Abstract
The G protein-coupled bradykinin B2 receptor (Bdkrb2) plays an important role in regulation of blood pressure under conditions of excess salt intake. Our previous work has shown that Bdkrb2 also plays a developmental role since Bdkrb2−/− embryos, but not their wild-type or heterozygous littermates, are prone to renal dysgenesis in response to gestational high salt intake. Although impaired terminal differentiation and apoptosis are consistent findings in the Bdkrb2−/− mutant kidneys, the developmental pathways downstream of gene-environment interactions leading to the renal phenotype remain unknown. Here, we performed genome-wide transcriptional profiling on embryonic kidneys from salt-stressed Bdkrb2+/+ and Bdkrb2−/− embryos. The results reveal significant alterations in key pathways regulating Wnt signaling, apoptosis, embryonic development, and cell-matrix interactions. In silico analysis reveal that nearly 12% of differentially regulated genes harbor one or more Pax2 DNA-binding sites in their promoter region. Further analysis shows that metanephric kidneys of salt-stressed Bdkrb2−/− have a significant downregulation of Pax2 gene expression. This was corroborated in Bdkrb2−/−;Pax2GFP+/tg mice, demonstrating that Pax2 transcriptional activity is significantly repressed by gestational salt-Bdkrb2 interactions. We conclude that gestational gene (Bdkrb2) and environment (salt) interactions cooperate to impact gene expression programs in the developing kidney. Suppression of Pax2 likely contributes to the defects in epithelial survival, growth, and differentiation in salt-stressed BdkrB2−/− mice.
Keywords: kidney development, bradykinin receptor-knockout mice, paired box transcription factor-2, gestational salt intake, microarray
congenital abnormalities of the kidney and urinary tract are the major cause of end-stage renal disease in infants and children younger than 4 yr of age (38). Depending on the study, genetic analysis has demonstrated that monogenic mutations account for 1.9–30% of cases of human renal dysgenesis (22, 36, 43, 47). These mutations usually involve key transcription factors (e.g., PAX2, EYA1, SALL1, HNF1β, WT1, HOXA11), secreted factors, extracellular matrix proteins, and receptor molecules (e.g., c-RET). Despite these advances, the etiology of the majority of cases of renal dysgenesis remains unknown. In this regard, environmental factors, such as vitamin A deficiency, gestational exposure to aminoglycosides, or nutritional protein deficiency, have been implicated (3, 7, 24, 45, 46). Another important cause of developmental defects is gene-environment interactions. In this case, neither the gene mutation nor the environmental stressor is capable of inducing a phenotypic change by itself; rather, a silent mutation requires an environmental stressor to reveal the abnormal phenotype. The cellular and molecular mechanism(s) by which gene-environment interactions induce organ dysgenesis and dysfunction are largely unknown, although DNA damage and epigenetic factors have been implicated (23).
In previous studies, we reported that mice lacking the G protein-coupled bradykinin B2 receptor gene (Bdkrb2) are prone to renal dysgenesis if exposed to gestational high salt via the maternal diet (13, 14). Importantly, neither Bdkrb2 gene disruption nor gestational salt stress alone is sufficient to cause renal dysgenesis, and both factors must interact to initiate metanephric apoptosis and impaired terminal epithelial cell differentiation (17, 21). The onset of renal dysgenesis is preceded by phosphorylation of the transcription factor p53 on serine 23 (Ser20 in human p53) by the checkpoint kinase 1 (Chk1) (17). In the presence of salt stress, P-Ser23-p53 activates promoters of proapoptotic genes (e.g., Bax), while repressing terminal differentiation genes (e.g., tubular transporters, cell adhesion molecules, and transcriptional regulators such as HNF1β) (17, 19). Germline deletion of p53 or Bax genes partially rescues the abnormal renal phenotype in salt-stressed Bdkrb2−/− mice (19). Similarly, germline (knock-in) substitution of Ser23 by Ala23 in p53 prevents the development of renal dysgenesis in salt-stressed Bdkrb2-null mutants (12).
The present study was designed to examine the effects of fetal salt-Bdkrb2 interactions on the overall gene expression profile of the midgestation developing kidney. The results reveal a wide-ranging effect on key cellular pathways such as Wnt signaling, calcium signaling, apoptosis, and patterning. Moreover, our studies uncover an important effect of fetal salt-Bdkrb2 interactions on Pax2 gene expression, a key transcriptional regulator of renal growth and differentiation.
MATERIALS AND METHODS
Animals.
All animal protocols utilized in this study were approved by and in strict adherence to guidelines established by the Tulane University Institutional Animal Care and Use Committee. Mice with disruption of the entire protein-coding region (exon 3) of Bdkrb2 bred on C57bl6 background have been described (4, 14). Pax2(GFP) BAC transgenic mice (Pax2gfp/+) carrying 30 kb of Pax2 genomic upstream regulatory sequences driving a GFP cassette were generously provided by the laboratory of Maxime Bouchard (30). These mice express the reporter faithfully mimicking the spatiotemporal developmental expression of endogenous Pax2. Bdkrb2+/−;Pax2-GFP+ were back-crossed to Bdkrb2−/− or Bdkrb2+/− mice, and pregnant mice were placed on 5% NaCl diet for the duration of pregnancy or until surgical dissection, as described (8, 14). Genotyping was performed by PCR of genomic DNA. Bdkrb2 wild-type and mutant alleles were amplified with the following primers: 435: GGTCCTGAACACCAACATGG, 434: TGTCCTCAGCGTGTTCTTCC, and 014: AGGTGAGATGACAGGAGATC, and 013: CTTGGGTGGAGAGGCTATTC; and GFP: forward ACTGGTGTGAGAGGCGGGTTCTT, reverse GCTGGCGAAAGGGGGATGTGCTG.
RNA extraction and RT-PCR.
Total RNA was isolated with the RNeasy Mini Kit (QIAGEN, Valencia, CA). Quantitative Real Time PCR assays were performed with the 1-Step Brilliant QRT-PCR Master Mix Kit (Stratagene) containing 100 nM forward primer, 100 nM reverse primer, and 20 ng total RNA. Relative mRNA levels were normalized to GAPDH. The following real-time primers were purchased from Applied Biosystems: GFP transgene forward: CCACATGAAGCAGCAGGACTT and GFP transgene reverse: GGTGCGCTCCTGGACGTA, and probe TTCAAGTCCGCCATGCCCGAA labeled on the 5′- and 3′-ends with 6-FAM and TAMRA, respectively; PAX2 TaqMan primer set Mm01217933mH; GAPDH TaqMan Rodent primer set number Mm4308313.
Microarray.
Heterozygous pairings were done to obtain Bdkrb2+/+ and Bdkrb2−/− litter-matched kidneys. RNA (500 ng) was obtained from each of three pairs of Bdkrb2+/+ and Bdkrb2+/+ embryonic day (E) 14.5 kidneys, amplified, and labeled, and the cDNA was hybridized to Agilent 4X44K Whole Mouse Genome Microarray slide from Agilent Technologies. To prevent bias from signaling detection on different colors (Cy5 red, Cy3 green), we applied dye-swap strategy to this experiment, in which identical pairs of Bdkrb2+/+ and Bdkrb2−/− cDNA samples were reversely labeled by Cy5 and Cy3 into two groups. Raw data were processed by GeneSpring software. Only genes showing a significant (P < 0.05) differential change in expression after Benjamini and Hochberg false discovery rate (FDR) correction in the three datasets were used for further analyses. The microarray results were deposited in the National Center for Biotechnology Information database; the Gene Expression Omnibus number is GSE26681.
Quantitative real-time PCR.
Validation of microarray data was done by RT-PCR on RNA from E14.5 Bdkrb2+/+ and Bdkrb2−/− kidneys. Exon-spanning primer-probes for TaqMan gene expression assay from Applied Biosystems were utilized. Reactions were prepared by TaqMan RNA-to-CT 1-Step Kit and amplified by Stratagene Mx3000P and Mx3005P. Expression was normalized against endogenous GAPDH mRNA levels. Experimental and biological triplicates were applied.
Metanephric organ culture.
Embryonic kidneys were aseptically microdissected from timed-pregnant mice at E12.5 and were cultured for the times indicated on polycarbonate Transwell filters (0.4 μm pore size; Corning Costar, Acton, MA) over medium (DMEM/F12 medium + 10% FBS) at 37°C and 5% CO2, as described (35).
Immunofluorescence staining.
Postnatal day (P) 1 and E14.5 kidneys were fixed in 10% formalin and processed for paraffin embedding and sectioning. Antibodies for the following proteins were used: Pax2 (1:200, Invitrogen), LTA (1:100, Vector Laboratories), AQP-2 (1:500, SC-9882; Santa Cruz Biotechnology, Santa Cruz, CA), and Cytokeratin (1:200, Sigma).
The spatiotemporal expression pattern of the Pax2-GFP transgene was recorded and analyzed by GFP Fluorescent Microscopy during 48 h ex vivo organ culture. Images were acquired at the same exposure with Slidebook 4 (Intelligent imaging innovations, Denver, CO) operating an Olympus Spinning Disc microscope fitted with a Hamamatsu CCD. Sum pixel intensity was measured with the “mask segment” function of Slidebook 4. The same mask parameters were assigned to all images. Relative GFP fluorescence pixel intensity mask data were then exported and subjected to statistical analysis by t-test or ANOVA.
RESULTS
BdkrB2-regulated transcriptome in the developing kidney.
We previously reported that gestational salt stress of BdkrB2-null mice causes renal dysgenesis in the homozygous but not heterozygous or wild-type littermates (12, 14, 17, 19). To identify differentially expressed genes in BdkrB2-null kidneys that might be responsible for the phenotype, we performed gene expression microarray on E14.5 BdkrB2+/+ and BdkrB2−/− litter-matched kidneys. We chose E14.5 embryonic age because the renal phenotype becomes histologically evident between E15.5 and E16.5 (14, 17, 19). By two-color dye-swap strategy, the microarrays (44,000 probes) detected 348 unique genes with significant changes in gene expression (>1.5-fold change, P < 0.05) (Fig. 1). Of these, 158 transcripts were upregulated, and 190 transcripts were downregulated.
Fig. 1.
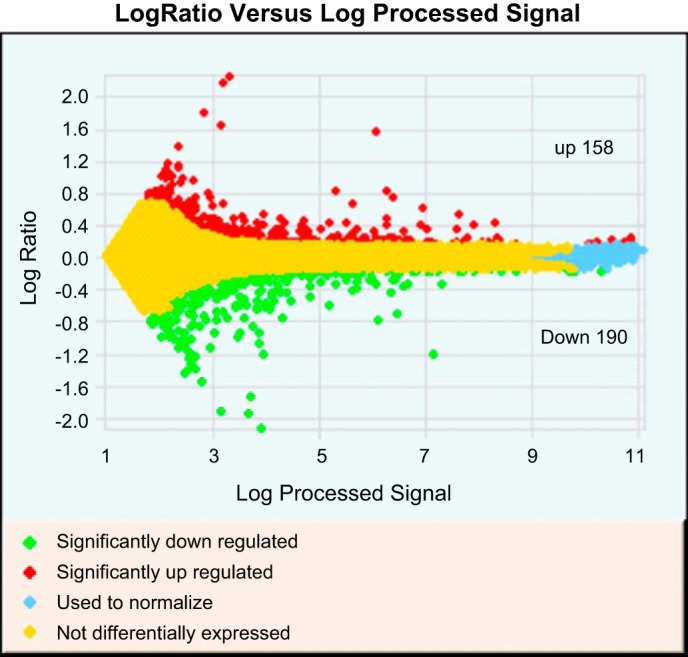
A scatterplot depicting the distribution of significantly altered genes between gestational salt-stressed Bdkrb2−/− and Bdkrb2+/+ kidneys at embryonic day (E) 14.5. Red, upregulated; yellow, unaltered; green, downregulated (P < 0.05, >1.5-fold).
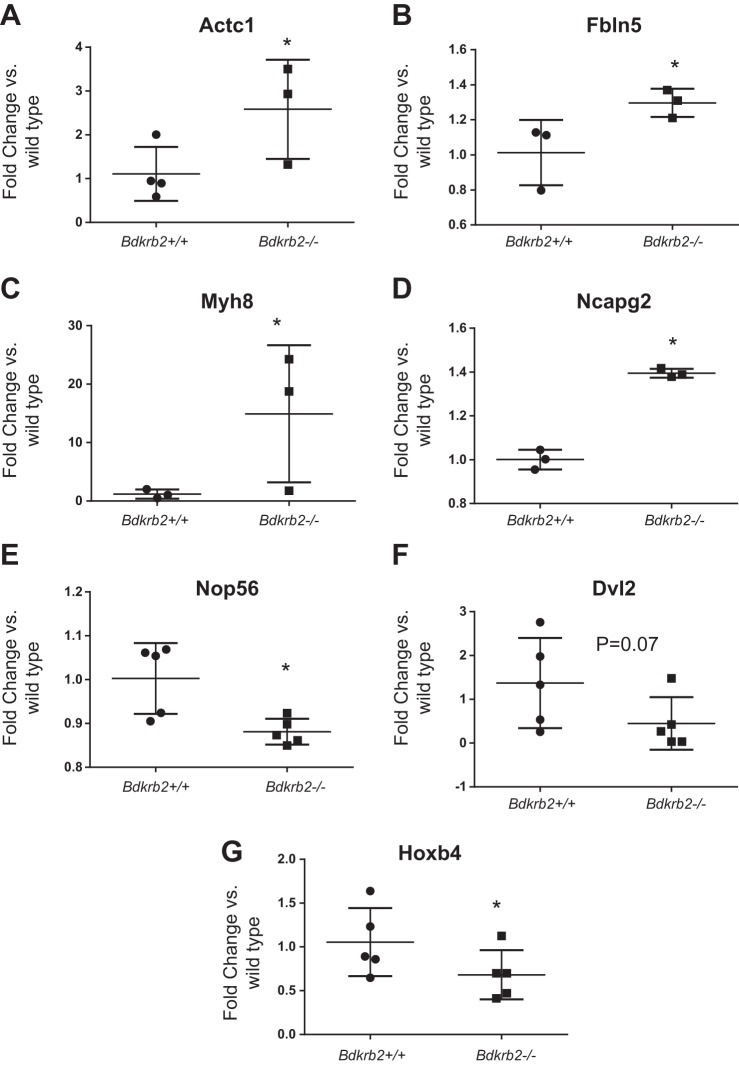
Gene Ontology (GO) functional annotation of differentially expressed genes in wild-type and BdkrB2 mutants revealed that the top functional categories comprise regulation of calcium transport, extracellular structure/matrix organization, cell communication, Wnt receptor signaling, pattern specification, apoptosis, embryonic development, and anatomical structure morphogenesis (Fig. 2). Further functional categorization revealed that the upregulated genes in BdkrB2 mutants play integral roles in cell adhesion, transcriptional regulation, muscle development, and cell proliferation and fate (Fig. 3A). Downregulated genes clustered in pathways pertaining to DNA, RNA and protein metabolism, transcriptional regulation, cell proliferation/fate, and chromatin modification (Fig. 3B). Table 1 lists a subset of differentially expressed transcripts with known expression pattern in the developing kidney based on the GUDMAP database. Interestingly, several transcription factors (e.g., Homeobox genes, GATA6, and RXR) and Wnt6 are among the downregulated genes in the mutant kidneys. A more complete list of significantly upregulated and downregulated genes is presented in Tables 2 and 3. Quantitative RT-PCR confirmed the directional changes in five randomly selected genes, as well as in Dvl2 and Hoxb4 (Fig. 4, A–G).
Fig. 2.
Gene ontology analysis of differentially regulated genes in E14.5 Bdkrb2−/− kidneys.
Fig. 3.

Functional categorization of upregulated (A) and downregulated (B) genes in gestational salt-stressed BdkrB2 mutant kidneys.
Table 1.
Selected differentially expressed transcripts with proven expression in GUDMAP database
| Probe Set ID | Gene Title | Gene Symbol | Fold Change | P Value |
|---|---|---|---|---|
| A_51_P402391 | activin A receptor, type 1B | Acvr1b | 2.47 | 0.00598 |
| A_51_P286301 | bradykinin receptor, beta 1 | Bdkrb1 | 2.30 | 0.0197 |
| A_51_P149469 | actin, alpha, cardiac muscle 1 | Actc1 | 1.99 | 0.0232 |
| A_51_P457196 | secreted frizzled-related protein 4 | Sfrp4 | 1.64 | 0.0204 |
| A_51_P116651 | dermatopontin | Dpt | 1.59 | 0.0362 |
| A_52_P480351 | lysyl oxidase | Lox | 1.56 | 0.0163 |
| A_51_P487518 | caspase 12 | Casp12 | 1.52 | 0.0113 |
| A_52_P299915 | mitogen-activated protein kinase kinase 6 | Map2k6 | 1.52 | 0.005 |
| A_51_P176365 | GTPase, IMAP family member 5 | Gimap5 | 1.52 | 0.0336 |
| A_52_P451073 | tumor necrosis factor receptor superfamily, member 21 | Tnfrsf21 | 1.50 | 0.000329 |
| A_52_P315155 | Eph receptor B2 | Ephb2 | −1.50 | 0.0488 |
| A_52_P415155 | wingless-related MMTV integration site 6 | Wnt6 | −1.50 | 0.00602 |
| A_52_P349939 | enolase 1, alpha nonneuron | Eno1 | −1.50 | 0.0371 |
| A_52_P285992 | ADP-ribosylation factor GTPase activating protein 2 | Zfp289 | −1.51 | 0.034 |
| A_51_P246471 | pre-B cell leukemia transcription factor 2 | Pbx2 | −1.51 | 0.0123 |
| A_51_P112319 | homeobox D11 | Hoxd11 | −1.52 | 0.0271 |
| A_51_P244892 | protein inhibitor of activated STAT 4 | Pias4 | −1.53 | 0.00837 |
| A_51_P360586 | homeobox A6 | Hoxa6 | −1.57 | 0.00555 |
| A_52_P519689 | Retinold X receptor beta | Rxrb | −1.59 | 0.0429 |
| A_52_P147070 | calcium channel alpha 2/delta subunit 2 | Cacna2d2 | −1.62 | 0.00131 |
| A_52_P565957 | homeobox B4 | Hoxb4 | −1.66 | 0.0055 |
| A_52_P595908 | ring finger protein 138 | Rnf138 | −1.68 | 0.0308 |
| A_52_P590175 | GATA binding protein 6 | Gata6 | −2.85 | 0.0205 |
| A_51_P301483 | homeobox C8 | Hoxc8 | −18.87 | 0.0357 |
Table 2.
Upregulated genes in gestational salt-stressed BdkrB2−/− relative to BdkrB2+/+ mice
| Gene Symbol | Entrez ID | Entrez Gene Name | Molecular Function | P Value |
|---|---|---|---|---|
| NCAPG2 | 76044 | nonSMC condensin II complex, subunit G2 | mitotic chromosome assembly and segregation | 3.22E-09 |
| MYH8 | 17885 | myosin, heavy chain 8, skeletal muscle, perinatal | structural constituent of muscle | 4.14E-02 |
| FBLN5 | 23876 | fibulin 5 | cell adhesion; cell-matrix adhesion | 4.03E-06 |
| PLAC9 | 100039175 | placenta-specific 9 | extracellular region | 7.04E-03 |
| ACTC1 | 11464 | actin, alpha, cardiac muscle 1 | actin filament-based movement | 1.84E-02 |
| MYOD1 | 17927 | myogenic differentiation 1 | myogenic factors, muscle regeneration | 2.34E-02 |
| RPL31 | 114641 | ribosomal protein L31 | Ribosomes, component of the 60S subunit | 3.32E-03 |
| FNDC1 | 68655 | fibronectin type III domain containing 1 | extracellular region | 4.47E-02 |
| CNN1 | 12797 | calponin 1, basic, smooth muscle | calmodulin binding, muscle contraction | 4.85E-02 |
| POP4 | 66161 | processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) | processing of precursor RNAs, alternative splicing | 4.85E-02 |
| TEX14 | 83560 | testis expressed 14 | protein amino acid phosphorylation | 2.99E-02 |
| CRIP1 | 12925 | cysteine-rich protein 1 (intestinal) | zinc ion binding, cell proliferation | 2.03E-02 |
| LUM | 17022 | lumican | regulate collagen fibril organization, epithelial cell migration | 3.44E-02 |
| PRELP | 116847 | proline/arginine-rich end leucine-rich repeat protein | extracellular matrix, skeletal system development | 2.51E-02 |
| NRXN1 | 18189 | neurexin 1 | cell adhesion molecules and receptors | 1.36E-03 |
| CD4 | 12504 | CD4 molecule | membrane glycoprotein of T lymphocytes | 1.65E-03 |
| KDM6A | 22289 | lysine (K)-specific demethylase 6A | demethylation of tri/dimethylated histone H3 | 4.47E-02 |
| ACTG2 | 11475 | actin, gamma 2, smooth muscle, enteric | muscle contraction | 4.42E-02 |
| CLEC2D | 232409 | C-type lectin domain family 2, member D | natural killer cell receptor C-type lectin family | 2.47E-02 |
| CLMN | 94040 | calmin (calponin-like, transmembrane) | actin binding | 6.17E-04 |
| WNT1 | 22408 | wingless-type MMTV integration site family, member 1 | signal transducer activity; frizzled binding, transcription activator activity | 2.47E-02 |
| MYH11 | 17880 | myosin, heavy chain 11, smooth muscle | smooth muscle myosin, muscle contraction | 4.06E-02 |
| COL12A1 | 12816 | collagen, type XII, alpha 1 | modify the interactions between collagen I fibrils | 3.62E-04 |
| FGF7 | 14178 | fibroblast growth factor 7 | type 2 fibroblast growth factor receptor binding | 1.94E-02 |
| GPRC5D | 93746 | G protein-coupled receptor, family C, group 5, member D | G protein-coupled receptor, undiscovered function | 3.45E-02 |
| KDM5C | 20591 | lysine (K)-specific demethylase 5C | chromatin modification; regulation of transcription | 2.84E-03 |
| SVEP1 | 64817 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | chromatin binding; calcium ion binding, cell adhesion | 1.53E-02 |
| AHNAK | 66395 | AHNAK nucleoprotein | protein binding, nervous system development | 4.75E-02 |
| ANXA8 | 11752 | annexin A8 | Anticoagulant, inhibits the thromboplastin-specific complex | 3.94E-02 |
| DCN | 13179 | decorin | component of connective tissue, binds to type I collagen fibrils | 2.35E-03 |
| CCBP2 | 59289 | chemokine binding protein 2 | beta chemokine receptor, development and growth of vascular tumors | 4.06E-02 |
| GPR116 | 224792 | G protein-coupled receptor 116 | G-protein coupled receptor | 1.02E-02 |
| CALM1 | 640703 | calmodulin 1 (phosphorylase kinase, delta) | calcium-binding protein, neurogenesis | 2.14E-02 |
| PRSS35 | 244954 | protease, serine, 35 | Trypsin, proteolysis, | 5.50E-03 |
| MATN2 | 17181 | matrilin 2 | formation of filamentous networks | 8.52E-04 |
| COL6A3 | 12835 | collagen, type VI, alpha 3 | type VI collagen, cell adhesion in muscle | 1.71E-02 |
| ELN | 13717 | elastin | elastic fibers, extracellular matrix | 3.09E-02 |
| F13A1 | 74145 | coagulation factor XIII, A1 polypeptide | blood coagulation; peptide cross-linking | 3.09E-03 |
| ARSI | 545260 | arylsulfatase family, member I | cell signaling, and degradation of macromolecules | 2.10E-02 |
| BCAT2 | 12036 | branched chain amino-acid transaminase 2, mitochondrial | branched chain family amino acid biosynthesis | 4.48E-02 |
| VWF | 22371 | von Willebrand factor | platelet-vessel wall mediator | 7.62E-03 |
| ADAMTS5 | 23794 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | metalloendopeptidase activity, proteolysis | 1.03E-03 |
| SPON2 | 100689 | spondin 2, extracellular matrix protein | cell adhesion; axon guidance | 1.55E-03 |
| AKAP12 | 83397 | A kinase (PRKA) anchor protein 12 | adenylate cyclase binding, cAMP biosynthesis | 6.99E-03 |
| CRIM1 | 50766 | cysteine rich transmembrane BMP regulator 1 (chordin-like) | interact with growth factors, implicated in motor neuron differentiation and survival | 6.42E-03 |
| EGFR | 13649 | epidermal growth factor receptor | activation of MAPKK activity; cell morphogenesis | 4.22E-02 |
| FLT1 | 14254 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | VEGFR family, receptor tyrosine kinases, angiogenesis and vasculogenesis | 1.62E-02 |
| WNT2 | 22413 | wingless-type MMTV integration site family member 2 | secreted signaling proteins, regulation of cell fate and patterning during embryogenesis | 2.07E-02 |
| ITPR2 | 16439 | inositol 1,4,5-triphosphate receptor, type 2 | ion channel activity, response to hypoxia | 2.50E-02 |
| SLIT3 | 20564 | slit homolog 3 (Drosophila) | calcium ion binding, organ morphogenesis | 4.88E-03 |
| CDK6 | 12571 | cyclin-dependent kinase 6 | G1 phase of mitotic cell cycle, cell-matrix adhesion | 1.12E-02 |
| MYLK | 107589 | myosin light chain kinase | phosphorylates myosin regulatory light chains | 1.65E-02 |
| COL1A2 | 12843 | collagen, type I, alpha 2 | extracellular matrix, skeletal system development | 4.80E-02 |
| CAV1 | 12389 | caveolin 1, caveolae protein, 22 kDa | coupling integrins to the Ras-ERK pathway and promoting cell cycle progression | 4.13E-02 |
| STARD9 | 668880 | StAR-related lipid transfer (START) domain containing 9 | microtubule motor activity, microtubule-based movement | 3.00E-02 |
| DSP | 109620 | desmoplakin | intercellular junctions, cell-cell adhesion | 5.78E-03 |
| LTBP4 | 108075 | latent transforming growth factor beta binding protein 4 | transforming growth factor beta receptor activity, regulation of cell growth | 1.36E-03 |
| RB1 | 19645 | retinoblastoma 1 | tumor suppressor gene | 4.78E-02 |
| FOXP2 | 114142 | forkhead box P2 | orkhead/winged-helix (FOX) family of transcription factors. | 2.47E-02 |
| FBLN1 | 14114 | fibulin 1 | incorporated into fibrillar extracellular matrix | 1.94E-02 |
| ITPR1 | 16438 | inositol 1,4,5-triphosphate receptor, type 1 | intracellular receptor for inositol 1,4,5-trisphosphate, response to hypoxia | 1.99E-02 |
| THBS1 | 21825 | thrombospondin 1 | adhesive glycoprotein, angiogenesis | 5.50E-03 |
| ABCC9 | 20928 | ATP-binding cassette, sub-family C (CFTR/MRP), member 9 | ATP-binding cassette (ABC) transporters, multi-drug resistance, | 3.01E-02 |
| NBEA | 26422 | neurobeachin | Target protein kinase A to specific subcellular sites | 4.85E-02 |
| LPP | 210126 | LIM domain containing preferred translocation partner in lipoma | protein binding, cell-cell adhesion and cell motility, transcriptional co-activator | 2.00E-02 |
| NFIA | 18027 | nuclear factor I/A | cellular transcription factors | 1.16E-02 |
| NAV3 | 260315 | neuron navigator 3 | ATPases associated with cellular activities | 6.52E-03 |
| COL3A1 | 12825 | collagen, type III, alpha 1 | type III collagen, cell-matrix adhesion | 1.02E-02 |
| RUNX2 | 12393 | runt-related transcription factor 2 | RUNX family of transcription factors, | 2.55E-02 |
| TNS1 | 21961 | tensin 1 | protein binding, focal adhesions, | 7.76E-03 |
| WISP1 | 22402 | WNT1 inducible signaling pathway protein 1 | a member of the WNT1 inducible signaling pathway (WISP) protein subfamily | 3.94E-02 |
| ADH1A | 0 | alcohol dehydrogenase 1A (class I), alpha polypeptide | alcohol metabolic process; oxidation reduction | 1.79E-02 |
| LAMA2 | 16773 | laminin, alpha 2 | mediate the attachment, migration, and organization of cells | 2.09E-06 |
| NID1 | 18073 | nidogen 1 | basement membrane glycoproteins, adhesion | 1.57E-03 |
| PCDH10 | 18526 | protocadherin 10 | cadherin superfamily, cell adhesion | 3.26E-02 |
| PPP2R2B | 72930 | protein phosphatase 2, regulatory subunit B, beta | negative control of cell growth and division, apoptosis | 2.96E-02 |
| SLC24A3 | 94249 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | intracellular calcium homeostasis, | 4.55E-02 |
| MANBA | 110173 | mannosidase, beta A, lysosomal | Lysosomal hydrolase activity, | 4.59E-02 |
| FBLN1 | 14114 | fibulin 1 | extracellular matrix structural constituent | 1.27E-02 |
| FIGF | 14205 | c-fos induced growth factor (vascular endothelial growth factor D) | angiogenesis, lymphangiogenesis, and endothelial cell growth | 4.49E-02 |
| FZD7 | 14369 | frizzled homolog 7 (Drosophila) | receptors for Wnt signaling proteins | 4.16E-03 |
| AGRN | 11603 | agrin | formation of the neuromuscular junction | 1.89E-02 |
| ITPR2 | 16439 | inositol 1,4,5-triphosphate receptor, type 2 | response to hypoxia, inositol 1,4,5-trisphosphate-sensitive calcium-release channel | 2.09E-03 |
| HIPK2 | 15258 | homeodomain interacting protein kinase 2 | interacts with homeodomain transcription factors | 4.45E-02 |
| KLF4 | 16600 | Kruppel-like factor 4 (gut) | transcription factor, mesodermal cell fate determination | 6.44E-03 |
| SIK3 | 70661 | SIK family kinase 3 | protein amino acid phosphorylation | 2.23E-03 |
| CHST3 | 53374 | carbohydrate (chondroitin 6) sulfotransferase 3 | catalyzes the sulfation of chondroitin, nvolved in cell migration and differentiation | 3.45E-02 |
| SLIT2 | 20563 | slit homolog 2 (Drosophila) | GTPase inhibitor activity, metanephros development, ureteric bud developmen | 4.35E-02 |
| COL15A1 | 12819 | collagen, type XV, alpha 1 | adhere basement membranes to underlying connective tissue stroma | 3.84E-02 |
| BACH2 | 12014 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 | transcription factor activity, regulation of transcription | 3.09E-02 |
| DMD | 13405 | dystrophin | muscle organ development | 1.19E-03 |
| EMX1 | 13796 | empty spiracles homeobox 1 | transcription factor activity | 3.22E-02 |
| ANTXR1 | 69538 | anthrax toxin receptor 1 | actin cytoskeleton reorganization | 1.27E-02 |
| EDNRA | 13617 | endothelin receptor type A | potent and long-lasting vasoconstriction | 1.37E-02 |
| CELSR1 | 12614 | cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) | establishment of planar polarity, cell adhesion | 4.45E-02 |
| LRP2BP | 67620 | LRP2 binding protein | protein binding | 8.84E-03 |
| PMP22 | 18858 | peripheral myelin protein 22 | major component of myelin | 1.71E-02 |
| DDX3X | 13205 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | alteration of RNA secondary structure | 3.38E-03 |
| POSTN | 50706 | periostin, osteoblast specific factor | heparin binding, cell adhesion, | 3.38E-02 |
| PLCG2 | 234779 | phospholipase C, gamma 2 (phosphatidylinositol-specific) | phosphoinositide phospholipase C activity | 4.75E-02 |
| ICAM1 | 15894 | intercellular adhesion molecule 1 | T cell activation, regulation of cell adhesion | 4.32E-02 |
| ITGB4 | 192897 | integrin, beta 4 | cell communication; cell adhesion | 4.62E-02 |
| COL6A1 | 12833 | collagen, type VI, alpha 1 | maintaining the integrity of various tissues | 3.46E-02 |
| TRPM5 | 56843 | transient receptor potential cation channel, subfamily M, member 5 | voltage-gated ion channel activity | 4.14E-02 |
| CALCRL | 54598 | calcitonin receptor-like | regulation of muscle contraction | 3.44E-02 |
| MLL5 | 69188 | myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila) | histone methyltransferase activity (H3-K4 specific) | 1.58E-02 |
| FAM46A | 212943 | family with sequence similarity 46, member A | N/A | 2.14E-02 |
| MYOF | 226101 | myoferlin | membrane regeneration and repair | 3.00E-02 |
| SFRP2 | 20319 | secreted frizzled-related protein 2 | putative Wnt-binding site of Frizzled proteins | 2.47E-02 |
| LHFP | 108927 | lipoma HMGIC fusion partner | member of the lipoma HMGIC fusion partner | 7.40E-03 |
| KCNK4 | 16528 | potassium channel, subfamily K, member 4 | voltage-gated ion channel activity | 3.00E-02 |
| ADAMTS15 | 235130 | ADAM metallopeptidase with thrombospondin type 1 motif, 15 | metalloendopeptidase activity, proteolysis | 4.13E-02 |
| TMCC3 | 319880 | transmembrane and coiled-coil domain family 3 | N/A | 4.12E-02 |
| G0S2 | 14373 | G0/G1 switch 2 | Apoptosis, cell cycle | 2.75E-02 |
| RBMS3 | 207181 | RNA binding motif, single stranded interacting protein 3 | c-myc gene single-strand binding protein | 4.06E-02 |
| CD248 | 70445 | CD248 molecule, endosialin | sugar binding | 4.06E-02 |
| FAM107A | 268709 | family with sequence similarity 107, member A | regulation of cell growth | 2.32E-02 |
| PDGFRB | 18596 | platelet-derived growth factor receptor, beta polypeptide | kidney development, positive regulation of cell proliferation | 4.45E-02 |
| NPPC | 18159 | natriuretic peptide precursor C | potent natriuretic, diuretic, and vasodilating activities | 1.84E-02 |
| CGNL1 | 68178 | cingulin-like 1 | motor activity | 3.73E-02 |
| B4GALT1 | 14595 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | type II membrane-bound glycoproteins, epithelial cell development | 2.70E-03 |
| MMRN2 | 105450 | multimerin 2 | extracellular matrix protein | 7.88E-03 |
| ATP7A | 11977 | ATPase, Cu2+ transporting, alpha polypeptide | copper transport across membranes, blood vessel development | 3.17E-03 |
| EMILIN3 | 280635 | elastin microfibril interfacer 3 | proteinaceous extracellular matrix | 2.67E-02 |
| SLC39A1 | 30791 | solute carrier family 39 (zinc transporter), member 1 | transmembrane transport | 6.44E-03 |
| PURB | 19291 | purine-rich element binding protein B | regulation of transcription | 2.84E-02 |
| SSH2 | 237860 | slingshot homolog 2 (Drosophila) | actin cytoskeleton organization | 6.46E-03 |
| FNDC3B | 72007 | fibronectin type III domain containing 3B | positive regulation of fat cell differentiation | 1.70E-02 |
| KCNJ8 | 16523 | potassium inwardly-rectifying channel, subfamily J, member 8 | inward rectifier potassium channel activity, kidney development | 6.80E-03 |
| ZFP36L1 | 12192 | zinc finger protein 36, C3H type-like 1 | nuclear-transcribed mRNA catabolic process, vasculogenesis | 2.50E-02 |
| CDS2 | 110911 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | hosphatidate cytidylyltransferase activity | 5.74E-03 |
| PBXIP1 | 229534 | preB-cell leukemia homeobox interacting protein 1 | transcription corepressor activity, cell differentiation | 4.93E-02 |
| PPP1R9A | 243725 | protein phosphatase 1, regulatory (inhibitor) subunit 9A | regulatory subunit of protein phosphatase I, controls actin cytoskeleton reorganization | 1.60E-02 |
| NID2 | 18074 | nidogen 2 (osteonidogen) | cell-adhesion protein that binds collagens I and IV | 1.08E-03 |
| PKD1 | 18763 | polycystic kidney disease 1 (autosomal dominant) | regulator of calcium permeable cation channels, calcium-independent cell-matrix adhesion | 2.47E-02 |
| SERTAD4 | 214791 | SERTA domain containing 4 | protein binding | 4.78E-02 |
| FBXO46 | 243867 | F-box protein 46 | protein binding | 4.01E-02 |
| SNX18 | 170625 | sorting nexin 18 | cell communication; protein transport | 4.85E-02 |
| PTPRF | 19268 | protein tyrosine phosphatase, receptor type, F | negative regulation of cytokine-mediated signaling pathway, cell adhesion | 2.30E-02 |
| SPCS2 | 66624 | signal peptidase complex subunit 2 homolog (S. cerevisiae) | peptidase activity, signal peptide processing | 2.67E-02 |
| NES | 18008 | nestin | central nervous system development | 5.50E-03 |
| SESN1 | 140742 | sestrin 1 | response to DNA damage stimulus, cell cycle arrest; negative regulation of cell proliferation | 3.94E-02 |
P < 0.05.
Table 3.
Downregulated genes in BdkrB2−/− relative to BdkrB2+/+ mice
| Gene Symbol | Entrez ID | Entrez Gene Name | Molecular Function | P value |
|---|---|---|---|---|
| KDM5D | 20592 | lysine (K)-specific demethylase 5D | chromatin modification | 6.52E-03 |
| NUDT16 | 75686 | nudix-type motif 16 | RNA binding; hydrolase activity; metal binding | 2.98E-02 |
| LTA | 16992 | lymphotoxin alpha | cytokine produced by lymphocytes | 4.75E-02 |
| STARD13 | 243362 | StAR-related lipid transfer (START) domain containing 13 | GTPase activator activity, signal transduction | 3.46E-02 |
| RAPGEF5 | 217944 | Rap guanine nucleotide exchange factor (GEF) 5 | small GTPase mediated signal transduction | 1.23E-05 |
| CREG2 | 263764 | cellular repressor of E1A-stimulated genes 2 | N/A | 1.27E-02 |
| DDX23 | 74351 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 | Embryogenesis and cellular growth and division | 4.49E-02 |
| NOP56 | 67134 | NOP56 ribonucleoprotein homolog (yeast) | rRNA processing | 3.51E-03 |
| RPS13 | 68052 | ribosomal protein S13 | negative regulation of RNA splicing | 3.53E-08 |
| ADAM23 | 23792 | ADAM metallopeptidase domain 23 | metalloendopeptidase activity | 1.27E-02 |
| MSX2 | 17702 | msh homeobox 2 | DNA binding; transcription factor activity | 3.43E-02 |
| TPCN1 | 252972 | two pore segment channel 1 | voltage-gated ion channel activity | 2.10E-02 |
| BCAT2 | 12036 | branched chain amino-acid transaminase 2, mitochondrial | Catalyzes production of the branched chain amino acids | 2.26E-02 |
| BSDC1 | 100383 | BSD domain containing 1 | protein binding | 2.47E-02 |
| GANC | 76051 | glucosidase, alpha; neutral C | carbohydrate metabolic process | 4.98E-02 |
| CHMP4B | 75608 | chromatin modifying protein 4B | interact with programmed cell death 6 interacting protein | 1.37E-02 |
| SCRIB | 105782 | scribbled homolog (Drosophila) | targeted to epithelial adherens junctions, cell polarization | 4.89E-02 |
| CLK1 | 12747 | CDC-like kinase 1 | governing splice site selection | 4.09E-03 |
| PABPN1 | 54196 | poly(A) binding protein, nuclear 1 | RNA processing; mRNA processing | 3.11E-02 |
| HOXD4 | 15436 | homeobox D4 | morphogenesis in all multicellular organisms | 3.51E-03 |
| TNKS2 | 74493 | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | NAD+ ADP-ribosyltransferase activity, regulation of multicellular organism growth | 1.66E-02 |
| DNASE1L2 | 66705 | deoxyribonuclease I-like 2 | DNA binding; endonuclease activity | 4.86E-02 |
| RAF1 | 110157 | v-raf-1 murine leukemia viral oncogene homolog 1 | MAP kinase kinase kinase (MAP3K), control of gene expression involved in the cell division cycle, apoptosis, cell differentiation and cell migration. | 1.70E-02 |
| GAS5 | 14455 | growth arrest-specific 5 (nonprotein coding) | N/A | 2.64E-02 |
| ZXDC | 80292 | ZXD family zinc finger C | positive regulation of transcription | 2.09E-03 |
| PNPLA6 | 50767 | patatin-like phospholipase domain containing 6 | deacetylates intracellular phosphatidylcholine | 4.85E-02 |
| BUB3 | 12237 | budding uninhibited by benzimidazoles 3 homolog (yeast) | spindle checkpoint function, mitotic sister chromatid segregation, cell division | 1.74E-02 |
| MTHFSD | 234814 | methenyltetrahydrofolate synthetase domain containing | folic acid and derivative biosynthetic process | 3.84E-02 |
| CLK4 | 12750 | CDC-like kinase 4 | peptidyl-tyrosine phosphorylation | 2.71E-02 |
| LSM7 | 66094 | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | nuclear mRNA splicing, via spliceosome | 3.88E-02 |
| RPL11 | 67025 | ribosomal protein L11 | RNA processing; translation; translational elongation | 9.14E-03 |
| NFATC2IP | 18020 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein | cytokine production; regulation of transcription | 4.45E-02 |
| IDI1 | 319554 | isopentenyl-diphosphate delta isomerase 1 | cholesterol biosynthetic process | 1.84E-02 |
| C16ORF70 | 234678 | chromosome 16 open reading frame 70 | Golgi to plasma membrane protein transport | 4.47E-02 |
| GRWD1 | 101612 | glutamate-rich WD repeat containing 1 | role in ribosome biogenesis | 1.25E-02 |
| KCTD5 | 69259 | potassium channel tetramerisation domain containing 5 | voltage-gated potassium channel activity | 1.89E-02 |
| DDX51 | 69663 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 | rRNA processing | 3.85E-02 |
| GTPBP3 | 70359 | GTP binding protein 3 (mitochondrial) | tRNA modification | 2.32E-02 |
| PCSK7 | 18554 | proprotein convertase subtilisin/kexin type 7 | proprotein convertases, peptide hormone processing | 4.06E-02 |
| RNF168 | 70238 | ring finger protein 168 | double-strand break repair; response to ionizing radiation; chromatin modification; positive regulation of DNA repair; histone H2A K63-linked ubiquitination | 1.27E-02 |
| EDC3 | 353190 | enhancer of mRNA decapping 3 homolog (S. cerevisiae) | associated with mRNA-decapping complex | 2.58E-02 |
| SNIP1 | 76793 | Smad nuclear interacting protein 1 | production of miRNAs involved in gene silencing | 3.39E-02 |
| DDX18 | 66942 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 | alteration of RNA secondary structure | 1.12E-04 |
| NANS | 94181 | N-acetylneuraminic acid synthase | lipopolysaccharide biosynthetic process | 3.73E-02 |
| MED29 | 67224 | mediator complex subunit 29 | a multiprotein coactivator of RNA transcription | 4.70E-03 |
| SUPV3L1 | 338359 | suppressor of var1, 3-like 1 (S. cerevisiae) | DNA duplex unwinding | 1.53E-02 |
| EXOSC7 | 66446 | exosome component 7 | 3¢-5¢-exoribonuclease activity, RNA processing | 2.47E-02 |
| TRIM39 | 79263 | tripartite motif-containing 39 | protein binding; zinc ion binding, apoptosis | 3.17E-02 |
| DOCK5 | 68813 | dedicator of cytokinesis 5 | guanyl-nucleotide exchange factor activity | 4.95E-02 |
| DNAJC11 | 230935 | DnaJ (Hsp40) homolog, subfamily C, member 11 | heat shock protein binding | 1.16E-02 |
| TBP | 21374 | TATA box binding protein | Initiation of transcription, regulation of transcription | 1.94E-02 |
| MOXD1 | 59012 | monooxygenase, DBH-like 1 | monooxygenase activity, catecholamine metabolic process | 1.02E-03 |
| DIDO1 | 23856 | death inducer-obliterator 1 | transcription; apoptosis | 4.06E-02 |
| NUP43 | 69912 | nucleoporin 43 kDa | Bidirectional transport of macromolecules | 8.52E-04 |
| KIAA1804 | 234878 | mixed lineage kinase 4 | protein phosphorylation, signal transduction | 4.85E-02 |
| RAD51AP1 | 19362 | RAD51 associated protein 1 | DNA recombination, DNA repair DNA binding | 3.85E-02 |
| HSPA14 | 50497 | heat shock 70 kDa protein 14 | ATP binding, nucleotide binding | 3.99E-03 |
| GPS1 | 1E +08 | G protein pathway suppressor 1 | catalyze deneddylation of proteins | 2.47E-02 |
| PRKAR2A | 19087 | protein kinase, cAMP-dependent, regulatory, type II, alpha | cAMP binding, cAMP-dependent protein kinase regulator activity | 3.91E-02 |
| GPKOW | 209416 | G patch domain and KOW motifs | ribosomal protein, nucleic acid binding | 1.37E-02 |
| TEX9 | 21778 | testis expressed 9 | testis-expressed sequence 9 protein | 3.01E-02 |
| GTPBP3 | 70359 | GTP binding protein 3 (mitochondrial) | GTP catabolic process, tRNA modification | 3.47E-02 |
| ASNS | 27053 | asparagine synthetase (glutamine-hydrolyzing) | asparagine biosynthetic process, cellular amino acid biosynthetic process | 1.65E-02 |
| KLHDC4 | 234825 | kelch domain containing 4 | peptidease activity, hydrolysis of peptide bond | 4.06E-02 |
| MED22 | 20933 | mediator complex subunit 22 | regulation of transcription, DNA directed RNA polymerase | 8.76E-03 |
| CCDC117 | 104479 | coiled-coil domain containing 117 | chromosomal protein | 4.13E-02 |
| HDAC10 | 170787 | histone deacetylase 10 | chromatin modification, histone deacetylation | 2.35E-03 |
| POLR3C | 74414 | polymerase (RNA) III (DNA directed) polypeptide C (62kD) | innate immune response, positive regulation of innate immune response | 1.98E-02 |
| MTRF1L | 108853 | mitochondrial translational release factor 1-like | translation release factor activity | 3.92E-02 |
| RNF34 | 80751 | ring finger protein 34 | ligase activity, metal ion binding, apoptosis, metabolic proces | 4.93E-04 |
| UBP1 | 22221 | upstream binding protein 1 (LBP-1a) | DNA binding, protein binding, angiogenesis, regulation of transcription | 3.94E-02 |
| BCCIP | 66165 | BRCA2 and CDKN1A interacting protein | kinase regulator activity, cell cycle, DNA repair | 4.85E-02 |
| LUC7L2 | 192196 | LUC7-like 2 (S. cerevisiae) | enzyme binding, metal ion binding | 1.89E-02 |
| NOC4L | 100608 | nucleolar complex associated 4 homolog (S. cerevisiae) | nucleolus protein, formation of ribosome | 8.76E-03 |
| PPHLN1 | 223828 | periphilin 1 | keratinization | 2.84E-03 |
| EIF4A3 | 192170 | eukaryotic translation initiation factor 4A3 | ATP-dependent RNA helicase activity, negative regulation of translation | 1.89E-02 |
| NIF3L1 | 65102 | NIF3 NGG1 interacting factor 3-like 1 (S. pombe) | transcription factor binding, positive regulation of transcription | 4.89E-02 |
| FASTKD5 | 380601 | FAST kinase domains 5 | protein kinase activity, apoptosis | 4.45E-02 |
| HAUS3 | 231123 | HAUS augmin-like complex, subunit 3 | cytoskeleton component, cell cycle, cell division | 3.53E-05 |
| PSEN1 | 19164 | presenilin 1 | beta-catenin binding, cadherin binding, activation of MAPKK activity | 1.66E-03 |
| PSMC1 | 19179 | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | hydrolase activity, negative regulation of ubiquitin- protein ligase activity involved in mitotic cell cycle | 4.06E-02 |
| EXO1 | 26909 | exonuclease 1 | 5′-3′ exodeoxyribonuclease activity, DNA recombination, DNA repair | 2.99E-02 |
| CBX2 | 12416 | chromobox homolog 2 | chromatin binding, DNA binding, chromatin assembly or disassembly, chromatin modification | 4.75E-02 |
| DIABLO | 66593 | diablo homolog (Drosophila) | activation of caspase activity, apoptosis | 4.42E-02 |
| EBNA1BP2 | 69072 | EBNA1 binding protein 2 | chromosome segregation, ribosome biogenesis | 1.25E-02 |
| FDPS | 110196 | farnesyl diphosphate synthase | dimethylallyltranstransferase activity, cholesterol biosynthetic proces | 3.00E-02 |
| METTL2B | 52686 | methyltransferase like 2B | methyltransferase activity, metabolic process, methylation | 2.45E-02 |
| PPP1R1A | 58200 | protein phosphatase 1, regulatory (inhibitor) subunit 1A | phosphoprotein phosphatase inhibitor activity, carbohydrate metabolic process | 4.85E-02 |
| SREBF2 | 20788 | sterol regulatory element binding transcription factor 2 | chromatin binding, DNA binding, cholesterol metabolic process | 2.10E-02 |
| RBM4 | 19653 | RNA binding motif protein 4 | mRNA 3′-UTR binding, circadian regulation of gene expression | 1.94E-02 |
| RPP38 | 227522 | ribonuclease P/MRP 38 kDa subunit | hydrolase activity, ribonuclease P activity, tRNA processing | 2.47E-02 |
| USP38 | 74841 | ubiquitin specific peptidase 38 | Process ubiquitin-dependent protein catabolic process | 4.95E-02 |
| TUBG1 | 103733 | tubulin, gamma 1 | GTPase activity, microtubule nucleation | 3.99E-03 |
| MAX | 17187 | MYC associated factor X | negative regulation of gene expression, protein complex assembly | 2.22E-02 |
| DGAT1 | 13350 | diacylglycerol O-acyltransferase 1 | acyltransferase activity, glycerolipid metabolic process | 3.99E-03 |
| POLR2G | 67710 | polymerase (RNA) II (DNA directed) polypeptide G | DNA-directed RNA polymerase activity, RNA splicing, transcription | 3.47E-02 |
| EIF3B | 27979 | eukaryotic translation initiation factor 3, subunit B | nucleic acid binding, regulation of translational initiation | 4.77E-02 |
| SC4MOL | 66234 | sterol-C4-methyl oxidase-like | C-4 methylsterol oxidase activity, iron ion binding, fatty acid biosynthetic process | 4.95E-02 |
| GLRX3 | 30926 | glutaredoxin 3 | electron carrier activity, iron-sulfur cluster binding, cell redox homeostasis | 3.01E-04 |
| TNFRSF19 | 29820 | tumor necrosis factor receptor superfamily, member 19 | receptor activity, positive regulation of I-kappaB kinase/NF-kappaB cascade | 4.85E-02 |
| DOHH | 1E +05 | deoxyhypusine hydroxylase/monooxygenase | oxidation-reduction process, peptidyl-lysine modification to hypusine | 5.49E-03 |
P < 0.05.
Fig. 4.
Quantitative RT-PCR validation of differentially expressed genes in the microarray. *P < 0.05.
Gestational salt stress suppresses epithelial Pax2 expression in BdkrB2−/− mice.
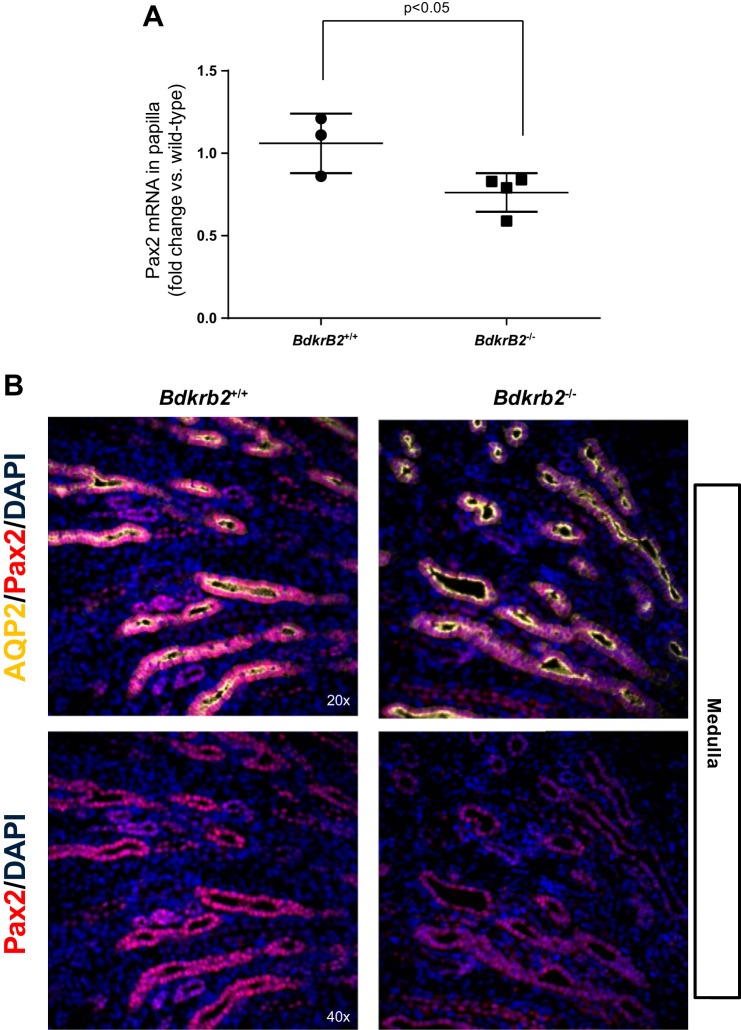
In silico analysis revealed that ∼12% of the altered genes in salt-stressed BdkrB2−/− embryonic kidneys (28 genes upregulated and 13 genes downregulated) harbor putative Pax2-binding sites in their promoter region (within −1 kb of the transcription start site) (Tables 4 and 5). Since Pax2-target genes in the E14.5 developing kidney are not known, it is not possible to discern if any of these genes are Pax2-regulated genes. Quantitative real-time RT-PCR performed on the renal medulla harvested from P1 salt-stressed Bdkrb2+/+ and Bdkrb2−/− kidneys validated expression changes of some genes (but not all). For example, we validated upregulation of Gpcr5d (+1.26 ± 0.10 fold vs. wild type, n = 5/group, P < 0.05) but failed to detect a statistically significant differential expression in Myh11, while there was tendency toward differential expression in Actg2 (+2.96 ± 1.15 P = 0.12). Although differential expression of Pax2 was not evident in the microarray, there was a significant yet modest (20%) reduction in Pax2 mRNA levels in mutants as compared wild-type mice (Fig. 5A). QPCR is more sensitive than hybridization-based technologies such as microarrays, which may explain why differential expression of Pax2 was not picked up by the microarray. Double immunofluorescence for Pax2 and the water channel AQP-2 confirmed the downregulation of Pax2 protein in collecting ducts of Bdkrb2 mice (Fig. 5B). Notably, downregulation of Pax2 is only observed in salt-stressed Bdkrb2 mutants indicating that Pax2 is responsive to gene-environment interactions and not to salt or gene mutations alone.
Table 4.
Upregulated genes in BdkrB2−/− with Pax2 binding sites
| Gene Symbol | Entrez ID | Entrez Name | Molecular Function | Expression in Kidney | Start Position | End Position | Inspecting Sequence ID |
|---|---|---|---|---|---|---|---|
| Myh8 | 17885 | myosin, heavy chain 8 | motor activity, constituent of muscle | trunk mesenchyme | 579 | 591 | GXP_92669 |
| Nrxn1 | 18189 | neurexin 1 | cell adhesion; axon guidance | trunk mesenchyme | 60 | 72 | GXP_185470 |
| Actg2 | 11468 | actin, gamma 2, smooth muscle, enteric | muscle contraction | ureter | 240 | 252 | GXP_303411 |
| Myh11 | 17880 | myosin, heavy chain 11, smooth muscle | muscle contraction | vasculature, pelvis, ureter | 238 | 250 | GXP_18305 |
| Gprc5d | 93746 | G protein-coupled receptor, family C, group 5, member D | G protein-coupled receptor activity | N/A | 449 | 461 | GXP_2537517 |
| Ahnak | 66395 | AHNAK nucleoprotein | nervous system development | ureteric tip and trunk, interstitium, vasculature | 341 | 353 | GXP_304725 |
| Ccbp2 | 59289 | chemokine binding protein 2 | chemokine receptor activity | kidney | 402 | 414 | GXP_1805682 |
| Col6a3 | 12835 | collagen, type VI, alpha 3 | cell adhesion; muscle organ development | diffused in metanephros | 652 | 664 | GXP_1457863 |
| Adamts5 | 23794 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | metalloendopeptidase activity, integrin binding, proteolysis | urogenital sinus, primitive bladder | 468 | 480 | GXP_1239277 |
| Sema5a | 20356 | sema domain, seven thrombospondin repeats | cell adhesion; cell-cell signaling; multicellular organismal development | ureteric tree and tips, vasculature, interstitium, mesenchyme, nephrons | 1098 | 1110 | GXP_89931 |
| Col1a2 | 12843 | collagen, type I, alpha 2 | extracellular matrix structural constituent | renal vasculature | 55 | 67 | GXP_821204 |
| Cav1 | 12389 | caveolin 1, caveolae protein | regulator of the Ras-p42/44 mitogen-activated kinase cascade | trunk mesenchyme | 32 | 44 | GXP_150594 |
| Rb1 | 19645 | retinoblastoma 1 | Cell cycle progression, apoptosis, proliferation regulator | kidney | 546 | 558 | GXP_1793835 |
| Itpr1 | 16438 | inositol 1,4,5-triphosphate receptor, type 1 | response to hypoxia, calcium ion transport, ion channel activity | renal vasculature | 537 | 549 | GXP_105230 |
| Thbs1 | 21825 | thrombospondin 1 | adhesive glycoprotein, cell-to-cell and cell-to-matrix interactions | metanephros, late tubules | 410 | 422 | GXP_1459831 |
| Tns1 | 21961 | tensin 1 | focal adhesions, cell attaches to the extracellular matrix | proximal and distal tubule, medullary collecting duct | 232 | 244 | GXP_2220687 |
| Manba | 110173 | mannosidase, beta A, lysosomal | hydrolase activity, glycoprotein catabolic process | N/A | 256 | 268 | GXP_1799746 |
| Figf | 14205 | c-fos induced growth factor | PDGF/VEGF family, active in angiogenesis | metanephros | 189 | 201 | GXP_1471435 |
| Spnb2 | 20742 | spectrin beta 2 | actin binding, calmodulin binding | trunk mesenchyme | 551 | 563 | GXP_118730 |
| Zfp26 | 22688 | Mus musculus zinc finger protein 26 | DNA binding, metal ion binding, transcription factor, glial development | mesenchyme, ureteric tip and trunk, interstitium, tubules and vasculature | 163 | 175 | GXP_826162 |
| Dmd | 13405 | dystrophin | dystrophin-glycoprotein complex, muscle organ development; skeletal muscle tissue development | metanephros | 338 | 350 | GXP_1470974 |
| Icam1 | 15894 | intercellular adhesion molecule 1 | receptor activity, integrin binding | N/A | 234 | 246 | GXP_445321 |
| Mll5 | 69188 | myeloid/lymphoid or mixed-lineage leukemia 5 | Histone methyltransferase, dimethylates 'Lys-4¢ of histone H3, myeloid differentiation | cap mesenchyme, interstitium, pelvic smooth muscle | 494 | 506 | GXP_239498 |
| Rbms3 | 207181 | RNA binding motif, single stranded interacting protein 3 | DNA replication, gene transcription, cell cycle progression and apoptosis | N/A | 267 | 279 | GXP_229583 |
| Nppc | 18159 | natriuretic peptide precursor C | vasodilating activities, body fluid homeostasis, blood pressure control | N/A | 218 | 230 | GXP_288775 |
| Atp7a | 11977 | ATPase, Cu++ transporting, alpha polypeptide | blood vessel development; blood vessel remodeling | N/A | 590 | 602 | GXP_187 |
| Fndc3b | 72007 | fibronectin type III domain containing 3B | positive regulation of fat cell differentiation | N/A | 168 | 180 | GXP_82518 |
| Nes | 18008 | nestin | neural tube intermediate filamen | drainage component | 198 | 210 | GXP_1455614 |
Table 5.
Downregulated genes in BdkrB2−/− with Pax2 binding sites
| Gene symbol | Entrez ID | Entrez Name | Molecular Function | Expression in Kidney | Start Position | End Position | Inspecting Sequence ID |
|---|---|---|---|---|---|---|---|
| Lta | 16992 | lymphotoxin alpha (TNF superfamily, member 1) | tumor necrosis factor, formation of secondary lymphoid organs, apoptosis | N/A | 325 | 337 | GXP_81886 |
| Stard13 | 243362 | StAR-related lipid transfer (START) domain containing 13 | GTPase activator activity, | N/A | 148 | 160 | GXP_1801980 |
| Rapgef5 | 217944 | Rap guanine nucleotide exchange factor (GEF) 5 | members of the RAS subfamily of GTPases function in signal transduction | N/A | 158 | 170 | GXP_413710 |
| Adam23 | 23792 | ADAM metallopeptidase domain 23 | proteolysis; cell adhesion; central nervous system development | N/A | 557 | 569 | GXP_132501 |
| D4Wsu53e | 27981 | DNA segment, Chr 4 | tagging genes with cassette-exchange sites | N/A | 170 | 182 | GXP_287874 |
| Ganc | 76051 | glucosidase, alpha; neutral C | carbohydrate metabolic process, hydrolase enzymes hydrolyse the glycosidic bond between two or more carbohydrates | N/A | 723 | 735 | GXP_2532326 |
| Raf1 | 110157 | v-raf-1 murine leukemia viral oncogene homolog 1 | MAP kinase kinase kinase, cell division cycle, apoptosis, cell differentiation and cell migration. | cap mesenchyme | 475 | 487 | GXP_2537320 |
| Clk4 | 12750 | CDC-like kinase 4 | CDC2-like protein kinase (CLK) family, regulation of alternative splicing | kidney | 605 | 617 | GXP_409562 |
| Igh-6 | 16019 | 2 days neonate thymus thymic cells cDNA | immunoglobulin heavy chain 6, antigen binding | N/A | 88 | 100 | GXP_1236446 |
| Supv3l1 | 338359 | suppressor of var1, 3-like 1 | DNA binding; DNA helicase activity, DNA duplex unwinding | kidney | 301 | 313 | GXP_115349 |
| Zfp108 | 54678 | Zinc finger protein 108 | Interacting selectively and noncovalently with any metal ion | N/A | 409 | 421 | GXP_1466602 |
| Tex9 | 21778 | testis expressed 9 | expressed in adult mouse testes | kidney | 182 | 194 | GXP_276197 |
| Ubp1 | 22221 | upstream binding protein 1 | transcription factor activity, angiogenesis | ureter | 169 | 181 | GXP_712 |
Gene list was generated by Genomatix Program with zero mismatches. The start position and end position of binding are relative to the inspecting sequence of Genomatix Program. Some of the genes in the list may have more than one binding sites; only one binding site is presented.
Fig. 5.
Pax2 mRNA and protein are suppressed in the medulla of salt-stressed Bdkrb2−/− mice. A: RNA was isolated from the medulla of P1 salt-stressed BdkrB2+/+ and Bdkrb2−/− kidneys. Pax2 and GAPDH mRNA were measured by real-time quantitative RT-PCR for the genes indicated. B: immunofluorescence staining of Pax2 in P1 kidney sections from postnatal day (P) 1 Bdkrb2+/+ and Bdkrb2−/− pups subjected to gestational high salt. AQP2 staining was used to mark the collecting ducts.
Gestational salt stress represses Pax2-driven reporter expression in mutant but not in wild-type mice.
To determine whether the reduction in Pax2 expression is mediated transcriptionally and is sensitive to gestational gene-environment interactions, we monitored GFP fluorescence as a surrogate of Pax2 promoter activity in vivo in salt-stressed Pax2-GFP;Bdkrb2−/− mice. At E12.5 and E14.5, GFP reporter activity was detected in the UB and its branches and in the metanephric mesenchyme, thus mimicking endogenous Pax2 gene expression. This expression pattern is similar in both genotypes, i.e., Bdkrb2+/+ and BdkrB2−/− embryos. However, GFP fluorescence is markedly lower in Bdkrb2−/− than Bdkrb2+/+ kidneys (Fig. 6, A and C). Quantitative real-time RT-PCR demonstrated that GFP mRNA is significantly lower in mutant than wild-type kidneys at E12.5 and E14.5 (Fig. 6, B and D).
Fig. 6.
Pax2 promoter-driven reporter activity is reduced in salt-stressed Bdkrb2−/− metanephroi. A, C: E12.5 and E14.5 metanephroi were harvested from genetic crosses of Bdkrb2−/−;Pax2-GFP mice subjected to gestational high salt. Photographs of GFP fluorescence (a surrogate of Pax2 promoter activity) were captured at the time of harvest. GFP fluorescence is visibly lower in Bdkrb2−/− than Bdkrb2+/+ metanephroi. B, D: GFP mRNA was measured by real-time quantitative RT-PCR in E12.5 and E14.5 Bdkrb2−/− and Bdkrb2+/+ metanephroi (n = 4–5/group).
Removal of the salt stressor fails to restore Pax2 reporter activity in ex vivo cultured BdkrB2 mutant explants.
To determine whether Pax2 gene repression is reversible upon removal of the intrauterine stress, salt-stressed E12.5 Pax2-GFP;Bdkrb2−/−, Bdkrb2+/−, and Bdkrb2+/+ metanephroi were grown in culture for 2 days, and the intensity of GFP recorded over time. As noted in Fig. 7A, at 0 h, GFP fluorescence intensity is higher in wild-type and heterozygous than mutant kidneys. More importantly, over the ensuing 48 h, although growth in size is observed in the three genotypes, GFP intensity remains low in the homozygous null mutants compared with wild-type and heterozygous kidneys. Measurement of GFP pixel intensity, shown in Fig. 7B, confirms that at E12.5, GFP fluorescence intensity is significantly lower in both heterozygous and homozygous Bdkrb2 mutants than wild-type controls.
Fig. 7.
Ex vivo culture of salt-stressed Bdkrb2−/− metanephroi. A: E12.5 metanephroi were harvested from Pax2-GFP;Bdkrb2+/+, Pax2-GFP;Bdkrb2+/−, and Pax2-GFP;Bdkrb2−/− littermates and grown in culture (n = 12–18/group). The intensity of GFP fluorescence, a surrogate of Pax2 promoter activity and expression, was recorded at times 0, 24, and 48 h. Bdkrb2−/− metanephroi are smaller than the heterozygous and homozygous null mutants and maintain lower GFP fluorescence intensity ex vivo. B: quantitative measurement of GFP fluorescence reveals that although GFP pixel intensity increases in all groups during 24–48 h in the absence of the intrauterine salt stressor, the reduction in GFP intensity remains significantly lower in the Bdkrb2−/− metanephroi.
DISCUSSION
We reported previously that BdkrB2−/− mice are predisposed to renal dysgenesis in the presence of embryonic salt stress (14, 17). This mouse model of a human disease offers a unique opportunity to uncover the genetic and epigenetic mechanisms underlying gene-environment interactions in congenital renal disease. The present study demonstrates that BdkrB2 mutant kidneys from salt-stressed embryos manifest gene expression changes involving transcription factors, Wnt, cell survival, and cell-matrix pathways. Moreover, we found that Pax2 gene expression is repressed in collecting ducts of salt-stressed Bdkrb2−/− embryos. Lastly, utilizing genetic crosses between Bdkrb2−/− mice and BAC transgenic mice carrying a Pax2-GFP reporter cassette, we show that Pax2 transcriptional activity is repressed in salt-stressed Bdkrb2−/− embryonic kidneys. To our knowledge, this is the first demonstration of a renal developmental regulator that is sensitive to gene-environment interactions.
Clinical studies have demonstrated that mutations in genes encoding renal developmental regulators account for a relatively small fraction of congenital abnormalities in renal and urinary tract development (22, 36). Examples of these genes include Pax2, Eya1, Six1, Sall1, HNF1β, Ret, Robo, WT1, and Notch. The etiology of the remaining cases is unknown, but it is possible that mutations in other developmental genes will be identified in the future. It is also conceivable that gestational exposure to environmental factors (e.g., drugs, toxins) or gene-environment interactions account for a significant proportion of sporadic renal dysgenesis.
The gene-environment interaction model implies that an inherited germline mutation is phenotypically silent, presumably because it is compensated for by genes with overlapping functions. The homeostatic function of the mutant gene is revealed following exposure to a stressor that overwhelms the capacity of compensatory mechanisms. An example of genetic susceptibility is a patient with a bleeding disorder who is asymptomatic but bleeds when exposed to a stressor that impairs hemostasis, such as an antiplatelet drug or physical trauma. In our animal model, renal dysgenesis is produced by defined gene-environment interactions (13, 14). Mice harboring a homozygous deletion of the Bdkrb2 are developmentally normal and fertile. However, Bdkrb2−/− embryos exposed to gestational salt stress via the maternal diet develop renal dysgenesis due to impaired collecting duct growth and differentiation (12, 14, 17, 19). Interestingly, adult Bdkrb2 mutants develop hypertension if fed a high-salt diet (1, 8, 25).
The combination of Bdkrb2 inactivation and embryonic salt stress results in transcriptional activation of proapoptotic and repression of terminal differentiation genes (17, 19). Germline deletion of p53 from the genome of Bdkrb2−/− mice protects against apoptosis (19). Since p53 is sensor and a hub for multiple intracellular pathways (15, 16, 40), it was important to determine the cellular and molecular pathways that are altered at the onset of renal dysgenesis. Our microarray analysis revealed that while apoptosis is one of the top 10 pathways that are altered in mutant kidneys, other important pathways included Wnt signaling, calcium signaling, extracellular matrix organization, and cell communication. The following discussion focuses on the major genetic networks downstream of gene-environment interactions and how they might affect kidney development.
Intracellular calcium signaling.
Several gene products involved in intracellular calcium removal (ATP2B2, decreased), sarcoplasmic calcium release (ryanodine receptor RYR1, increased), or calcium entry/release into the cell (bradykinin receptor type 1 BdkrB1, increased) suggest that salt-stressed BdkrB2−/− cells accumulate excess intracellular calcium. The latter effect may have significant effects on membrane protein function, cell shape and mobility, gene expression, and activation of apoptosis (33).
Extracellular matrix metabolism.
Enhanced expression of fibulin 5 (Fbln5), a secreted extracellular matrix protein, and lysyl oxidase (Lox), which catalyzes cross-linking of collagen fibers, is observed in dysplastic BdkrB2−/− kidneys, which manifest expanded interstitium. Mutations in Fbln5 and Lox are implicated in various vasculopathies and inherited musculoskeletal disorders.
Regulation of cellular communication and signaling.
Several key cell membrane-associated or cell surface protein-encoding genes, e.g., small G protein binding and Rho-dependent signaling (ARHGEF1), cell-matrix signaling (integrin B3, ITGB3), and ephrin B2 (EPHB2) are downregulated, whereas genes involved in TGF-β (activin A type 1B receptor, ACVR1B) and NF-κB (IKBKG) signaling are upregulated. This imbalance may contribute to cellular injury in the dysplastic BdkrB2 null kidneys.
Pattern specification process.
Several Homeobox genes (HoxB4, HoxC8, HoxA6, and HoxD11) are downregulated in salt-stressed BdkrB2 mutant kidneys. Hox genes are known to play a key role in anterio-posterior axis specification (26), and within the kidney they may play a role in defining the segmental specification of the nephron (29, 44). Hoxd11 paralogs and Pbx2 genes are expressed in the kidney and their mutations cause renal dysgenesis.
Wnt receptor signaling.
Both canonical and noncanonical Wnts play important roles in kidney development (39). BdkrB2−/− exhibit reduced expression of Wnt6, Dvl2, and Axin1 and upregulation of the soluble receptor for Wnt, Sfrp4, suggesting impaired Wnt signaling.
Apoptosis.
Enhanced cell death is a prominent feature in BdkrB2−/− mutant kidneys (19). This is partly mediated by p53 activation via Chk1-mediated serine 23 phosphorylation. Indeed, multiple proapoptotic genes in the tumor necrosis factor pathway (TNFRSF21, LTA, EIF5A), caspase 12, and mitochondrial membrane protein Mtch1 are dysregulated in BdkrB2 null kidneys.
Downregulation of Pax2.
The present study demonstrates reduced Pax2 protein and mRNA abundance in embryonic and newborn kidneys of salt-stressed Bdkrb2−/− mice and reduced reporter (GFP) expression in kidneys of Bdkrb2−/− mice carrying a BAC Pax2-GFP transgene. Pax2 is a transcription factor that is highly expressed in proliferating epithelial progenitors and is rapidly downregulated during terminal differentiation (2, 11, 41). Pax2 plays multiple and sequential roles in renal development, including specification of the metanephric blastema, and survival and differentiation of nephron progenitors and collecting ducts (5, 6, 9, 11, 27, 34, 42). Deregulated expression of Pax2 results in cystic kidneys (10, 28), whereas Pax2 gene dosage reduction causes renal hypoplasia in humans and mice (31, 32, 37). Based on these data, we suggest that reduced Pax2 activity in salt-stressed BdkrB2−/− mice is an important contributing factor in the pathogenesis of the renal phenotype. Interestingly, allowing metanephric growth ex vivo failed to normalize Pax2 reporter activity, suggesting that the Pax2 promoter retains a repressive memory even after removal of the embryonic stressor or that a longer period outside the intrauterine environment may be necessary to reverse the repression. Epigenetic therapy that favors a permissive chromatin environment (e.g., HDAC or Dnmt inhibitors) may be of therapeutic benefit.
In summary, the present study demonstrates that gestational gene-environment interactions alter the metanephric transcriptome prior to the onset of the renal phenotype. Moreover, the Pax2 gene is sensitive to gestational gene-environment interactions.
GRANTS
This work was supported by National Institutes of Health Grants DK-56264 and Center Of Biomedical Research Excellence 1P20 RR-017659.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: L.Y., X.Y., and D.B. performed experiments; L.Y., D.B., Z.S., and S.S.E.-D. analyzed data; L.Y., D.B., Z.S., and S.S.E.-D. interpreted results of experiments; L.Y. prepared figures; L.Y. drafted manuscript; L.Y., X.Y., D.B., Z.S., and S.S.E.-D. approved final version of manuscript; Z.S. and S.S.E.-D. edited and revised manuscript; S.S.E.-D. conception and design of research.
ACKNOWLEDGMENTS
We thank Dr. Maxime Bouchard for the BAC Pax2-GFP transgenic mice and the Tulane Renal and Hypertension Center and Tulane Center for Stem Cell Research and Regenerative Medicine for the use of molecular and imaging cores.
REFERENCES
- 1.Alfie ME, Sigmon DH, Pomposiello SI, Carretero OA. Effect of high salt intake in mutant mice lacking bradykinin-B2 receptors. Hypertension 29: 483–487, 1997 [DOI] [PubMed] [Google Scholar]
- 2.Bates CM. Kidney development: regulatory molecules crucial to both mice and men. Mol Genet Metab 71: 391–396, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Batourina E, Gim S, Bello N, Shy M, Clagett-Dame M, Srinivas S, Costantini F, Mendelsohn C. Vitamin A controls epithelial/mesenchymal interactions through Ret expression. Nat Genet 27: 74–78, 2001 [DOI] [PubMed] [Google Scholar]
- 4.Borkowski JA, Ransom RW, Seabrook GR, Trumbauer M, Chen H, Hill RG, Strader CD, Hess JF. Targeted disruption of a B2 bradykinin receptor gene in mice eliminates bradykinin action in smooth muscle and neurons. J Biol Chem 270: 13706–13710, 1995 [DOI] [PubMed] [Google Scholar]
- 5.Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev 16: 2958–2970, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128: 4747–4756, 2001 [DOI] [PubMed] [Google Scholar]
- 7.Burrow CR. Retinoids and renal development. Exp Nephrol 8: 219–225, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Cervenka L, Harrison-Bernard LM, Dipp S, Primrose G, Imig JD, El-Dahr SS. Early onset salt-sensitive hypertension in bradykinin B(2) receptor null mice. Hypertension 34: 176–180, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Dressler GR. Epigenetics, development, and the kidney. J Am Soc Nephrol 19: 2060–2067, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Dressler GR, Wilkinson JE, Rothenpieler UW, Patterson LT, Williams-Simons L, Westphal H. Deregulation of Pax-2 expression in transgenic mice generates severe kidney abnormalities. Nature 362: 65–67, 1993 [DOI] [PubMed] [Google Scholar]
- 11.Eccles MR, He S, Legge M, Kumar R, Fox J, Zhou C, French M, Tsai RW. PAX genes in development and disease: the role of PAX2 in urogenital tract development. Int J Dev Biol 46: 535–544, 2002 [PubMed] [Google Scholar]
- 12.El-Dahr SS, Aboudehen K, Dipp S. Bradykinin B2 receptor null mice harboring a Ser23-to-Ala substitution in the p53 gene are protected from renal dysgenesis. Am J Physiol Renal Physiol 295: F1404–F1413, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Dahr SS, Dipp S, Meleg-Smith S, Pinna-Parpaglia P, Madeddu P. Fetal ontogeny and role of metanephric bradykinin B2 receptors. Pediatr Nephrol 14: 288–296, 2000 [DOI] [PubMed] [Google Scholar]
- 14.El-Dahr SS, Harrison-Bernard LM, Dipp S, Yosipiv IV, Meleg-Smith S. Bradykinin B2 null mice are prone to renal dysplasia: gene-environment interactions in kidney development. Physiol Genomics 3: 121–131, 2000 [DOI] [PubMed] [Google Scholar]
- 15.El-Dahr S, Hilliard S, Aboudehen K, Saifudeen Z. The MDM2-p53 pathway: multiple roles in kidney development. Pediatr Nephrol 29: 621–627, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Dahr SS, Saifudeen Z. Interactions between BdkrB2 and p53 genes in the developing kidney. Biol Chem 394: 347–351, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Fan H, Harrell JR, Dipp S, Saifudeen Z, El-Dahr SS. A novel pathological role of p53 in kidney development revealed by gene-environment interactions. Am J Physiol Renal Physiol 288: F98–F107, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Fan H, Stefkova J, El-Dahr SS. Susceptibility to metanephric apoptosis in bradykinin B2 receptor null mice via the p53-Bax pathway. Am J Physiol Renal Physiol 291: F670–F682, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Harrison-Bernard LM, Dipp S, El-Dahr SS. Renal and blood pressure phenotype in 18-mo-old bradykinin B2R(-/-)CRD mice. Am J Physiol Regul Integr Comp Physiol 285: R782–R790, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Kerecuk L, Schreuder MF, Woolf AS. Renal tract malformations: perspectives for nephrologists. Nat Clin Pract Nephrol 4: 312–325, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Latham KE, Sapienza C, Engel N. The epigenetic lorax: gene-environment interactions in human health. Epigenomics 4: 383–402, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lelievre-Pegorier M, Vilar J, Ferrier ML, Moreau E, Freund N, Gilbert T, Merlet-Benichou C. Mild vitamin A deficiency leads to inborn nephron deficit in the rat. Kidney Int 54: 1455–1462, 1998 [DOI] [PubMed] [Google Scholar]
- 25.Madeddu P, Emanueli C, El-Dahr S. Mechanisms of disease: the tissue kallikrein-kinin system in hypertension and vascular remodeling. Nat Clin Pract Nephrol 3: 208–221, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol 344: 7–15, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narlis M, Grote D, Gaitan Y, Boualia SK, Bouchard M. Pax2 and pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J Am Soc Nephrol 18: 1121–1129, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Ostrom L, Tang MJ, Gruss P, Dressler GR. Reduced Pax2 gene dosage increases apoptosis and slows the progression of renal cystic disease. Dev Biol 219: 250–258, 2000 [DOI] [PubMed] [Google Scholar]
- 29.Patterson LT, Potter SS. Hox genes and kidney patterning. Curr Opin Nephrol Hypertens 12: 19–23, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Pfeffer PL, Payer B, Reim G, di Magliano MP, Busslinger M. The activation and maintenance of Pax2 expression at the mid-hindbrain boundary is controlled by separate enhancers. Development 129: 307–318, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Porteous S, Torban E, Cho NP, Cunliffe H, Chua L, McNoe L, Ward T, Souza C, Gus P, Giugliani R, Sato T, Yun K, Favor J, Sicotte M, Goodyer P, Eccles M. Primary renal hypoplasia in humans and mice with PAX2 mutations: evidence of increased apoptosis in fetal kidneys of Pax2(1Neu) +/- mutant mice. Hum Mol Genet 9: 1–11, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Quinlan J, Lemire M, Hudson T, Qu H, Benjamin A, Roy A, Pascuet E, Goodyer M, Raju C, Zhang Z, Houghton F, Goodyer P. A common variant of the PAX2 gene is associated with reduced newborn kidney size. J Am Soc Nephrol 18: 1915–1921, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Rao A. Signaling to gene expression: calcium, calcineurin and NFAT. Nat Immunol 10: 3–5, 2009 [DOI] [PubMed] [Google Scholar]
- 34.Reidy KJ, Rosenblum ND. Cell and molecular biology of kidney development. Sem Nephrol 29: 321–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saifudeen Z, Dipp S, Stefkova J, Yao X, Lookabaugh S, El-Dahr SS. p53 regulates metanephric development. J Am Soc Nephrol 20: 2328–2337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saisawat P, Tasic V, Vega-Warner V, Kehinde EO, Gunther B, Airik R, Innis JW, Hoskins BE, Hoefele J, Otto EA, Hildebrandt F. Identification of two novel CAKUT-causing genes by massively parallel exon resequencing of candidate genes in patients with unilateral renal agenesis. Kidney Int 81: 196–200, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salomon R, Tellier AL, Attie-Bitach T, Amiel J, Vekemans M, Lyonnet S, Dureau P, Niaudet P, Gubler MC, Broyer M. PAX2 mutations in oligomeganephronia. Kidney Int 59: 457–462, 2001 [DOI] [PubMed] [Google Scholar]
- 38.Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genet 8: 791–802, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Ott KM, Barasch J. WNT/beta-catenin signaling in nephron progenitors and their epithelial progeny. Kidney Int 74: 1004–1008, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen N, Satija YK, Das S. p53 and metabolism: old player in a new game. Transcription 3: 119–123, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terzic J, Muller C, Gajovic S, Saraga-Babic M. Expression of PAX2 gene during human development. Int J Dev Biol 42: 701–707, 1998 [PubMed] [Google Scholar]
- 42.Torban E, Eccles MR, Favor J, Goodyer PR. PAX2 suppresses apoptosis in renal collecting duct cells. Am J Pathol 157: 833–842, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber S, Moriniere V, Knuppel T, Charbit M, Dusek J, Ghiggeri GM, Jankauskiene A, Mir S, Montini G, Peco-Antic A, Wuhl E, Zurowska AM, Mehls O, Antignac C, Schaefer F, Salomon R. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol 17: 2864–2870, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Wellik DM. Hox genes and kidney development. Pediatr Nephrol 26: 1559–1565, 2011 [DOI] [PubMed] [Google Scholar]
- 45.Woods LL, Ingelfinger JR, Nyengaard JR, Rasch R. Maternal protein restriction suppresses the newborn renin-angiotensin system and programs adult hypertension in rats. Pediatr Res 49: 460–467, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Woods LL, Rasch R. Perinatal ANG II programs adult blood pressure, glomerular number, and renal function in rats. Am J Physiol Regul Integr Comp Physiol 275: R1593–R1599, 1998 [DOI] [PubMed] [Google Scholar]
- 47.Woolf AS, Price KL, Scambler PJ, Winyard PJ. Evolving concepts in human renal dysplasia. J Am Soc Nephrol 15: 998–1007, 2004 [DOI] [PubMed] [Google Scholar]