Abstract
IL-13-induced epithelial gene and protein expression changes are central to the pathogenesis of multiple allergic diseases. Herein, using human esophageal squamous and bronchial columnar epithelial cells, we identified miRNAs that were differentially regulated after IL-13 stimulation. Among the IL-13-regulated miRNAs, miR-375 showed a conserved pattern of down-regulation. Furthermore, miR-375 was down regulated in the lung of IL-13 lung transgenic mice. We subsequently analyzed miR-375 levels in a human disease characterized by IL-13 overproduction – the allergic disorder eosinophilic esophagitis (EE), and observed down-regulation of miR-375 in EE patient samples compared to control patients. MiR-375 expression levels reflected disease activity, normalized with remission, and inversely correlated to the degree of allergic inflammation. Using a lentiviral strategy and whole-transcriptome analysis in epithelial cells, miR-375 over-expression was sufficient to markedly modify IL-13-associated immunoinflammatory pathways in epithelial cells in vitro, further substantiating interactions between miR-375 and IL-13. Taken together, our results support a key role of miRNAs, particularly miR-375, in regulating and fine-tuning IL-13 mediated responses.
Introduction
IL-13 is an adaptive immune cytokine that is involved in mediating the effector functions of T helper type 2 (Th2) responses. The central role of IL-13 in allergic disorders has been demonstrated by the attenuation of experimental allergic diseases in animals with blockade and/or gene deletion of IL-13 and/or its receptor signaling components.1–5 Furthermore, one of the critical functions of IL-13 is to modify epithelial gene expression at sites of inflammation. Notably, IL-13-induced gene expression changes in epithelial cells in vitro have been shown to significantly overlap with the gene expression changes seen in patients in vivo.6–13 Recent early clinical studies with IL-13 neutralizing agents provide evidence that anti-IL-13 holds promise for the treatment of allergic disorders, especially in patient subgroups, based on various gene expression profiles (especially periostin).14 Therefore, a better understanding of IL-13-mediated responses and the pathways that regulate IL-13-induced gene expression are likely to provide insight into therapeutic strategies, especially for allergic disorders characterized by IL-13 overproduction, such as eosinophilic esophagitis (EE).
MicroRNAs (miRNAs) are short single-stranded RNA molecules that regulate post-transcriptional gene silencing of target genes.15 In animals, miRNAs base pair with the complementary regions in the 3’ untranslated regions of mRNA and induce translational repression and/or mRNA degradation depending on the degree of complementarity of the base pairing.16 Recently, the miRNA let-7 has been shown to target IL-13 directly and miR-155 has been shown to target IL-13Rα1.17–19 However, whether IL-13-induced miRNAs could regulate or fine tune IL-13 mediated-responses has not been extensively explored. The epithelial cell is a particular attractive model to investigate in this area since it has been shown to be a key target cell type for IL-13 mediated responses. For example, epithelial cells are required for IL-13 induced airway hyper-reactivity and mucus production20 and IL-13-induced epithelial cell gene expression changes have a critical role in the pathogenesis of EE.9
Here, we used miRNA array analysis to determine the differentially expressed miRNAs after IL-13 stimulation in two distinct human epithelial cell types: esophageal squamous cells and bronchial columnar cells. Among the IL-13-regulated miRNAs, miR-375 showed a conserved pattern of down-regulation between these two epithelial cell types. Direct examination of human allergic tissue (esophageal biopsies from patients with EE) indicated that miR-375 was inversely related to the degree of allergic inflammation including esophageal eosinophil levels and gene expression levels of Th2 cytokine and mast cell specific proteases. Functionally, miR-375 over-expression was sufficient to markedly modify IL-13-associated immunoinflammatory pathways in epithelial cells in vitro.
Results
Expression profiling of miRNA in IL-13-stimulated epithelial cells
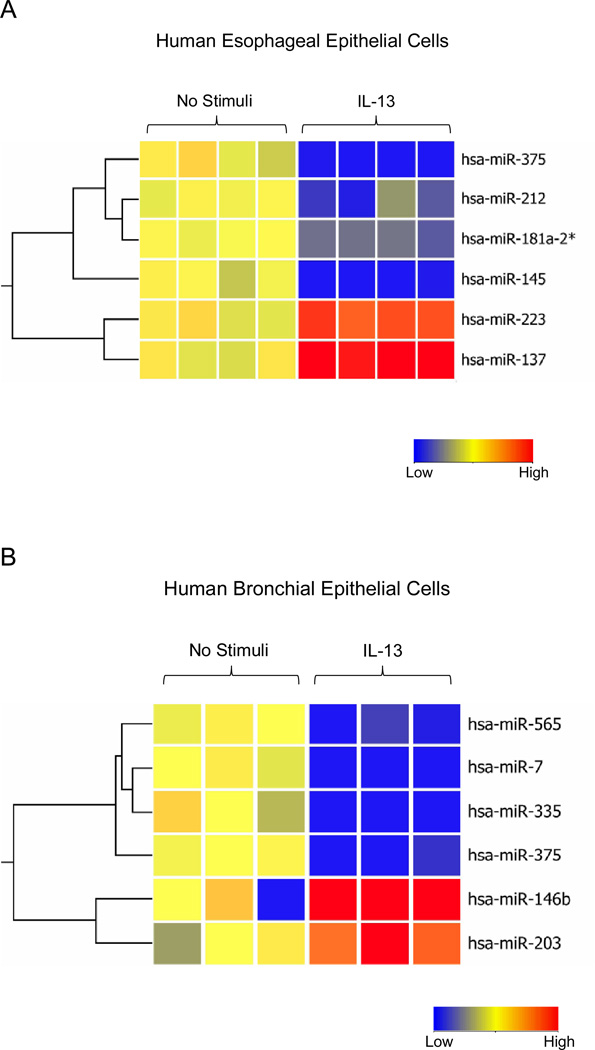
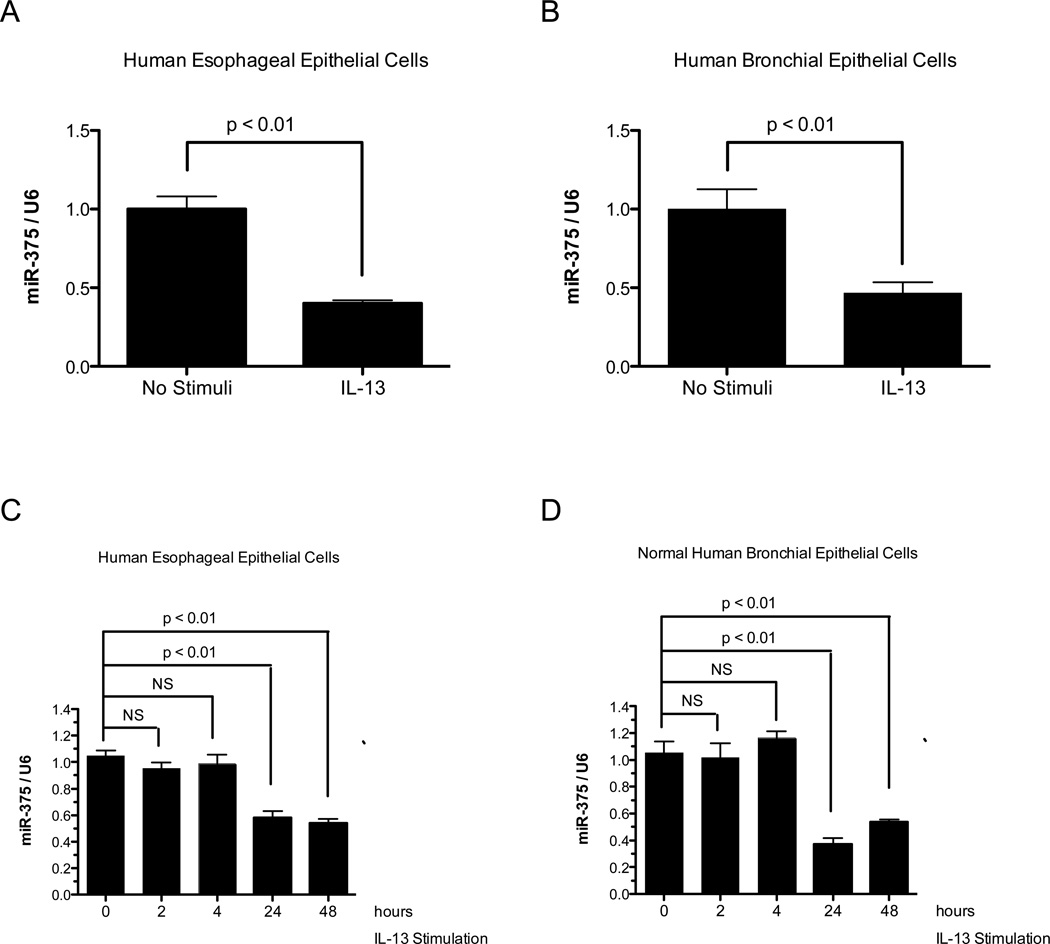
To identify miRNAs differentially expressed in epithelial cells in response to IL-13 stimulation, we profiled miRNA expression in IL-13-stimulated human bronchial and esophageal epithelial cells using miRNA microarrays. Comparing IL-13-treated and untreated esophageal epithelial cells, we found 6 miRNAs that were differentially regulated in response to IL-13 (Fig. 1A). These include four down-regulated miRNAs – miR-375, miR-212, miR-181a-2*, miR-145 and 2 up-regulated miRNAs – miR-223, miR-137. A similar analysis of IL-13 treated human bronchial epithelial cells found four down-regulated miRNAs – miR-565, miR-7, miR-335, miR-375 and 2 up-regulated miRNAs – miR-146b, miR-203 (Fig. 1B). MiR-375 was the only miRNA that was differentially regulated in both epithelial cell types after IL-13 treatment. We subsequently focused on miR-375 and validated its down-regulation in both the human esophageal and human bronchial epithelial cells after IL-13 stimulation by qPCR (Fig. 2 A-B). Kinetic analysis of miR-375 expression in human esophageal epithelial cells and normal human primary bronchial epithelial cells indicated that miR-375 was down-regulated after 24 and 48 hours of IL-13 stimulation (Fig. 2C–D).
Figure 1. MiRNA expression profile in human esophageal epithelial cells and human bronchial epithelial cells after 24 hrs of IL-13 stimulation.
(A) Heat map of 4 down-regulated and 2 up-regulated miRNAs in IL-13 stimulated human esophageal epithelial cells compared to controls. (B) Heat map of 4 down-regulated and 2 up-regulated miRNAs in IL-13 stimulated human bronchial epithelial cells compared to controls. Red: Up-regulated in IL-13 stimulated cells compared to controls; blue: down-regulated in IL-13 stimulated cells compared to controls.
Figure 2. Quantitative RT-PCR verification of miR-375 expression in IL-13 stimulated human esophageal epithelial cells and human bronchial epithelial cells.
Expression of miR-375 was determined in (A) IL-13 stimulated human esophageal epithelial cells compared to controls and (B) IL-13 stimulated human bronchial epithelial cells compared to controls. (C) Kinetic analysis of miR-375 expression in IL-13 stimulated primary human esophageal epithelial cells. (D) Kinetic analysis of miR-375 expression in IL-13 stimulated normal human primary bronchial epithelial cells. The relative expression levels were normalized to U6 small nuclear RNA. N = 4 per group; data are represented as mean ± S.E.M. NS: not significant.
Expression of miR-375 in IL-13 lung transgenic mice and in EE patients
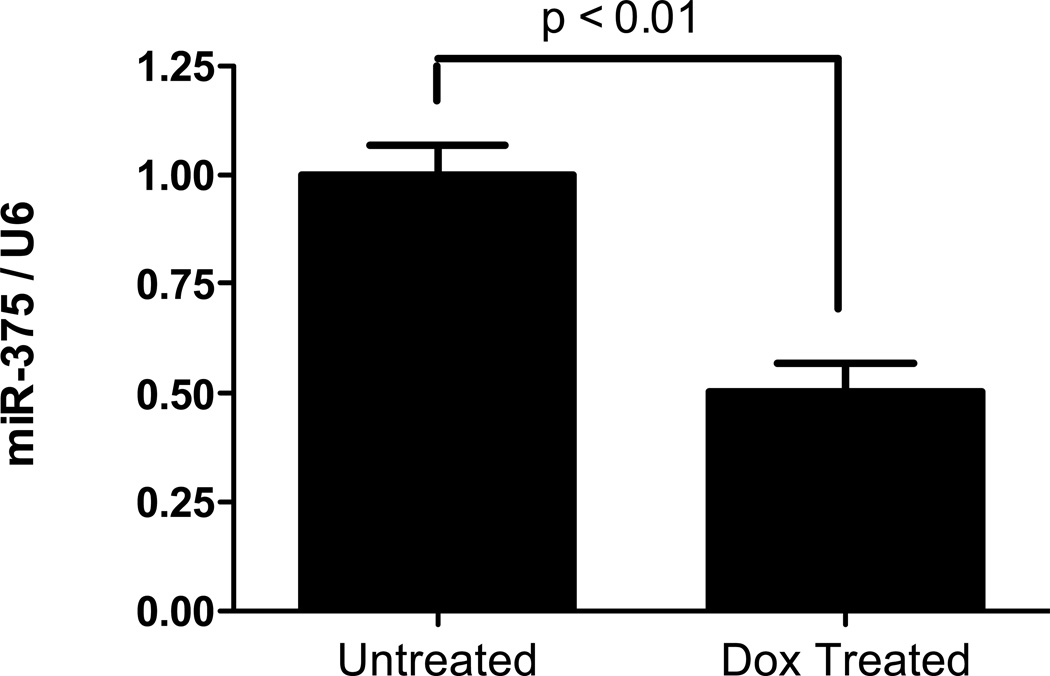
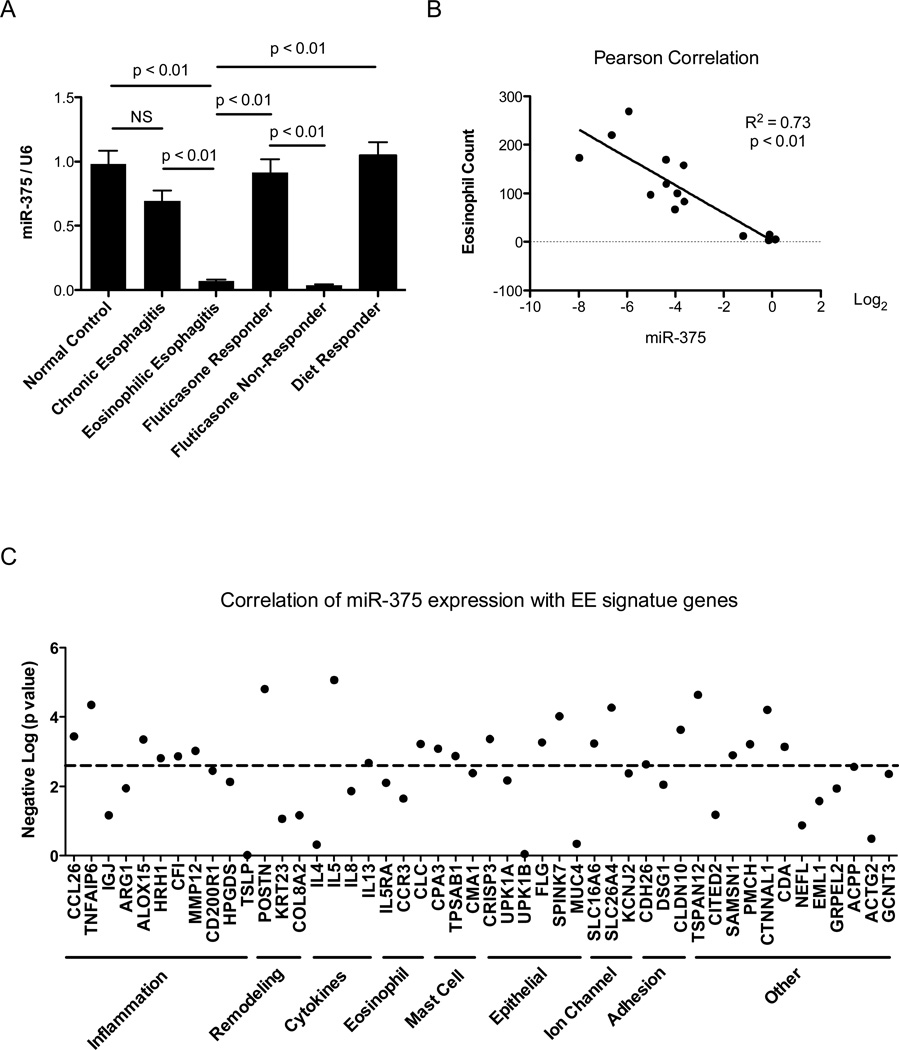
To investigate the long-term effect of IL-13 exposure on miR-375 expression, we utilized an IL-13 lung transgenic mouse model where experimental asthma was induced by 4 weeks of doxycycline induced IL-13 transgene expression. Compared to control mice that received no doxycycline, doxycycline treated IL-13 transgenic mice had significant down-regulation of miR-375 in the lungs (Fig. 3). We also investigated whether miR-375 was down-regulated in EE patients since EE has been reported to be a Th2 associated disease with IL-13 having a major role in its pathogenesis.21, 22 Compared to normal healthy controls, EE patients had significant down-regulation of miR-375 in esophageal tissue. This down-regulation was not evident in patients with chronic (non-eosinophilic) esophagitis which has a distinct etiology and pathogenesis from EE (Fig. 4A).23 We next investigated whether the expression levels of miR-375 were normalized in EE patients in remission. EE patients that responded to either fluticasone propionate therapy or diet modification had miR-375 levels comparable to normal healthy controls; whereas, patients that did not respond to therapy continued to have repressed miR-375 levels (Fig. 4A). Since miR-375 down-regulation fluctuated with disease activity, we further investigated the correlation of miR-375 expression with other markers of EE disease activity including the high level of eosinophil infiltration observed in the esophageal biopsies of EE patients23, 24 and the expression of previously identified EE signature genes. Esophageal miR-375 expression exhibited a significant inverse correlation with the level of eosinophil infiltration in the esophageal biopsies as well as esophageal expression of genes involved in inflammation including CCL26 (eotaxin-3),23 remodeling including POSTN (periostin),25 Th2 cytokines including IL-5 and IL-13,26 and cell specific markers for eosinophils (CLC),27 mast cells (CPA3 and TPSAB1)28 and epithelial cells (FLG) (Fig. 4 B–C).29
Figure 3. Expression of miR-375 in doxycycline induced IL-13 lung transgenic experimental asthma model.
Relative expression level of miR-375 determined by qPCR normalized to U6. N = 6 mice per group; Data are represented as mean ± S.E.M.
Figure 4. Expression of miR-375 in esophageal biopsies from EE patients and its correlation with esophageal eosinophil counts and EE signature genes.
(A) Expression of miR-375 in normal control, EE patients, chronic esophagitis patients, EE patients responsive to glucocorticoid therapy (fluticasone proprionate), EE patients unresponsive to glucocorticoid therapy and EE patients responsive to diet modification. Expression levels were determined by qPCR normalized to U6 small nuclear RNA. N = 8–15 patients per group; data are represented as mean ± S.E.M. (B) Correlation between miR-375 expression and esophageal eosinophil counts. (C) Correlation between miR-375 expression and EE signature genes. The significance of the correlation was plotted as the negative log of p value for each gene. The dashed line represents significance level after false discovery rate correction.
MiR-375 is predominately expressed in the esophageal and bronchial epithelial cells
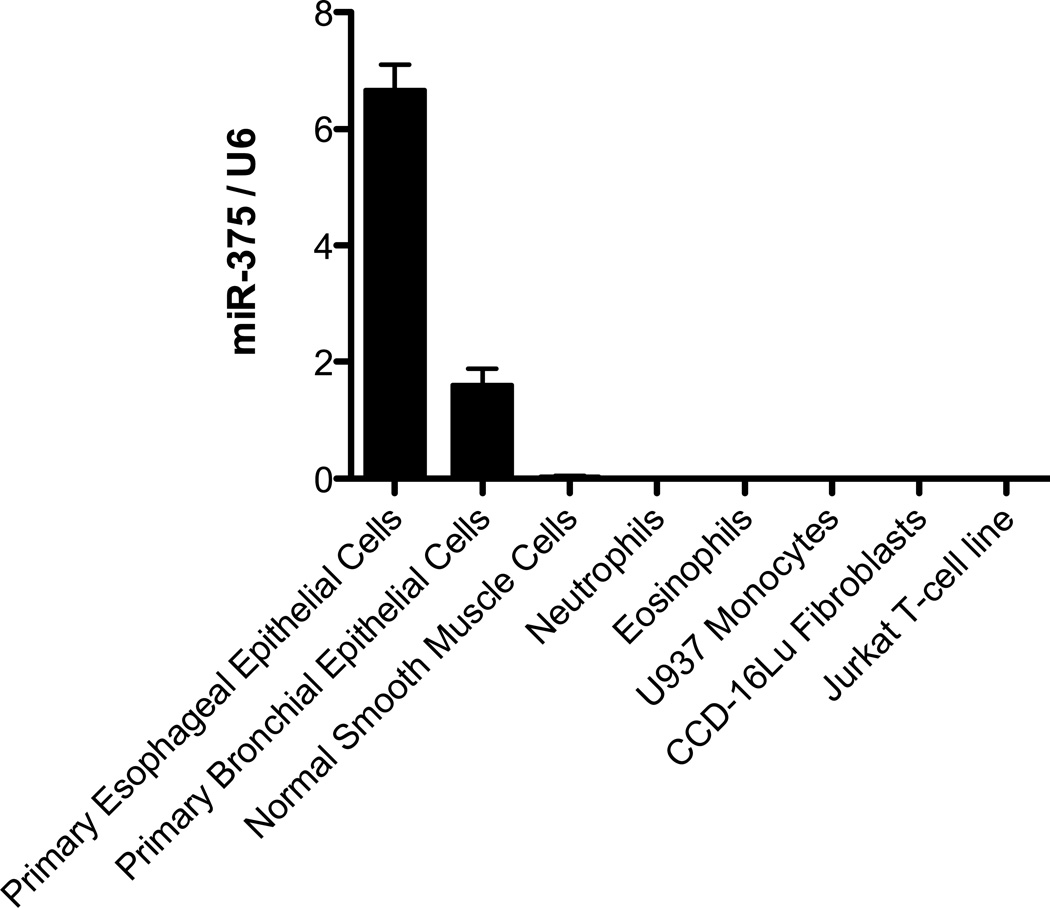
To determine the levels of miR-375 expression in different cell types, we performed qPCR analysis of miR-375 levels in primary esophageal epithelial cells, normal human primary bronchial epithelial cells, normal smooth muscle cells, neutrophils, eosinophils, monocytes, fibroblasts and T-cells. The esophageal epithelial cells had the highest miR-375 expression (Fig. 5). We subsequently focused on the role of miR-375 in the esophageal epithelial cells.
Figure 5. Expression level of miR-375 in different cell types.
Relative expression level of miR-375 in different cell types determined by qPCR normalized to U6. N = 3 per group; Data are represented as mean ± S.E.M.
MiR-375 regulates IL-13-regulated signature genes
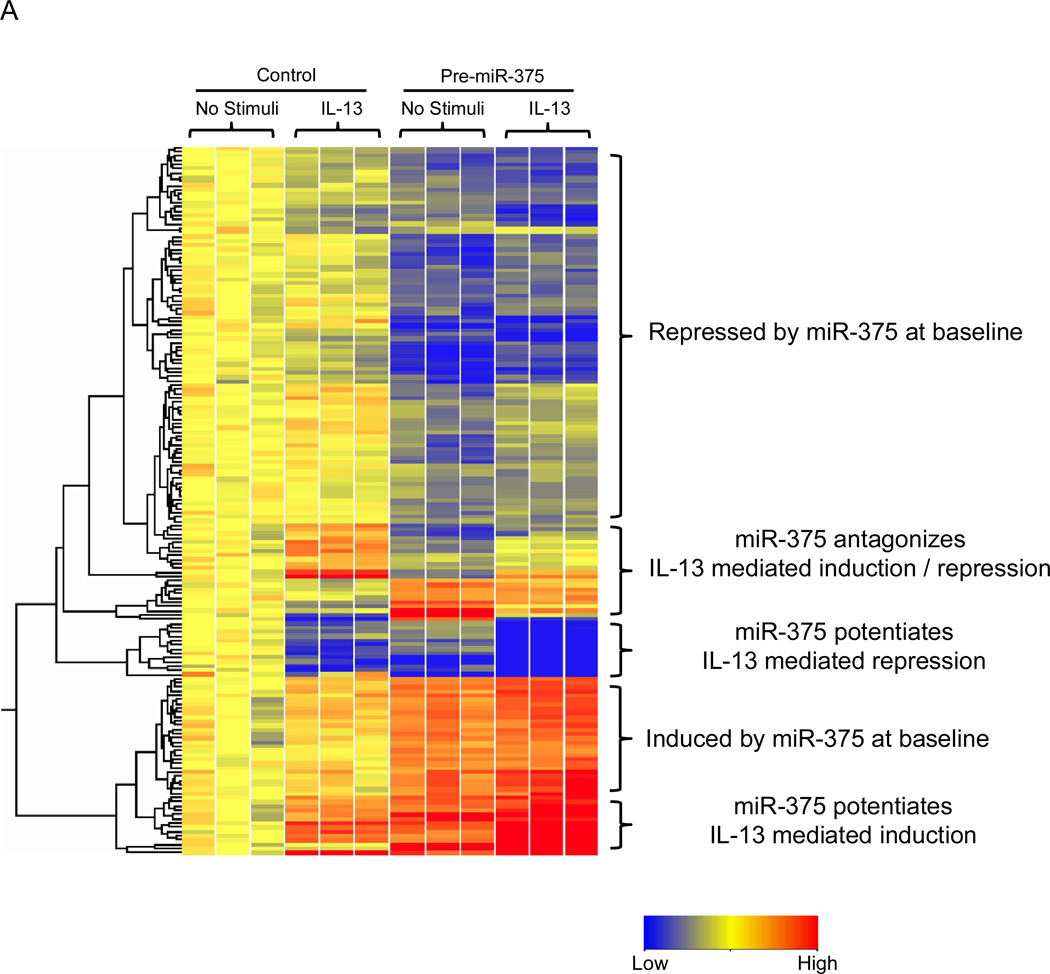
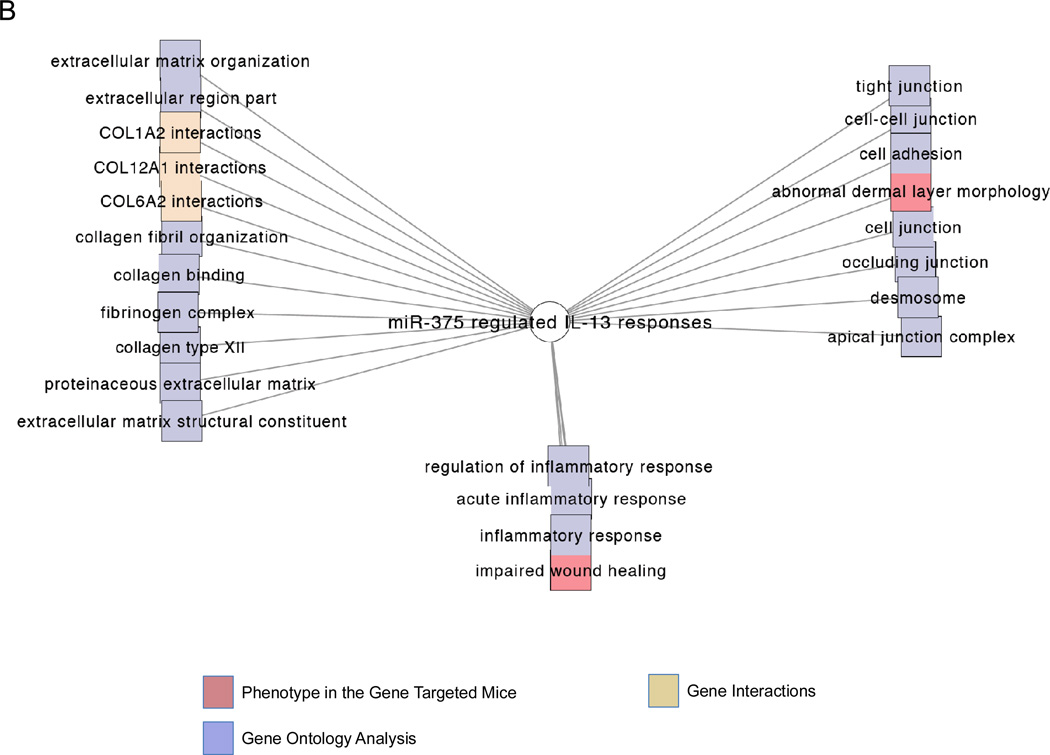
Using a lentiviral vector, we stably over-expressed miR-375 in the esophageal epithelial cell line TE-7. We then treated the control-transduced and miR-375-transduced TE-7 cells with IL-13 to determine the effect of miR-375 on IL-13 induced-esophageal epithelial transcriptome. MiR-375 was able to repress a large set of genes at baseline, consistent with the function of miRNAs as repressors of gene expression (Fig. 6A). We also noted a smaller set of genes that were induced at baseline, most likely though miR-375 mediated repression of transcriptional repressors (Fig. 6A). Interestingly, miR-375 was able to both potentiate and antagonize a subset of IL-13 mediated gene signatures, indicating a complex interaction between miR-375 and effects of IL-13 (Fig. 6A). Functional analysis indicated that the pathways affected by miR-375 under IL-13-stimulated conditions were enriched for processes involved in extracellular matrix organization, cellular junctions and inflammation (Fig. 6B). The differentially regulated genes are listed in Supplemental Table 1. Analysis of all miR-375-regulated genes indicated that inflammatory diseases and immunological diseases are the two most significantly over-represented disease states (Supplemental Fig. 1). The miR-375 regulated genes in each of these disease processes are listed in Supplemental Table 2.
Figure 6. Genes differentially regulated by miR-375 in esophageal epithelial cells before and after IL-13 stimulation.
(A) Heatmap showing genes differentially expressed in esophageal epithelial cell line transduced with either control vector or pre-miR-375 expression vector before and after IL-13 stimulation. Red: Up-regulated compared to control transduced unstimulated cells; blue: down-regulated compared to control transduced unstimulated cells. (B) Functional enrichment analysis of pathways affected by miR-375 under IL-13 stimulated conditions. The networks are shown as Cytoscape graph networks generated from ToppCluster network analysis.
Discussion
Herein, we have identified miRNA changes induced by IL-13 stimulation in human bronchial and esophageal epithelial cells. In particular, we found that miR-375 was the only miRNA that was down-regulated in both epithelial cell types after IL-13 stimulation. Analysis of different human cell types involved in allergic inflammation identified the highest expression of miR-375 in epithelial cells, strengthening the relevance of our findings. In addition, we found that miR-375 was inversely correlated with the level of esophageal eosinophils and expression of the mast cell specific genes CPA3 and TPSAB1. The down-regulation of miR-375 was specific to EE patients since the chronic esophagitis patients have miR-375 expression levels comparable to normal controls. Disease remission with either fluticasone therapy or diet modification was associated with normalization of miR-375 levels, likely due to reduced IL-13; whereas, patients that did not respond to fluticasone therapy continued to have repressed miR-375 levels. Further, modulation of miR-375 levels was sufficient to regulate IL-13 mediated gene expression, particularly with pathways involved in immunoinflammatory processes. Indeed, levels of miR-375 markedly inversely correlated with a large set of immunoinflammatory genes (including IL-13) in the esophagus of patients with EE. Collectively, these findings provide multiple lines of evidence for a key role of miR-375 in epithelial cell driven allergic inflammation.
Using a genome wide transcriptome based approach, we demonstrated that miR-375 could potentiate and repress IL-13-mediated effects, underlining the complex interaction between cytokine and miRNA mediated gene regulation. The inflammatory diseases and immunological diseases are the two most significantly over-represented disease states regulated by miR-375. These include allergy-associated genes such as MMP12 and MUC4 (Supplemental Table 2).30–33 Our results differed from those recently reported by Biton et al. in that their results demonstrated up-regulation of miR-375 after 2 hours of IL-13 stimulation; whereas, we found that miR-375 expression was unchanged after 2 hours. However, they did demonstrate that miR-375 levels were at or below baseline after 16 hours of IL-13 stimulation.34 This latter finding corresponds with our data that miR-375 is down-regulated after 24 and 48 hours of IL-13 stimulation. The disparity between our results at the 2-hour time point could potentially be due to the different cell types used in the studies; they used the HT-29 human colon adenocarcinoma cell line, and we used human esophageal squamous cells and bronchial columnar cells. Notably, while miR-375 down-regulation has been reported in patient samples from multiple Th2-associated diseases [e.g. atopic dermatitis and ulcerative colitis35, 36] and hyperproliferative diseases [e.g. esophageal squamous carcinoma37], an up-regulation of miR-375 in a Th2-associated disease in humans has yet to be reported. Thus, the long-term effect of IL-13 is likely to down-regulate miR-375 expression.
MiR-375 has been previously shown to enhance goblet cell differentiation by repressing KLF5 expression.34 It has also been shown to attenuate cell proliferation by targeting IGF1R, PDK1 and YWHAQ.37, 38 However, neither of these pathways was affected in the EE patients or in our analysis of miR-375-regulated genes in the esophageal epithelial cells 23. This suggests that the activity of miR-375 may be dependent on the cellular context, as indicated by previous reports.38, 39 MiR-375 has been previously reported to regulate TSLP expression in HT-29 human colonic adenocarcinoma cell line. TSLP and miR-375 were concomitantly induced by IL-13 in HT-29 cells and knockdown of miR-375 inhibited TSLP production. In addition, over-expression of miR-375 induced TSLP expression in HT-29 cells.34 Since TSLP has been reported to have an important role in EE pathogenesis,40, 41 we analyzed whether miR-375 was able to regulate TSLP expression in esophageal epithelial cells. We did not find any effect of miR-375 on TSLP production (Supplemental Figure 1) and there was no correlation between miR-375 and TSLP in the esophageal samples (Fig. 3C). Our control transduced cells and pre-miR-375 transduced cells expressed TSLP at similar levels without stimulation and have similar levels of induction after polyI:C stimulation. This disparity could be due to the different cell types used in our studies and/or different mechanisms in TSLP induction in these cells since IL-13 induced TSLP expression in HT-29 cells but not in esophageal epithelial cells according to previous reports.9, 34
While IL-13 down-regulates miR-375 and miR-375 and could modulate IL-13 regulated gene expression, whether the over-expression of miR-375 could correct the allergic phenotype in asthma and EE remains to be investigated. This will likely be resolved in future studies utilizing miR-375 lung and/or esophageal epithelial specific transgenic mice. In addition to miR-375, we identified 10 other miRNAs that were differentially regulated in either the human esophageal epithelial cells or the human bronchial epithelial cells. These likely reflect cell type specific effects of IL-13 stimulation. Notably, previous reports indicated that the miRNAs miR-203 and miR-223 were differentially regulated in Th2-associated diseases.35, 36
In summary, we report miRNA signatures of human esophageal and bronchial epithelial cells after IL-13 stimulation. We demonstrated that one epithelial derived miRNA, miR-375, was down-regulated in both epithelial cells types after IL-13 stimulation and was sufficient to regulate an IL-13-induced epithelial transcriptome. MiR-375 expression levels reflected disease activity, normalized with remission, and inversely correlated to the degree of allergic inflammation. It is notable that miR-375 was strongly associated with parameters germane to allergic responses including eosinophil levels, gene expression levels of the Th2 cytokines IL-5 and IL-13, the mast cell-specific enzymes CPA3 and TPSAB1, and POSTN (the gene that encodes periostin). It is notable that periostin has been demonstrated to have a key role in IL-13 associated remodeling responses42 and its level predicts responsiveness to anti-IL-13 therapy in humans14 highlighting the potential importance of our findings as we have found that miR-375 strongly correlates with human POSTN levels in vivo. Using a lentiviral strategy and whole-transcriptome analysis in epithelial cells, miR-375 over-expression was sufficient to markedly modify IL-13-associated immunoinflammatory pathways in epithelial cells in vitro, further substantiating interactions between miR-375 and IL-13. Taken together, our results support a key role of miRNAs in regulating and fine-tuning IL-13 mediated responses; we propose miR-375 is a key downstream mediator of IL-13-induced responses.
Material and Methods
Human esophageal tissues
Patients were selected without regard to age, race, or sex. Normal patients presented to the clinic with symptoms consistent with gastroesophageal reflux disease or EE but the endoscopic and histologic findings were normal. The active EE patients have a clinical diagnosis of EE and eosinophil counts of ≥24 per 400× high power field in the esophageal biopsies. The active chronic esophagitis patients have eosinophil counts of 1–15 per 400x high power field in the esophageal biopsies. Patients with systemic or swallowed topical glucocorticoid use were excluded from the selection of active EE or active chronic esophagitis patients. The EE remission patients responding to steroid treatment have a clinical history of EE, treatment with swallowed topical glucocorticoids, and responsiveness as indicated by an eosinophil count of ≤1 per 400x high power field and normalization of histological features of the disease. The EE remission patients responding to diet treatment have a clinical history of EE, treatment with diet modification, and responsiveness as described above. The EE patients not responding to glucocorticoid treatment have a clinical history of EE, treatment with swallowed topical glucocorticoid, and non-responsiveness as indicated by an eosinophil count of ≥24 per 400× high power field. This study was approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
Cell culture
Human esophageal epithelial cells derived from human patient biopsies were cultured as previously described.9 The human bronchial epithelial cell line HBEC was cultured as previously described.43 The normal human primary bronchial epithelial cells were purchased from Lonza (Catalog #: CC-2540) and cultured as previously described.44 The U937 monocytes and Jurkat T cells were cultured in RPMI 1640 medium supplemented with 10%FBS, 100 U/mL penicillin, and 100 µg/ml streptomycin. The CCD-16Lu fibroblasts were cultured in Eagle’s Minimum Essential Medium supplemented with 10%FBS, 100 U/mL penicillin, and 100 µg/ml streptomycin.
RNA extraction and miRNA microarray analysis
Human esophageal epithelial cells and bronchial epithelial cells were stimulated with media or 100 ng/mL IL-13 for 24 hours. Total RNA including miRNA was isolated using miRNeasy Mini Kit according to manufacturer’s instructions (Qiagen). RNA quality was assessed using the Agilent 2100 bioanalyzer (Agilent Technologies) and only samples with RNA integrity number >8 were used. MiRNA expression from human bronchial epithelial cells was profiled using TaqMan Human MicroRNA Array v1.0 (Early Access), which includes probes for 365 human miRNAs, according to manufacturer’s protocols (Applied Biosystems). Data analysis was carried out using GeneSpring software (Agilent Technologies). To identify miRNAs differentially regulated between unstimulated and IL-13 stimulated samples, the expression data were normalized to the average of two endogenous control probes RNU44 and RNU48, then filtered on cycle threshold values < 35 and at least a 2 fold change between unstimulated and IL-13 stimulated samples. Statistical significance was determined at p < 0.05 with Benjamini Hochberg false discovery rate correction. The list of differentially expressed miRNAs was clustered using hierarchical clustering and a heatmap was generated. A similar analysis was carried out comparing unstimulated and IL-13 stimulated human esophageal epithelial cells, except that the TaqMan Human MicroRNA Array v2.0 was used, which includes probes for 667 human miRNAs. The microarray data have been deposited into the Array Express database (www.ebi.ac.uk/arrayexpress) with accession number E-MEXP-3351 and E-MEXP-3353 in compliance with minimum information about microarray experiment (MIAME) standards.
Experimental Asthma Induction in IL-13 bitransgenic mice
Bitransgenic mice bearing CCSP-rtTA and (tetO)7CMV-IL-13 transgenes were previously described.45 Experimental asthma was induced in IL-13 bitransgenic mice by feeding bi-transgenic mice doxycycline-impregnated food for 4 weeks as previously described.45 All animals were housed under specific pathogen free conditions in accordance with institutional guidelines. The use of animals in these experiments was approved by the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital Medical Center.
qRT-PCR for miRNA
Levels of miRNA expression were measured quantitatively by using TaqMan MicroRNA Assays (Applied Biosystems) following manufacturer’s protocol. The expression levels were normalized to the U6 endogenous control. Relative expression was calculated as previously described.45
qRT-PCR for mRNA
Total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). All primer/probe sets were obtained from Applied Biosytems. Samples were analyzed by TaqMan qRT-PCR for CPA3 (Assay ID: Hs00157019_m1) and normalized to HPRT1 (Assay ID: Hs01003267_m1). Relative expression was calculated as previously described.45
Correlation of miR-375 with major EE signature genes
Esophageal mRNA from EE patients was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer’s protocol. The TaqMan reagents for amplification of EE signature genes 9, 23, 24 were obtained from Applied Biosystems. TaqMan real-time PCR amplification was performed on an Applied Biosystems 7900HT Real-Time PCR System. The expression correlation study between miR-375 and 48 EE genes was performed in GraphPad Prism software. Negative log of p values from Pearson correlation analysis were plotted to demonstrate correlation significance with EE genes. To control for the increased risk of false positives due to the number of statistical tests performed, we applied a Bonferroni correction based on the number of gene expression profiles compared. Because the average pairwise correlation between gene expression profiles was 0.54, we applied principal components analysis to determine the effective number of independent comparisons as previously described.46 Using this approach, a p-value of 0.002 was required to achieve a family wise error rate of 0.05.
Lentiviral transdcution
The human esophageal epithelial cell line TE-7 cells were transduced with pmiRNA1-Pre-miR-375 vector or pmiRNA1-Control vector (System Biosciences). The vectors include GFP and puromycin resistance genes as selection markers. Three days after transduction, cells were selected by FACS sorting for GFP+ cells and further cultured in media containing 4 µg/mL puromycin for 1 week. The cells were > 99% GFP+ after selection.
Human genome-wide mRNA microarray
The Affymetrix human Gene 1.0ST array was used to compare gene expression profile of control transduced TE-7 cells and pre-miR-375 transduced TE-7 cells before and after IL-13 treatment. Microarray data were analyzed using the GeneSpring software (Agilent Technologies) as previously described.47 Global scaling was performed to compare genes from chip to chip, and a base set of probes was generated by requiring a minimum raw expression level of 20th percentile out of all probes on the microarray. The resulting probe sets were then baseline transformed and filtered on at least 1.2 fold difference between control transduced and pre-miR-375 transduced cells with or without IL-13 treatment to identify miR-375 regulated genes. Statistical significance was determined at p < 0.05 with Benjamini Hochberg false discovery rate correction. The resulting list of genes was clustered using hierarchical clustering and a heatmap was generated. Biological functional enrichment analysis was carried out using Ingenuity Pathway Analysis (Ingenuity Systems) and Toppgene/Toppcluster.46, 48 The microarray data have been deposited into the Array Express database (www.ebi.ac.uk/arrayexpress) with accession number E-MEXP-3345 in compliance with MIAME standards.
Statistical Analysis
Student’s t-test was used to determine the significance between two groups. One-way ANOVA with Tukey post-hoc test was used to determine the significance between more than 2 groups. Statistical significance and the p values were indicated on the figures where appropriate. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by the NHLBI Ruth L. Kirschstein National Research Service Award for individual predoctoral MD/PhD fellows F30HL104892 (T.X.L), the Ryan Fellowship from Albert J. Ryan Foundation (T.X.L.), and the Organogenesis Training Grant (NIH T32HD046387 supporting T.X.L.). Additionally, this work was supported by NIH R01DK076893 (M.E.R), the Campaign Urging Research for Eosinophilic Disease (CURED), the Buckeye Foundation, and the Food Allergy Project / Food Allergy Initiative.
Footnotes
Disclosures: M.E.R. has an equity interest in reslizumab, a drug being developed by Cephalon, and is a consultant for Immune Pharmaceuticals. The other authors have declared that they have no conflict of interest.
Author Contributions: T.X.L. designed experiments, performed experiments, analyzed data, interpreted all results, and wrote the manuscript. E.J.L and T.W. performed experiments and analyzed data. A.J.P. and B.J.A. performed bioinformatics analysis. L.J.M. performed biostatistics analysis. M.E.R. designed experiments, interpreted all results, wrote the manuscript and coordinated overall research efforts.
References
- 1.Leigh R, Ellis R, Wattie JN, Hirota JA, Matthaei KI, Foster PS, et al. Type 2 cytokines in the pathogenesis of sustained airway dysfunction and airway remodeling in mice. Am J Respir Crit Care Med. 2004;169(7):860–867. doi: 10.1164/rccm.200305-706OC. [DOI] [PubMed] [Google Scholar]
- 2.Yang M, Rangasamy D, Matthaei KI, Frew AJ, Zimmmermann N, Mahalingam S, et al. Inhibition of arginase I activity by RNA interference attenuates IL-13-induced airways hyperresponsiveness. J Immunol. 2006;177(8):5595–5603. doi: 10.4049/jimmunol.177.8.5595. [DOI] [PubMed] [Google Scholar]
- 3.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282(5397):2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junttila IS, Mizukami K, Dickensheets H, Meier-Schellersheim M, Yamane H, Donnelly RP, et al. Tuning sensitivity to IL-4 and IL-13: differential expression of IL-4Ralpha, IL-13Ralpha1, and gammac regulates relative cytokine sensitivity. J Exp Med. 2008;205(11):2595–2608. doi: 10.1084/jem.20080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee PJ, Zhang X, Shan P, Ma B, Lee CG, Homer RJ, et al. ERK1/2 mitogen-activated protein kinase selectively mediates IL-13-induced lung inflammation and remodeling in vivo. J Clin Invest. 2006;116(1):163–173. doi: 10.1172/JCI25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 7.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128(1):3–20. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 8.Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137(4):1238–1249. doi: 10.1053/j.gastro.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Kaminski N, Dolganov G, Grunig G, Koth L, Solomon C, et al. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am J Respir Cell Mol Biol. 2001;25(4):474–485. doi: 10.1165/ajrcmb.25.4.4522. [DOI] [PubMed] [Google Scholar]
- 11.Laprise C, Sladek R, Ponton A, Bernier MC, Hudson TJ, Laviolette M. Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics. 2004;5(1):21. doi: 10.1186/1471-2164-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104(40):15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, Paquet AC, et al. IL-13 and epidermal growth factor receptor have critical but distinct roles in epithelial cell mucin production. Am J Respir Cell Mol Biol. 2007;36(2):244–253. doi: 10.1165/rcmb.2006-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab Treatment in Adults with Asthma. N Engl J Med. 2011;365(12):1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 15.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 16.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286(3):1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128(5):1077–1085. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 19.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285(39):30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med. 2002;8(8):885–889. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 21.Blanchard C, Rothenberg ME. Basic pathogenesis of eosinophilic esophagitis. Gastrointest Endosc Clin N Am. 2008;18(1):133–143. doi: 10.1016/j.giec.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulder DJ, Justinich CJ. Understanding eosinophilic esophagitis: the cellular and molecular mechanisms of an emerging disease. Mucosal immunol. 2011;4(2):139–147. doi: 10.1038/mi.2010.88. [DOI] [PubMed] [Google Scholar]
- 23.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):140–149. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stansfield WE, Andersen NM, Tang RH, Selzman CH. Periostin is a novel factor in cardiac remodeling after experimental and clinical unloading of the failing heart. Ann Thorac Surg. 2009;88(6):1916–1921. doi: 10.1016/j.athoracsur.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiko GE, Foster PS. New insights into the generation of Th2 immunity and potential therapeutic targets for the treatment of asthma. Curr Opin Allergy Clin Immunol. 2011;11(1):39–45. doi: 10.1097/ACI.0b013e328342322f. [DOI] [PubMed] [Google Scholar]
- 27.Ackerman SJ, Liu L, Kwatia MA, Savage MP, Leonidas DD, Swaminathan GJ, et al. Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion. J Biol Chem. 2002;277(17):14859–14868. doi: 10.1074/jbc.M200221200. [DOI] [PubMed] [Google Scholar]
- 28.Xing W, Austen KF, Gurish MF, Jones TG. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc Natl Acad Sci U S A. 2011;108(34):14210–14215. doi: 10.1073/pnas.1111048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay S, Sypek J, Tavendale R, Gartner U, Winter J, Li W, et al. Matrix metalloproteinase-12 is a therapeutic target for asthma in children and young adults. J Allergy Clin Immunol. 2010;126(1):70–76. doi: 10.1016/j.jaci.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Lavigne MC, Thakker P, Gunn J, Wong A, Miyashiro JS, Wasserman AM, et al. Human bronchial epithelial cells express and secrete MMP-12. Biochemical and biophysical research communications. 2004;324(2):534–546. doi: 10.1016/j.bbrc.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 32.Fahy JV. Remodeling of the airway epithelium in asthma. Am J Respir Crit Care Med. 2001;164(10 Pt 2):S46–S51. doi: 10.1164/ajrccm.164.supplement_2.2106066. [DOI] [PubMed] [Google Scholar]
- 33.Pouladi MA, Robbins CS, Swirski FK, Cundall M, McKenzie AN, Jordana M, et al. Interleukin-13-dependent expression of matrix metalloproteinase-12 is required for the development of airway eosinophilia in mice. Am J Respir Cell Mol Biol. 2004;30(1):84–90. doi: 10.1165/rcmb.2003-0051OC. [DOI] [PubMed] [Google Scholar]
- 34.Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12(3):239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 35.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135(5):1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 36.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126(3):581–589. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 37.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61(1):33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 38.Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Nguyen LT, Uchida T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70(6):2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 39.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70(22):9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 40.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–291. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):160–165. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal immunol. 2008;1(4):289–296. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64(24):9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 44.Kariyawasam HH, Pegorier S, Barkans J, Xanthou G, Aizen M, Ying S, et al. Activin and transforming growth factor-beta signaling pathways are activated after allergen challenge in mild asthma. J Allergy Clin Immunol. 2009;124(3):454–462. doi: 10.1016/j.jaci.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182(8):4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 Limits In Vivo Immune Response-Mediated Activation of the IL-12/IFN-{gamma} Pathway, Th1 Polarization, and the Severity of Delayed-Type Hypersensitivity. J Immunol. 2011;187(6):3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–W102. doi: 10.1093/nar/gkq418. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.