Abstract
Spliceosomal snRNAs are extensively 2'-O-methylated and pseudouridylated. The modified nucleotides are relatively highly conserved across species, and are often clustered in regions of functional importance in pre-mRNA splicing. Over the past decade, the study of the mechanisms and functions of spliceosomal snRNA modifications has intensified. Two independent mechanisms behind these modifications, RNA-independent (protein-only) and RNA-dependent (RNA-guided), have been discovered. The role of spliceosomal snRNA modifications in snRNP biogenesis and spliceosome assembly has also been verified.
Introduction
The removal of intervening sequences, introns, from pre-messenger RNA (pre-mRNA) is of fundamental importance to gene expression. In eukaryotic organisms, the majority of introns are removed by the spliceosome, a massively large and equally dynamic complex consisting of five small nuclear (sn) RNAs (U1, U2, U4, U5 and U6) and numerous protein components.1-4 snRNAs participate in the pre-mRNA splicing reaction as a small nuclear ribonucleoprotein (snRNP) complex, which includes a single spliceosomal snRNA in complex with a number of proteins.
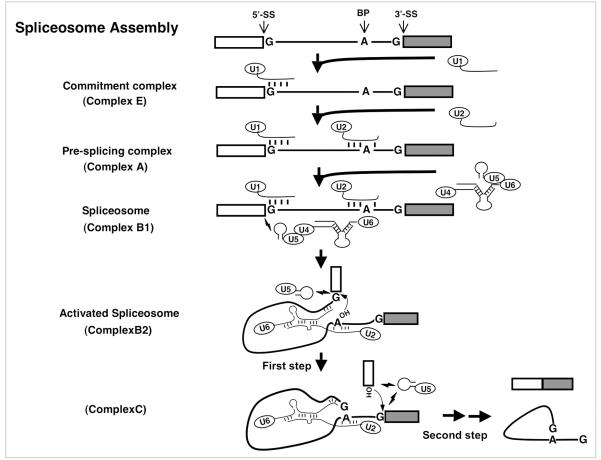
Spliceosomal snRNPs are key components of the spliceosome and are absolutely required for pre-mRNA splicing. In the classical view of pre-mRNA splicing there is a step-wise assembly of the spliceosome initiated by recognition of the 5' splice-site (5' SS) by complementary base-pairing interactions with the 5'-end of U1 snRNA (Fig. 1).5-14 Subsequently, the branch-site sequence (BSS) is engaged by the U2 snRNP, resulting in the formation of a pre-splicing complex, namely complex A.11,14-24 The U2 snRNA-BSS interaction, which is mediated through base-pairing interactions, bulges out the branch point nucleotide (typically an adenosine). Addition of the U4/U6.U5 tri-snRNP, in which U4 and U6 are extensively base-paired, to complex A results in the formation of complex B1, and initiates a series of RNA-RNA rearrangements, resulting in the destabilization and release of the U1 and U4 snRNPs.19,25-28 The result of these rearrangements is the formation of complex B2 and concomitant activation of the spliceosome, leading to the first step of splicing, in which the 2'-OH group of the bulged out branch point adenosine nucleophilically attacks the 5' SS. Upon the completion of the first step of splicing, Complex B2 is converted into complex C. After additional conformational changes, the second step of splicing occurs, resulting in the production of mature mRNA and the release of the excised intron and the U2, U5 and U6 snRNPs, which are recycled for further rounds of pre-mRNA splicing.
Figure 1.
Major spliceosome assembly and catalysis of pre-mRNA splicing. The thick lines represent the intron and the boxes are exons. The 5' splice site (5'-SS), the 3' splice site (3'-SS) and the branch point adenosine (BP) are indicated in the pre-mRNA. The conserved residues at the 5' and 3' splice sites and the branch site are shown. The headed thin lines are snRNAs with their names in the ellipses. The short thick lines between RNA strands represent Watson-Crick base-pairing interactions. The lightning symbols depict non-Watson-Crick base-pairing interactions. The 2'-OH groups of branch point adenosine and the cut-off 5' exon are pictured in the activated spliceosome. The small arrows near those 2'-OH group indicate the nucleophilic chemical reactions also known as trans-esterification reactions.
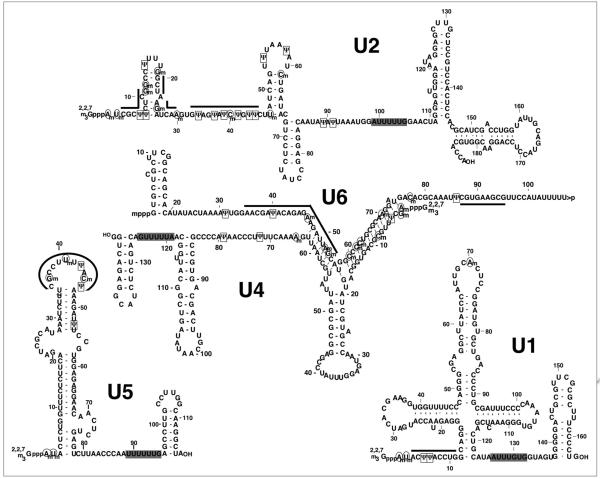
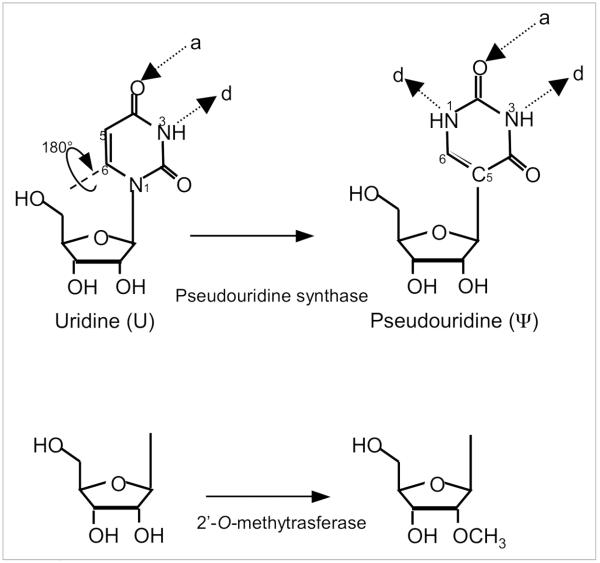
Interestingly, all five spliceosomal snRNAs are extensively posttranscriptionally modified (Fig. 2).29,30 Aside from the 5' cap modification, there are essentially two types of internal modifications, namely, 2'-O-methylation and pseudouridylation. Pseudouridylation is a uridine-specific modification that results in the formation of the 5-ribosyl isomer of uridine, pseudouridine (Ψ), while 2'-O-methylation (2'-Ome) is an RNA backbone modification that introduces a methyl group at the 2'-O position of the sugar ring (Fig. 3). Analysis of the distribution of modified nucleotides in spliceosomal snRNAs from various organisms has demonstrated conservation in the location of modifications throughout evolution. Strikingly, the majority of modified nucleotides are present in regions known to be functionally important for pre-mRNA splicing, including the regions of RNA-RNA interactions described above.29,31
Figure 2.
Pseudouridines and 2'-O-methylated residues in human spliceosomal snRNAs. Primary and secondary structures of human major spliceosomal snRNAs (U1, U2, U4, U5 and U6) are shown. Pseudouridines (Ψ) are surrounded by rectangles; 2'-O-methylations are circled. The thick lines indicate the nucleotides participating in RNA-RNA interactions or involved in catalysis during pre-mRNA splicing. The gray boxes highlight the Sm-binding sites. The 5' caps (2,2,7 trimethylated guanosine cap for U1, U2, U4, U5 and γ-methylated guanosine cap for U6) are also depicted.
Figure 3.
Schematic depiction of the two most abundant modified nucleotides in spliceosomal snRNA. (Top) Pseudouridine is a rotational isomer of uridine, in which the N-C glycosidic bond is broken to form an C-C bond. This results in the presence of an extra hydrogen bond donor (d), while the number of hydrogen bond acceptors (a) is unchanged. (Bottom) Schematic representation of a 2'-O-methylated ribose.
Over the years, great efforts have been made toward understanding the mechanisms and functions of spliceosomal snRNA modifications. It is now clear that two distinct molecular mechanisms exist that are capable of site-specifically introducing modified residues within spliceosomal snRNAs. It has also become increasingly clear that modified residues are not just bystanders in the process of pre-mRNA splicing, but actually participate in and influence snRNP and spliceosome assembly. In this review we discuss the mechanisms and functions of spliceosomal snRNA modifications.
Mechanisms of Spliceosomal snRNA Modifications
RNA-dependent mechanism
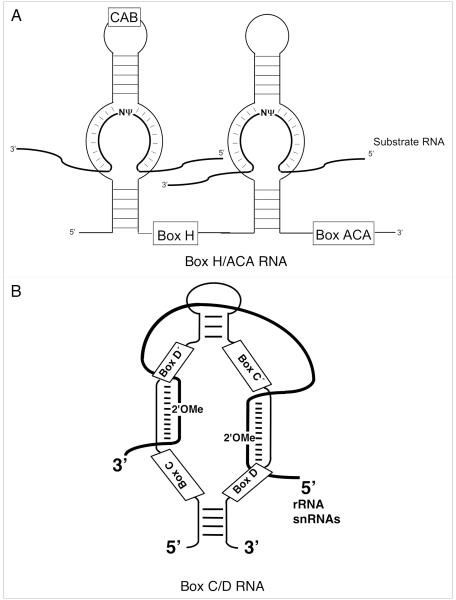
Posttranscriptional modification of spliceosomal snRNAs can occur via two distinct pathways, known as RNA-dependent and RNA-independent mechanisms (see below). In the RNA-dependent mechanism, small noncoding RNAs, namely Box H/ACA or Box C/D RNAs, are responsible for the site-specific posttranscriptional pseudouridylation and 2'-O-methylation of substrate RNAs, respectively (Fig. 4A and B). Both RNAs assemble with an evolutionarily conserved, yet distinct set of four core proteins [C/D RNAs: Nop1p, Nop56p, Nop58p and Snu13p; H/ACA RNAs: Cbf5p (Dyskerin in humans), Nhp2p, Nop10p and Gar1p].31-47 While the RNA component is responsible for dictating site-specificity through complementary base pairing interactions with the substrate RNA, the catalytic activity, i.e., modification activity, is provided by one of the core protein components (Nop1p for 2'-O-methylation, and Cbf5p for pseudouridylation).48-51
Figure 4.
Schematic depiction of box H/ACA and C/D RNAs. (A) Secondary structure of a eukaryotic pseudouridylation guide box H/ACA RNA. The RNA adapts a Hairpin-Hinge-Hairpin-Tail structure. Present within the hinge region is the box H (5'-ANANNA-3'), the box ACA (5'-ACA-3') motif typically lies three nucleotides from the 3'-end of the RNA. A CAB box (5'-ugAG-3'), responsible for Cajal body localization, may be present in the apical loop of either hairpin. Pseudouridylation is targeted to substrate RNAs by complementary base-pairing interactions between the internal loop (pseudouridylation pocket) and nucleotides adjacent to the target uridine. The thick lines denote substrate RNAs. (B) Secondary structure of a box C/D RNA. Boxes C, D, C’ and D’ are shown. The 2'OMe represents the target 2'-O-methylation site that is always the fifth nucleotide from box D or D’. The thick line represents substrate RNA.
Analyses of the subnuclear localization of guide-RNAs directing snRNA modification have revealed that they primarily reside within Cajal bodies, a subnuclear compartment present in eukaryotic cells.52-54 Thus, these RNAs have been referred to as small Cajal body-specific RNAs (scaRNA).54 Recently, the mechanism behind Cajal body localization has received significant attention, and it has been demonstrated that Cajal body retention of H/ACA RNAs in mammalian cells is mediated by a 4-nucleotide (nt) sequence (5'-ugAG-3', refereed to as the CAB box: lower case letters are less conserved) located within the apical loop of either hairpin (see Fig. 4B).55 Interestingly, the Sm proteins, SmB and SmD3, have been shown to specifically interact with the CAB box of both H/ACA and telomerase RNAs by immunoprecipitation and northern blot analysis.56 However, whether these interactions are necessary for Cajal body localization has not been addressed. More recently, the Steitz group has shed light on the mechanism of Cajal body retention and identified a CAB box for Drosophila C/D RNAs (5'-cgaGUUAnUg-3': lower case letters are less conserved).57 Using a UV crosslinking approach, a Drosophila WD40 repeat protein, p70, was identified which recognizes the Drosophila C/D RNA CAB box. In addition, both p70 and its human homologue, WDR79, were shown to interact with both human and Drosophila C/D, and H/ACA RNA CAB boxes. Importantly, this interaction was shown to be required for Cajal body retention.57
While Cajal bodies are considered to be the site of spliceosomal snRNA modification, there is growing evidence suggesting that RNA-guided modification is not restricted to Cajal bodies. For example, nuclear fractionation and northern blot analysis indicate that pugU2-34/44, a Xenopus H/ACA RNA that directs U2 snRNA pseudouridylation at two different positions, resides within the nucleoplasm.58 Furthermore, it has recently been shown that while flies null for coilin lack detectable Cajal bodies, their spliceosomal snRNAs are efficiently posttranscriptionally modified.59,60 Analysis of scaRNA localization by fluorescent in situ hybridization failed to detect any sites of scaRNA accumulation.59 Taken together, these results strongly suggest that the modification machinery is dispersed throughout the nucleoplasm (rather than being present exclusively in the Cajal bodies). In addition, our lab has recently shown that in Saccharomyces. cerevisiae artificial C/D RNAs are capable of site-specifically modifying pre-mRNA.61 These data strongly suggest that the guide-RNA mechanism of modification is functional in the nucleoplasm and raises the possibility that other nuclear RNAs, i.e., mRNA, may be natural targets of the RNA-dependent modification scheme. The apparent lack of detectable H/ACA and C/D RNAs in the nucleoplasm maybe a result of being too dilute within this compartment.
RNA-independent mechanism
To date, the majority of spliceosomal snRNA modifications have either been predicted or proven to be catalyzed by the RNA-dependent mechanism (Tables 1 and 2). However, while investigating whether S. cerevisiae spliceosomal snRNAs are posttranscriptionally modified, Massenet et al. demonstrated that Ψ44 of U2 snRNA is catalyzed by a single polypeptide enzyme known as Pus1p, demonstrating the existence of a second mechanism for spliceosomal snRNA modification, the RNA-independent or protein-only mechanism.62 In this mechanism, an enzyme is responsible for both substrate recognition and catalysis. While this was an interesting finding, the protein-only mechanism had been known for decades to catalyze the modifications of tRNA (both in prokaryotes and eukaryotes) and rRNA (in prokaryotes and in 5S rRNA of S. cerevisiae).63,64 In fact, Pus1p had already been shown to be responsible for eight different uridine-to-pseudouridine conversions in tRNA.62 Additionally, using a yeast GST-ORF genomic library, Ma et al. identified the previously uncharacterized ORF YOR243c as being responsible for Ψ35 formation in U2 snRNA.65 ORF YOR243c was subsequently renamed as PseudoUridine Synthase 7, PUS7. Surprisingly, when the amino acid sequence of Pus7p was compared with those of other known pseudouridine synthases, namely those of the TruA, TruB, RluA and RsuA families, no significant homology was identified. Thus, Pus7p represented a novel family of pseudouridine synthases. Furthermore, a BLAST search of available databases indicated the presence of Pus7p homologues in many organisms, including Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis and humans. The Xenopus and human Pus7p homologues have been cloned, and both of them are capable of catalyzing U2 pseudouridylation at position 34, which is equivalent to position 35 of yeast U2 (Ma and Yu YT, unpublished data). Shortly following the identification of Pus7p, an E. coli pseudouridine synthase, TruD, was identified which contained homology to Pus7p.66 Pus7p has since been classified as a member of the TruD family of pseudouridine synthases. Analysis of Pus7p substrates has revealed the importance of a 7 nucleotide-sequence flanking the target uridine in substrate recognition and catalysis.67
Table 1.
Sites of pseudouridylation within yeast and human spliceosomal snRNAs
| Organism | snRNA | Position | Mechanism | Enzyme | Verified/Predicted | Reference |
|---|---|---|---|---|---|---|
| U1 | Ψ5 | Cbf5 dependent | Cbf5 | NR | 62, Unpublished data Yu Lab | |
| Ψ6 | Cbf5 dependent | Cbf5 | NR | 62, Unpublished data Yu lab | ||
| Yeast | Ψ35 | Protein only | Pus 7 | Verified | 62, 65 | |
| U2 | Ψ42 | H/ACA RNP | snR81 | Verified | 62, 68 | |
| Ψ44 | Protein only | Pus 1 | Verified | 62 | ||
| U5 | Ψ99 | NR | NR | NR | 62 | |
| U1 | Ψ5 | H/ACA RNP | ACA47 | Verified | 132, 133 | |
| Ψ6 | H/ACA RNP | U109 | Predicted | 132, 134 | ||
| Ψ6 | NR | NR | NR | |||
| Ψ7 | H/ACA RNP | U100 | Predicted | 135, 136 | ||
| Ψ15 | NR | NR | NR | |||
| Ψ34 | H/ACA RNP | U92 | Predicted | 54, 137 | ||
| Ψ37 | H/ACA RNP | ACA45 | Predicted | 133, 137 | ||
| Ψ39 | H/ACA RNP | ACA26 | Predicted | 133, 137 | ||
| U2 | Ψ41 | H/ACA RNP | ACA26 | Predicted | 133, 137 | |
| Ψ43 | NR | NR | NR | |||
| Ψ44 | H/ACA RNP | U92 | Predicted | 54, 137 | ||
| Ψ54 | H/ACA RNP | U93 | Predicted | 29, 136-138 | ||
| Ψ88 | NR | NR | NR | |||
| Ψ89 | H/ACA RNP | ACA35 | Predicted | 133, 137 | ||
| Human | Ψ91 | NR | NR | NR | ||
| Ψ4 | NR | NR | NR | |||
| U4 | Ψ72 | NR | NR | NR | ||
| Ψ79 | NR | NR | NR | |||
| Ψ43 | H/ACA RNP | ACA57 | Predicted | 133, 139 | ||
| U5 | Ψ46 | H/ACA RNP | U85 | Verified | 139, 140 | |
| Ψ53 | H/ACA RNP | U93 | Predicted | 29, 137, 138 | ||
| Ψ31 | H/ACA RNP | ACA65 | Predicted | 136 | ||
| U6 | Ψ40 | H/ACA RNP | ACA12 | Predicted | 133, 141 | |
| HBI-100 | Predicted | 135 | ||||
| Ψ86 | H/ACA RNP | ACA65 | Predicted | 135 | ||
| U4atac | Ψ12 | NR | NR | NR | 117 | |
| U6atac | Ψ83 | NR | NR | NR | 117 | |
| U12 | Ψ19 | H/ACA RNP | ACA68 | Predicted | 117, 136 | |
| Ψ28 | H/ACA RNP | ACA66 | Predicted | 117, 136 |
Note: NR is for Not Reported.
Table 2.
Sites of 2'-0-methylation within human spliceosomal snRNAs
| Organism | snRNA | Position | Mechanism | Enzyme | Verified/Predicted | Reference |
|---|---|---|---|---|---|---|
| U1 | A70 | Box C/D RNP | U90 | Predicted | 54, 135 | |
| G11 | Box C/D RNP | HBII-382 | Predicted | 54, 135 | ||
| G19 | Box C/D RNP | mgU2-19/30 | Predicted | 54, 118 | ||
| U2 | G25 | Box C/D RNP | mgU2-25/61 | Predicted | 118, 135 | |
| A30 | Box C/D RNP | mgU2-19/30 | Predicted | 54, 118 | ||
| C61 | Box C/D RNP | mgU2-25/61 | Predicted | 118, 135 | ||
| HBII-382 | Predicted | 54, 135 | ||||
| C8 | Box C/D RNP | mgU12-22/U4-8 | Predicted | 54, 118 | ||
| U4 | U91 | Predicted | 54, 118, 135 | |||
| Human | A65 | Box C/D RNP | U87 | Predicted | 54, 142, 143 | |
| U41 | Box C/D RNP | U87 | Predicted | 54, 139, 143 | ||
| U5 | U88 | Predicted | 54, 139 | |||
| C45 | Box C/D RNP | U85 | Predicted | 54, 55, 140 | ||
| A47 | Box C/D RNP | mgU6-47 | Predicted | 144 | ||
| A53 | Box C/D RNP | mgU6-53 | Predicted | 145 | ||
| U6 | mgU6-53B | Predicted | 145 | |||
| C60 | Box C/D RNP | HBII-166 | Predicted | 135 | ||
| C62 | Box C/D RNP | U94 | Predicted | 146 | ||
| C77 | Box C/D RNP | mgU6-77 | Predicted | 144 | ||
| U12 | G22 | Box C/D RNP | mgU12-22/U4-8 | Predicted | 54, 118 |
RNA-dependent mechanism versus RNA-independent mechanism: which came first?
It appears that both RNA-dependent and RNA-independent mechanisms co-exist in various organisms. In higher eukaryotes, while spliceosomal snRNA modifications appear to be predominantly catalyzed by the RNA-dependent mechanism, at least one such modification (the pseudouridylation of human and Xenopus U2 at position 34) appears to be catalyzed by the RNA-independent mechanism as well. Thus, two mechanisms, RNA-dependent and RNA-independent, act at the same site. However, in S. cerevisiae, spliceosomal snRNA pseudouridylation at a given site can only be catalyzed by one of the two mechanisms, either RNA-dependent or RNA-independent mechanism, but not by both. For instance, snR81, a H/ACA RNA, directs pseudouridylation of position 42, and Pus1p and Pus7p, protein-only enzymes, pseudouridylate position 44 and 35, respectively.62,65,68
The coexistence of the two mechanisms and their distinct usage in S. cerevisiae and other organisms is rather interesting from an evolutionary point of view. It is possible that the RNA-dependent mechanism evolved from the protein-only mechanism in both higher eukaryotes and S. cerevisiae. However, in yeast, while some modifying enzymes evolved (e.g., snR81), others (e.g., Pus1p and Pus7p) may have remained unchanged (or evolved but were subsequently lost from the genome; e.g., via chromosomal deletion). Conversely, it is equally possible that the RNA-dependent mechanism is the most ancient mechanism, given that archaeal H/ACA RNPs, C/D RNPs, and ribosomes share a common core protein, L7 (homologous with Nhp2p (H/ACA RNP) and Snu13p (C/D RNP) in Eukarya).69 Thus, it is possible that the RNA-dependent mechanism evolved from the ancient translation apparatus, rather than from the RNA-independent mechanism. While the issue of which came first remains controversial, the preservation of the putatively ancient RNA-dependent (or RNA-independent) modifying mechanism throughout evolution implies that such modifications are functionally important.
Localization of snRNA-specific modifying enzymes in S. cerevisiae
With regard to subnuclear localization, it is worth noting that a coilin homologue has thus far escaped detection in S. cerevisiae. While it is possible that S. cerevisiae lacks a coilin homologue, it does however possess a structure analogous to Cajal bodies, the nucleolar body.70 The notion that the nucleolar body is the functional homologue of the Cajal body comes from several lines of evidence. For instance, when the human-Cajal body specific protein, survival of motor neuron (SMN), is ectopically expressed in S. cerevisiae, it concentrates to the nucleolar body.70 Furthermore, the 5'-cap hypermethylase, Tgs1p, responsible for spliceosomal snRNA 5'-cap hypermethylation, specifically localizes to the nucleolar body.71,72 However, analysis of spliceosomal snRNA localization through indirect immunofluorescence using an anti-m3G antibody has proposed that the nucleolus is the site of snRNA accumulation.73 While this would stand in direct opposition to the localization of spliceosomal snRNAs in mammalian cells, this interpretation of the data relies on the assumption that there are no other m3G-capped small nuclear RNAs. In fact, numerous yeast snoRNAs, including members of the H/ACA and C/D RNA families, are m3G-capped.74 Thus, a detailed and systematic analysis of spliceosomal snRNA localization in S. cerevisiae has yet to be carried out. Likewise, the specific subnuclear sites, to which the spliceosomal snRNA-specific modification enzymes (RNA-dependent and RNA-independent) localize, are yet to be determined. Thus, whether nucleolar bodies are the premier sites of spliceosomal snRNA modification remains uncertain.
Functions of Spliceosomal snRNA Modifications
Posttranscriptional modification provides a means to expand the vocabulary of nucleotides in the genetic code. Importantly, it is clear that modified nucleotides have distinct chemical properties from their unmodified counterparts. Thus, they have the potential to impact numerous aspects of the modified RNA, including structure, thermal stability and biochemical interactions.75 In each case, the structural, thermodynamic and biochemical contributions imparted by the modified nucleotide depend on the structural context and can extend beyond the site of modification. Indeed, 1H NMR, UV, and CD (circular dichroism) spectroscopy have demonstrated that short RNA fragments containing pseudouridine are more stable than if the same RNA contained uridine.76 Conformational stabilization appears to be an intrinsic property of pseudouridine at the nucleotide level, and is mediated by both an increase in base stacking and the ability to coordinate a water molecule through the extra hydrogen bond present.76-78 Similarly, 2'-O-methylation promotes increased stability in RNA conformations. For instance, 2'-O-methylation alters the hydration sphere around the oxygen resulting in the blockage of sugar edge interactions.79-81 In addition, methylation of the 2'-OH alters the ability of the ribose to participate in hydrogen bonding interactions.
In the context of the spliceosome, posttranscriptional modifications have the potential to influence numerous aspects of pre-mRNA splicing, including (1) RNA-RNA interactions, (2) interactions of spliceosomal snRNAs with spliceosomal proteins, and (3) directly participating in the catalytic reactions. To date, not much data has been generated regarding the latter two. Thus, we will primarily focus on the role of the modified nucleotides engaged in RNA-RNA interactions.
U1 snRNA
Within the initial step of pre-mRNA splicing, recognition of the 5'SS by the 5'-end of U1 snRNA, there are four modified nucleotides that can influence the U1 snRNA-pre-mRNA base pairing interaction (Fig. 2). Interestingly, however, U1 snRNA-depleted mammalian splicing extracts can be successfully reconstituted using in vitro transcribed U1 snRNA (presumably lacking modifications).82 Taken at face value these results suggest that modified nucleotides within the 5'-end of U1 snRNA are not necessary for pre-mRNA splicing; however, this relies on the assumption that the spliceosomal snRNAs are not capable of being modified in the splicing extracts. In fact, based on our own experience and that of others, in vitro transcribed U2 snRNA is readily modified when added to yeast splicing extracts (perhaps mammalian extracts as well). Furthermore, as only a strong splicing substrate was analyzed, whether modified nucleotides are required for the splicing of a suboptimal 5'SS was not addressed. Indeed, in vitro splicing assays in which two 5'SS are in competition with each other suggest that the presence of pseudouridine within the 5' end of U1 snRNA provides an advantage in 5'SS discrimination.83 Furthermore, a Ψ-G base pair was shown to contribute to the stability of the U1 snRNA interaction with the 5'SS of HIV-1 SD4 RNA.84 However, a complete functional dissection of the role of U1 snRNA posttranscriptional modification will have to wait for the identification of the enzymes responsible for their formation.
U2 snRNA
Of the spliceosomal snRNAs, U2 snRNA has the most posttranscriptional modifications. Human U2 snRNA contains ten 2'-O-methylated residues and 13 pseudouridines within the 5' half of the molecule (Fig. 2). It is perhaps for this reason that the functions of U2 snRNA posttranscriptional modifications have been the most extensively studied.
U2 snRNA modifications, snRNP biogenesis and spliceosome assembly
The initial experiments of Patton in the early 1990s provided the first functional analysis of U2 snRNA modification.85,86 Using HeLa cell S100 and nuclear extracts he demonstrated that the incorporation of 5-fluororidine (5-FU) in to U2 snRNA blocked U2 snRNA pseudouridylation. In addition, while it was observed that 5-FU-substituted U2 snRNA was able to form an U2 snRNP, the snRNP was overwhelmingly susceptible to salt dissociation.85 A more detailed and systematic analysis of the effects of U2 snRNA pseudouridylation on pre-mRNA splicing eventually established a nice correlation between modification status, pre-mRNA splicing competency, and U2 snRNP biogenesis.87 Using Xenopus oocytes, Yu et al. demonstrated that while in vitro transcribed U2 snRNA was unable to rescue splicing in U2 snRNA-depleted oocytes, upon longer reconstitution periods in vitro transcribed U2 snRNA gained the ability to reconstitute splicing activity. Strikingly, the pseudouridylation status of U2 snRNA mirrored the ability of U2 snRNA to reconstitute pre-mRNA splicing, that is, in vitro transcribed U2 snRNA became pseudouridylated following the longer reconstitution periods. Further analyses using anti-snRNP immunoprecipitation in conjunction with glycerol gradient sedimentation demonstrated that while U2 snRNA lacking pseudouridine is able to form nonfunctional 12S U2 snRNP particles, it is unable to detectably form functional 17S particles.87 Consequently, U2 snRNA lacking pseudouridine is unable to participate in spliceosome assembly. Furthermore, Zhao and Yu (2004) were able to show that pseudouridine residues within the branch site recognition region of Xenopus U2 snRNA are essential for U2 snRNP assembly and spliceosome assembly. Interestingly, the rate at which in vitro transcribed U2 snRNA is modified within the branch site recognition region is significantly faster than within the 5' region of U2 snRNA, when injected into Xenopus oocytes.88
In 2004 the Lührmann group carried out an extensive analysis on the role of modified nucleotides in the first 24-nt of human U2 snRNA.89 Interestingly, 2'-O-methylations at positions 1, 2, 12 and 19, were individually shown to be required for pre-mRNA splicing, while pseudouridines located within this region were shown to have a cumulative effect on splicing, as none were absolutely required for pre-mRNA splicing.89 Interestingly, their study demonstrated that the internal modifications are required for E complex formation. Taken together, the data accumulated thus far have clearly demonstrated that most modified nucleotides in U2 snRNA, including those residing within the 5'-end region and the branch site recognition region, are functionally important.
U2 modification and splicing efficiency
In addition to playing a role in the assembly of catalytically competent snRNPs and splicing complexes, posttranscriptional modifications, in particular pseudouridylation within the branch site recognition region of U2 snRNA, has been demonstrated to influence, directly or indirectly, the catalytic phase of pre-mRNA splicing. For instance, deletion of the gene encoding Pus7p, responsible for Ψ35 formation in S. cerevisiae U2 snRNA (see above), although viable, displayed reduced fitness under conditions of high salt or when in competition with wildtype yeast.65 Further analysis demonstrated that loss of Ψ35 in conjunction with U40G or U40Δ mutations in U2 snRNA severely reduced the organism’s fitness.90 Analysis of pre-mRNA splicing by semi-quantitative RT-PCR indicated an accumulation of pre-mRNA in the pus7Δ U2-U40G and pus7Δ U2-U40Δ strains, while any single mutation resulted in minimal if any accumulation of pre-mRNA. In line with the notion of pseudouridylation within the branch site recognition region of U2 snRNA affecting the catalytic phase of pre-mRNA splicing, the change of a single uridine (U35) to pseudouridine (Ψ35) significantly enhances the production of X-RNA, a product generated by a splicing related reaction in a cell- and protein-free system.91,92 It should be noted that Ψ35 is the nucleotide nearly opposite the branch-point adenosine.
Nucleophile positioning via U2 snRNA pseudouridylation
In recent years, the role of U2 snRNA pseudouridylation has been extensively investigated using various biophysical techniques. In this regard, the crystal structure of a self-complementary RNA designed to mimic the S. cerevisiae U2 snRNA-branch point interaction was determined in the absence of pseudouridine.93 Surprisingly, the adenosine 5' of the expected branch point adenosine was bulged out. Subsequently, the Greebaum group determined solution structures of the S. cerevisiae U2 snRNA-branch point interaction either in the presence or absence of pseudouridine (Ψ34, corresponding to Ψ35 in mammals).94,95 Interestingly, NMR data coupled with 2-aminopurine fluorescence titration data indicated that the presence of the pseudouridine was required for the bulging out of the expected branch point adenosine.95 However, in the NMR structure, the bulged adenine base was inserted in the minor groove, burying the 2'-OH, the nucleophile in the reaction, making it unlikely to participate in the splicing reaction. More recently, Lin and Kielkopf determined the crystal structure of the U2 snRNA-branch point interaction in the presence of pseudouridine and observed an extra-helical branch point adenosine in which its 2'-OH was prominently exposed and available for attack on the 5'SS.96 Thus, the structure proposed by Lin and Kielkopf is more likely to be functionally relevant for catalysis.
Taken together, U2 snRNA modification is required for pre-mRNA splicing. Biochemical and molecular data have clearly established links between spliceosomal snRNP biogenesis, spliceosome assembly, and splicing efficiency with the status of U2 snRNA modification. In addition, biophysical data have provided detailed structural information indicating that U2 snRNA pseudouridylation is required for proper positioning of the 2'-OH of the branch-point adenosine so that it is accessible and exposed for recognition and nucleophilic activity.
U5 snRNA
The U5 snRNA plays a critical role in juxtaposing the 5' SS, 3' SS and the BSS during pre-mRNA splicing. To date all U5 snRNAs examined possess an 11-nt loop, called loop 1, which contains the conserved 9-nt sequence 5'-G1C2C3U4U5U6U7A8C9-3'.97 Loop 1 engages the 5' exon before the first step of splicing and this interaction is maintained throughout the second step of splicing. In contrast, the loop 1-3' exon interaction can not be detected until the second step.98-100 Strikingly, there are 4 post-transcriptional modifications within the conserved 9-nt sequence of loop 1 (2'-Ome at positions 1, 5 and 9; Ψ at position 7; Fig. 2). Furthermore, nucleotides at positions 5 and 7 have been shown to interact with the splicing substrate by photochemical crosslinking and genetic suppression analyses.14,99,101-106 Unfortunately, the mechanism of U5 snRNA modification has not been elucidated, thus precluding a functional analysis of these modifications in pre-mRNA splicing. However, based on the known function of loop 1 in pre-mRNA splicing, it is reasonable to speculate a role for these modifications in influencing the stability of the pre-mRNA-U5 snRNA interaction. Alternatively, posttranscriptional modification of U5 snRNA loop 1 may be required for the Prp8p-mediated stabilization of exon-U5 snRNA loop 1 interactions during pre-mRNA splicing.107-109
U4 and U6 snRNAs
U4 and U6 snRNAs enter the pre-mRNA splicing reaction as the U4/U6.U5 tri-snRNP. Following tri-snRNP addition, a series of RNA-RNA rearrangements proceed, resulting in the exclusion of the U1 and U4 snRNAs from the spliceosome (see above). U6 snRNA is one of three snRNAs present in active spliceosomes (U2 and U5 are also present). However, evidence suggests that it is U2 and U6 that are directly involved in the catalytic steps (see above).98,110-112
Within the tri-snRNP, there are extensive base-pairing interactions between U4 and U6. Strikingly, this interaction is particularly strong with an experimentally determined Tm for affinity purified yeast U4/U6 di-snRNP of 55°C; on the other hand, human U4/U6 di-snRNP has two Tms, a lower Tm of 37°C and a higher Tm of 55°C.113 It should be noted that the human di-snRNP also presented with two alternate mobilities by native gel analysis.113 Interestingly, between human U4 and U6 snRNAs there are six Ψs and 12 2'-Ome residues (Fig. 2 and Tables 1 and 2), and half of them map to the regions involved in U4/U6 snRNA base-pairing interactions (Fig. 2). It has previously been proposed that the ATP-dependent unwinding of this duplex provides a proofreading step to ensure proper positioning of the tri-snRNP on the spliceosome,114 and timely formation of the catalytic center (U2-U6 duplexes). Within this context one could imagine that the extra thermodynamic stability afforded to the base-pairing interactions by the posttranscriptional modifications influences the rate of unwinding. Alternatively, as the binding positions of the helicases responsible for di-snRNP unwinding have not been mapped on to the snRNAs, modifications may induce a structure conducive to recognition by helicases.
Minor Spliceosomal snRNAs are Pseudouridylated
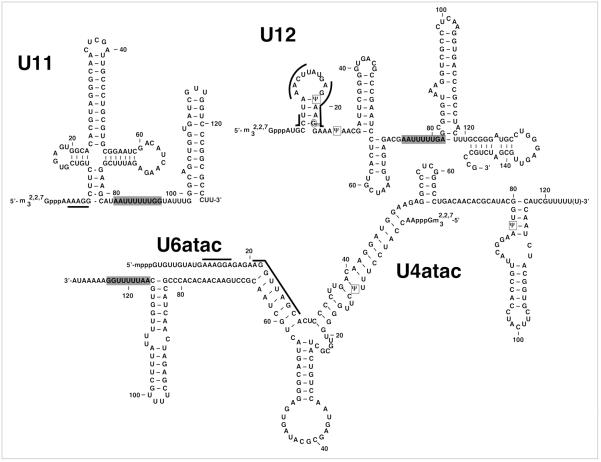
While the majority of introns are removed by the aforementioned spliceosome (or the major spliceosome), there exists a rare class of introns (~1–300) that are removed by a functionally similar, yet structurally distinct spliceosome, which is of much lower abundance (~104 copies per cell) relative to components of the major spliceosome.115,116 Thus, this spliceosome is referred to as the minor spliceosome. The activity of the minor spliceosome requires four distinct spliceosomal snRNAs, namely U11, U12, U4atac and U6atac, while sharing the U5 snRNA with the major spliceosome (Fig. 5).115 Analysis of minor spliceosomal snRNAs from HeLa cells has demonstrated that they too are posttranscriptionally modified (Fig. 5). To date, four pseudouridines have been identified in the minor spliceosomal snRNAs, two within U12, and one each within U4atac and U6atac.117 A single 2'-O-methylation has been detected in U12.118 While all of the modifications present in the minor class spliceosomal snRNAs have been predicted to be guided via the RNA-dependent mechanism, none have had their mechanism of formation experimentally determined (Table 2).
Figure 5.
Shown are primary and secondary structures of human minor spliceosomal snRNAs, U11, U12, U4atac and U6atac. U5 snRNA is shared by both the major and minor spliceosomes. Pseudouridines (Ψ) are surrounded by rectangles; 2'-O-methylations are circled. Pseudouridines within U12 and U4atac are believed to function analogously to their homologous modifications within U2 and U4 snRNAs, respectively. The thick lines indicate the nucleotides participating in RNA-RNA interactions or involved in catalysis during pre-mRNA splicing. The gray boxes highlight the Sm-binding sites.
Although fewer pseudouridine residues are present in the minor spliceosomal snRNAs when compared to the major spliceosomal snRNAs, the positions of pseudouridylation for U12 and U4atac are homologous to those within U2 and U4, respectively, thus suggesting that these pseudouridines are important for the splicing of minor introns. In fact, Ψ19 of U12 snRNA, which is adjacent to the branch point adenosine, is present in equivalent positions in U2 snRNA in human (Ψ34), plant (Ψ34) and S. cerevisiae (Ψ35). Interestingly, introns removed by the minor spliceosome contain more constrained consensus sequences at the 5' end of the intron and BSS.119-121 Thus, it is reasonable to hypothesize that the increased amount of modified nucleotides present in the major spliceosomal snRNAs, relative to the amount present in the minor spliceosomal snRNAs, is necessitated by the fact that major class (U2-type) introns contain less conserved consensus splice site sequences than the minor class (U12-type) introns. In support of this hypothesis, the introns of S. cerevisiae contain highly conserved consensus splice site sequences, while the spliceosomal snRNAs contain relatively few modified residues.
Spliceosomal snRNA Pseudouridylation as a Therapeutic Target
5-FU is commonly used in the treatment of a variety of solid tumors such as colorectal, breast, and liver carcinomas.122,123 Although nearly six decades have passed since the initial uses of 5-FU as a chemotherapeutic agent, its mechanism of action is one of debate. Initially it was hypothesized that 5FU affects DNA metabolism through inhibition of the enzyme thymidylate synthase, which is required for the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP).124,125 A reduction in the amount of dTMP, in turn, results in an inhibitory effect on the production of its downstream product deoxythymidine triphosphate (dTTP). Consequently, dUMP accumulates, resulting in elevated synthesis of its downstream product, deoxyuridine triphosphate (dUTP), and the incorporation of dUTP into DNA, thus resulting in DNA damage.126,127 Paradoxically, however, when 5FU-exposed cells are treated with thymidine, which can be converted to dTMP through the action of thymidine kinase (a pathway independent of the thymidylate synthase pathway), 5FU-mediated cytotoxic and apoptotic effects remain, suggesting that DNA metabolism is not the primary target of 5FU.128,129
Given that 5FU can be readily converted into 5-fluorouridine triphosphate (5FUTP), a ribonucleotide analog that can be incorporated into RNA, it has been proposed that 5FU may directly affect RNA metabolism.122,123 Indeed, 5FU-treated HeLa cells show a dramatic accumulation of pre-mRNA.130 In addition, thin-layer chromatography analysis of U2 snRNA isolated from 5FU-treated HeLa cells demonstrated the presence of 5FU and a reduction in the amount of pseudouridine present. Furthermore, while U2 snRNA isolated from uracil-treated HeLa cells can efficiently reconstitute pre-mRNA splicing in U2 snRNA-depleted Xenopus oocytes, U2 snRNA purified from 5FU-treated HeLa cells failed to reconstitute pre-mRNA splicing.130 Thus, some of the therapeutic effect of 5FU can be attributed to the inhibition of pre-mRNA splicing as a result of precluding pseudouridine formation.
Conclusions
The past decade has seen remarkable progress towards elucidating the mechanism and function of spliceosomal snRNA modification. However, relative to the progress made on DNA and protein modifications, research on RNA modifications has lagged behind. The key to addressing the in vivo role of spliceosomal snRNA modifications is to identify all the gene products responsible for their formation. While in vitro studies and studies using small molecule inhibitors of modification, i.e., 5FU, can offer insight in to the function of modifications, they are no substitute to a clean loss of that particular modification. To date, only 16 of the 24 known sites of pseudouridylation within the major spliceosomal snRNAs (U1, U2, U4, U5 and U6), and two of four for the minor spliceosomal snRNAs (U11, U12, U4atac and U6atac) of mammals have had the enzymes responsible for their formation proven or predicted. In addition, only three of six pseudouridines have had their modifying enzyme identified in S. cerevisiae (Table 1). Thus, a daunting task of identifying the enzymes responsible for the remaining modifications lies ahead.
It should be reiterated that the only spliceosomal snRNA which has had its modifications subjected to a detailed and systematic experimental analysis is U2 snRNA. Thus, whether modified nucleotides in U1, U4, U5 and U6 play any roles in splicing is an open question. In addition, it is our hope that as structural techniques continue to advance and become more powerful we will see detailed images of the spliceosome at defined functional stages. In fact, we are already beginning to see high-resolution electron microscopy images.131 With the growing attention given to RNA modification and pre-mRNA splicing, we expect a clear picture, regarding whether and how spliceosomal snRNA modifications contribute to function, to emerge soon.
Acknowledgements
We would like to thank the members of the Yu laboratory for discussion and inspiration. Our work was supported by grant GM62937 (to Yi-Tao Yu) from the National Institute of Health. J.K. was supported by a NIH Institutional Ruth L. Kirschstein National Research Service Award GM068411.
References
- 1.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell. 1998;92:315–26. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 2.Yu YT, Scharl EC, Smith CM, Steitz JA. The RNA World. 2nd Cold Spring Harbor Laboratory Press; Cold Spring Harbor NY: 1999. pp. 487–524. [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 4.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–8. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Kramer A, Keller W, Appel B, Luhrmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- 6.Bindereif A, Green MR. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987;6:2415–24. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruby SW, Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988;242:1028–35. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- 8.Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–60. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–58. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 10.Seraphin B, Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991;10:1209–16. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–46. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 12.Mount SM, Steitz JA. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981;9:6351–68. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827–35. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 14.Wassarman DA, Steitz JA. Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–25. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang Y, Weiner AM. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–52. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 16.Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–55. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 17.Pikielny CW, Rymond BC, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature. 1986;324:341–5. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- 18.Frendewey D, Kramer A, Keller W. Different small nuclear ribonucleoprotein particles are involved in different steps of splicing complex formation. Cold Spring Harb Symp Quant Biol. 1987;52:287–98. doi: 10.1101/sqb.1987.052.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–27. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 20.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–74. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang YA, Goldstein AM, Weiner AM. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci USA. 1989;86:2752–6. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–39. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 23.Black DL, Chabot B, Steitz JA. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–50. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 24.Krainer AR, Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985;42:725–36. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 25.Lesser CF, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–8. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 26.Moore MJ, Query CC, SPA . The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor NY: 1993. Splicing of precursors to mRNAs by the splicesome. [Google Scholar]
- 27.Ruby SW, Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991;7:79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- 28.Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci USA. 1992;89:11269–73. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy R, Busch H. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag Press; Heidelberg: 1988. pp. 1–37. [Google Scholar]
- 30.Massenet S, Mougin A, Branlant C. Modification and Editing of RNA. ASM Press; Washington DC: 1998. pp. 201–28. [Google Scholar]
- 31.Yu YT, Terns RM, Terns MP. Fine-Tuning of RNA Functions by Modification and Editing. Springer-Verlag; Berlin heidelberg: 2005. Mechanisms and Functions of RNA-guided RNA Modification; pp. 223–62. [Google Scholar]
- 32.Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54:123–33. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 33.Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–98. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henras A, et al. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–90. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins NJ, et al. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACAmotif snoRNPs and constitute a common bipartite structure. Rna. 1998;4:1549–68. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lafontaine DL, Tollervey D. Nop58p is a common component of the box C + D snoRNPs that is required for snoRNA stability. Rna. 1999;5:455–67. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028–40. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watkins NJ, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–66. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe Y, Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28:2342–52. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafontaine DL, Tollervey D. Synthesis and assembly of the box C + D small nucleolar RNPs. Mol Cell Biol. 2000;20:2650–9. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dragon F, Pogacic V, Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol. 2000;20:3037–48. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn JF, Tran EJ, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–41. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galardi S, et al. Purified box C/D snoRNPs are able to reproduce site-specific 2'-O-methylation of target RNA in vitro. Mol Cell Biol. 2002;22:6663–8. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci USA. 2002;99:5289–94. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozhdestvensky TS, et al. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–77. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–67. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–24. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–57. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 50.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–37. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–72. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–22. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–8. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 54.Darzacq X, et al. Cajal body-specific small nuclear RNAs: a novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–56. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richard P, et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–93. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu D, Collins K. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 2006;20:531–6. doi: 10.1101/gad.1390306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X, Li ZH, Terns RM, Terns MP, Yu YT. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. Rna. 2002;8:1515–25. [PMC free article] [PubMed] [Google Scholar]
- 59.Deryusheva S, Gall JG. Small Cajal Body-Specific RNAs (scaRNAs) of Drosophila Function in the Absence of Cajal Bodies. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu JL, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–70. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao X, Yu YT. Targeted pre-mRNA modification for gene silencing and regulation. Nat Methods. 2008;5:95–100. doi: 10.1038/nmeth1142. [DOI] [PubMed] [Google Scholar]
- 62.Massenet S, et al. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–54. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grosjean H. Springer-Verlag; Berlin Heidelberg: 2005. Modification and editing of RNA: historical overview and important facts to remember; pp. 223–62. [Google Scholar]
- 64.Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol Cell Biol. 2008;28:3089–100. doi: 10.1128/MCB.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–97. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea and eukarya. RNA. 2003;9:711–21. doi: 10.1261/rna.5230603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urban A, Behm-Ansmant I, Branlant C, Motorin Y. RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J Biol Chem. 2009;284:5845–58. doi: 10.1074/jbc.M807986200. [DOI] [PubMed] [Google Scholar]
- 68.Ma X, et al. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–13. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tran E, Brown J, Maxwell ES. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem Sci. 2004;29:343–50. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Verheggen C, et al. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001;20:5480–90. doi: 10.1093/emboj/20.19.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 72.Verheggen C, et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–45. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Potashkin JA, Derby RJ, Spector DL. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol Cell Biol. 1990;10:3524–34. doi: 10.1128/mcb.10.7.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 75.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 76.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 78.Charette M, Gray MW. Pseudouridine in RNA: what, where, how and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 79.Auffinger P, Westhof E. Rules governing the orientation of the 2'-hydroxyl group in RNA. J Mol Biol. 1997;274:54–63. doi: 10.1006/jmbi.1997.1370. [DOI] [PubMed] [Google Scholar]
- 80.Auffinger P, Westhof E. Hydration of RNA base pairs. J Biomol Struct Dyn. 1998;16:693–707. doi: 10.1080/07391102.1998.10508281. [DOI] [PubMed] [Google Scholar]
- 81.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–33. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Will CL, Rumpler S, Klein Gunnewiek J, van Venrooij WJ, Luhrmann R. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24:4614–23. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5' splice sites. Rna. 2005;11:683–98. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Freund M, et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–75. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patton JR. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry. 1993;32:8939–44. doi: 10.1021/bi00085a027. [DOI] [PubMed] [Google Scholar]
- 86.Patton JR. Multiple pseudouridine synthase activities for small nuclear RNAs. Biochem J. 1993;290:595–600. doi: 10.1042/bj2900595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–95. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. Rna. 2004;10:681–90. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5' end of human U2 snRNA are required for spliceosomal E-complex formation. Rna. 2004;10:1925–33. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280:6655–62. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 91.Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–7. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 92.Valadkhan S, Manley JL. Characterization of the catalytic activity of U2 and U6 snRNAs. Rna. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. Rna. 2001;7:682–91. doi: 10.1017/s1355838201002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. Rna. 2001;7:833–45. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–65. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 96.Lin Y, Kielkopf CL. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry. 2008;47:5503–14. doi: 10.1021/bi7022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frank DN, Roiha H, Guthrie C. Architecture of the U5 small nuclear RNA. Mol Cell Biol. 1994;14:2180–90. doi: 10.1128/mcb.14.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–96. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 99.Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5' and 3' splice sites monitored by photoactivated crosslinking in yeast spliceosomes. Rna. 1995;1:968–80. [PMC free article] [PubMed] [Google Scholar]
- 100.McGrail JC, Tatum EM, O’Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–22. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chiara MD, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3' splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–59. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991;65:115–23. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 103.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992;68:743–54. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 104.Cortes JJ, Sontheimer EJ, Seiwert SD, Steitz JA. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5' splice sites in vivo. EMBO J. 1993;12:5181–9. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alvi RK, Lund M, Okeefe RT. ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5' splice site. Rna. 2001;7:1013–23. doi: 10.1017/s135583820101041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McGrail JC, O’Keefe RT. The U1, U2 and U5 snRNAs crosslink to the 5' exon during yeast pre-mRNA splicing. Nucleic Acids Res. 2008;36:814–25. doi: 10.1093/nar/gkm1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–16. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5' and 3' splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–12. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. Rna. 1998;4:1239–50. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–4. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. Rna. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- 112.Yu YT, Maroney PA, Nilsen TW. Functional reconstitution of U6 snRNA in nematode cis- and trans-splicing: U6 can serve as both a branch acceptor and a 5' exon. Cell. 1993;75:1049–59. doi: 10.1016/0092-8674(93)90315-h. [DOI] [PubMed] [Google Scholar]
- 113.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–8. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 114.Raghunathan PL, Guthrie C. RNA unwinding in U4/ U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–55. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 115.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:801–11. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 116.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–70. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 117.Massenet S, Branlant C. A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnRNAs. Rna. 1999;5:1495–503. doi: 10.1017/s1355838299991537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5' caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol. 2004;14:1985–95. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 119.Dietrich RC, Incorvaia R, Padgett RA. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol Cell. 1997;1:151–60. doi: 10.1016/s1097-2765(00)80016-7. [DOI] [PubMed] [Google Scholar]
- 120.Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell. 1997;91:875–9. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 121.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2:773–85. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 122.Ghoshal K, Jacob ST. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem Pharmacol. 1997;53:1569–75. doi: 10.1016/s0006-2952(97)00040-3. [DOI] [PubMed] [Google Scholar]
- 123.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 124.Sommer H, Santi DV. Purification and amino acid analysis of an active site peptide from thymidylate synthetase containing covalently bound 5-fluoro-2'-de-oxyuridylate and methylenetetrahydrofolate. Biochem Biophys Res Commun. 1974;57:689–95. doi: 10.1016/0006-291x(74)90601-9. [DOI] [PubMed] [Google Scholar]
- 125.Santi DV, McHenry CS, Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorode-oxyuridylate. Biochemistry. 1974;13:471–81. doi: 10.1021/bi00700a012. [DOI] [PubMed] [Google Scholar]
- 126.Aherne GW, Hardcastle A, Raynaud F, Jackman AL. Immunoreactive dUMP and TTP pools as an index of thymidylate synthase inhibition; effect of tomudex (ZD1694) and a nonpolyglutamated quinazoline antifolate (CB30900) in L1210 mouse leukaemia cells. Biochem Pharmacol. 1996;51:1293–301. doi: 10.1016/0006-2952(96)00035-4. [DOI] [PubMed] [Google Scholar]
- 127.Mitrovski B, Pressacco J, Mandelbaum S, Erlichman C. Biochemical effects of folate-based inhibitors of thymidylate synthase in MGH-U1 cells. Cancer Chemother Pharmacol. 1994;35:109–14. doi: 10.1007/BF00686631. [DOI] [PubMed] [Google Scholar]
- 128.Pritchard DM, Watson AJ, Potten CS, Jackman AL, Hickman JA. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci USA. 1997;94:1795–9. doi: 10.1073/pnas.94.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–47. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 130.Zhao X, Yu YT. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res. 2007;35:550–8. doi: 10.1093/nar/gkl1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fabrizio P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 132.Reddy R, Henning D, Busch H. Pseudouridine residues in the 5'-terminus of uridine-rich nuclear RNA I (U1 RNA) Biochem Biophys Res Commun. 1981;98:1076–83. doi: 10.1016/0006-291x(81)91221-3. [DOI] [PubMed] [Google Scholar]
- 133.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24:5797–807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gu AD, Zhou H, Yu CH, Qu LH. A novel experimental approach for systematic identification of box H/ACA snoRNAs from eukaryotes. Nucleic Acids Res. 2005;33:194. doi: 10.1093/nar/gni185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huttenhofer A, et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–53. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. Rna. 2006;12:15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shibata H, et al. The primary nucleotide sequence of nuclear U-2 ribonucleic acid. The 5'-terminal portion of the molecule. J Biol Chem. 1975;250:3909–20. [PubMed] [Google Scholar]
- 138.Kiss AM, et al. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–9. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Krol A, Gallinaro H, Lazar E, Jacob M, Branlant C. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 1981;9:769–87. doi: 10.1093/nar/9.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2'-O-ribose methylation and pseudouri-dylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–51. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Epstein P, Reddy R, Henning D, Busch H. The nucleotide sequence of nuclear U6 (4.7 S) RNA. J Biol Chem. 1980;255:8901–6. [PubMed] [Google Scholar]
- 142.Krol A, Branlant C, Lazar E, Gallinaro H, Jacob M. Primary and secondary structures of chicken, rat and man nuclear U4 RNAs. Homologies with U1 and U5 RNAs. Nucleic Acids Res. 1981;9:2699–716. doi: 10.1093/nar/9.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reddy R, Henning D, Busch H. The primary nucleotide sequence of U4 RNA. J Biol Chem. 1981;256:3532–8. [PubMed] [Google Scholar]
- 144.Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2:629–38. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 145.Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2'-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol. 1999;19:6906–17. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vitali P, et al. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–51. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]