ABSTRACT
The miR-17∼92 cluster family is composed of three members encoding microRNAs that share seed sequences. To assess their role in cerebellar and medulloblastoma (MB) development, we deleted the miR-17∼92 cluster family in Nestin-positive neural progenitors and in mice heterozygous for the Sonic Hedgehog (SHH) receptor Patched 1 (Ptch1+/−). We show that mice in which we conditionally deleted the miR-17∼92 cluster (miR-17∼92floxed/floxed; Nestin-Cre+) alone or together with the complete loss of the miR-106b∼25 cluster (miR-106b∼25−/−) were born alive but with small brains and reduced cerebellar foliation. Remarkably, deletion of the miR-17∼92 cluster abolished the development of SHH-MB in Ptch1+/− mice. Using an orthotopic transplant approach, we showed that granule neuron precursors (GNPs) purified from the cerebella of postnatal day 7 (P7) Ptch1+/−; miR-106b∼25−/− mice and overexpressing Mycn induced MBs in the cortices of naïve recipient mice. In contrast, GNPs purified from the cerebella of P7 Ptch1+/−; miR-17∼92floxed/floxed; Nestin-Cre+ animals and overexpressing Mycn failed to induce tumors in recipient animals. Taken together, our findings demonstrate that the miR-17∼92 cluster is dispensable for cerebellar development, but required for SHH-MB development.
Keywords: MicroRNA, miR-17∼92 and miR-106b∼25 clusters, Cerebellum, Development, Nestin, Medulloblastoma, Granule neuron progenitors (GNPs)
INTRODUCTION
The cerebellum develops in the mouse from embryonic day 9 (E9) with the cerebellar anlage forming from the roof (the alar plates) of the metencephalon. It is composed of different types of neurons that arise from the ventricular zone (VZ) of the cerebellar neuroepithelium localized on the roof of the fourth ventricle, and from the rostral rhombic lip (rRL), localized at the posterior edge of the cerebellar anlage (Hatten and Roussel, 2011). In the mouse, granule neuron progenitors (GNPs) are born in the rRL (E11–E16) and migrate along the surface of the developing cerebellum over the Purkinje cells, which are born in the VZ (E11–E13), to form the external granule layer (EGL) (E13–E16). After birth, Sonic Hedgehog (SHH), secreted by the Purkinje cells, promotes the proliferation of GNPs, which peaks between postnatal days 5 (P5) and P7. Subsequently, GNPs exit the cell cycle, migrate inwardly through the Purkinje cell layer and settle as postmitotic neurons in the internal granular layer (IGL). By 3 weeks, the mouse cerebellum is fully formed consisting of ten folia separated by fissures (Hatten and Roussel, 2011). Constitutive activation of the SHH signaling pathway leads to defects in cell cycle exit, migration and differentiation, which, in turn, induce medulloblastoma (MB). This SHH-subgroup of MBs (SHH-MB) represents ∼25% of all human cases (Taylor et al., 2012).
MicroRNAs (miRNAs) are ∼22 nucleotides long non-coding RNAs. They are derived from pri-miRNAs that are processed by Drosha and DGCR8 into pre-miRNAs in the nucleus, and then translocated into the cytoplasm where they are converted into mature miRNAs by the processing enzyme Dicer. In turn, single-stranded miRNAs are loaded into the RNA-induced silencing complex (RISC) to bind the 3′-untranslated region (3′-UTR) of mRNAs to inhibit their translation or degradation (Carmell and Hannon, 2004; Kim, 2005).
Abnormal expression of miRNAs is often seen in cancers. MicroRNAs encoded by the miR-17∼92 cluster, also called oncomiR-1, are overexpressed in various cancers (Concepcion et al., 2012; Mogilyansky and Rigoutsos, 2013) including mouse and human medulloblastomas with constitutively activated SHH signaling (Uziel et al., 2009; Northcott et al., 2009). The miR-17∼92 cluster encoded by chromosome 14 in the mouse (13 in humans) has two paralogs, the miR-106b∼25 and miR-106a∼363 clusters, each of which is located on different chromosomes (Fig. 1A). The miR-106b∼25 cluster is encoded on chromosome 5 in the mouse (7 in humans) while the miR-106a∼363 cluster maps to chromosome X in mice and humans (Concepion et al., 2012; Mogilyansky and Rigoutsos, 2013). Mice lacking the miR-17∼92 cluster die shortly after birth from lung and heart defects while mice lacking each of its two paralogs do not show any obvious phenotypes (Ventura et al., 2008). However, the miR-17∼92 and miR-106b∼25 clusters share overlapping functions since mice with combined deletion exhibit a more profound phenotype than those lacking miR-17∼92 alone (Ventura et al., 2008). The miR-17∼92 cluster is a downstream target of Myc (c-Myc) (O'Donnell et al., 2005) and Mycn (Northcott et al., 2009; de Pontual et al., 2011). Mice lacking one copy of miR-17∼92 show skeletal and growth defects recapitulating the Feingold syndrome observed in patients harboring MYCN mutations or hemizygous deletion of MIR-17∼92 (de Pontual et al., 2011). The miR-17∼92 cluster is expressed in proliferative GNPs but not in post-mitotic granule neurons. Overexpression of the miR-17∼92 cluster in GNPs heterozygous for the SHH receptor, Patched 1 (Ptch1+/−), induces early onset of SHH-MB formation after orthotopic transplant in the cortices of naïve recipient animals (Uziel et al., 2009). Similarly when overexpressed in wild-type GNPs, the miR-17∼92 cluster collaborates with SHH signaling to provide GNPs with a proliferative advantage (Northcott et al., 2009). These results suggested that, besides its role in SHH-MBs, the miR-17∼92 cluster might play a role in cerebellar development. We here show that the miR-17∼92 cluster and its paralog the miR-106b∼25 cluster, are differently required for cerebellar and medulloblastoma development.
Fig. 1. Expression of miR-19a and miR-106b encoded by the miR-17∼92 cluster family in cerebella during embryogenesis.
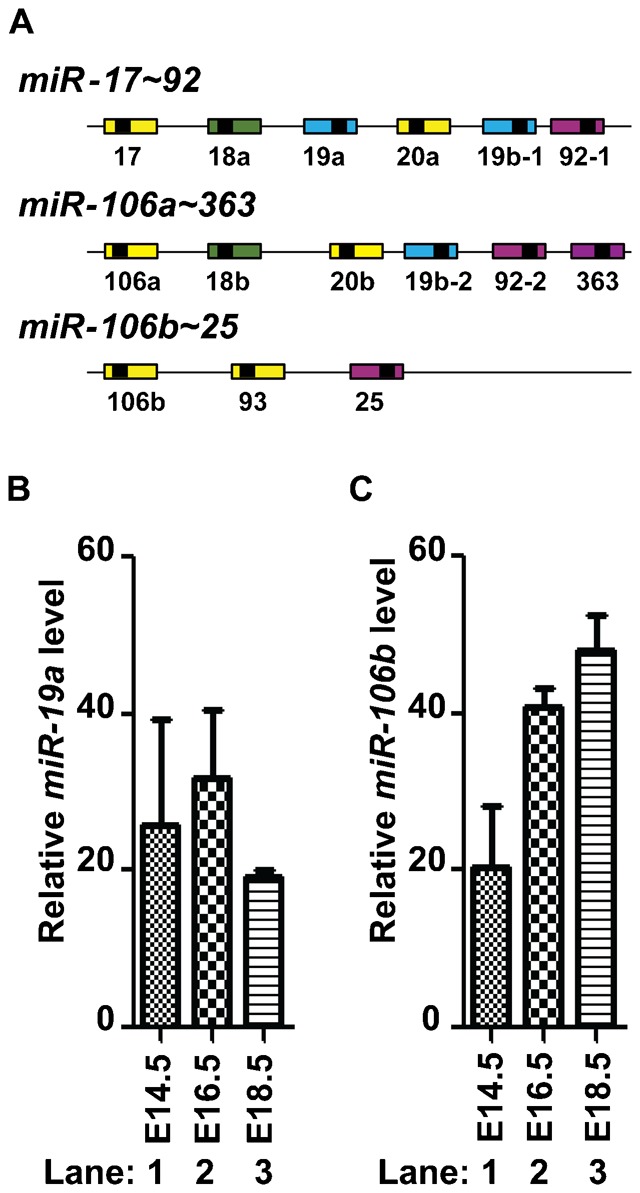
(A) Schematic representation of the three microRNA clusters. MiR-17∼92, miR-106a∼363 and miR-106b∼25 clusters encode 6, 6 and 3 mature microRNAs (colored boxes), respectively. MicroRNAs sharing the same seed sequence are represented by boxes of the same color. Relative levels of mature microRNAs miR-19a (B) and miR-106b (C) were determined by Q-RT-PCR on total RNA extracted from total cerebella of wild-type mice (lanes 1–3) at E14.5 (lanes 1), E16.5 (lanes 2) and E18.5 (lanes 3).
MATERIALS AND METHODS
Mice
Mouse lines carrying conditional alleles of miR-17∼92 (miR-17∼92floxed/floxed) (Ventura et al., 2008), or lacking miR-106b∼25 (miR-106b∼25−/−) (Ventura et al., 2008) were generously provided by Dr Tyler Jacks (Boston, MA, USA). The transgenic line in which the Cre recombinase is expressed under the promoter of the rat Nestin gene (Nestin-Cre) (stock number 003771) (Tronche et al., 1999) and C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Ptch1+/− mice in a Cdkn2c-null background (Ptch1+/−; Cdkn2c−/−) were previously described (Uziel et al., 2005). All mice were maintained on a mixed 129×C57BL/6 background. CD-1 nu/nu mice were obtained from Charles River (Wilmington, MA, USA). Mice were housed in an accredited facility of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) in accordance with National Institute of Health guidelines. The Animal Care and Use Committee (ACUC) of St Jude Children's Research Hospital approved all procedures.
Histology, immunohistochemistry, proliferation assays and X-gal staining
Heads of embryos or whole brains from mice were harvested, fixed overnight in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) at 4°C, soaked in 30% sucrose in PBS until the tissues sank to the bottom of the tube and embedded in Optimal Cutting Temperature (OCT) compound. Frozen 12 µm sections were collected on Fisherbrand superfrost plus slides using a cryostat. Slides were stained with Hematoxylin and Eosin (H&E) or antibodies, as previously described (Uziel et al., 2005). The following antibodies used were: 5-Bromo-2-Deoxy-Uridine (BrdU) (SC-32323; Santa Cruz, Dallas, TX, USA; 1/1000 dilution), p27Kip1 (610242; BD Biosciences, San Jose, CA; 1/200 dilution), cyclin D2 (SC-593; Santa Cruz; 1/50 dilution), NeuN (MAB377; EMD Millipore, Billerica, MA, USA; 1/200 dilution), Ki67 (NLC-Ki67p; Leica microsystems, Buffalo Grove, IL; 1/1000 dilution) and GABA(A) receptor α6 subunit (AB5610; EMD Millipore; 1/200 dilution). BrdU (100 mg/kg) was injected intra-peritoneally two hours before harvesting the brain. For X-gal staining, whole brains from 3-week-old animals were fixed in 2% PFA in PBS at 4°C for 3 hours. Fixed brains were washed twice in PBS and then incubated in the “Rinse” buffer (100 mM sodium phosphate pH 7.3, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% IGEPAL CA-360) for 5 minutes at room temperature. Brains were stained with X-gal staining solution (“Rinse” buffer containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-galactoside 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside (X-gal) (Invitrogen, Life technologies, Grand Island, NY, USA), at 37°C for 4 hours.
Quantitative-reverse transcriptase-polymerase chain reaction (Q-RT-PCR) and Affymetrix microarrays analysis
Purification of GNPs from P7 cerebella, RNA extraction from dissected embryonic and postnatal (P4 and P7) cerebella or from purified GNPs, and Q-RT-PCR for miR-19a and miR-106b were performed, as previously described (Uziel et al., 2005; Uziel et al., 2009). For comparative gene expression analysis, RNAs from cerebella of P4 and P7 mice were subjected to hybridization using Affymetrix Mouse GeneChip MG430PM (Affymetrix, Santa Clara, CA). Principal component analysis (PCA) and gene set enrichment analysis (GSEA) were performed using Partek Genomics Suite 6.6 software (St Louis, MO) and GSEA software (Broad Institute, Cambridge, MA), respectively.
Orthotopic transplantation
GNPs were purified from P7 cerebella and infected with retroviruses encoding Mycn and the red fluorescent protein (RFP), as previously described (Kawauchi et al., 2012). 48 hours after infection, 2×106 infected GNPs were injected into the cortices of naïve recipient CD-1 nu/nu mice. The rate of infection was assessed by fluorescence activated cell sorting (FACS). The percentage of RFP positive (RFP+) cells ranged from 30 to 50%.
Statistics
Statistical significance was determined using GraphPad Prism software (version 5.0). Data were shown as mean ± s.e.m. P-values <0.05 were used as significance threshold from unpaired two-tailed Student's t test. For the survival curves, p-values were determined with a log-rank (Mantel Cox) test.
RESULTS
Co-inactivation of the miR-17∼92 and miR-106b∼25 clusters reduced cerebellar size and foliation by limiting proliferation
We previously found that microRNAs encoded by the miR-17∼92 and miR-106b∼25, but not the miR-106a∼363, clusters are expressed in proliferating GNPs in the postnatal cerebellum (Uziel et al., 2009). Because these miRNAs were expressed in developing cerebella from E14.5 to birth (Fig. 1B,C, lanes 1–3), we assessed their role during cerebellar development. Since miR-17∼92-null mice die shortly after birth (Ventura et al., 2008), the miR-17∼92 cluster was conditionally deleted in Nestin-positive neural progenitors using a Nestin-Cre transgenic mouse (miR-17∼92floxed/floxed; Nestin-Cre+) in a wild-type miR-106b∼25 (referred as miR-17∼92cKO) or in a miR-106b∼25-null (referred as miR-17∼92cKO; miR-106b∼25KO) background. We also generated miR-17∼92floxed/floxed; Nestin-Cre−; miR-106b∼25−/− (referred as miR-106b∼25KO) and miR-17∼92floxed/floxed; Nestin-Cre−; miR-106b∼25+/+ (referred as control) littermates. Mice of all genotypes were born at a Mendelian ratio.
At E18.5, cerebella of all 4 genotypes were indistinguishable displaying the 5 cardinal lobes separated by four principal fissures (Fig. 2A–D) despite complete Cre-mediated recombination of the two miR-17∼92 alleles (supplementary material Fig. S1A) and reduced levels of miR-19a in cerebella of miR-17∼92cKO mice (supplementary material Fig. S1B, lane 2).
Fig. 2. Progressive foliation defects in cerebella of mice lacking the miR-17∼92 and miR-106b∼25 clusters.
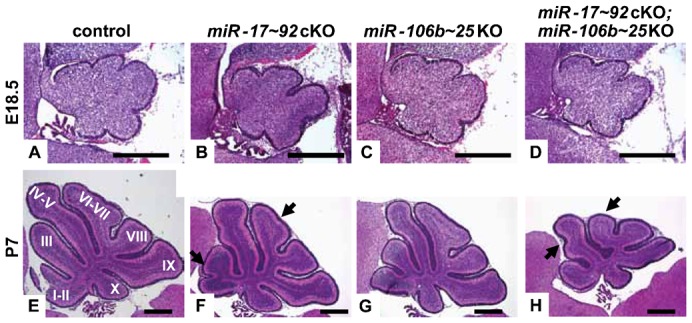
Mid-sagittal sections through the cerebellum of a control (A,E), miR-17∼92cKO (B,F), miR-106b∼25KO (C,G) and miR-17∼92cKO; miR-106b∼25KO (D,H) mouse at E18.5 (A–D) and P7 (E–H) of age were stained by H&E. The ten vermis lobules are indicated by Roman numbers. Arrows indicate the area of abnormal foliation. Scale bars: 500 µm.
At P7, the ten folia were observed in the cerebella of control mice (Fig. 2E). However, cerebella of miR-17∼92cKO; miR-106b∼25KO mice and, to a lesser extent, cerebella of miR-17∼92cKO mice, demonstrated foliation defects mainly in folia VI–VII and folia I–V (compare Fig. 2H and Fig. 2F with Fig. 2E, respectively, see arrows), while the folia in the cerebella of miR-106b∼25KO mice appeared similar to those of control mice (compare Fig. 2G with Fig. 2E).
At one month of age, miR-17∼92cKO and miR-17∼92cKO; miR-106b∼25KO mice showed statistically smaller body weight (Fig. 3A, lanes 2 and 4 versus lane 1, respectively), brains and cerebella (Fig. 3B,C, lanes 2 and 4 versus lane 1, respectively) compared to control mice. In contrast, the size of miR-106b∼25KO mice was similar to control animals (Fig. 3A–C, lane 3 versus lane 1). This demonstrated that, despite the small brain size of the miR-17∼92cKO; miR-106b∼25KO mice, their cerebella was significantly smaller than expected. The cerebellar folia from miR-17∼92cKO; miR-106b∼25KO and miR-17∼92cKO mice were misshapen with shallow fissures. Folia I–V and VI–VII were under developed when compared to cerebella of control mice (compare Fig. 3G and Fig. 3E with Fig. 3D, respectively, arrows). However, folia from the cerebella of miR-106b∼25KO animals appeared similar to those of controls (compare Fig. 3F with Fig. 3D). Scattered ectopic clusters of mature granule neurons were found on the surface of the molecular layer in miR-17∼92cKO; miR-106b∼25KO mice (Fig. 3G–I). The rest of the cerebellum cortical architecture appeared normal including a proper IGL, a monolayer of Purkinje cells with normal arborization, and Bergmann glia with normal fibers (negative data not shown). In spite of their small cerebella with foliation defects, miR-17∼92cKO; miR-106b∼25KO mice were asymptomatic with no evidence of gross neurological symptoms including motor coordination or balance defects measured on a Rotarod (negative data not shown).
Fig. 3. Mice lacking the miR-17∼92 and miR-106b∼25 clusters have small brains and small cerebella.
(A) Body weight, (B) ratio (expressed in %) of brain versus body weight and (C) ratio (expressed in %) of cerebellum versus brain weight of control (lanes 1; n = 23), miR-17∼92cKO (lanes 2; n = 45), miR-106b∼25KO (lanes 3; n = 11) and miR-17∼92cKO; miR-106b∼25KO (lanes 4; n = 31) mice at 1 month of age. (* and +) p-values <0.05, (**) p-values <0.01, (***, +++ and ###) p-values <0.001. P-values were calculated by comparing lanes 2–4 to lane 1 (*), lanes 3 and 4 to lane 2 (+) and lane 4 to lane 3 (#). Mid-sagittal sections through the cerebellum of control (D), miR-17∼92cKO (E), miR-106b∼25KO (F) and miR-17∼92cKO; miR-106b∼25KO (G) mouse at 1 month of age stained by H&E. The ten vermis lobules are indicated by Roman numbers. Arrows indicate foliation abnormalities. Enlarged view of boxed area in panel G is presented in panels H and I. Clusters of mature granule neurons (white arrows) on the surface of the molecular layer were stained with DAPI (H) and an antibody against GABA(A) receptor α6 subunit (GABARa6) (I). Scale bars: 500 µm.
Cerebellar development of miR-17∼92cKO; miR-106b∼25KO mice appeared normal during embryogenesis until birth, a time when GNPs respond to SHH to rapidly proliferate with maximal proliferation between P5 and P7. At P7, we found a significant reduction in BrdU incorporation in the EGL of cerebella from miR-17∼92cKO; miR-106b∼25KO mice compared to controls (Fig. 4D versus Fig. 4A and Fig. 4E) consistent with the requirement for massive proliferation of GNPs for proper foliation from birth until P7 (Lauder et al., 1974). The miR-17∼92 cluster not only regulates proliferation but also apoptosis (Concepcion et al., 2012; Mogilyansky and Rigoutsos, 2013). We observed no significant differences in apoptosis by Terminal deoxynucleotidyl transferase dUTP nick end labeling staining between the cerebella of all 4 genotypes at P7 (negative data not shown). These results suggested that decreased proliferation of GNPs from cerebella of miR-17∼92cKO; miR-106b∼25KO mice might account for the diminished pool of GNPs during post-natal development. Consistent with this possibility, we observed premature exhaustion of proliferative GNPs at P14 in miR-17∼92cKO; miR-106b∼25KO mice. The EGL did not uniformly cover the surface of the cerebellum in contrast to that of control mice (compare Fig. 5G and Fig. 5I with Fig. 5A and Fig. 5C). The remaining cells in the EGL were mainly negative for BrdU and cyclin D2 but positive for p27Kip1, suggesting premature exit from the cell cycle and migration defects (Fig. 5H and Fig. 5J–L versus Fig. 5B and Fig. 5D–F).
Fig. 4. Reduced proliferation in cerebella of mice lacking the miR-17∼92 and miR-106b∼25 clusters.
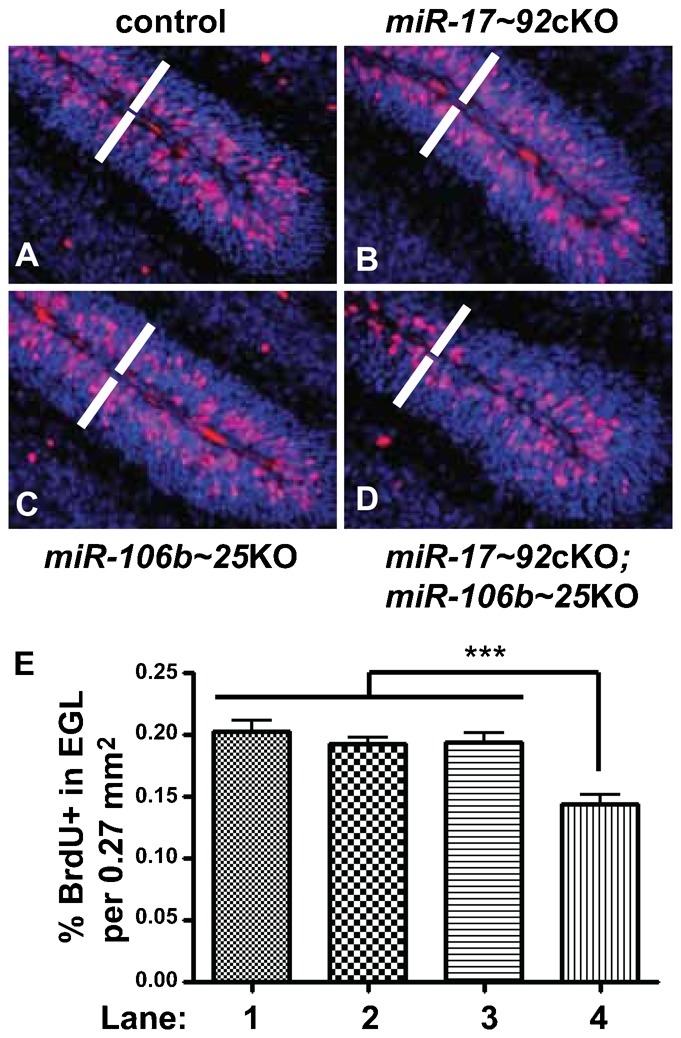
(A–D) Proliferation assessed by BrdU staining (red) on mid-sagittal sections through P7 cerebellum. Nuclei were counterstained with propidium iodide (blue). White bars indicate the EGL. (E) The percentage of BrdU+ cells in the EGL was quantified per 0.27 mm2. BrdU+ cells were counted in the EGL of lobule I and II from 2 sections per animal. Three mice for each genotype were analyzed. (***) p-values <0.001. (A and E, lane 1) control, (B and E, lane 2) miR-17∼92cKO, (C and E, lane 3) miR-106b∼25KO and (D and E, lane 4) miR17∼92cKO; miR-106b∼25KO.
Fig. 5. Premature termination of EGL proliferation in mice lacking the miR-17∼92 and miR-106b∼25 clusters.
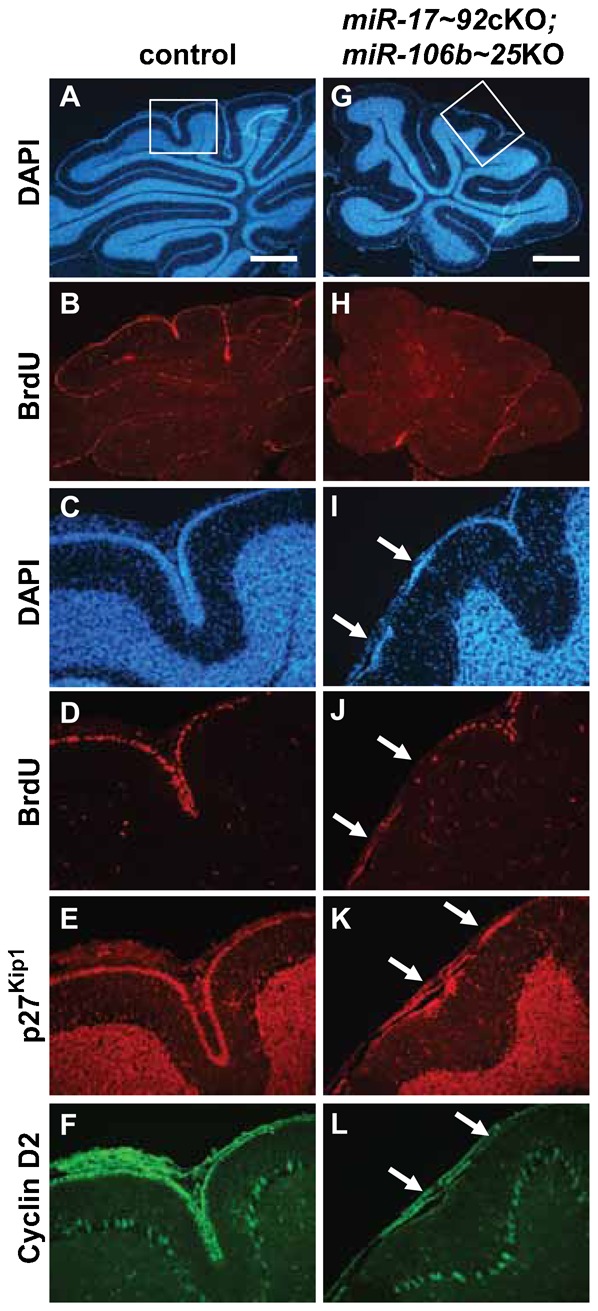
Mid-sagittal sections through the cerebellum of P14 control (A–F) and miR-17∼92cKO; miR-106b∼25KO (G–L) mice stained with DAPI (A,C,G,I), or antibodies raised against BrdU (B,D,H,J), p27Kip1 (E,K) and cyclin D2 (F,L). Enlarged views of boxed areas in panels A and G are presented in panels C–F and I–L, respectively. Arrows indicate 2 clusters of granule neurons out of cycle, on the surface of the cerebellum, in miR-17∼92cKO; miR-106b∼25KO mice. Scale bars: 500 µm.
The miR-17∼92 cluster is required for medulloblastoma formation
We previously reported that GNPs purified from Ptch1+/−; Cdkn2c−/− mice and overexpressing the miR-17∼92 cluster induced SHH-MB after transplant in the cortex of naïve CD-1 nu/nu recipient mice (Uziel et al., 2009). However, unlike Mycn, enforced expression of the miR-17∼92 cluster in GNPs purified from the cerebella of Trp53−/−; Cdkn2c−/− mice failed to induce SHH-MBs after orthotopic transplantation demonstrating that activation of the Patched signaling pathway was required for miR-17∼92 induction of SHH-MBs (Uziel et al., 2009). To assess whether the miR-17∼92 cluster was required for MB formation, we bred miR-17∼92floxed/+; Nestin-Cre+ with Ptch1+/−; Cdkn2c−/− mice. While, as expected, 45.5% (10/22) of miR-17∼92+/+; Ptch1+/; Cdkn2c+/− mice succumbed to SHH-MBs, none (0/20) of the miR-17∼92cKO; Ptch1+/−, Cdkn2c+/− mice developed tumors over a period of 300 days (Fig. 6A).
Fig. 6. The mir-17∼92 cluster is required for medulloblastoma formation.
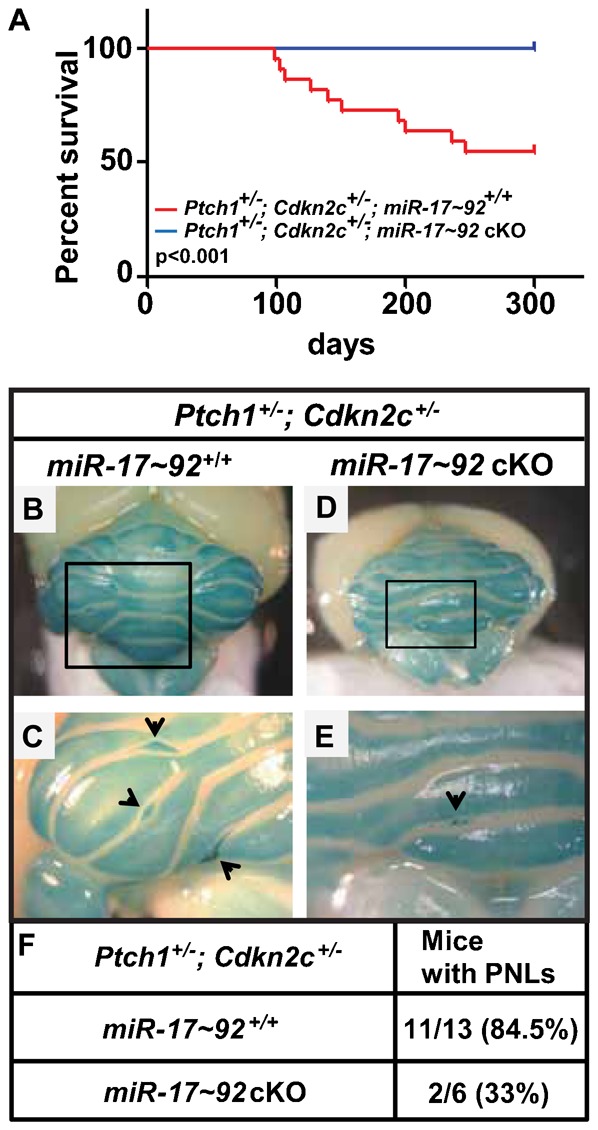
(A) Survival curves for Ptch1+/−; Cdkn2c+/− mice with different miR-17∼92 and Nestin-Cre genotypes. MiR-17∼92cKO (blue line, n = 20) and miR-17∼92+/+ (red line, n = 22) mice. P-value <0.001. (B–E) In situ X-gal staining of cerebella from a 3 week old Ptch1+/−; Cdkn2c+/− mouse wild type (B,C) or null (D,E) for the miR-17-92 cluster. Enlarged views of boxed areas in panels B and D are presented in panels C and E, respectively. (C) Arrows indicate 3 PNLs composed of densely packed cells. (E) Arrow indicates a region composed of scattered ectopic cells localized on the surface of the cerebellum. (F) Number of 3 week old mice revealing PNLs on the surface of their cerebella.
In wild-type mice, by 3 weeks after birth, all GNPs have exited the cell cycle and migrated from the surface of the cerebellum into the IGL. In contrast, in Ptch1+/− mice, several GNPs continue to divide and remain on the surface of the molecular layer (ML), to form clusters of densely packed cells called pre-neoplastic lesions (PNLs) (Goodrich et al., 1997; Kim et al., 2003; Oliver et al., 2005). In 6 weeks old mice, the majority of PNLs have regressed, while only few progress to MBs. To visualize PNLs, we stained the entire brains of 3 week old miR-17∼92cKO; Ptch1+/−, Cdkn2c+/− and miR-17∼92+/+; Ptch1+/−, Cdkn2c+/− mice with X-gal, since a portion of the wild-type allele of Patched is replaced by LacZ in the Ptch1+/− mice (Goodrich et al., 1997). We detected PNLs in 84.5% (11/13) miR-17∼92+/+; Ptch1+/−, Cdkn2c+/− mice (Fig. 6B,C,F). As expected, those PNLs were Ki67 positive but negative for NeuN and GABA (A) receptor α6 subunit (Fig. 7C–E, arrows). However, only 33% (2/6) of cerebella from miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− mice showed PNLs (p = 0.0248) (Fig. 6F). Scattered foci of non-proliferating, differentiated neurons (negative for Ki67 but positive for NeuN and GABA (A) receptor α6 subunit) were observed on the surface of the cerebellar ML of miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− mice (Fig. 6D,E, Fig. 7H–J, arrows). These results suggest that the miR-17∼92 cluster is required for SHH-induced PNL formation and for SHH-MB development.
Fig. 7. Foci of differentiated neurons at the cerebellar surface of 3 week old miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− mice.
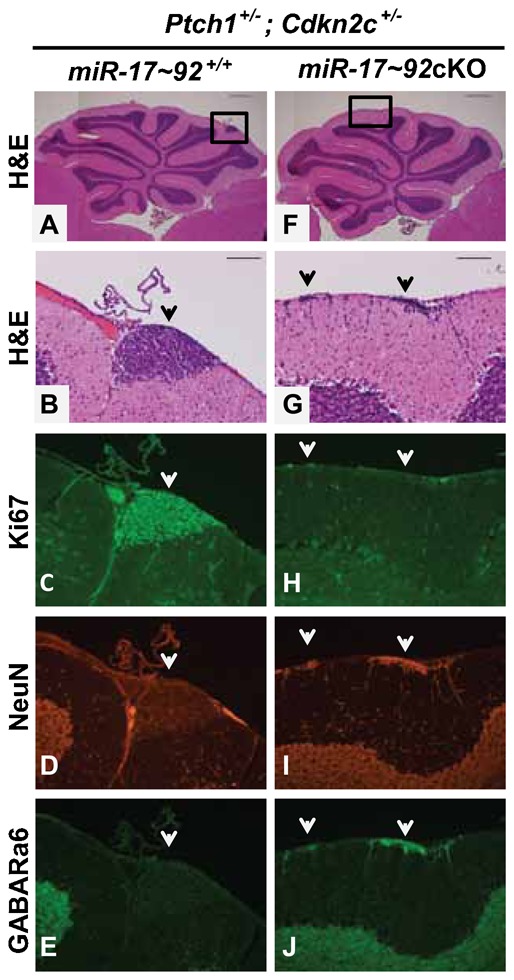
Cerebella from 3 week old Ptch1+/−; Cdkn2c+/− mice wild type (A–E) or null (F–J) for the miR-17∼92 cluster were stained with H&E (A,B,F,G), an antibody against Ki67 (C,H), NeuN (D,I), and GABA(A) receptor α6 subunit (GABARa6) (E,J). Enlarged views of boxed areas (A,F) are presented in panels B–E and G–J, respectively. PNL (arrow, B–E) is composed of densely packed undifferentiated and proliferating neurons. Foci of differentiated, non-proliferating neurons (arrows, G–J). Scale bars: 500 µm (A,F), 100 µm (B,G).
Mycn is a direct target of SHH signaling and is required for SHH-MB formation (Kenney et al., 2003; Hatton et al., 2006). Using an orthotopic transplantation approach, we previously showed that enforced expression of Mycn in GNPs purified from P7 cerebella of Ptch1+/−, Cdkn2c−/− mice induces SHH-MBs (Zindy et al., 2007; Kawauchi et al., 2012). GNPs purified from the cerebella of P7 miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− and miR-17∼92+/+; Ptch1+/−; Cdkn2c+/− mice were infected with retroviruses encoding Mycn and the red fluorescence protein (RFP), and stereotactically implanted 2 days later into the cortices of naïve recipient animals. Mycn expression did not increase apoptosis or cell cycle arrest in infected GNPs after three days in culture (negative data not shown). As expected, 100% (9/9) of the animals transplanted with GNPs purified from the cerebella of P7 miR-17∼92+/+; Ptch1+/−; Cdkn2c+/− mice and overexpressing Mycn developed SHH-MBs (Fig. 8A) (Zindy et al., 2007; Kawauchi et al., 2012). In contrast, none of the 12 mice transplanted with GNPs purified from the cerebella of P7 miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− mice and expressing Mycn developed medulloblastoma, 180 days post-implantation. However, 5/5 mice transplanted with GNPs purified from the cerebella of P7 miR-106b∼25KO; Ptch1+/−; Cdkn2c+/− mice in which we enforced Mycn expression developed MBs (Fig. 8B). Pathological analysis of the tumors of all genotypes confirmed that they were MBs of the SHH subtype with classic morphology of round cells (Fig. 8C–H), as previously described (Zindy et al., 2007; Kawauchi et al., 2012).
Fig. 8. Enforced Mycn expression in GNPs from Ptch1+/−; Cdkn2c+/− but not from miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− mice induces SHH-MBs.
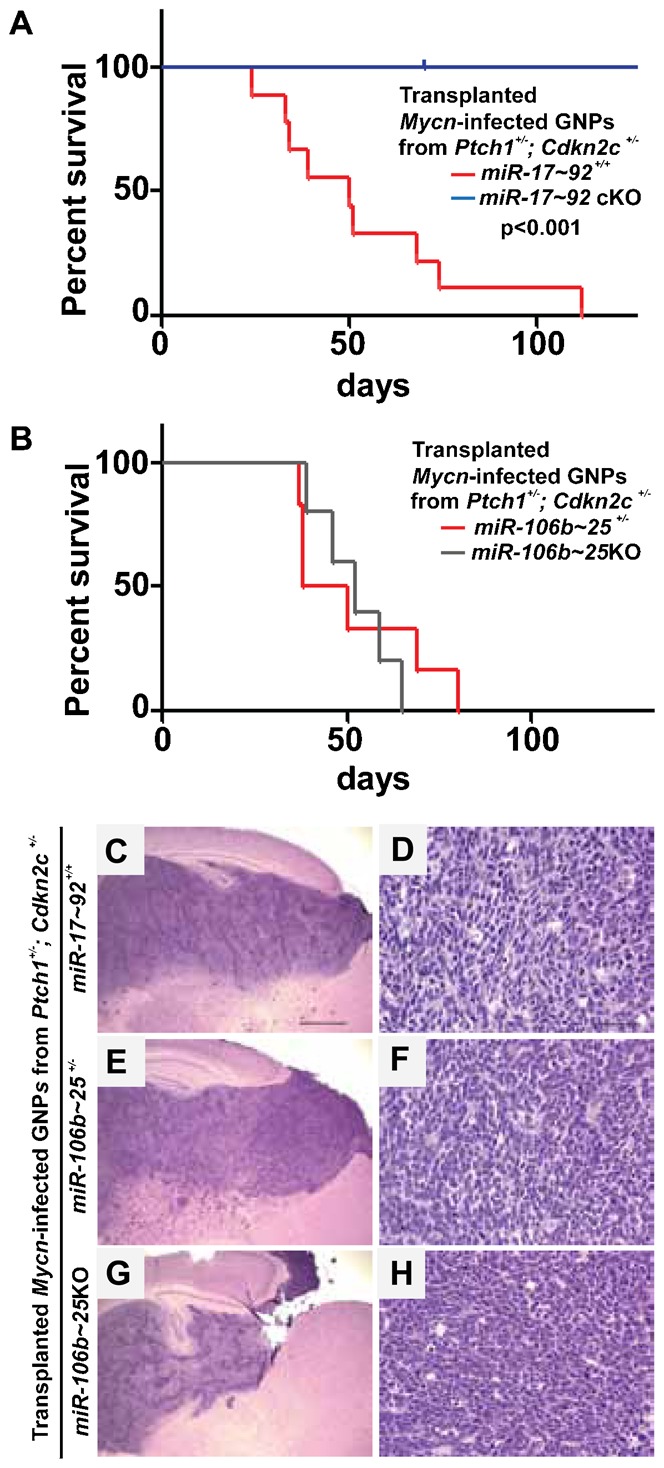
(A) Survival curves for naïve CD-1 nu/nu mice transplanted with GNPs purified from the cerebella of P7 miR-17∼92cKO; Ptch1+/−; Cdkn2c+/− (blue line, n = 12) or from miR-17∼92+/+; Ptch1+/−; Cdkn2c+/− (red line, n = 9) mice and infected with retroviruses expressing Mycn. P-value <0.001. (B) Survival curves for naïve CD-1 nu/nu mice transplanted with GNPs purified from the cerebella of P7 miR-106b∼25KO; Ptch1+/−; Cdkn2c+/− (grey line, n = 5) or from miR-106b∼25+/−; Ptch1+/−; Cdkn2c+/− (red line, n = 6) mice and infected with retroviruses expressing Mycn. (C–H) H&E staining of medulloblastoma after cortical implants of GNPs purified from the cerebella of P7 miR-17∼92+/+; Ptch1+/−; Cdkn2c+/− (C,D) or miR-106b∼25+/−; Ptch1+/−; Cdkn2c+/− (E,F) or miR-106b∼25KO; Ptch1+/−; Cdkn2c+/− mice (G,H), infected with retroviruses expressing Mycn. Scale bars: 1 mm (C,E,G) and 50 µm (D,F,H).
These results point to an absolute requirement for the miR-17∼92, but not for the miR-106b∼25, cluster in SHH-MB initiation.
Targets of the miR-17∼92 family cluster
We recently reported that the miR-17∼92 cluster down-regulates bone morphogenic protein (Bmp) receptor type 2 (Bmpr2), the receptor for Bmp-2, -4 and -7 and that higher levels of Bmpr2 are detected in cerebella of P7 miR-17∼92cKO; miR-106b∼25KO mice compared to those of controls (Murphy et al., 2013). To identify potential targets of the miR-17∼92 cluster family, we compared the gene expression profile of cerebella from control and miR-17∼92cKO; miR-106b∼25KO mice at P4, and P7, times at which difference in cerebellar size was detectable. Principal component analysis revealed that cerebella from control mice clustered together but independently from those of miR-17∼92cKO; miR-106b∼25KO mice for each time point (Fig. 9A). To gain insights into the pathways affected by the deletion of miR-17∼92 and miR-106b∼25 clusters we performed GSEA. Using the Biocarta gene sets, we found that the Transforming Growth Factor (TGF)-β responsive signaling pathway was enriched in cerebella from P7 miR-17∼92cKO; miR-106b∼25KO mice relatively to controls (Fig. 9B). Although these results were not statistically significant, the TGF-β responsive gene set was significantly enriched when we combined P4 and P7 cerebella (Fig. 9C) suggesting that this pathway was responsible in part for the reduced proliferation, as previously reported (Aref et al., 2013).
Fig. 9. The miR-17∼92 cluster suppresses TGF-β signaling.
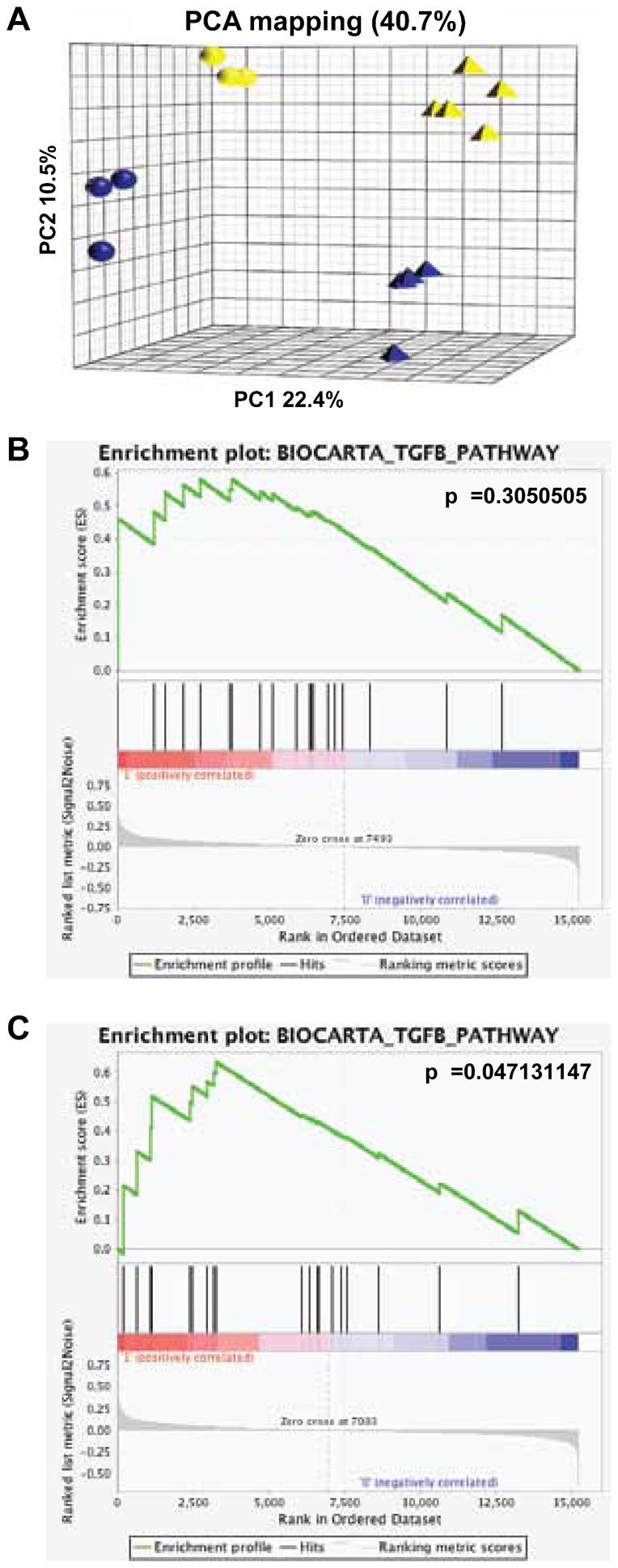
(A) Principal component analysis (PCA) of cerebella from P4 (blue spheres, n = 3) and P7 (blue pyramids, n = 4) control mice, and from P4 (yellow spheres, n = 3) and P7 (yellow pyramids, n = 5) miR-17∼92cKO; miR-106b∼25KO mice. (B,C) Gene set enrichment analysis plot for Biocarta TGF-β responsive genes set between cerebella from P7 miR-17∼92cKO; miR-106b∼25KO and P7 control mice (B; p-value = 0.3050) and between cerebella from P4 and P7 miR-17∼92cKO; miR-106b∼25KO and P4 and P7 control mice (C; p-value = 0.04713).
DISCUSSION
Here we analyzed the role of the miR-17∼92 cluster family during cerebellar development by conditional deletion the miR-17∼92 cluster alone or together with the miR-106b∼25 cluster (miR-17∼92cKO; miR-106b∼25KO) in neural progenitors. While lack of the miR-106b∼25 cluster had no obvious phenotype, loss of the miR-17∼92 cluster induced a reduction in cerebellar size and foliation. Loss of both clusters induced a more severe phenotype. Deletion of the miR-17∼92 cluster was sufficient to completely abolish tumor development, pointing to the absolute requirement for the miR-17∼92 cluster, and to the lack of compensation by the miR-106b∼25 cluster, for MB development in collaboration with constitutively activated SHH signaling.
The miR-17∼92 and miR106b∼25 clusters control the number of GNPs during post-natal cerebellar development
We previously reported that microRNAs from the miR-106a∼363 cluster are not expressed in wild-type cerebella (Uziel et al., 2009). We found no significant up-regulation of microRNAs encoded by this cluster in cerebella lacking both miR-17∼92 and miR-106b∼25 clusters (data not shown). This suggested that the miR-106a∼363 cluster is unlikely to contribute to the phenotype seen in miR-17∼92cKO; miR-106b∼25KO mice.
Co-deletion of the two clusters miR-17∼92 and miR-106b∼25 led to the exhaustion of progenitor neurons in the cerebellum resulting in reduction in cerebellar size and the number of folia. The miR-17∼92 cluster controls the number of oligodendroglial cells (Budde et al., 2010), renal tubular cells (Patel et al., 2013), neural stem cells (Bian et al., 2013) and cardiomyocytes (Chen et al., 2013). However, in contrast, the miR-17∼92 cluster alone or in combination with miR-106b∼25 is dispensable during retinal development (Conkrite et al., 2011; Nittner et al., 2012), mammary development (Feuermann et al., 2012) and Langerhans cells (Zhou et al., 2014). Thus, the requirement for microRNAs encoded by the miR-17∼92 cluster is cell context specific.
The miR-17∼92 and miR-106b∼25 clusters are direct targets of Myc and Mycn (O'Donnell et al., 2005; Northcott et al., 2009; de Pontual et al., 2011). In the central nervous system, the loss of Mycn in Nestin-positive cells induces a reduced size and misformed cerebellum (Knoepfler et al., 2002). However, in these mice, the proliferation of the residual GNPs is driven by the up-regulation of Myc that normally is not expressed in these neuronal progenitors (Zindy et al., 2006). Expansion of GNPs in postnatal cerebella of mice deficient for both Mycn and Myc is severely reduced leading to a cerebellum lacking all folia (Wey et al., 2010). The fact that the loss of the miR-17∼92 and miR-106b∼25 clusters attenuated proliferation of GNPs but still caused folia formation suggests that both clusters are required but not sufficient to induce the cerebellar phenotype of Mycn; Myc double-null mice.
The miR-17∼92 cluster is absolutely required for SHH-MB development
MiRs encoded by the miR-17∼92 cluster are overexpressed in both mouse and human tumors (Uziel et al., 2009; Northcott et al., 2009). We previously found that enforced expression of the miR-17∼92 cluster collaborates with SHH signaling to induce SHH-MB (Uziel et al., 2009). Strikingly, deletion of the miR-17∼92 cluster in Nestin-positive cells from Ptch1+/−; Cdkn2c+/− mice completely abolished SHH-MB development. This was associated with a significant decrease in the number of 3 week old Ptch1+/−; Cdkn2c+/− mice lacking the miR-17∼92 cluster that exhibit PNLs on the surface of their cerebella. Therefore, the miR-17∼92 cluster plays a critical role in GNPs during SHH-MB initiation as was shown in retinoblastoma (Nittner et al., 2012).
Our data also showed that the miR-106b∼25 did not compensate for the loss of the miR-17∼92 cluster in tumor initiation. Analysis of the microRNAs expressed by the two clusters revealed that miR-19a and miR-19b-1 that share the same seed sequence are encoded only by the miR-17∼92 but not the miR-106b∼25 cluster (Fig. 10). This suggests that these two microRNAs might be sufficient for tumor development and that they might recapitulate the oncogenic function of the miR-17∼92 cluster, as reported previously for the development of Eμ-Myc B cell lymphoma (Mu et al., 2009; Olive et al., 2009). Treatment of SHH-MB with tiny LNAs directed against miR-17/20a/106b/93 and miR-19a/b-1 from the miR-17∼92 cluster family inhibits proliferation of SHH-MB in vitro and in vivo (Murphy et al., 2013). Therefore the miR-19a/b-1 might not be the only microRNA required for MB progression, although this will need further evaluation.
Fig. 10. Schematic representation of the miR-17∼92 cluster family and its regulation by SHH signaling.
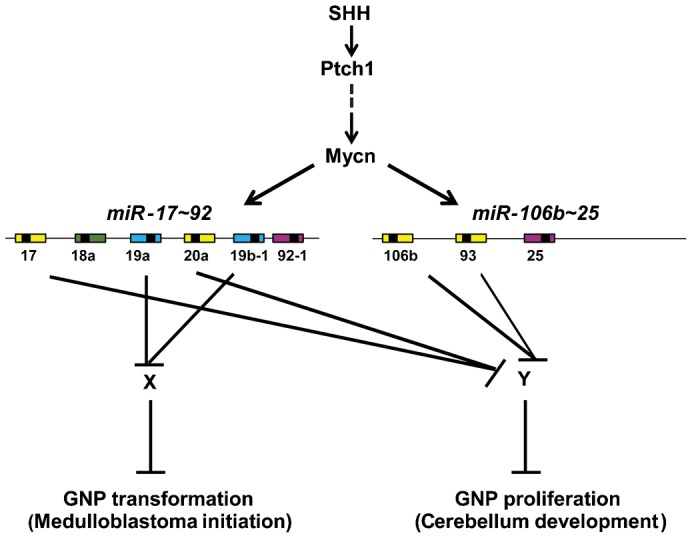
SHH signaling induces the transcription of Mycn, which, in turn, induces the expression of the miR-17∼92 and miR-106b∼25 clusters that encode 6 and 3 microRNAs, respectively. MicroRNAs sharing the same seed sequence are represented by boxes of the same color. X and Y represent putative targets. Loss of the miR-17∼92 but not the miR-106b∼25 cluster inhibits Ptch1+/−-induced SHH-MBs. Loss of both clusters reduces cerebellar size.
Given the absolute requirement of the miR-17∼92 cluster for Ptch1+/− induced SHH-MB formation, the identification of bone fide miR-17∼92 targets is clearly warranted. Analysis of previously published targets (Brock et al., 2009; Concepcion et al., 2012; Mogilyansky and Rigoutsos, 2013; Sun et al., 2013) revealed that Bmpr2, but not PTEN and p21Cip1, is regulated by the miR-17∼92 cluster (Murphy et al., 2013). We found that Bmpr2 was upregulated in the cerebella of miR-17∼92cKO; miR-106b∼25KO mice, which might be responsible, in part, for the cerebellar anomalies. Constitutive activation of Bmpr1a in cerebella induced a simplified foliation pattern (Ming et al., 2002). These data are consistent with ours and others findings showing that BMP signaling antagonizes the SHH pathway (Rios et al., 2004) and that genes in the BMP pathway are downregulated in SHH-MB (Zhao et al., 2008). It has been previously reported that the miR-17∼92 cluster down-regulates multiple components of the TGF-β pathway (Tagawa et al., 2007; Petrocca et al., 2008; Dews et al., 2010; Mestdagh et al., 2010; Concepion et al., 2012; Li et al., 2012; Mogilyansky and Rigoutsos, 2013). In agreement with these reports, we found, by GSEA analysis, that the TGF-β pathway was upregulated in the cerebella of miR-17∼92cKO; miR-106b∼25KO mice. TGF-β pathway is associated with SHH-MB pathogenesis with high levels connoting a good prognosis (Aref et al., 2013). Our results are in agreement with a study suggesting that miR-17∼92 cluster downregulates two different signaling pathways, Bmp and TGF-β pathways (Brock et al., 2009). Further analysis using Clip-Seq approaches will be required to identify the bona fide miR-17∼92 targets in mouse and human MBs (Darnell, 2010).
In summary, while the miR-17∼92 and miR-106b∼25 clusters are essential for cerebellum development and homeostasis, the miR-17∼92, but not the miR-106b∼25, cluster is required for tumor initiation in Ptch1+/− mice.
Supplementary Material
Acknowledgments
We are indebted to Tyler Jacks for graciously providing the conditional miR-17∼92 mice as well as the miR-106b∼25-null animals. We thank Shelly Wilkerson, Sarah Robinson, Dana Farmer and Jose Grenet for mouse genotyping, immunohistochemistry experiments, RNA extraction and mouse colony management. We thank Melissa Johnson and Shantel Brown for help with orthotopic transplants, Jerold Rehg for pathology analysis, Brandon Cox for performing Rotarod testing, and David Finkelstein and Marie Morfouace for microarray analysis. We also thank Charles J. Sherr, David Solecki, Brandon Wainwright and all members of the laboratory for helpful discussions during the course of these experiments.
Footnotes
Author Contributions: M.F.R., F.Z. and A.V. developed concepts and approaches; F.Z., D.K., Y.L., O.A. and L.B.M. performed experiments; F.Z., D.K. and M.F.R. performed data analysis; M.F.R. and F.Z. wrote the manuscript; M.F.R., P.J.M. and A.V. edited the manuscript; all authors reviewed the manuscript.
Competing interests: The authors have no competing interests to declare.
Funding
This work was supported in part by National Institutes of Health Grant CA-096832 (to M.F.R.), Core Grant CA-21765 (to M.F.R.), and the American Lebanese-Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital.
References
- Aref D., Moffatt C. J., Agnihotri S., Ramaswamy V., Dubuc A. M., Northcott P. A., Taylor M. D., Perry A., Olson J. M., Eberhart C. G. et al. (2013). Canonical TGF-β pathway activity is a predictor of SHH-driven medulloblastoma survival and delineates putative precursors in cerebellar development. Brain Pathol. 23, 178–191 10.1111/j.1750-3639.2012.00631.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S., Hong J., Li Q., Schebelle L., Pollock A., Knauss J. L., Garg V., Sun T. (2013). MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406 10.1016/j.celrep.2013.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M., Trenkmann M., Gay R. E., Michel B. A., Gay S., Fischler M., Ulrich S., Speich R., Huber L. C. (2009). Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ. Res. 104, 1184–1191 10.1161/CIRCRESAHA.109.197491 [DOI] [PubMed] [Google Scholar]
- Budde H., Schmitt S., Fitzner D., Opitz L., Salinas-Riester G., Simons M. (2010). Control of oligodendroglial cell number by the miR-17-92 cluster. Development 137, 2127–2132 10.1242/dev.050633 [DOI] [PubMed] [Google Scholar]
- Carmell M. A., Hannon G. J. (2004). RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 11, 214–218 10.1038/nsmb729 [DOI] [PubMed] [Google Scholar]
- Chen J., Huang Z. P., Seok H. Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. et al. (2013). mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566 10.1161/CIRCRESAHA.112.300658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion C. P., Bonetti C., Ventura A. (2012). The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 18, 262–267 10.1097/PPO.0b013e318258b60a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkrite K., Sundby M., Mukai S., Thomson J. M., Mu D., Hammond S. M., MacPherson D. (2011). miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 25, 1734–1745 10.1101/gad.17027411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R. B. (2010). HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1, 266–286 10.1002/wrna.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L., Yao E., Callier P., Faivre L., Drouin V., Cariou S., Van Haeringen A., Geneviève D., Goldenberg A., Oufadem M. et al. (2011). Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat. Genet. 43, 1026–1030 10.1038/ng.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M., Fox J. L., Hultine S., Sundaram P., Wang W., Liu Y. Y., Furth E., Enders G. H., El-Deiry W., Schelter J. M. et al. (2010). The myc-miR-17∼92 axis blunts TGFbeta signaling and production of multiple TGFbeta-dependent antiangiogenic factors. Cancer Res. 70, 8233–8246 10.1158/0008-5472.CAN-10-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y., Robinson G. W., Zhu B. M., Kang K., Raviv N., Yamaji D., Hennighausen L. (2012). The miR-17/92 cluster is targeted by STAT5 but dispensable for mammary development. Genesis 50, 665–671 10.1002/dvg.22023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L. V., Milenković L., Higgins K. M., Scott M. P. (1997). Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 10.1126/science.277.5329.1109 [DOI] [PubMed] [Google Scholar]
- Hatten M. E., Roussel M. F. (2011). Development and cancer of the cerebellum. Trends Neurosci. 34, 134–142 10.1016/j.tins.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton B. A., Knoepfler P. S., Kenney A. M., Rowitch D. H., de Alborán I. M., Olson J. M., Eisenman R. N. (2006). N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 66, 8655–8661 10.1158/0008-5472.CAN-06-1621 [DOI] [PubMed] [Google Scholar]
- Kawauchi D., Robinson G., Uziel T., Gibson P., Rehg J., Gao C., Finkelstein D., Qu C., Pounds S., Ellison D. W. et al. (2012). A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 21, 168–180 10.1016/j.ccr.2011.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A. M., Cole M. D., Rowitch D. H. (2003). Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130, 15–28 10.1242/dev.00182 [DOI] [PubMed] [Google Scholar]
- Kim V. N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Nelson A. L., Algon S. A., Graves O., Sturla L. M., Goumnerova L. C., Rowitch D. H., Segal R. A., Pomeroy S. L. (2003). Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev. Biol. 263, 50–66 10.1016/S0012-1606(03)00434-2 [DOI] [PubMed] [Google Scholar]
- Knoepfler P. S., Cheng P. F., Eisenman R. N. (2002). N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 16, 2699–2712 10.1101/gad.1021202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder J. M., Altman J., Krebs H. (1974). Some mechanisms of cerebellar foliation: effects of early hypo- and hyperthyroidism. Brain Res. 76, 33–40 10.1016/0006-8993(74)90511-3 [DOI] [PubMed] [Google Scholar]
- Li L., Shi J. Y., Zhu G. Q., Shi B. (2012). MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J. Cell. Biochem. 113, 1235–1244 10.1002/jcb.23457 [DOI] [PubMed] [Google Scholar]
- Mestdagh P., Boström A. K., Impens F., Fredlund E., Van Peer G., De Antonellis P., von Stedingk K., Ghesquière B., Schulte S., Dews M. et al. (2010). The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell 40, 762–773 10.1016/j.molcel.2010.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming J. E., Elkan M., Tang K., Golden J. A. (2002). Type I bone morphogenetic protein receptors are expressed on cerebellar granular neurons and a constitutively active form of the type IA receptor induces cerebellar abnormalities. Neuroscience 114, 849–857 10.1016/S0306-4522(02)00348-2 [DOI] [PubMed] [Google Scholar]
- Mogilyansky E., Rigoutsos I. (2013). The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20, 1603–1614 10.1038/cdd.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P., Han Y. C., Betel D., Yao E., Squatrito M., Ogrodowski P., de Stanchina E., D'Andrea A., Sander C., Ventura A. (2009). Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 23, 2806–2811 10.1101/gad.1872909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B. L., Obad S., Bihannic L., Ayrault O., Zindy F., Kauppinen S., Roussel M. F. (2013). Silencing of the miR-17∼92 cluster family inhibits medulloblastoma progression. Cancer Res. 73, 7068–7078 10.1158/0008-5472.CAN-13-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittner D., Lambertz I., Clermont F., Mestdagh P., Köhler C., Nielsen S. J., Jochemsen A., Speleman F., Vandesompele J., Dyer M. A. et al. (2012). Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat. Cell Biol. 14, 958–965 10.1038/ncb2556 [DOI] [PubMed] [Google Scholar]
- Northcott P. A., Fernandez-L A., Hagan J. P., Ellison D. W., Grajkowska W., Gillespie Y., Grundy R., Van Meter T., Rutka J. T., Croce C. M. et al. (2009). The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 69, 3249–3255 10.1158/0008-5472.CAN-08-4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 10.1038/nature03677 [DOI] [PubMed] [Google Scholar]
- Olive V., Bennett M. J., Walker J. C., Ma C., Jiang I., Cordon-Cardo C., Li Q. J., Lowe S. W., Hannon G. J., He L. (2009). miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 23, 2839–2849 10.1101/gad.1861409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T. G., Read T. A., Kessler J. D., Mehmeti A., Wells J. F., Huynh T. T., Lin S. M., Wechsler-Reya R. J. (2005). Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439 10.1242/dev.01793 [DOI] [PubMed] [Google Scholar]
- Patel V., Williams D., Hajarnis S., Hunter R., Pontoglio M., Somlo S., Igarashi P. (2013). miR-17∼92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc. Natl. Acad. Sci. USA 110, 10765–10770 10.1073/pnas.1301693110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F., Vecchione A., Croce C. M. (2008). Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 68, 8191–8194 10.1158/0008-5472.CAN-08-1768 [DOI] [PubMed] [Google Scholar]
- Rios I., Alvarez-Rodríguez R., Martí E., Pons S. (2004). Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development 131, 3159–3168 10.1242/dev.01188 [DOI] [PubMed] [Google Scholar]
- Sun Q., Mao S., Li H., Zen K., Zhang C. Y., Li L. (2013). Role of miR-17 family in the negative feedback loop of bone morphogenetic protein signaling in neuron. PLoS ONE 8, e83067 10.1371/journal.pone.0083067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa H., Karube K., Tsuzuki S., Ohshima K., Seto M. (2007). Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 98, 1482–1490 10.1111/j.1349-7006.2007.00531.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. D., Northcott P. A., Korshunov A., Remke M., Cho Y. J., Clifford S. C., Eberhart C. G., Parsons D. W., Rutkowski S., Gajjar A. et al. (2012). Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123, 465–472 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., Schütz G. (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Uziel T., Zindy F., Xie S., Lee Y., Forget A., Magdaleno S., Rehg J. E., Calabrese C., Solecki D., Eberhart C. G. et al. (2005). The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev. 19, 2656–2667 10.1101/gad.1368605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T., Karginov F. V., Xie S., Parker J. S., Wang Y. D., Gajjar A., He L., Ellison D., Gilbertson R. J., Hannon G. et al. (2009). The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. USA 106, 2812–2817 10.1073/pnas.0809579106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R. et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey A., Martinez Cerdeno V., Pleasure D., Knoepfler P. S. (2010). c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum 9, 537–547 10.1007/s12311-010-0190-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Ayrault O., Zindy F., Kim J. H., Roussel M. F. (2008). Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 22, 722–727 10.1101/gad.1636408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Qi R. Q., Liu M., Xu Y. P., Li G., Weiland M., Kaplan D. H., Mi Q. S. (2014). microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun. 15, 57–61 10.1038/gene.2013.61 [DOI] [PubMed] [Google Scholar]
- Zindy F., Knoepfler P. S., Xie S., Sherr C. J., Eisenman R. N., Roussel M. F. (2006). N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc. Natl. Acad. Sci. USA 103, 11579–11583 10.1073/pnas.0604727103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Uziel T., Ayrault O., Calabrese C., Valentine M., Rehg J. E., Gilbertson R. J., Sherr C. J., Roussel M. F. (2007). Genetic alterations in mouse medulloblastomas and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res. 67, 2676–2684 10.1158/0008-5472.CAN-06-3418 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



