 [DOI] [PubMed] [Google Scholar]
- 53. Domené HM, Hwa V, Argente J, et al. Human acid-labile subunit deficiency: clinical, endocrine and metabolic consequences. Horm Res. 2009;72:129–141 [DOI] [PubMed] [Google Scholar]
- 54. Woods KA, Camacho-Hübner C, Savage MO, Clark AJ. Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N Engl J Med. 1996;335:1363–1367 [DOI] [PubMed] [Google Scholar]
- 55. Abuzzahab MJ, Schneider A, Goddard A, et al. IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N Engl J Med. 2003;349:2211–2222 [DOI] [PubMed] [Google Scholar]
- 56. Warman ML, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kant SG, Grote F, de Ru MH, et al. Radiographic evaluation of children with growth disorders. Horm Res. 2007;68:310–315 [DOI] [PubMed] [Google Scholar]
- 58. Veeramani AK, Higgins P, Butler S, et al. Diagnostic use of skeletal survey in suspected skeletal dysplasia. J Clin Res Pediatr Endocrinol. 2009;1:270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rao E, Weiss B, Fukami M, et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 1997;16:54–63 [DOI] [PubMed] [Google Scholar]
- 60. Binder G, Ranke MB, Martin DD. Auxology is a valuable instrument for the clinical diagnosis of SHOX haploinsufficiency in school-age children with unexplained short stature. J Clin Endocrinol Metab. 2003;88:4891–4896 [DOI] [PubMed] [Google Scholar]
- 61. Chen J, Wildhardt G, Zhong Z, et al. Enhancer deletions of the SHOX gene as a frequent cause of short stature: the essential role of a 250 kb downstream regulatory domain. J Med Genet. 2009;46:834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huber C, Rosilio M, Munnich A, Cormier-Daire V. High incidence of SHOX anomalies in individuals with short stature. J Med Genet. 2006;43:735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jorge AA, Souza SC, Nishi MY, et al. SHOX mutations in idiopathic short stature and Leri-Weill dyschondrosteosis: frequency and phenotypic variability. Clin Endocrinol (Oxf). 2007;66:130–135 [DOI] [PubMed] [Google Scholar]
- 64. Rappold G, Blum WF, Shavrikova EP, et al. Genotypes and phenotypes in children with short stature: clinical indicators of SHOX haploinsufficiency. J Med Genet. 2007;44:306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rappold GA, Fukami M, Niesler B, et al. Deletions of the homeobox gene SHOX (short stature homeobox) are an important cause of growth failure in children with short stature. J Clin Endocrinol Metab. 2002;87:1402–1406 [DOI] [PubMed] [Google Scholar]
- 66. Benito-Sanz S, Barroso E, Heine-Suñer D, et al. Clinical and molecular evaluation of SHOX/PAR1 duplications in Leri-Weill dyschondrosteosis (LWD) and idiopathic short stature (ISS). J Clin Endocrinol Metab. 2011;96:E404–E412 [DOI] [PubMed] [Google Scholar]
- 67. Benito-Sanz S, Royo JL, Barroso E, et al. Identification of the first recurrent PAR1 deletion in Léri-Weill dyschondrosteosis and idiopathic short stature reveals the presence of a novel SHOX enhancer. J Med Genet. 2012;49:442–450 [DOI] [PubMed] [Google Scholar]
- 68. Rosilio M, Huber-Lequesne C, Sapin H, Carel JC, Blum WF, Cormier-Daire V. Genotypes and phenotypes of children with SHOX deficiency in France. J Clin Endocrinol Metab. 2012;97:E1257–E1265 [DOI] [PubMed] [Google Scholar]
- 69. Moreno-García M, Fernández-Martínez FJ, Barreiro Miranda E. Chromosomal anomalies in patients with short stature. Pediatr Int. 2005;47:546–549 [DOI] [PubMed] [Google Scholar]
- 70. Richter-Unruh A, Knauer-Fischer S, Kaspers S, Albrecht B, Gillessen-Kaesbach G, Hauffa BP. Short stature in children with an apparently normal male phenotype can be caused by 45,X/46,XY mosaicism and is susceptible to growth hormone treatment. Eur J Pediatr. 2004;163:251–256 [DOI] [PubMed] [Google Scholar]
- 71. Bober MB, Bellus GA, Nikkel SM, et al. Hypochondroplasia. GeneReviews [Internet] In: Pagon RA, Adam MP, Ardinger HH, et al., eds. Seattle, WA: University of Washington, Seattle; http://www.ncbi.nlm.nih.gov/books/NBK1477/. Published July 15, 1999. Updated September 26, 2013 [Google Scholar]
- 72. Song SH, Balce GC, Agashe MV, et al. New proposed clinico-radiologic and molecular criteria in hypochondroplasia: FGFR 3 gene mutations are not the only cause of hypochondroplasia. Am J Med Genet A. 2012;158A:2456–2462 [DOI] [PubMed] [Google Scholar]
- 73. Prinster C, Carrera P, Del Maschio M, et al. Comparison of clinical-radiological and molecular findings in hypochondroplasia. Am J Med Genet. 1998;75:109–112 [DOI] [PubMed] [Google Scholar]
- 74. Vasques GA, Amano N, Docko AJ, et al. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) gene as a cause of short stature in patients initially classified as idiopathic short stature. J Clin Endocrinol Metab. 2013;98:E1636–E1644 [DOI] [PubMed] [Google Scholar]
- 75. Amano N, Mukai T, Ito Y, et al. Identification and functional characterization of two novel NPR2 mutations in Japanese patients with short stature. J Clin Endocrinol Metab. 2014;99:E713–E718 [DOI] [PubMed] [Google Scholar]
- 76. Nilsson O, Guo MH, Dunbar N, et al. Short stature, accelerated bone maturation, and early growth cessation due to heterozygous aggrecan mutations [published online April 24, 2014]. J Clin Endocrinol Metab. doi:10.1210/jc.2014–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang SR, Carmichael H, Andrew SF, et al. Large-scale pooled next-generation sequencing of 1077 genes to identify genetic causes of short stature. J Clin Endocrinol Metab. 2013;98:E1428–E1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Romano AA, Allanson JE, Dahlgren J, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2010;126:746–759 [DOI] [PubMed] [Google Scholar]
- 79. Caliebe J, Broekman S, Boogaard M, et al. IGF1, IGF1R and SHOX mutation analysis in short children born small for gestational age and short children with normal birth size (idiopathic short stature). Horm Res Paediatr. 2012;77:250–260 [DOI] [PubMed] [Google Scholar]
- 80. Wit JM, van Duyvenvoorde HA, Scheltinga SA, et al. Genetic analysis of short children with apparent growth hormone insensitivity. Horm Res Paediatr. 2012;77:320–333 [DOI] [PubMed] [Google Scholar]
- 81. Clayton P, Bonnemaire M, Dutailly P, et al. Characterizing short stature by insulin-like growth factor axis status and genetic associations: results from the prospective, cross-sectional, epidemiogenetic EPIGROW study. J Clin Endocrinol Metab. 2013;98:E1122–E1130 [DOI] [PubMed] [Google Scholar]
- 82. Renes JS, Willemsen RH, Wagner A, Finken MJ, Hokken-Koelega AC. Bloom syndrome in short children born small for gestational age: a challenging diagnosis. J Clin Endocrinol Metab. 2013;98:3932–3938 [DOI] [PubMed] [Google Scholar]
- 83. Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Edery P, Marcaillou C, Sahbatou M, et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science. 2011;332:240–243 [DOI] [PubMed] [Google Scholar]
- 87. He H, Liyanarachchi S, Akagi K, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rauch A, Thiel CT, Schindler D, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319:816–819 [DOI] [PubMed] [Google Scholar]
- 89. O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501 [DOI] [PubMed] [Google Scholar]
- 90. Ogi T, Walker S, Stiff T, et al. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel syndrome. PLoS Genet. 2012;8:e1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. J Med Genet. 2010;47:411–414 [DOI] [PubMed] [Google Scholar]
- 92. Kalay E, Yigit G, Aslan Y, et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43:23–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qvist P, Huertas P, Jimeno S, et al. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet. 2011;7:e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Griffith E, Walker S, Martin CA, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bicknell LS, Bongers EM, Leitch A, et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011;43:356–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Guernsey DL, Matsuoka M, Jiang H, et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43:360–364 [DOI] [PubMed] [Google Scholar]
- 97. Alatzoglou KS, Dattani MT. Genetic causes and treatment of isolated growth hormone deficiency-an update. Nat Rev Endocrinol. 2010;6:562–576 [DOI] [PubMed] [Google Scholar]