SUMMARY
Appropriate DNA double-strand break (DSB) repair factor choice is essential for ensuring accurate repair outcome and genomic integrity. The factors that regulate this process remain poorly understood. Here, we identify two repressive chromatin components, the macrohistone variant macroH2A1 and the H3K9 methyltransferase and tumor suppressor PRDM2, which together direct the choice between the antagonistic DSB repair mediators BRCA1 and 53BP1. The macroH2A1/PRDM2 module mediates an unexpected shift from accessible to condensed chromatin that requires the ataxia telangiectasia mutated (ATM)-dependent accumulation of both proteins at DSBs in order to promote DSB-flanking H3K9 dimethylation. Remarkably, loss of macroH2A1 or PRDM2, as well as experimentally induced chromatin decondensation, impairs the retention of BRCA1, but not 53BP1, at DSBs. As a result, mac-roH2A1 and/or PRDM2 depletion causes epistatic defects in DSB end resection, homology-directed repair, and the resistance to poly(ADP-ribose) polymerase (PARP) inhibition—all hallmarks of BRCA1-deficient tumors. Together, these findings identify dynamic, DSB-associated chromatin reorganization as a critical modulator of BRCA1-dependent genome maintenance.
INTRODUCTION
Eukaryotic DNA double-strand breaks (DSBs) occur in the context of a highly organized chromatin environment. DSB repair, therefore, requires the reorganization and structural modification of break-proximal chromatin to facilitate and regulate access for repair factors and DNA damage response (DDR) mediators (Price and D’Andrea, 2013; Smeenk and van Attikum, 2013). Numerous, often functionally distinct, DSB-associated chromatin alterations have been identified over the past decade (Polo and Jackson, 2011; Shi and Oberdoerffer, 2012), suggesting that a balanced and dynamic sequence of remodeling events is critical for accurate genome maintenance.
Chromatin reorganization generally involves the covalent modification of histone tails as well as histone (variant) exchange, which together affect nucleosome density and DNA accessibility (Zentner and Henikoff, 2013). Both processes have been implicated in the cellular response to DSBs and can be broadly separated into modifications associated with accessible or repressive chromatin (Polo and Jackson, 2011; Shi and Oberdoerffer, 2012). Increased chromatin accessibility counteracts physical restraints that would otherwise impede DDR initiation, and chromatin relaxation can be observed within seconds of DSB induction (Altmeyer and Lukas, 2013; Kruhlak et al., 2006; Price and D’Andrea, 2013; Soria et al., 2012). The latter temporally coincides with and largely depends on the activation of poly(ADP-ribose)polymerase1(PARP1), which promotes therecruitment of various chromatin remodeling factors to facilitate phosphatidylinositol 3-kinase-related kinase (PIKK)-mediated DNA damage signaling (Altmeyer et al., 2013; Smeenk et al., 2013; Young et al., 2013). Activation of the PIKK ataxia telangiectasia mutated (ATM) further depends on the sensing of DSB-induced chromatin perturbations via the KAT5 (or Tip60) acetyltransferase (Bakkenist and Kastan, 2003; Kaidi and Jackson, 2013; Murr et al., 2006; Sun et al., 2009). In addition, DDR activation has been linked to the incorporation of the histone H2A variant H2A.Z, which is thought to help destabilize DSB-flanking nucleosomes and thereby regulate repair factor access (Xu et al., 2012a).
Although the formation of an accessible chromatin environment is a critical step in DDR initiation, several factors associated with repressive or transcriptionally inactive chromatin have now been linked to DSB repair, including polycomb group proteins, histone deacetylases (HDACs), the macrohistone variant macroH2A1.1, and HP1 proteins (Ayoub et al., 2008; Baldeyron et al., 2011; Chou et al., 2010; Ismail et al., 2010; Lee et al., 2013; Luijsterburg et al., 2009; Miller et al., 2010; Soria and Almouzni, 2013; Timinszky et al., 2009; Xu et al., 2012b). Moreover, PIKK activation was shown to promote transcriptional silencing in cis to DSBs (Kruhlak et al., 2007; Pankotai et al., 2012; Shanbhag et al., 2010), thus challenging the view of uniformly accessible chromatin as an optimal environment for DNA damage signaling and repair.
Consistent with this notion, recent work suggests the existence of functionally distinct chromatin domains associated with a single DNA lesion. Specifically, the DDR mediators BRCA1 and 53BP1 were found to occupy large and often mutually exclusive DSB-flanking regions, in agreement with their opposing roles in DSB repair (Chapman et al., 2012). 53BP1 is a negative regulator of DNA end resection at DSBs, thereby suppressing unscheduled homology-directed repair, which in turn promotes nonhomologous end joining (NHEJ) (Bouwman et al., 2010; Bunting et al., 2010). BRCA1 antagonizes 53BP1 to facilitate end resection and homologous recombination (HR) in the presence of a sister chromatid. As a result, 53BP1 accounts for the HR defects in BRCA1-deficient cells and the concomitant increase in cancer susceptibility (Bouwman et al., 2010; Bunting et al., 2010). Several, often functionally opposed chromatin alterations, including histone acetylation, HP1 recruitment, and H2A.Z histone variant exchange, have been linked to the recruitment of both 53BP1 and BRCA1 (Lee et al., 2013; Murr et al., 2006; Soria and Almouzni, 2013; Tang et al., 2013; Xu et al., 2012b), raising the intriguing possibility that DSB repair pathway choice may be regulated through spatially and/or temporally controlled chromatin reorganization.
Using a chromatin-focused RNAi screen for HR modulators, we uncover a repressive chromatin module that links the dynamic condensation of DSB-proximal chromatin to BRCA1-dependent genome maintenance, with implications for cancer predisposition associated with defective BRCA1 function (Silver and Livingston, 2012).
RESULTS
MacroH2A1 and Its Splice Variant MacroH2A1.2 Promote DSB Repair by HR
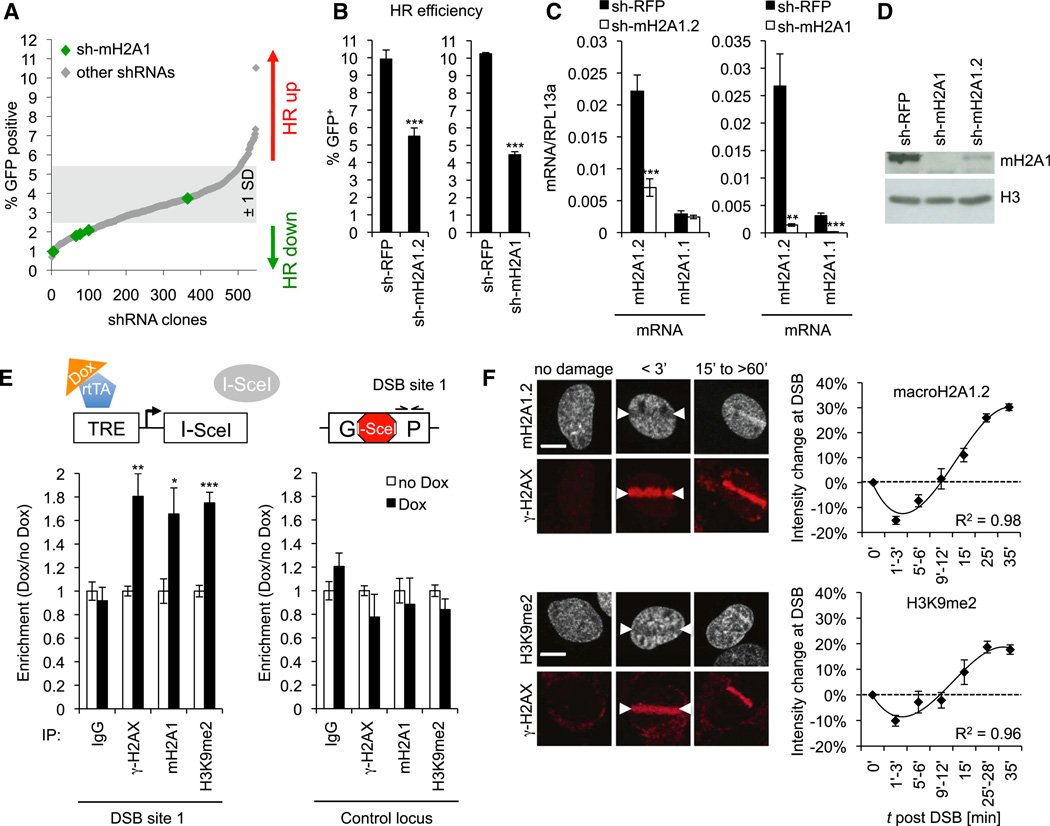
To systematically dissect the contribution of chromatin to DSB repair by HR, we analyzed over 400 Gene Ontology-annotated chromatin modifiers using high-throughput small hairpin RNA (shRNA)-based RNAi screening of an HR reporter cell line (DRGFP-U2OS) (Weinstock et al., 2006). Based on the number of scoring hairpins combined with RNAi gene enrichment ranking of a total of ~2,000 shRNAs, we identified the repressive histone variant macroH2A1 as one of the top-five HR-promoting candidates, together with two known mediators of HR, RBBP8/CtIP and TRRAP (Figure 1A; Table S1) (Murr et al., 2006; Sartori et al., 2007). The macroH2A1-encoding H2AFY gene produces two splice variants, macroH2A1.1 and macroH2A1.2, which differ in 32 aa within the ~30 kDa carboxy-terminal macrodomain, resulting in the presence of a poly-ADP-ribose (PAR)-binding pocket in the 1.1 but not the 1.2 variant (Timinszky et al., 2009). To dissect the effect of macroH2A1 isoforms on HR, we measured HR frequency following depletion of either macroH2A1.2 alone or both macroH2A1 isoforms simultaneously, using a DRGFP reporter cell line that allows for doxycycline (Dox)-inducible DSB formation via the I-SceI endonuclease (Figure 1E). Both mac-roH2A1 and macroH2A1.2 knockdown caused a reduction in HR efficiency that correlated with the extent of macroH2A1.2 depletion and had no major impact on DSB formation or the frequency of HR-permissive S/G2 cells (Figures 1B–1D, 2F, and S1). Our results, thus, identify macroH2A1 and its predominant macroH2A1.2 splice variant as mediators of HR.
Figure 1. RNAi Screen Identifies a Role for MacroH2A1/MacroH2A1.2 in HR.
(A) HR efficiency in DRGFP-U2OS cells stably transduced with shRNAs from a chromatin-focused RNAi library. HR was measured as percent (%) GFP+ cells; green diamonds represent macroH2A1-specific shRNAs.
(B) HR efficiency (percent [%] GFP+ cells) in a Dox-inducible DRGFP gene conversion assay (see E). Samples were analyzed in triplicate. Values are expressed as mean and SD. Unless noted otherwise, p values are based on Student’s two-tailed t test: *p < 0.05; **p < 0.01; ***p < 0.001.
(C) MacroH2A1.1-and macroH2A1.2-encoding mRNA levels of samples in (B) relative to RPL13a. Samples were analyzed in triplicate. Values are expressed as mean and SD.
(D) Western blot analysis of macroH2A1 expression following macroH2A1 or macroH2A1.2 knockdown.
(E) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox-induced I-SceI expression. I-SceI DSB site-flanking primer locations are indicated (DSB site 1). A non-DSB-associated genomic locus served as control. Enrichment was normalized to no Dox. Values are expressed as mean and SEM (n ≥ 5).
(F) Immunofluorescence analysis of macroH2A1.2 (top) or H3K9me2 (bottom) at laser-induced DSBs. Arrows depict the site of laser microirradiation; γ-H2AX served as a marker for DSBs. Arrows depict site of laser microirradiation. Scale bars, 10 µm. DSB-associated intensity changes were measured as the ratio of γ-H2AX+ over γ-H2AX− nuclear areas (7–18 cells per time point). Values are expressed as mean and SEM (n ≥ 3). R2 values are based on a third-order polynomial regression. See also Figures S1-S3 and Table S1.
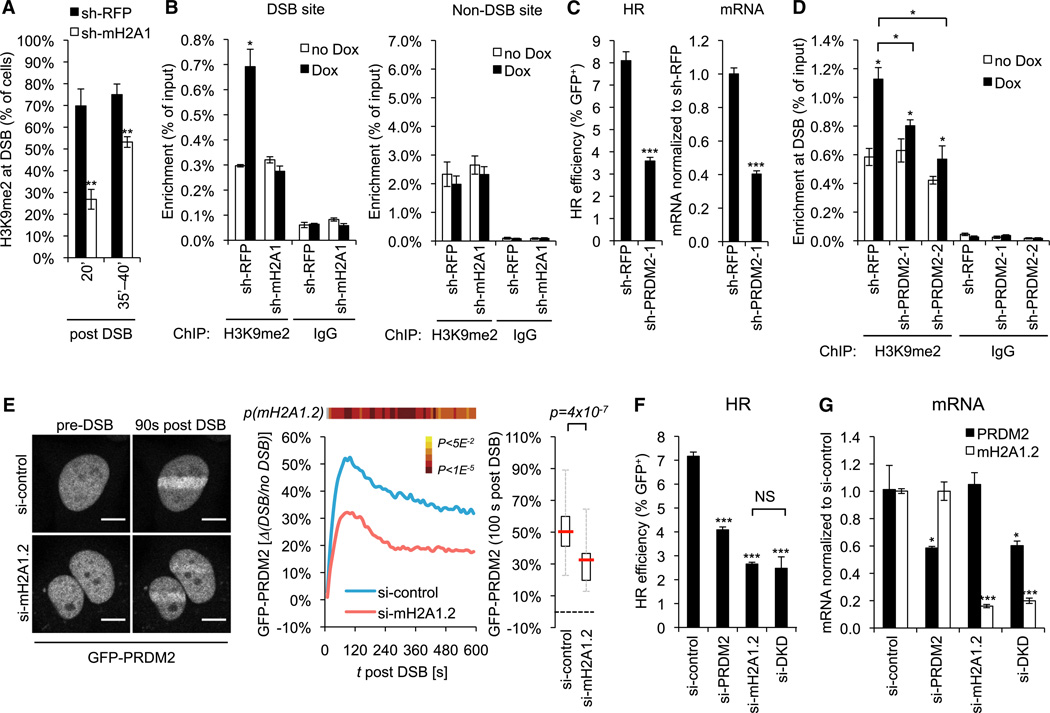
Figure 2. PRDM2 Is a MacroH2A1.2-Dependent Regulator of HR.
(A) Frequency of cells with H3K9me2 enrichment at laser-induced DSBs at the indicated time points after laser microirradiation. Values are expressed as mean and SEM (n = 4).
(B) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox. Enrichment relative to input is shown at the I-SceI DSB site and a non-DSB control locus. Values are expressed as mean and SEM (n = 3).
(C) HR efficiency and PRDM2 mRNA levels following PRDM2 knockdown. Samples were analyzed in triplicate. mRNA levels are relative to sh-RFP and were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), RPL13a, and RPS16. Values are expressed as mean and SD.
(D) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox. Enrichment is shown relative to input. Values are expressed as mean and SEM (n = 3).
(E) GFP-PRDM2 recruitment to laser-induced DSBs in cells expressing si-control (n = 29) or si-macroH2A1.2 (n = 30). Representative images are shown. Scale bars, 10 µm. Two independent experiments were combined. Data sets were subjected to Student’s two-tailed t test at each imaging time point. The p(mH2A1.2) heatmap depicts the p value distribution over time. gray indicates nonsignificance (ns). The right panel shows a representative box plot for data sets acquired 100 s post DSB. The red line indicates the median. The box shows the 25th–75th percentile. Whiskers show range between minimum and maximum values. (F and G) HR efficiency (F) and mRNA levels normalized to GAPDH, RPL13a, and RPS16 (G) in the presence of the indicated siRNAs. si-DKD, combined knockdown of macroH2A1.2 and PRDM2. Samples were analyzed in triplicate. Values are expressed as mean and SD.
See also Figures S1, S4, and S5 and Movie S1.
MacroH2A1 Recruitment to DSBs Coincides with H3K9 Dimethylation
We next asked if macroH2A1 functions directly at DSBs. Using chromatin immunoprecipitation (ChIP), we found that I-SceI-mediated DSB induction resulted inarobust, DSB-specific accumulation of macroH2A1 that coincided with known markers of DSB repair, such as H2AX phosphorylation (γ-H2AX) and BRCA1 recruitment, and was detectable following both S phase and G0/G1 arrest (Figures 1E and S2). For a kinetic dissection of the recruitment of macroH2A1, and specifically the macroH2A1.2 splice variant, to DSBs, we performed laser microirradiation in combination with macroH2A1.2-specific immunostaining (Sporn et al., 2009). Consistent with the DSB-associated chromatin relaxation reported previously (Kruhlak et al., 2006; Murr et al., 2006; Smeenk and van Attikum, 2013), we detected an immediate but transient depletion of macroH2A1.2 at DSBs. The latter was followed by prolonged macroH2A1.2 reaccumulation that initiated within minutes of DSB induction and resulted in discern-able, DSB-associated enrichment in >88% ± 10% of cells within 30–40 min (Figure 1F). Dynamic depletion and reaccumulation of macroH2A1.2 at DSBs were further observed in breast epithelial (MCF7) or skin-derived tumor cell lines (WM-115), although macroH2A1.2 levels did not accumulate beyond predamage levels in the former (Figure S3).
Because macroH2A1 is frequently associated with chromatin silencing (Gamble and Kraus, 2010), we next examined if the accumulation of macroH2A1.2 at DSBs coincides with other repressive chromatin marks. Indeed, we observed DSB-proximal dimethylation of histone H3 on lysine 9 (H3K9me2), a histone mark associated with silent chromatin (Barski et al., 2007), following DSB induction by either I-SceI expression or laser microirradiation (Figures 1E and 1F). In analogy to macroH2A1.2, H3K9me2 accumulation at laser-induced DSBs was observed in 81% ± 14% of cells within 30–40 min, following a brief phase of depletion. Consistent with this, accumulation at I-SceI-induced DSBs occurred both in S/G2 and in G1 phase cells, albeit at moderately lower levels in the latter (Figures 1E and Figure S2). Together, these findings reveal the dynamic depletion and reaccumulation of repressive chromatin marks at DSBs.
MacroH2A1 Mediates Recruitment of the H3K9 Methyltransferase PRDM2 to DSBs
Based on the kinetic similarities in macroH2A1.2 and H3K9me2 accumulation at DSBs, we next asked if the two processes are mechanistically linked. Indeed, depletion of macroH2A1 or macroH2A1.2 resulted in reduced DSB-associated H3K9me2 accumulation following both laser microirradiation and I-SceI-mediated DSB induction (Figures 2A, 2B, and S4). However, macroH2A1 does not exhibit known methyltransferase activity and cannot directly account for the latter. When revisiting the top 10%of hits that downregulated HR in our RNAi screen (Table S1), we identified three genes with annotated lysine methyltransferase activity, one of which, PRDM2 (or RIZ1), was previously shown to modify H3K9 (Kim et al., 2003). PRDM2 is a PR/SET domain protein and has been implicated in tumor suppression in mice and humans (Kim et al., 2003; Steele-Perkins et al., 2001). However, no direct role in DSB repair has been reported. Confirming our RNAi screen results, partial knockdown of PRDM2 with two independent shRNAs resulted in a decrease in HR efficiency following Dox-induced I-SceI expression without reducing S/G2 frequencies (Figures 2C, S1D, S1E, and S5A). To determine if PRDM2 is responsible for DSB-proximal H3K9 dimethylation, we performed H3K9me2 ChIP at I-SceI-induced DSBs. Following both shRNA- and small interfering RNA (siRNA)-mediated PRDM2 depletion, DSB-induced H3K9 dimethylation was significantly reduced, despite comparable DSB formation (Figures 2D, S1A–S1C, and S5B). PRDM2 depletion also reduced the frequency of cells with H3K9me2 accumulation at laser-induced DSBs (Figure S5C). Finally, GFP-tagged PRDM2 was recruited to DSBs, peaking shortly after H3K9me2 depletion and remaining enriched for the duration of the experiment (Figure 2E; Movie S1). In agreement with the finding that H3K9 dimethylation is at least in part dependent on macroH2A1, PRDM2 recruitment was impaired following depletion of either macroH2A1 or macroH2A1.2, whereas PRDM2 knockdown did not significantly alter macroH2A1.2 accumulation at laser-induced DSBs (Figures 2E, S5D, and S5E). Moreover, siRNA-mediated codepletion of both macroH2A1.2 and PRDM2 caused no additive reduction in HR, supporting the notion that macroH2A1.2 and PRDM2 function in the same DSB repair pathway (Figures 2F and 2G). Loss of either protein did not result in major changes in the expression of 11 key DSB repair factors, further indicating that the observed HR defect is due to DSB site- specific chromatin perturbations rather than a global transcriptional deregulation of DDR mediators (Figure S5F). We have thus identified macroH2A1 and PRDM2 as components of a DSB repair pathway that links HR to the deposition of DSB-proximal repressive chromatin marks.
MacroH2A1 and H3K9me2 Accumulation at DSBs Is ATM Dependent
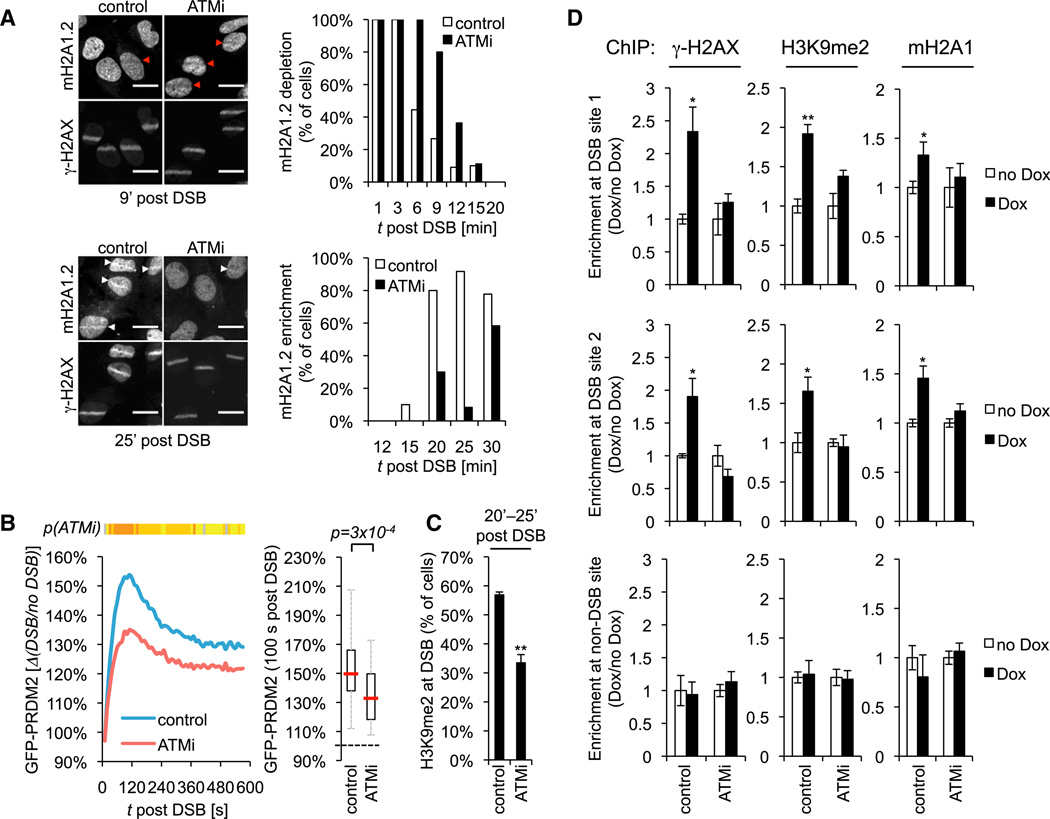
Given that changes in DSB-proximal chromatin structure are tightly linked to ATM activation (Bakkenist and Kastan, 2003; Kaidi and Jackson, 2013; Sun et al., 2009), we asked if ATM signaling is implicated in the dynamic accumulation of macroH2A1 and H3K9me2 at DSBs. Using a specific small molecule inhibitor of ATM kinase (ATMi), we detected a prolonged depletion and impaired reaccumulation of macroH2A1.2 at laser-induced DSB sites (Figure 3A). Similarly, laser damage-induced PRDM2 recruitment and H3K9 dimethylation were reduced following ATM inhibition (Figures 3B and 3C). Accordingly, the accumulation of macroH2A1 and H3K9me2 at I-SceI-induced DSBs was blunted in the presence of ATMi (Figure 3D). Together, these findings demonstrate that macroH2A1 and PRDM2 function at DSBs is ATM dependent.
Figure 3. MacroH2A1, PRDM2, and H3K9me2 Accumulation at DSBs Is Dependent on ATM Kinase.
(A) Coimmunostaining for macroH2A1.2 and γ-H2AX in the presence or absence of ATMi. Scale bars, 20 µm. The frequency of depletion (red arrows) or enrichment (white arrows) is shown for the indicated time points post laser microirradiation.
(B) GFP-PRDM2 recruitment to sites of laser-induced DSBs in the absence (n = 60) or presence (n = 26) of ATMi. At least three independent experiments were combined. p(ATMi) heatmap and box plot were generated as described in Figure 2E.
(C) Frequency of cells with laser damage-associated H3K9me2 in the presence or absence of ATMi (n = 3).
(D) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox. Enrichment relative to no Dox is shown for DSB site 1, DSB site 2 (see Figure 4B), and a non-DSB control locus. Values are expressed as mean and SEM (n = 3).
MacroH2A1 Promotes DNase I Resistance in DSB-Flanking Chromatin
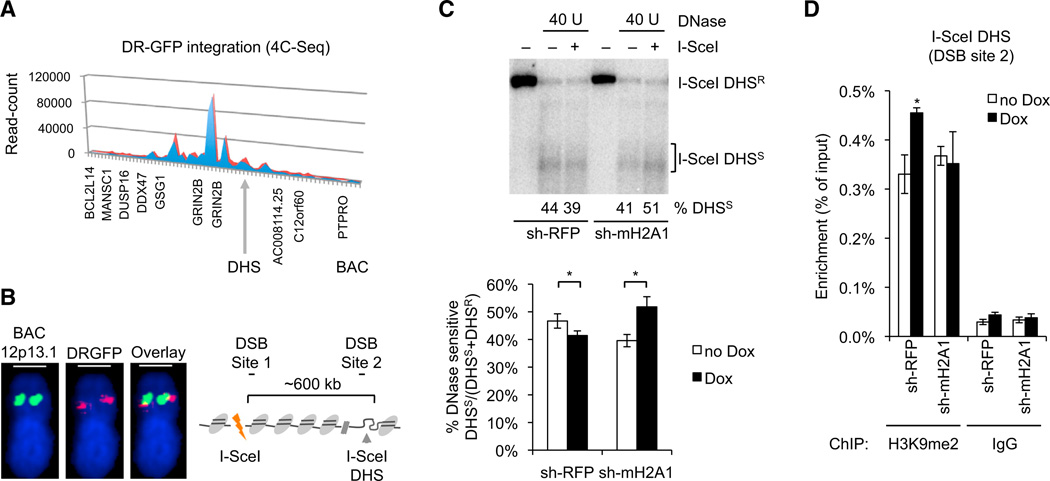
We next sought to investigate if the accumulation of the repressive macroH2A1 variant has functional consequences for DSB-proximal chromatin accessibility. Using circular chromosome conformation capture (4C) sequencing followed by DNA fluorescence in situ hybridization (FISH), we mapped the I-SceI containing DRGFP transgene to an intronic region of the GRIN2B gene on chromosome 12p13.1 in DRGFP-U2OS cells (Figures 4A and 4B; Table S4). Based on UCSC Genome Browser data, we identified a conserved DNase I-hypersensitive site (DHS) located within ~600 kb of the I-SceI site (Figure 4B), which displayed DSB-dependent accumulation of macroH2A1 and H3K9me2 repressive chromatin marks (Figure 3D, DSB site 2). Consistent with the latter, DSB induction caused a moderate decrease in nuclease sensitivity in sh-red fluorescent protein (RFP) control cells (Figure 4C). In contrast, depletion of macroH2A1 resulted in a significant shift toward DNase I-hypersensitive, accessible chromatin and a concomitant loss of DSB-induced, DHS-proximal H3K9 dimethylation (Figures 4C and 4D). Our findings, therefore, indicate that DSB-associated chromatin relaxation, and the associated increase in nuclease sensitivity (Ziv et al., 2006), is followed by macroH2A1-dependent condensation, resulting in the reestablishment of nuclease-resistant chromatin.
Figure 4. MacroH2A1 Promotes DSB-Induced DNase I Resistance.
(A) 4C mapping of the I-SceI DSB site-containing DRGFP transgene. Normalized read counts are shown. Red and blue graphs represent independent experiments; bins are in 50 kb intervals.
(B) Validation of the DRGFP integration site by DNA FISH. Green indicates GRIN2B–proximal BAC, and red indicates DRGFP probe. Scale bars, 1 mm. Colocalization was observed in ~25% of GRIN2B alleles, consistent with U2OS cell polyploidy.
(C) Southern blot analysis of an endogenous DHS downstream of DRGFP. DHSR, DNase resistant; DHSS, DNase sensitive. A representative experiment is shown. DNase hypersensitivity was determined as the ratio of DHSS over total (DHSR + DHSS) signal intensities in the presence or absence of Dox. Values are expressed as mean and SEM (n = 3).
(D) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox. Enrichment relative to input is shown for the DSB-proximal DHS (DSB site 2). Values are expressed as mean and SEM (n = 3).
See also Table S4.
MacroH2A1/PRDM2 Promote ATM-Dependent Chromatin Recondensation at DSBs
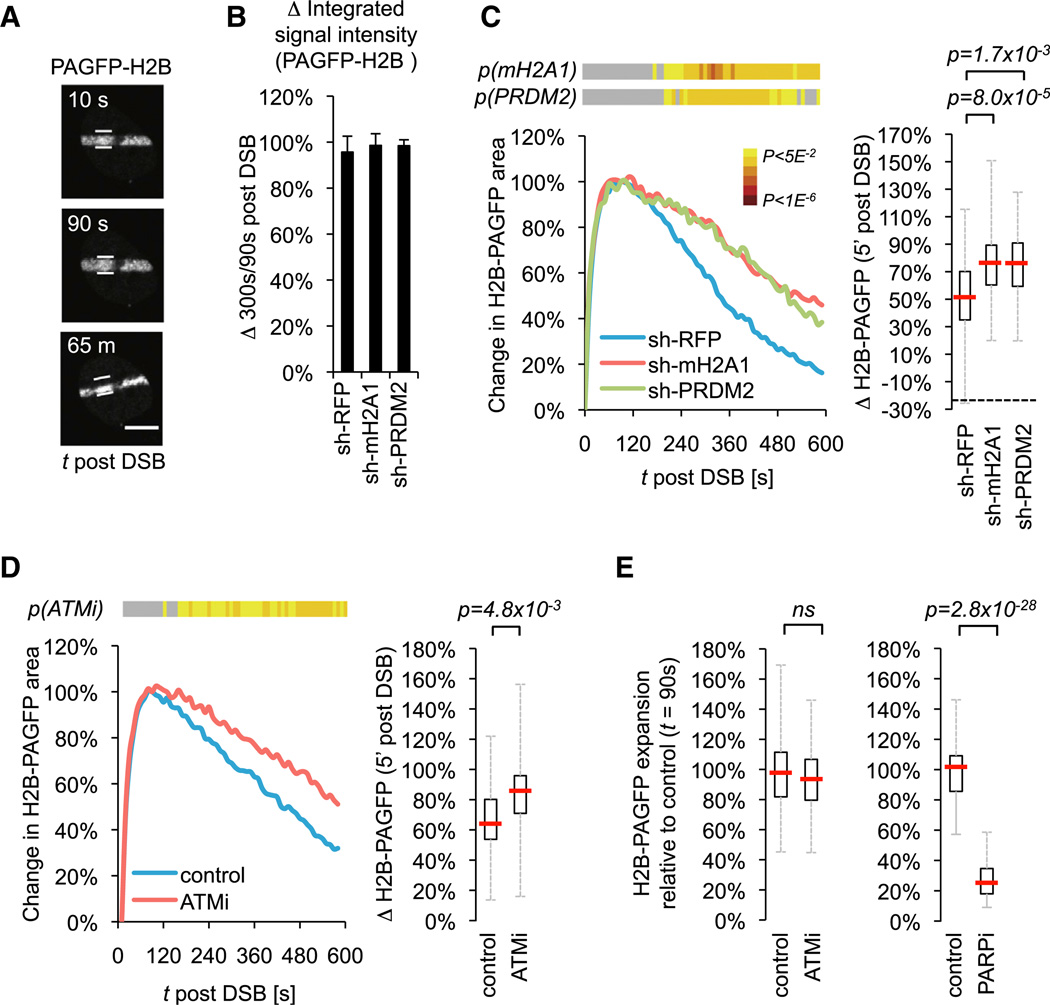
To visualize DSB-associated chromatin reorganization over time in living cells, we took advantage of U2OS cells expressing his-tone H2B fused to a photoactivatable GFP (PAGFP-H2B). Laser microirradiation of PAGFP-H2B–expressing nuclei resulted in the simultaneous introduction of DSBs and photoactivation of PAGFP-H2B, thus allowing us to track changes in the nuclear area occupied by DNA damage-associated nucleosomes (Figure 5A) (Kruhlak et al., 2006). Integrated PAGFP-H2B signal intensities did not vary significantly over time, or between control and knockdown samples, indicating that PAGFP-H2B remained stably associated with damaged chromatin (Figure 5B).
Figure 5. MacroH2A1 and PRDM2 Promote ATM-Dependent Chromatin Condensation.
(A) PAGFP-H2B imaging at the indicated time points after laser microirradiation. White lines depict maximal expansion (90 s). Scale bar, 10 µm.
(B) Integrated PAGFP-H2B signal intensities normalized to t = 90 s.
(C) Change in PAGFP-H2B nuclear area following laser microirradiation in sh-macroH2A1 (n = 39), sh-PRDM2–2 (n = 29), or sh-RFP cells (n = 32). Three independent experiments were combined. p(mH2A1) and p(PRDM2) heatmaps and box plot were generated as described in Figure 2E.
(D) Change in PAGFP-H2B nuclear area in control (n = 42) and ATMi-treated cells (n = 39). Three independent experiments were pooled and analyzed as in (C).
(E) Maximal expansion of PAGFP-H2B nuclear area following laser microirradiation in control (n = 36) and ATMi- (n = 35), or control (n = 47) and PARPi-treated cells (n =30). Nuclear area changes were normalized to the mean of controls.
In agreement with previous work (Kruhlak et al., 2006), we observed a phase of rapid chromatin expansion immediately after DSB induction (Figure 5C). Strikingly, however, DSB-proximal chromatin was found to recondense within minutes after the initial opening. DSB-induced chromatin compaction was first observed when PRDM2-GFP accumulation at DSBs was maximal and continued for the duration of the experiment (>40 min) (Figures 2E and 5C). Knockdown of either macroH2A1 or PRDM2 significantly impaired chromatin recondensation without affecting the kinetics of expansion (Figure 5C; Movies S2 and S3).Consistent with the ATM dependence of macroH2A1 and PRDM2 recruitment to DSBs, ATM inhibition resulted in a similar delay in condensation (Figure 5D). Chromatin expansion, on the other hand, was independent of ATM signaling but significantly impaired following inhibition of PARP, in agreement with previous reports (Figure 5E) (Kruhlak et al., 2006; Smeenk et al., 2013). These findings identify a biphasic change in DSB-proximal chromatin that is mediated by two distinct DDR signaling arms and involves the recruitment of the repressive chromatin components macroH2A1 and PRDM2.
MacroH2A1 and PRDM2 Promote BRCA1 Recruitment
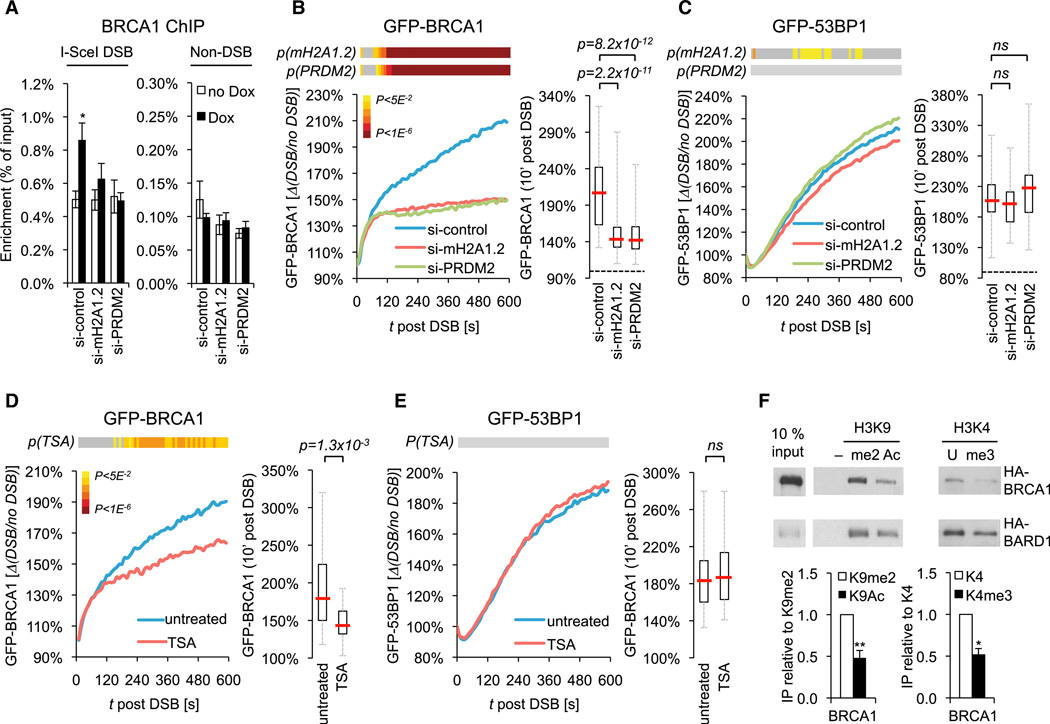
Having established a role for macroH2A1 and PRDM2 as modulators of DSB-proximal chromatin, we next asked how the depletion of these proteins accounts for the observed HR defects. Notably, the HR-associated repair factor BRCA1 was previously reported to accumulate at condensed DNA elements, such as pericentromeric repeats and the inactive × chromosome, and was further found to partially colocalize with macroH2A1 (Silver et al., 2007; Zhu et al., 2011). We, thus, sought to determine if macroH2A1 and/or PRDM2 can modulate BRCA1 accumulation at DSBs. Indeed, both shRNA- and siRNA-mediated depletion of macroH2A1, macroH2A1.2, or PRDM2 significantly impaired BRCA1 recruitment to I-SceI-induced DSBs (Figures 6A and S6A). Moreover, we observed a defect in the recruitment of a GFP-BRCA1 fusion protein to laser-induced DSBs that coincided with the initiation of macroH2A1/PRDM2-dependent chromatin recondensation (Figures 6B, S6B, and S6C). Impaired BRCA1 recruitment was not the result of a uniformly impaired DDR because accumulation of a GFP-53BP1 fusion protein showed little or no change compared to controls (Figures 6C, S6B, and S6D) (Mailand et al., 2007). Together, these findings implicate the macroH2A1.2/PRDM2 module in BRCA1 repair factor choice.
Figure 6. Depletion of MacroH2A1/PRDM2 and Chromatin Decondensation Promote BRCA1 Loss at DSBs.
(A) ChIP analysis 8 hr after release from double-thymidine block in the presence or absence of Dox. Enrichment relative to input is shown. Values are expressed as mean and SEM (n = 3).
(B and C) Recruitment kinetics of GFP-BRCA1 (B) or GFP-53BP1 (C) to laser-induced DSBs in si-macroH2A1.2, si-PRDM2, and si-control cells. Two independent experiments were combined (n > 50 cells per sample). p(mH2A1.2) and p(PRDM2) heatmaps and box plot were generated as described in Figure 2E.
(D) GFP-BRCA1 recruitment in the absence (n = 56) or presence of TSA (n = 50). At least three independent experiments were pooled and analyzed as described in (B).
(E) GFP-53BP1 recruitment in the absence (n = 22) or presence of TSA (n = 31). Two independent experiments were combined and analyzed as in (B).
(F) Peptide immunoprecipitation (IP) assays of HA-BRCA1 and HA-BARD1 with modified or unmodified histone H3 N-terminal peptides or beads alone (−). H3K9 peptide IPs were normalized to H3K9me2, H3K4 peptide IPs to the unmodified peptide (U). Values are expressed as mean and SEM (n ≥ 3).
See also Figure S6.
Notably, depletion of both macroH2A1.2 and PRDM2 impaired BRCA1 recruitment to a similar degree, which is consistent with their epistatic function in HR and points to a role for coordinated, macroH2A1.2/PRDM2-associated chromatin reorganization in this process. To determine if perturbed chromatin condensation is sufficient to modulate BRCA1 recruitment, we experimentally increased chromatin accessibility in macroH2A1.2/PRDM2-proficient cells by inducing histone hyperacetylation via trichostatin A (TSA)-mediated HDAC inhibition (Tóth et al., 2004). In direct agreement with the effects of macroH2A1.2 or PRDM2 depletion, TSA treatment resulted in reduced GFP-BRCA1 retention at DSBs, whereas GFP-53BP1 recruitment remained unaltered (Figures 6D, 6E, S6E, and S6F). Consistent with this, we observed decreased BRCA1 binding to histone H3 peptides carrying marks associated with open chromatin, such as acetylated lysine 9 or trimethylated lysine 4, when compared to K9-dimethylated or unmodified peptides, respectively (Figure 6F). No detectable interaction with either H3K9ac or H3K9me2 was observed for 53BP1 (Figure S6G). Together, these results suggest that the macroH2A1/PRDM2-mediated chromatin reorganization is functionally linked to BRCA1 accumulation at DSBs.
MacroH2A1.2/PRDM2 Direct Repair Pathway Choice by Promoting End Resection
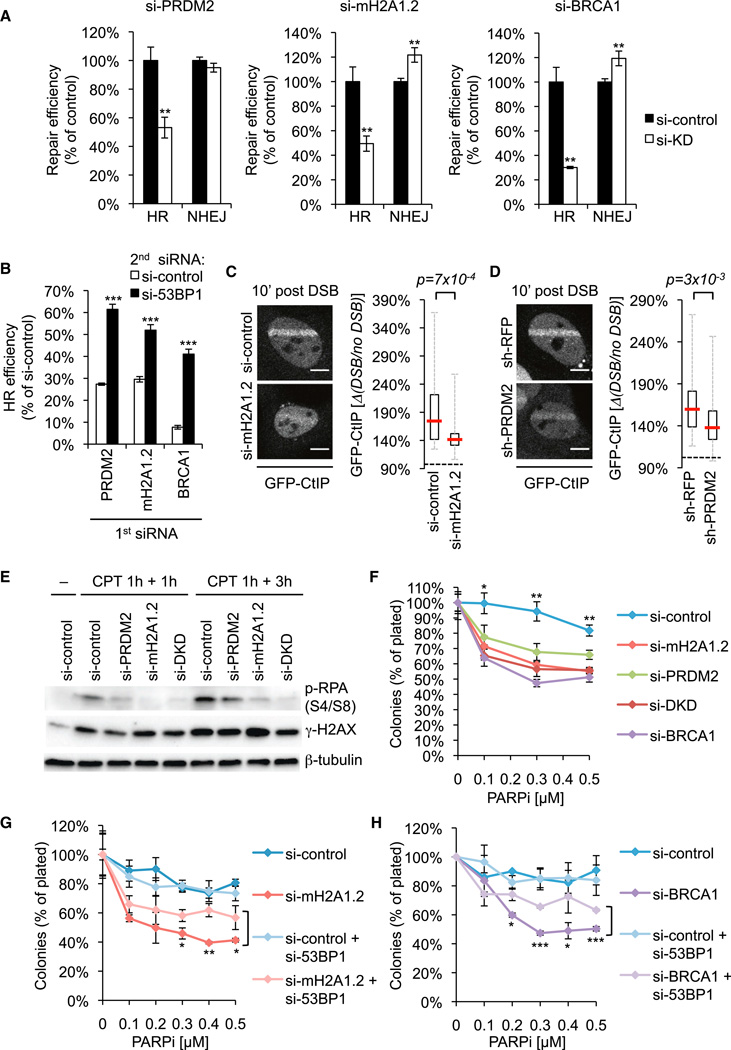
The selective effect of macroH2A1/macroH2A1.2 and PRDM2 on BRCA1 recruitment points to a role in DSB repair pathway choice. In analogy to BRCA1 loss, we found that, following siRNA-mediated depletion of macroH2A1.2 or PRDM2, HR was significantly impaired, whereas NHEJ was unaltered or moderately increased (Figure 7A). Notably, HR defects were partially restored following co-depletion of 53BP1, indicating that 53BP1 antagonizes HR following depletion of macroH2A1.2 or PRDM2 (Figure 7B). Mechanistically, 53BP1 was found to oppose HR in the absence of BRCA1 by interfering with the recruitment of CtIP, which mediates the resection of DNA ends to single-stranded DNA (ssDNA) (Bouwman et al., 2010; Bunting et al., 2010; Sartori et al., 2007). Consistent with this, we observed a significant reduction of GFP-CtIP as well as the ssDNA-binding protein RPA at laser-induced DSBs in S phase cells following depletion of either macroH2A1.2 or PRDM2 (Figures 7C, 7D, and S7A). Moreover, the DNA damage-induced phosphorylation of RPA was impaired following treatment with the topoisomerase I inhibitor camptothecin (CPT), which causes replication-dependent DSBs that initiate HR (Figure 7E). The reduction in RPA phosphorylation in macroH2A1.2/PRDM2-depleted cells was comparable to that observed following BRCA1 loss and less pronounced when 53BP1 was depleted simultaneously (Figure S7B). Impaired RPA phosphorylation was also apparent in MCF7 and WM-115 cells, two cell lines that showed dynamic changes in macroH2A1.2 occupancy at DSBs (Figures S7C and S7D). In agreement with the epistatic function of macroH2A1.2 and PRDM2 during HR (Figure 2F), no major additive or synergistic effects were observed following codepletion of macroH2A1.2 and PRDM2 (Figures 7E and S7C). These findings demonstrate that macroH2A1.2 and PRDM2 cooperate to promote HR, at least in part, through CtIP-dependent end resection.
Figure 7. MacroH2A1.2 and PRDM2 Direct Repair Pathway Choice by Promoting End Resection.
(A) HR and NHEJ efficiency in stable U2OS reporter cell lines. Repair efficiency was normalized to si-control (black). Samples were analyzed in triplicate. Values are expressed as mean and SD.
(B) HR efficiency in the presence or absence of si-53BP1. Samples were analyzed in triplicate. Values are expressed as mean and SD.
(C) GFP-CtIP recruitment in S phase cells (1–2 hr post double-thymidine block). MacroH2A1.2 knockdown (n = 32) and control cells (n = 34) were analyzed 10 min post DSB. Two independent experiments were combined. Representative images are shown. Scale bars, 10 µm.
(D) GFP-CtIP recruitment in sh-PRDM2–1 (n = 47) and sh-RFP control cells (n = 55). Three independent experiments were combined and analyzed as in (C). Representative images are shown. Scale bars, 10 µm.
(E) Western blot analysis in U2OS cells treated with CPT for 1 hr followed by a 1 or 3 hr release. si-DKD, combined knockdown of macroH2A1.2 and PRDM2. (F–H) Clonogenic survival assays in response to treatment with PARPi. Samples were analyzed in triplicate. Values are expressed as mean and SD (Fand G) or as mean and SEM (n = 2) (H).
See also Figure S7.
HR-deficient cells exhibit an acute sensitivity to killing by PARP inhibitors (PARPis). PARPi cytotoxicity is predominantly attributed to a defect in the repair of ssDNA breaks in G1, which are then converted into DSBs during DNA replication. The latter cannot be faithfully repaired in the absence of BRCA1 due to 53BP1-mediated end protection (Bouwman et al., 2010; Bryant et al., 2005; Bunting et al., 2010; Farmer et al., 2005). Similar to BRCA1-deficient cells, cells depleted for either macroH2A1.2 or PRDM2 showed increased sensitivity to PARPi, and no additive effect was observed following codepletion of both proteins (Figure 7F). Moreover, simultaneous loss of 53BP1 was able to partially rescue PARPi sensitivity both in BRCA1-deficient and macroH2A1.2- or PRDM2-deficient cells (Figures 7G, 7H, and S7E). Together, these findings establish the repressive macroH2A1/PRDM2 chromatin module as a selective modulator of BRCA1-dependent DSB repair.
DISCUSSION
DSBs Induce Biphasic Chromatin Reorganization
The relaxation of damage-proximal chromatin is widely considered a critical aspect of the eukaryotic response to DNA breaks (Price and D’Andrea, 2013; Smeenk and van Attikum, 2013; Soria et al., 2012). Here, we show that chromatin expansion at DSBs is followed by prolonged chromatin recondensation, which is at least in part promoted by the coordinated recruitment of the repressive H2A variant macroH2A1 and the H3K9 methyl-transferase PRDM2 (Figures 1 and 2). Supporting the biphasic nature of DSB-induced chromatin reorganization, expansion and recondensation are mediated by distinct arms of the DDR. Chromatin expansion occurs independently of ATM signaling and instead involves PARP-induced chromatin remodeling (Figure 5) (Kruhlak et al., 2006; Smeenk et al., 2013). Consistent with this, PARP promotes the recruitment of the histone demethylase KDM4B to DSBs, which accounts for the transient reduction in DSB-proximal H3K9 methylation observed by us and others (Young et al., 2013). PARP1 further facilitates the temporary recruitment of the chromatin scaffold protein SAFB1 to render DSB-surrounding chromatin permissive for repair (Altmeyer et al., 2013). Efficient macroH2A1/PRDM2 recruitment and concomitant chromatin recondensation, on the other hand, are dependent on ATM kinase (Figures 3 and 5D). This finding is consistent with previous reports demonstrating that ATM can act as a sensor of DSB-associated chromatin changes (Bakkenist and Kastan, 2003; Kaidi and Jackson, 2013; Sun et al., 2009) and suggests that a central aspect of ATM-mediated DNA damage signaling may be to control and/or contain DSB-induced chromatin reorganization.
Notably, the biphasic nature of DSB-induced chromatin reorganization is also reflected by the recruitment of the macroH2A1.1 and macroH2A1.2 splice variants to DSBs. Accumulation of macroH2A1.1 is transient, depends on PARP, and coincides with the early phase of macroH2A1.2 depletion described here (Figure 1) (Timinszky et al., 2009). In contrast, recruitment of macroH2A1.2, which lacks the macroH2A1.1 PAR-binding domain, is ATM dependent and temporally delayed compared to macroH2A1.1. These findings point to kinetically distinct but possibly complementary roles for macroH2A1 splice variants in the biphasic reorganization of DSB-proximal chromatin that are functionally linked to the respective signaling pathways.
A Link between Repressive Chromatin and HR
Our data suggest that DSB-induced chromatin condensation may have direct implications for DSB repair outcome: both macroH2A1/macroH2A1.2 and PRDM2 are required for efficient BRCA1 but not 53BP1 recruitment to DSBs. In addition, and consistent with macroH2A1/PRDM2-dependent, DSB-proximal H3K9 dimethylation, BRCA1 was found to associate with H3K9-dimethylated histone tails, whereas histone acetylation interfered with BRCA1 recruitment in vitro and in vivo (Figure 6). A link between repressive chromatin and BRCA1 recruitment is further suggested by recent reports demonstrating that the HP1 variants HP1-α and HP1-β can selectively recruit BRCA1, facilitate end resection, and in turn promote HR (Lee et al., 2013; Soria and Almouzni, 2013). Notably, the HP1-interacting KAP1 protein was also found to be required for efficient homology-directed repair (Geuting et al., 2013). However, the impact of both HP1 and KAP1 on DSB-proximal chromatin structure remains to be elucidated. In striking analogy to the biphasic chromatin reorganization identified here, both HP1 and KAP1 were found to be dispersed from as well as recruited to DSB sites (Ayoub et al., 2008; Baldeyron et al., 2011; Goodarzi et al., 2008; Lee et al., 2013; Soria and Almouzni, 2013; Ziv et al., 2006). It is, therefore, tempting to speculate that HP1 and/or KAP1 may cooperate with macroH2A1/PRDM2-mediated H3K9 dimethylation to promote DSB-associated chromatin condensation and BRCA1 repair factor choice. The establishment of a repressive chromatin environment at DSBs was further proposed to facilitate homology search in reduced spatial distance (Sonoda et al., 2006), a notion consistent with the limited mobility of broken DNA ends in mammalian cells (Soutoglou et al., 2007).
Interestingly, and in contrast to mammalian cells, DSBs in yeast are surprisingly mobile in their search for homologous DNA templates (Dion et al., 2012). Moreover, DSBs that occur in yeast heterochromatin require relocalization to the outside of the condensed chromatin domain to complete the HR process (Torres-Rosell et al., 2007), and a similar phenomenon has recently been described in flies by Chiolo et al. (2011). Notably, macrohistone variants are absent in flies and yeast, raising the intriguing possibility that DSB-induced repressive chromatin formation may have evolved to protect the genomes of longer-lived organisms from aberrant DSB repair, translocations, and ultimately, malignant transformation.
Chromatin Dynamics and Repair Factor Choice
The biphasic chromatin reorganization described here implies that (experimentally induced) changes in chromatin structure can have distinct consequences for repair factor recruitment, depending on which phase of the repair process is affected. Consistent with this, increased histone acetylation as well as macroH2A1.2/PRDM2 loss resulted in reduced BRCA1 retention at a time when DSB-induced chromatin condensation would normally occur, whereas the same perturbations did not interfere with BRCA1 recruitment during the initial phase of chromatin expansion (Figures 6B and 6D). Indeed, recent work suggests that increased histone acetylation can revert BRCA1 recruitment defects early in the DDR when chromatin relaxation is impaired as a result of KAT5 depletion (Tang et al., 2013). Similarly, increased histone acetylation can have diverse consequences for the recruitment of 53BP1 to DSBs, ranging from impaired binding to nucleosomes to unaltered or increased 53BP1 retention at DSBs (Figure 6E) (Miller et al., 2010; Murr et al., 2006; Tang et al., 2013). Together, these findings underscore the potential impact of kinetically as well as functionally distinct chromatin alterations on DSB repair factor choice.
MacroH2A1.2 Controls End Resection
Consistent with a role for macroH2A1.2 and PRDM2 as modulators of DSB repair outcome, depletion of either factor impaired HR without reducing NHEJ. HR defects were partially rescued upon depletion of 53BP1, pointing to a defect in the BRCA1-mediated inhibition of 53BP1-dependent end protection (Figure 7) (Bouwman et al., 2010; Bunting et al., 2010). In further support of the latter, we show that macroH2A1.2 and PRDM2 facilitate both CtIP recruitment and end resection. Although macroH2A1.2 recruitment and PRDM2-mediated H3K9 dimethylation at DSBs were observed throughout the cell cycle (Figures 1 and S2), their impact on DSB repair is likely to be most relevant in S/G2 because CtIP-dependent end resection was found to depend on S/G2-specific CtIP phosphorylation events (Huertas and Jackson, 2009; Yun and Hiom, 2009).
Notably, CtIP function does not appear to depend on its direct interaction with BRCA1, and resection can occur independently of BRCA1 when 53BP1 is absent (Bouwman et al., 2010; Bunting et al., 2010; Polato et al., 2014; Reczek et al., 2013). Moreover, depletion of the chromatin-binding protein LEDGF, which mediates CtIP recruitment to sites of DSBs, impairs end resection without affecting BRCA1 recruitment (Daugaard et al., 2012). Interestingly, LEDGF recruitment, end resection, and HR were recently found to occur preferentially at transcriptionally active regions (Aymard et al., 2014; Daugaard et al., 2012). Together with our work, these findings point to a dynamic role for chromatin in the control of end resection, where active chromatin can facilitate CtIP recruitment, which is then maintained by stabilizing BRCA1, and thereby opposing 53BP1, via the recruitment of the repressive macroH2A1.2/PRDM2 module. This model is further consistent with the previously described, ATM-dependent repression of actively transcribed genes in response to DNA damage (Kruhlak et al., 2007; Shanbhag et al., 2010). Nevertheless, we cannot formally rule out distinct, locus-specific contributions of the macroH2A1.2/PRDM2 module as well as LEDGF in controlling resection and HR.
In contrast to macroH2A1.2, the H2A variant H2A.Z was recently shown to restrict ssDNA production through end resection processes, while promoting chromatin relaxation and DSB-proximal H4 acetylation (Xu et al., 2012a). The differential use of histone variants, which may further involve macroH2A1.1 versus macroH2A1.2 splice variant choice, is, thus, emerging as a critical modulator of DSB repair outcome. The latter may further help explain why macroH2A1.2 was not identified as a modulator of HR in previous, pooled RNAi screens (Adamson et al., 2012; Słabicki et al., 2010), whereas PRDM2 depletion was found to reduce HR in at least one of these screens (Adamson et al., 2012).
The identification of factors that differentially control the recruitment of BRCA1 and 53BP1 has significant implications for the regulation of genome maintenance during malignant transformation. Cells deficient in BRCA1 but not 53BP1 are particularly sensitive to PARP inhibition, and PARPis are used in clinical trials to treat tumors with HR defects (Bouwman et al., 2010; Bryant et al., 2005; Bunting et al., 2010; Farmer et al., 2005). Our findings, thus, raise the intriguing possibility that, by modulating BRCA1 versus 53BP1 retention, macroH2A1 and PRDM2 may provide molecular targets for therapeutic intervention.
EXPERIMENTAL PROCEDURES
Cell Culture
Cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in the presence of 5% CO2. To generate a Dox-inducible I-SceI/DRGFP cell line (TRI-DR-U2OS), DRGFP-U2OS cells (Weinstock et al., 2006) were stably transfected with pTet-ON and a pTRE-tight-regulated I-SceI cDNA (Clontech Laboratories). Stable, I-SceI-dependent NHEJ-U2OS reporter cells were generated using the pEJ5 construct (Bennardo et al., 2008). HR/NHEJ efficiencies were analyzed by fluorescence-activated cell sorting. The GFP-PRDM2 pcDNA 3.1 was generated using full-length human PRDM2 (Open Biosystems). Stable knockdown was performed by spin infection, lentiviral particles were generated using 293T cells, and transient transfections were performed following standard procedures. For transient knockdown, cells were transfected with the indicated siRNAs (50 nM) using DharmaFect-1 (Thermo Scientific). See Table S2 for shRNA/siRNA target sequences and the Supplemental Experimental Procedures for drug treatments.
RNAi Screen
A custom lentiviral shRNA library directed against 412 Gene Ontology-annotated chromatin modifiers was used to determine their function in HR using the DRGFP-U2OS reporter system (see the Supplemental Experimental Procedures for details).
Laser Microirradiation and Imaging
Laser microirradiation and PAGFP photoactivation were performed using a Zeiss LSM510 META confocal microscope with a 364 nm UVA laser (Coherent).
Image Analysis
Changes in chromatin structure were measured using MetaMorph (v.7.7.9) or Imaris (v.7.4) image processing and analysis software. Recruitment of GFP-tagged proteins in laser-microirradiated cells was quantified using MIPAV software (v.5.1). See the Supplemental Experimental Procedures for details.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS and permeabilized with 10% methanol. Following two-step immunostaining, images were acquired using a Zeiss LSM510 or LSM710 META confocal microscope (Zeiss). See the Supplemental Experimental Procedures for a list of antibodies.
DNase I Hypersensitivity
TRI-DR-U2OS cells were treated with Dox for 12 hr or left untreated. Isolation of nuclei, DNase I treatment (40 U/ml), and DNA purification were performed as described by Lu and Richardson (2004). DNase I-treated DNA was digested with HindIII (New England Biolabs), subjected to Southern blotting, and probed with a PCR product specific to an I-SceI-proximal DHS (see Table S3 for primer sequences). Band intensities were quantified using a Typhoon Phosphorimager (GE Healthcare).
ChIP Analysis
For ChIP analysis, TRI-DR-U2OS cells were either left untreated or treated with Dox for the indicated time points following double-thymidine block or serum starvation. Crosslinked chromatin was fragmented by MNase digestion and immunoprecipitated overnight (see the Supplemental Experimental Procedures). Purified ChIP DNA was analyzed by quantitative PCR using a LightCycler 480 II (Roche) (see Table S3 for primer sequences).
4C Sequencing
4C was performed as previously described with minor modifications, using HindIII/Csp6I restriction digests followed by religation (Simonis et al., 2006). 4C DNA libraries were PCR amplified using DRGFP-specific primers (see Table S3) and subjected to Illumina HiSeq 2000 paired-end sequencing. HindIII-proximal reads were trimmed to remove bait sequence and low-quality bases and mapped to the human genome (hg18) using the Illumina single-end algorithm. The quality of mapped reads was assessed using FastQC (Babraham Bioinformatics). Read counts were normalized based on total read counts for each sample and binned into 50 kb nonoverlapping windows using SeqMonk software (Babraham Bioinformatics).
Peptide-Binding Assays
U2OS cells transfected with HA-BRCA1/HA-BARD1 or GFP-53BP1 expression vectors were lysed 48 hr posttransfection in NETN buffer (100 mM NaCl, 1 mM EDTA, 10 mM Tris-Cl [pH 8.0], 0.1% Nonidet P-40,10% glycerol, and 1 mM dithiothreitol). Lysates were incubated at 4°C with biotin-conjugated unmodified or modified histone peptides immobilized on streptavidin agarose beads (Millipore; AnaSpec).
Western Blotting
Whole-cell extracts were prepared using radioimmunoprecipitation assay buffer with protease inhibitors followed by SDS-PAGE. For phospho-RPA western blot analyses, cells were lysed and sonicated in high-salt buffer (20 mM Tris-HCl, 400 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40). Antibodies are listed in the Supplemental Experimental Procedures.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using the RNeasy Mini Kit according to the manufacturer’s instructions (QIAGEN). cDNA was synthesized from 0.2–1 µg of total RNA using the ThermoScript RT-PCR system (Invitrogen), and expression of the indicated genes was analyzed by quantitative RT-PCR using a LightCycler 480 II (Roche) (see Table S3 for primer sequences).
Ligated-Mediated PCR
Genomic DNA was extracted using the QIAGEN DNeasy Blood and Tissue kit and sheared in an ultrasonicator water bath (Bioruptor; Diagenode). Blunt ends were generated using the End-It Repair kit (Epicenter), and A tails were added using the NEB Next dA-Tailing protocol. Between reactions, DNA was purified with QIAquick PCR Purification (QIAGEN). DNA was ligated to adaptors followed by three rounds of nested PCR amplification using one DRGFP-specific and one adaptor-specific primer (see Table S3).
DNA FISH
Chromosome spreads were generated as described previously by Singh et al. (2013). For dual-color FISH, a bacterial artificial chromosome (BAC) probe (clone RP11–164B11, chromosome 12p13.1) or a DRGFP-derived 3.5 kb NotI fragment was labeled by nick translation with biotin-16–2’-deoxyuri-dine-5’-triphosphate (dUTP) or digoxigenin-11-dUTP, respectively (Roche). FISH was performed following standard procedures, using fluorescein isothiocyanate-conjugated avidin (Vector Laboratories) and rhodamine-conjugated anti-digoxigenin (Roche); DNA was counterstained with DAPI. FISH images were acquired on an Axioplan 2 fluorescence microscope (Zeiss) using FISHView 5.5 software (Applied Spectral Imaging).
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Dalal, T. Misteli, A. Nussenzweig, and S. Oberdoerffer for critical reading of the manuscript; S. Silver, T Nieland, and the Broad RNAi platform for assistance with RNAi screening; A. Mazumder for help with Cellomics high-content imaging; B. Tran and J. Shetty for Illumina sequencing; S. Burkett for FISH analysis; and C. Lukas, Y. Galanty, S. Jackson, D. Livingston, and A. Sartori for reagents. This work was supported by federal funds from the National Cancer Institute, NIH, and the NIH intramural research program.
Footnotes
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus accession number for the 4C sequencing data presented in this paper is GSE58563.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, four tables, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.07.024.
REFERENCES
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Lukas J. Guarding against collateral damage during chromatin transactions. Cell. 2013;153:1431–1434. doi: 10.1016/j.cell.2013.05.044. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Toledo L, Gudjonsson T, Grøfte M, Rask MB, Lukas C, Akimov V, Blagoev B, Bartek J, Lukas J. The chromatin scaffold protein SAFB1 renders chromatin permissive for DNA damage signaling. Mol. Cell. 2013;52:206–220. doi: 10.1016/j.molcel.2013.08.025. [DOI] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iaco-voni JS, Daburon V, Miller KM, Jackson SP, Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J. Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiácovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive poly-comb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sørensen CS, Petersen NH, Sorensen PH, Lukas C, Bartek J, et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Kraus WL. Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle. 2010;9:2568–2574. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- Geuting V, Reul C, Löbrich M. ATM release at resected double-strand breaks provides heterochromatin reconstitution to facilitate homologous recombination. PLoS Genet. 2013;9:e1003667. doi: 10.1371/journal.pgen.1003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim KC, Geng L, Huang S. Inactivation of a histone methyl-transferase by mutations in human cancers. Cancer Res. 2003;63:7619–7623. [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montaño C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013;41:5784–5798. doi: 10.1093/nar/gkt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Richardson B. DNaseI hypersensitivity analysis of chro-matin structure. Methods Mol. Biol. 2004;287:77–86. doi: 10.1385/1-59259-828-5:077. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J. Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- Polato F, Callen E, Wong N, Faryabi R, Bunting S, Chen HT, Kozak M, Kruhlak MJ, Reczek CR, Lee WH, et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J. Exp. Med. 2014;211:1027–1036. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek CR, Szabolcs M, Stark JM, Ludwig T, Baer R. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 2013;201:693–707. doi: 10.1083/jcb.201302145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription incis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Oberdoerffer P. Chromatin dynamics in DNA double-strand break repair. Biochim. Biophys. Acta. 2012;1819:811–819. doi: 10.1016/j.bbagrm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Livingston DM. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2012;2:679–684. doi: 10.1158/2159-8290.CD-12-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Dimitrov SD, Feunteun J, Gelman R, Drapkin R, Lu SD, Shestakova E, Velmurugan S, Denunzio N, Dragomir S, et al. Further evidence for BRCA1 communication with the inactive × chromosome. Cell. 2007;128:991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered bychromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Ober-doerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J. Exp. Med. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Jun-queira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, Romeijn RJ, Pastink A, Mailand N, Vermeulen W, van Attikum H. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 2013;126:889–903. doi: 10.1242/jcs.109413. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Soria G, Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell Cycle. 2013;12:422–429. doi: 10.4161/cc.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol. Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J, Buyse IM, Fletcher JA, Liu J, Bronson R, et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyl-transferase superfamily. Genes Dev. 2001;15:2250–2262. doi: 10.1101/gad.870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. HistoneH3 methylation links DNA damage detection toactivation of the tumour suppressor Tip60. Nat. Cell Biol. 2013;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, et al. A macro-domain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragón L, Lisby M. The Smc5-Smc6 complex and SUMO modification ofRad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2004;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- Tóth KF, Knoch TA, Wachsmuth M, Frank-Stöhr M, Stöhr M, Bacher CP, Müller G, Rippe K. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 2004;117:4277–4287. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012a;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Xu Y, Gursoy-Yuzugullu O, Price BD. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012b;586:3920–3925. doi: 10.1016/j.febslet.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LC, McDonald DW, Hendzel MJ. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 2013;288:21376–21388. doi: 10.1074/jbc.M113.491514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via hetero-chromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.