Abstract
We studied the effect of inactivation of genes, which control biosynthesis of inosine monophosphate (IMP)de novo and purine salvage and interconversion pathways, on sensitivity of yeast Saccharomyces cerevisiae to the mutagenic and toxic action of 6-hydroxylaminopurine (HAP) and 2-amino-6-hydroxylaminopurine(AHA). It was shown that the manifestation of HAP and AHA mutagenic properties depends on the action of enzyme adenine phosphoribosyltransferase encoded in yeast by APT1 gene. A blockade of any step of IMP biosynthesis, with the exception of the block mediated by inactivation of genes ADE16 and ADE17 leading to the accumulation of 5-aminoimidazole-4carboxamide ribonucleotide (AICAR), was shown to enhanceyeast cell sensitivity to the HAP mutagenic effect; however, it does not affect the sensitivity to AHA. A block of conversion of IMP into adenosine monophosphate (AMP) causes hypersensitivity of yeast cells to the mutagenic action of HAP and to the toxic effect of HAP, AHA, and hypoxanthine. It is possible that this enhancement of sensitivity to HAP and AHA is due to changes in the pool of purines. We conclude that genes ADE12, ADE13, AAH1, and HAM1 controlling processes of purine salvage and interconversion in yeast, make the greatest contribution to the protection against the toxic and mutagenic action of the examined analogs. Possible mechanisms of HAP detoxication in bacteria, yeast, and humans are discussed.
Introduction
Synthetic purine analogs 6-hydroxylaminopurine (HAP) and 2-amino-6-hydroxylaminopurine (AHA) are strong mutagens for a wide spectrum of pro- and eukaryotic organisms and possess toxic activity [1, 2]. HAP was shown to cause teratogenic effects [3], malignant transformation of mammalian cells [4] and chromosome fragmentation [5]. Some experimental data suggest the possibility of endogenic HAP generation [6–8]. Endogenic purine analogs may be a reason for hereditary and oncologic diseases [9], and this determines the importance of studying their metabolism in detail and elucidating mechanisms responsible for the mutagenic action.
Materials and Methods
In this work, we used a genetic approach to the study of HAP and AHA metabilism in yeast Saccharomyces cerevisiae.The previously conducted screening for S. cerevisiae deletion mutants allowed the identification of 18 various genes, the inactivation of which enhances yeast sensitivity to the mutagenic or toxic action of HAP (three of these genes also control sensitivity to AHA) [10]. Half of the identified genes were found to control purine metabolism: three genes (AAH1, ADE12 and HAM1) control purine salvage and interconversion pathway and six ADE genes controlling IMP biosynthesis de novo (Fig. 1). This approach used in this screening did not allow us to examine the effect of inactivation of the remaining four genes (ADE4, ADE13, ADE16, and ADE17) controlling IMP biosynthesis along with key genes (FCY2, APT1, XPT1, andHPT1) governing the transport of purine bases and their conversion into nucleoside monophosphates (NMP). In this work, the impact of inactivation of these genes on sensitivity of yeast cells to HAP and AHA was studied. To test mutagenic activity and toxicity of these compounds, we used quantitative and qualitative methods developed earlier in our laboratory [10].
Fig. 1.

Genetic control of IMP biosynthesis de novo (genes are underlined) and purine salvage and interconversion pathways in yeast S. cerevisiae (Ade, adenine; Hyp, hypoxanthine; Gua, guanine; X, xanthine; SAMP, succinyl-AMP; AICAR, 5-aminoimidazole-4-carboxamide-5′-phosphoribonucleotide; IMP, inosine-5′-monophosphate; HAPMP, HAP-5′-monophosphate; HAPTP, HAP-5′-triphosphate; dHAPTP, HAP-2′-deoxyriboside-5′-triphosphate. * Gene inactivation leads to an increase in sensitivity to mutagenic or toxic effect of HAP; ** gene inactivation increases resistance to HAP and AHA.
Results and Discussion
We established that mutant ade4 as all earlier studied ade mutants [10] with the block in IMP iosynthesis, is more sensitive to HAP than the wild-type strain and insensitive to AHA (Table 1). It should be noted that various ade mutants have different HAP sensitivity [10]. Possibly, intermediate products ccumulated in cells upon a blockade of various stages of IMP biosynthesis differentially affect the expression of the ADE genes [11, 12] and other genes for purine metabolism, thus modifying the sensitivity of yeast cells to purine analogs. An exception to this rule are genes ADE16 and ADE17. Single mutants for these genes do not display enhanced sensitivity to HAP and AHA [10]. Genes ADE 16 and ADE 17 encode isozymes and the complete block of purine biosynthesis (leading to adenine auxotrophy) is observed only in double mutants ade16 ade17[11]. However, the double mutant ade16 ade17 constructed in our study, does not show enhanced sensitivity to the examined analogs (Table 1), unlike other adenine auxotrophs. A unique property of mutant ade16 ade17 may be related to the accumulation in its cells of a low-molecular-weight regulator 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) participating in regulation of the expression of genes for yeast purine and histidine biosynthesis [12]. We suggest that one possible echanism responsible for AICAR action is the competition of this compound with AMP, as an activator of AMP-dependent protein kinases regulating the expression of many eukaryotic genes [16]. It is possible that AICAR is directly involved in the competition with mutagenic AMP analogs for enzymes of purine metabolism, decreasing their mutagenic effect. In addition, mutant ade16 ade17 requires not only adenine but also histidine [11, 12], because AICAR accumulated in cells seems to block biosynthesis of histidine. It is known that histidine is formed from the purine ring of ATP molecule. Thus, one can assume that the double mutation ade16 ade17 may increase the pool of adenine nucleotides (since ATP is not utilized for histidine biosynthesis), which may be the reason for ensuring the enhanced resistance of mutant ade16 ade17 to the adenine analog HAP. This assumption is in good agreement with the data showing the nhibition of HAPinduced mutagenesis in yeast and Escherichia coli with high amount of adenine. Thus, we showed that the block of IMP biosynthesis (inactivation of any of ADE genes, except for genes ADE16 and ADE17) leads to an increase in yeast cell sensitivity to the mutagenic effect of HAP (but does not affect AHA sensitivity). Most probably, the enhanced sensitivity of adenine auxotrophs to HAP is mediated by changes in the pool of purines, because it is known that the level of yeast mutagenesis strongly depends on the qualitative and quantitative ratio of deoxynucleoside triphosphates (dNTP) in the cell [17].
Table 1. Influence of mutations in the genes of purine metabolism on sensitivity of yeast cells to the mutagenic action of HAP and AHA.
| Strain (inactivated gene) | The median value (×10–7) of the frequency of forward mutations to canavanine resistance (confidence interval is given in brackets) |
||
|---|---|---|---|
| YPD | YPD + HAP (25 mg/l) |
YPD + AHA (100 mg/l) |
|
| PLY122 (wt)1 | 3 (1-10) | 540 (430-735) | 31 (24-53) |
| Y513 (fcy2)1 | 2 (0,7-10) | 503 (96-653) | 50 (22-80) |
| Y508 (hpt1)1 | 4 (2-7) | 480 (280-620) | 34 (29-79) |
| YD-16931 (xpt1)2 | 1 (0,5-5) | 600 (380-750) | 28 (17-40) |
| Y511 (apt1)1 | 2 (1-11) | 6 (1-39) | 10 (5-26) |
| PLY122-HLAM (ham1)3 | 3 (1-8) | 10000 (8000-1300) | 40 (18-65) |
| Y520 (aah1)1 | 2 (0.5-6) | 5500 (3500-7000) | 150 (100-220) |
| Y511-HLAM (ham1 apt1)3 | 2 (0.7-6) | 128 (80-210) | 43 (10-80) |
| Y550 (aah1 apt1)1 | 3 (1-7) | 245 (132-686) | 75 (30-115) |
| YD-10888 (ade4)2 | 4 (1-8) | 1131 (1050-1309) | 28 (15-48) |
| 16-17-BY4741 (ade16, 17)3 | 3 (1-10) | 563 (418-688) | 60 (42-80) |
Note:
Isogenic strains are described in [13].
Strains isogenic with strain BY4742 (see Table 2) are described in [14]. Strains 3PLY122-HLAM (MATα leu2-3, 112 lys2-Δ201 ura3–52 ham1::LEU2), Y511-HLAM (MATα leu2-3, 112 lys2-Δ201 ura3-52 apt1::URA3 ham1::LEU2) and 16-17-BY4741 (MATα leu2Δ lys2Δ ura3Δ his3Δ ade16Δ ade17Δ) are obtained in this work.
The values that significantly differed (P = 0.05) from the values for strain PLY122 are underlined (the comparison was performed using Wilcoxon's nonparametric rank test and standard methods of calculating confidence intervals for medians [15]).
Genes ADE12 and ADE13 control the conversion of IMP into AMP. Furthermore, gene ADE13 participates in IMP biosynthesis de novo (Fig. 1). Mutation ade13 is a conditional lethal: ade13 mutants are unable to grow on a medium with glucose, but they can grow on the medium containing glycerol as the sole source of carbon. This phenotype of mutation ade13 is partially suppressed by additional mutations at earlier stages of IMP biosynthesis [18]. Therefore, we constructed the double mutant ade4 ade13. Strain ade4 ade13, like mutant ade12, proved to grow poorly on the complete YPD medium containing glucose (Fig. 2). We have found that this phenotype is caused by an insufficient amount of exogenous adenine required for growth of mutants: the addition of adenine at a concentration of 100 mg/l to YPD medium led to the recovery of normal cell growth in strains ade4 ade13and ade12 (Fig. 2). Interestingly, hypoxanthine was toxic for mutants ade4 ade13 and ade12 (Fig. 2, Table 2). It is possible that the toxic effect of hypoxanthine is connected with the fact that IMP (produced from exogenous hypoxanthine due to the action of gene зPT1 product) cannot be converted to AMP as a result of the block mediated by the inactivation of gene ADE12 or ADE13. When there is limited amount of AMP, the accumulation of IMP seems to cause the toxic effect in cells. Mutant ade4 ade13 proved to be far more sensitive to HAP than the wild-type strain or single mutant ade4 (Fig. 2, Tables 1 and 2). At the same time, mutants ade4 ade13 and ade12 did not manifest enhanced sensitivity to the mutagenic effect of AHA, but, unlike other ade mutants, they were sensitive to the toxic effect of this analog (Fig. 2, Tables 1 and 2). As previously shown [10], inactivation of gene AAH1 also markedly enhances cell sensitivity to HAP and AHA, whereas inactivation of HAM1 enhances sensitivity to only HAP (see Table 1). According to our preliminary data obtained for cell-free extracts of strains PLY122 (wt), BY4742 (wt), and Y520 (aah1), HAP and AHA are converted, by adenine aminohydrolase encoded by gene AAH1, to hypoxanthine and guanine, respectively. Deoxyribonucleoside triphosphate pyrophosphohydrolase encoded by gene HAM1 catalyzes conversion of dHAPTP to dHAPMP [19] (Fig. 1) and is probably inactive for dAHAPTP. Thus, inactivation of genes ADE12, ADE13, AAH1, and HAM1 responsible for salvage and interconversion of purines caused the strongest increase in the toxic and mutagenic effect of HAP and AHA suggesting the largest contribution of these genes to the protection against the action of the examined analogs.
Fig. 2.
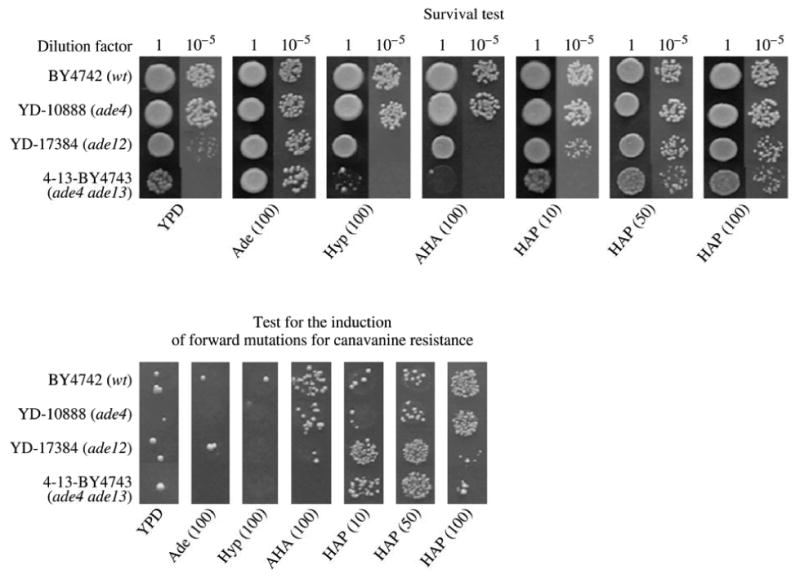
Results of the qualitative test for induction of forward mutations for canavanine resistance in yeast strains carrying mutations at various genes of the AMP biosynthesis pathway. The test was performed as in [10]. Below photos, concentrations of purine bases (mg/l) in complete YPD medium are given in brackets.
Table 2. Influence of inactivation of ADE12 and ADE13 genes on sensitivity of yeast cells to the mutagenic and toxic effect of HAP, AHA, and hypoxanthine.
| Strain (inactivated gene) | The median value (×10–7) of the frequency of forward mutations for canavanine resistance (confidence interval is given in brackets) and percent of survival4 |
|||
|---|---|---|---|---|
| YPD | YPD + Гип (100 Mг/л) |
YPD + AПAΠ (100 Mг/л) |
YPD + ГAП ( 25 Mг/л) |
|
| BY4742 (wt)1 | 3 (1-10) 100% |
4 (1-10) 100% |
31 (24-53) 100% |
540 (430-735) 100% |
| YD-17384 (ade12) 1 | 6 (2-10) 100% |
5 (0-18) 13% |
68 (48-106) 17% |
3722 (2627-6117) 11% |
| 4-13-BY4743 (ade4 ade13)2 | 17 (26-43) 100% |
-3 1,3% |
93 (72-806) 8% |
6168 (3691-8909) 24% |
Note:
Isogenic strains are described in [13].
Strain 4-13-BY4743 (MATα leu2Δ lys2Δ ura3Δ his3Δ ade4 ade13Δ) was provided by A.M. Zekhnov.
Mutation rate was not determined because of low survival. The survival on YPD medium was taken as 100%. The values that significantly differed (P = 0.05) from the values for strain BY4742 are underlined (an estimation was performed using Wilcoxon's nonparametric rank test and standard methods of calculating confidence intervals [15]).
By analogy to adenine metabolism, we assumed that HAP and AHA are ransported into the cell via the purine cytosine permease encoded by gene FCY2 [20] and converted, under the action of purine phosphoribosyltransferases, to the corresponding nucleosidmonophosphates, which is the critical condition for their further conversion to dNTP and incorporation into DNA. However, we found that the inactivation of gene FCY2does not affect the sensitivity to HAP and AHA (Table 1). In addition to Fcy2 permease, seven more potential permeases of nucleobases were detected in the yeast genome, two of which (Fcy21p and Fcy22p) have a high level of similarity (74 and 88%, respectively) with Fcy2 [21]. Genes FCY21 and FCY22, like FCY2 gene, are located in chromosome 5. Inactivation of these genes causes enhanced resistance to the adenine analog 8-azaadenine [21], which allows one to consider these proteins as potential purine transporters, activating HAP and AHA. Inactivation of gene APT1 encoding adenine phosphoribosyltransferase leads to a drastic decrease in HAP- and AHA-induced mutagenesis (by a factor of 90 and 3, see Table 1). The introduction of an additional mutation apt1 into HAP-hypersensetive strains (mutants aah1 and ham1) revealed that, in comparison with the single mutant apt1, double mutants apt1 aah1 show a rather high level of mutagenesis under the action of HAP and AHA, whereas the double mutants apt1 ham1, only when exposed to HAP (because ham1 mutants are insensitive to AHA). On the other hand inactivation of genes зPT1 and XPT1 encoding guanine-hypoxanthine and xanthine phosphoribosyltransferases, respectively, does not markedly affect the level of mutagenesis induced by the indicated analogs (Table 1). In summary, we can conclude that the principal pathway of HAP and AHA activation is regulated by gene APT1. Moreover, a less efficient APT1-independent pathway of conversion of HAP and AHA into corresponding NMP, which is probably governed by genes HPT1/XPT1, also exists in yeasts (Fig. 1).
A comparative analysis of our data on Saccharomyces yeast and the data describing the action of HAP on bacteria E. coli revealed both similarity and significant differences between these organisms. Three systems play a leading role in cell protection against HAP. The first system depends on the molybdenum cofactor [22] and is able to effectively inactivate HAP and AHA [23, 24]. The second system is mediated by the rdgB gene, an ortholog of the yeast gene зДel [19]. The third system, involving endonuclease V (encoded by gene nfi), provides HAP repair in DNA [25]. The operation of this endonuclease at high doses of HAP may cause cell death resulting from the accumulation of unrepaired single-stranded breaks in DNA leading to the formation of lethal double-stranded breaks during replication [25]. As S. cerevisiae bacteria lack both the molybdenum cofactor and the nfi-dependent protection system, we think that the main mechanism underlying yeast protection against purine analogs is the maintenance of the normal pool of nucleotides and its sanitization from mutagenic analogs via enzymes of purine metabolism. A similar mechanism of protection against mutagenic and toxic purine analogs may be also realized in other eukaryotes, including humans. This idea is consistent with the fact that the patients with a decreased activity of the product of gene ITPA (homolog of gene HAM1) show enhanced sensitivity to a number of base analogs used as drug preventing organ rejection after transplantation [26]. Moreover, it has been established recently that enzymes encoded by gene HAM1 and its orthologs in pro- and eukaryotes, including humans, in addition to the natural substrates ITP and dITP, effectively catalyze conversion of dHAPTP to dHAPMP [19].
Orthologs of bacterial genes of the molybdenum cofactor-dependent system [27] and of gene nfi (gene hNFI [28]) encoding endonuclease V were found in humans. Therefore, the HAPinduced chromosome fragmentation observed in human carcinoma cell cultures [5] (which has long been known and the reason of which remains unclear) can be explained by the formation of double-stranded DNA breaks during HAP repair by endonuclease V. We propose that various strategies used to protect organisms against the mutagenic and toxic action of synthetic analogs HAP and AHA in various organisms appear in evolution to avoid the incorporation into DNA of certain natural mutagenic purine bases. Most probably, these systems appeared to protect organisms against hypoxanthine and its nucleotides. Indeed, enzymes encoded by gene HAM1 and its orthologs from otherorganisms hydrolyze ITP and dITP to the corresponding monophosphates as effectively as dHAPTP and HAPTP [19]. Synthetic hypoxanthine analogs HAP and AHA proved to be useful tools for studying mechanisms responsible for the maintenance of genome stability.
Acknowledgments
We are grateful to A.M. Zekhnov for providing yeast strains and to N.N. Khromov-Borisov for discussing the results.
This study was supported by NCI grant RO1CA129925 to YIP, grant BRHE, presented by CRDF to EIS, and the Ministry of Education of the Russian Federation to EIS.
References
- 1.Khromov-Borisov NN. Naming the Mutagenic Nucleic Acid Base Analogs: The Galatea Syndrome. Mutat Res. 1997;379(no. 1):95–103. doi: 10.1016/s0027-5107(97)00112-7. [DOI] [PubMed] [Google Scholar]
- 2.Pavlov YI, Noskov VN, Lange EK, et al. The Genetic Activity of N6-Hydroxyadenine and 2-Amino-N6-Hydroxyadenine in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae. Mutat Res. 1991;253:33–46. doi: 10.1016/0165-1161(91)90343-7. [DOI] [PubMed] [Google Scholar]
- 3.Chaube S, Murphy ML. Teratogenic Effects of 6-Hydroxylaminopurine in the Rat—Protection by Inosine. Biochem Pharmacol. 1969;18(no. 5):1147–1156. doi: 10.1016/0006-2952(69)90118-x. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC. Induction of Gene Mutation in and Cell Transformation of Mammalian Cells by Modified Purines: 2-Aminopurine and 6-N-Hydroxylaminopurine. Proc Natl Acad Sci USA. 1981;78(no. 9):5685–5689. doi: 10.1073/pnas.78.9.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesele JJ. Some Morphological Effects of Alkylating Agents. Exp Cell Res. 1963;(suppl 9):525–534. doi: 10.1016/0014-4827(63)90293-3. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman I. Enzymatic Synthesis of Adenosine-5′-Phosphate from Inosine-5′-Phosphate. J Biol Chem. 1956;223:327–339. [PubMed] [Google Scholar]
- 7.Clement B, Kunze T. Hepatic Microsomal N-Hydroxylation of Adenine to 6-N-Hydroxylaminopurine. Biochem Pharmacol. 1990;39:925–933. doi: 10.1016/0006-2952(90)90209-4. [DOI] [PubMed] [Google Scholar]
- 8.Simandan T, Sun J, Dix TA. Oxydation of DNA Bases, Deoxyribonucleosides and Homopolymers by Peroxyl Radicals. Biochem J. 1998;335:233–240. doi: 10.1042/bj3350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekiguchi M, Tsuzuki T. Oxidative Nucleotide Damage: Consequences and Prevention. Oncogene. 2002;16(no. 21(58)):8895–8904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- 10.Stepchenkova EI, Kozmin SG, Alenin VV, Pavlov YI. Genome-Wide Screening for Genes Whose Deletions Confer Sensitivity to Mutagenic Purine Base Analogs in Yeast. BMC Genet. 2005;6(no. 31):1–6. doi: 10.1186/1471-2156-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tibbetts AS, Appling DR. Characterization of Two 5-Aminoimidazole-4-Carboxamide Ribonucleotide Transformylase/Inosine Monophosphate Cyclohydrolase Isozymes from Saccharomyces cerevisiae. J Biol Chem. 2000;275(no. 27):20920–20927. doi: 10.1074/jbc.M909851199. [DOI] [PubMed] [Google Scholar]
- 12.Rebora K, Laloo B, Daignan-Fornier B. Revisiting Purine–Histidine Cross-Pathway Regulation in Saccharomyces cerevisiae: A Central Role for a Small Molecule. Genetics. 2005;170(no. 1):61–70. doi: 10.1534/genetics.104.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guetsova ML, Lecoq K, Daignan-Fornier B. The Isolation and Characterization of Saccharomyces cerevisiae Mutants That Constitutively Express Purine Biosynthetic Genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winzeler EA, Shoemaker DO, Astromoff A, et al. Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science. 1999;258:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 15.Glotov NV, Zhivotovsky LA, Khovanov NV, Khromov-Borisov NN. Biometriya (Biometry) eningrad: Leningrad Gos. Univ.; 1982. [Google Scholar]
- 16.Leclerc I, Viollet B, da Silva Xavier G, et al. Role of AMP-Activated Protein Kinase in the Regulation of Gene Transcription. Biochem Soc Trans. 2002;30(no. 2):307–311. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 17.Chabes A, Georgieva B, Domkin V, et al. Survival of DNA Damage in Yeast Directly Depends on ncreased dNTP Levels Allowed by Relaxed Feedback Inhibition of Ribonucleotide Reductase. Cell. 2003;112(no. 3):391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 18.Zekhnov AM, Domkin VD, Dembereliin O, et al. Mutation of ade13–1 of the Yeasts Saccharomyces cerevisiae Leads to the Absence of Growth on a Complete Medium with Glucose and Epistatically Interacts with Mutations in Other Genes of Purine Biosyntheses. RussJ Genet. 1995;31(no. 1):15–23. [PubMed] [Google Scholar]
- 19.Burgis NE, Cunningham RP. Substrate Specificity of RdgB Protein, a Deoxyribonucleoside Triphosphate Pyrophosphohydrolase. J Biol Chem. 2007;282(no. 6):3531–3538. doi: 10.1074/jbc.M608708200. [DOI] [PubMed] [Google Scholar]
- 20.Weber E, Rodriguez C, Chevallier MR, Jund R. The Purine Cytosine Permease of Saccharomyces cerevisiae: Primary Structure and Deduced Protein Sequence of the FCY2 Gene Product. Mol Microbiol. 1990;4:585–596. doi: 10.1111/j.1365-2958.1990.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 21.Paluszynski JP, Klassen R, Rohe M, Meinhardt F. Various Cytosine/Adenine Permease Homologues Are Involved in the Toxicity of 5-Fluorocytosine in Saccharomyces cerevisiae. Yeast. 2006;23(no. 9):707–715. doi: 10.1002/yea.1387. [DOI] [PubMed] [Google Scholar]
- 22.Rajagopalan KV. Biosyntheses of the Molybdenum Cofactor. In: Neidhardt FC, editor. Escherichia coli and Salmonella, Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 674–679. [Google Scholar]
- 23.Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. Hypersensitivity of Escherichia coli (uvrB-bio) Mutants to 6-Hydroxylaminopurine and Other Base Analogs Is Due to a Defect in Molybdenum Cofactor Biosynthesis. J Bacteriol. 2000;182:3361–3367. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozmin SG, Schaaper RM. Molybdenum Cofactor-Dependent Resistance to N-Hydroxylated Base Analogs in E. coli Is Independent of MobA Function. Mutat Res. 2007;619:9–15. doi: 10.1016/j.mrfmmm.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgis NE, Brucker JJ, Cunningham RP. Repair System for Noncanonical Purines in Escherichia coli. J Bacteriol. 2003;185:3101–3110. doi: 10.1128/JB.185.10.3101-3110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinaki AM, Ansari A, Duley JA, et al. Adverse Drug Reactions to Azathioprine Therapy Are Associated with Polymorphism in the Gene Encoding Inosine Triphosphate Pyrophosphatase (ITPase) Pharmacogenetics. 2004;14(no. 3):181–187. doi: 10.1097/00008571-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz G. Molybdenum Cofactor Biosynthesis and Deficiency. Cell Mol Life Sci. 2005;62(no. 23):2792–2810. doi: 10.1007/s00018-005-5269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moe A, Ringvoll J, Nordstrand L, et al. Incision at Hypoxanthine Residues in DNA by a Mammalian Homologue of the Escherichia coli Antimutator Enzyme Endonuclease V. Nucleic Acids Res. 2003;31(no. 14):3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]


