Abstract
Accuracy in the flow of genetic information from DNA to protein, or gene expression, is essential to an organism’s viability. Pre-mRNA splicing and protein translation are two major steps in eukaryotic gene expression that necessitate the production of accurate gene products. Both processes occur in large complexes, consisting of both proteins and noncoding RNAs. Interestingly, the RNA components contain a large number of posttranscriptional modifications, including 2′-O-methylation and pseudouridylation, which are functionally important. In this chapter, we highlight the functional aspects of the modifications of spliceosomal snRNA and rRNA and provide a framework for understanding how posttranscriptional modifications are capable of influencing gene expression.
Keywords: Pre-mRNA splicing, RNA modifications, 2′-O-methylation, pseudouridylation
1. Introduction
Inherent to the expression of the majority of higher eukaryotic genes is the processing of pre-messenger RNA (pre-mRNA) and synthesis of proteins (translation). Pre-mRNA splicing is an RNA-processing reaction in which two successive transesterification reactions result in the removal of noncoding sequences, introns, and ligation of coding sequences, exons (including the 5′ and 3′ untranslated sequences), ensuing in the production of a mature mRNA. The mature mRNA can then be exported to the cytoplasm where it directs the synthesis of protein.
Both pre-mRNA splicing and protein synthesis are multistep processes coordinated by large and highly dynamic ribonucleoprotein (RNP) complexes (1). In the case of the spliceosome, the machinery responsible for intron removal, five uridyl-rich small nuclear RNAs (snRNA), namely U1, U2, U4, U5, and U6, as well as a number of proteins, orchestrate the splicing of pre-mRNAs (2-5). On the other hand, the ribosome, the machinery that directs protein synthesis, is composed of four ribosomal RNAs (rRNAs), namely 5S, 5.8S, 18S, 28S (in high eukaryotic organisms), and numerous protein factors. Over the years, the mechanism of both processes has been extensively studied, and it is now clear that in each case, the RNA components play indispensable roles in orchestrating their respective processes.
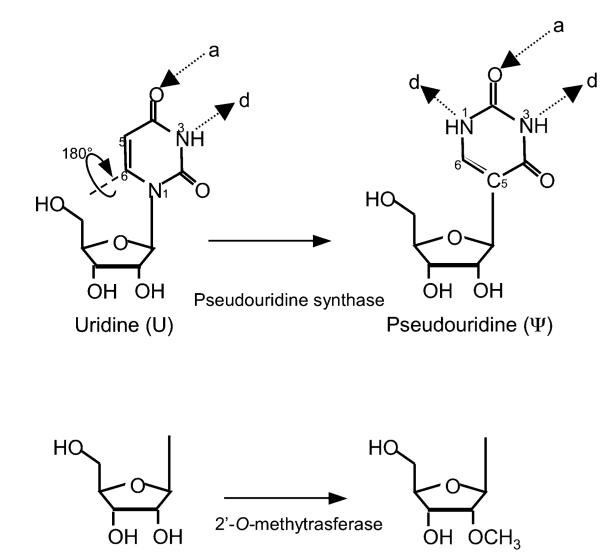
Interestingly, both spliceosomal snRNAs and rRNAs contain a variety of posttranscriptionally modified nucleotides. By far, the two most abundant modifications are 2′-O-methylation and pseudouridylation. While 2′-O-methylation is an RNA backbone-targeting reaction that results in the introduction of a methyl group at the 2′-O position of the sugar ring, pseudouridylation is a uridine-specific modification, which results in the formation of the 5-ribosyl isomer of uridine, pseudouridine (ψ) (Fig. 1.1). Over the last few decades, considerable effort has been directed at elucidating the mechanism by which these modifications are introduced as well as their molecular functions. To date, far progress has been most significant in the area regarding the introduction of modifications (see Chapter 1). In contrast, knowledge regarding the function of posttranscriptionally modified nucleotides has lagged behind. This has been in part due to the lack of proper assays and experimental systems. Recently, especially over the past 10 years, there has been a considerable growth in the field of RNA modification function, resulting in the accumulation of a large amount of data. This review discusses the functional aspects of the modifications of spliceosomal snRNA and rRNA and provides a framework for understanding how posttranscriptional modifications are capable of influencing gene expression.
Fig. 1.1.
Schematic depiction of the two most abundant modified nucleotides in snRNA and rRNA. (Top) Pseudouridine is a rotational isomer of uridine, in which the C–C glycosidic bond is broken to form an N–C bond, resulting in the presence of an extra hydrogen bond donor (d) while the number of hydrogen bond acceptors (a) is unchanged. (Bottom) Schematic representation of a 2′-O-methylated ribose.
2. Spliceosomal snRNA Modification
Prior to 1951, RNA was believed to consist of the four classical ribonucleosides (i.e., adenosine, guanosine, uridine, and cytidine). The field of RNA modification has its birth in 1951 when Cohn and Volkin reported the identification of an unknown nucleoside, subsequently identified as 5-ribosyluracil (pseudouridine), present in the RNA hydrolysates of calf liver (6). Shortly thereafter, additional modified ribonucleosides were discovered including various 2′-o-methylribose derivatives.
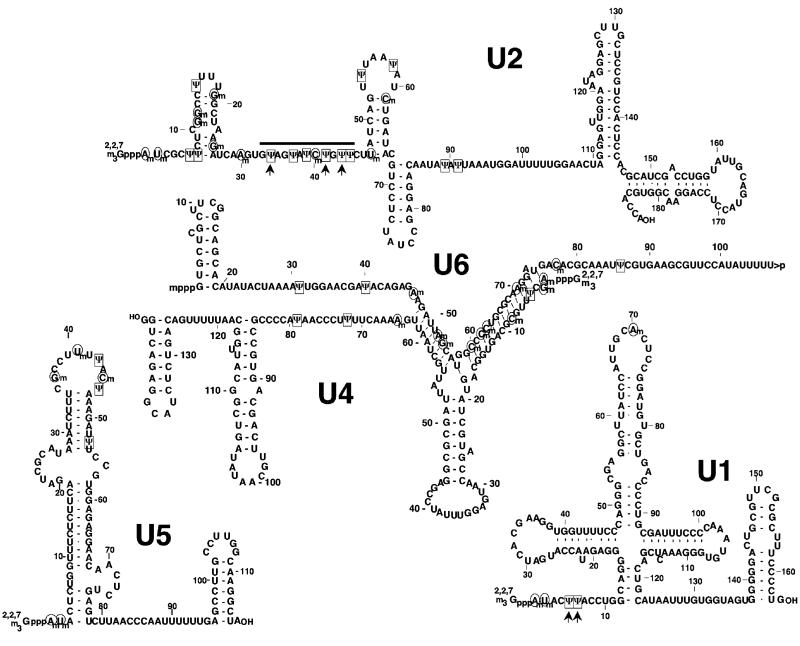
Spliceosomal snRNAs were initially identified as U-rich small nuclear RNAs (7-11). Surprisingly, however, when the exact nucleotide composition was finally determined, it became clear that these RNAs contained a number of pseudouridines and 2′-O-methylated residuals (Fig. 1.2). Whether these modified nucleotides are important for function in pre-mRNA splicing became a central question.
Fig. 1.2.
Primary sequences and secondary structures of the vertebrate major spliceosomal snRNAs. Positions of pseudouridine residues are marked in boxes and 2′-O-methylated nucleotides are circled. The 5′ caps (2,2,7-trimethylated guanosine cap for U1, U2, U4, and U6, and a γ-methylated guanosine for U6) are also shown. The pseudouridine residues in U2 and U1 snRNAs that are conserved in yeast are marked with arrows. The thick line indicates the U2 branch site recognition region.
2.1. Experimental Clues that Spliceosomal snRNA Modifications Might Affect Pre-mRNA Splicing
Although the spliceosomal U snRNAs had been known to be extensively posttranscriptionally modified since their initial identification in the 1960s and 1970s, the exploration for a functional role of these modifications was not fruitful until the early 1990s. However, as early as 1988 Reddy and Busch did make note of the fact that the modified nucleotides were distributed nonrandomly throughout the snRNAs (11) (Fig. 1.2). In fact, modifications are particularly clustered in regions of functional significance, such as regions engaged in RNA–RNA interactions (4). Furthermore, analysis of the distribution of modified nucleotides in spliceosomal snRNAs from various organisms demonstrated conservation in the location of modifications throughout evolution.
In the early 1990s, a number of functional reconstitution systems were developed, permitting a direct assessment of the function of spliceosomal snRNAs and their modifications in pre-mRNA splicing. In particular, Patton developed an in vitro U snRNA assembly-modification system from HeLa cell S100 and nuclear extracts that was capable of both pseudouridylating U snRNAs and assembling functional U snRNPs (12). Interested in addressing the effect of 5-fluorouridine (5FU) incorporation on snRNP assembly, he demonstrated that the presence of 5FU in U2 snRNA prevented the formation of salt-resistant complexes when analyzed by cesium-sulfate buoyant density gradient centrifugation. This data, coupled with previous data demonstrating the inhibition of pseudouridylation on 5FU-containing snRNA, was suggestive of a role for pseudouridylation in U2 snRNP biogenesis (13).
Reconstitution systems were also developed that involved the specific depletion of one of the endogenous spliceosomal snR-NAs followed by the supplementation of that respective snRNA synthesized in vitro (14-28). As the in vitro synthesized snRNAs are void of modifications, the ability (or lack thereof) of the RNA to reconstitute pre-mRNA splicing would indicate whether the modifications were required for pre-mRNA splicing. Surprisingly, in vitro synthesized U1 (15), U4 (16), and U6 (21-24) were shown to be effective in reconstituting pre-mRNA splicing in mammalian extracts depleted of the respective endogenous snRNA. Furthermore, in vitro synthesized U6 snRNA was successful in rescuing pre-mRNA splicing in U6-depleted cell-free extracts prepared from Ascaris embryos (25, 26). In addition, in vitro synthesized U2 and U6 were capable of restoring pre-mRNA splicing in yeast cell extracts (14, 18-20, 28). These initial data created rather a conundrum, as a lack of U snRNP assembly would have been expected to negatively impact pre-mRNA splicing. However, it is important to note that none of the above-mentioned reconstitution experiments monitored modification of the in vitro transcribed snRNA. In fact, based on the results of others (D. S. McPheeters, personal communication) and our own (unpublished data), in vitro transcribed U2 snRNA is readily modified when added to yeast splicing extracts. Thus, whether modified nucleotides contributed to pre-mRNA splicing was still an open question.
In 1995, the Luhrmann group studied the function of spliceosomal snRNAs in a HeLa reconstitution system (17). Interestingly, while in vitro synthesized U2 snRNA completely failed to reconstitute pre-mRNA splicing in U2 snRNA-depleted extracts, cellularly derived U2 snRNA successfully restored splicing, suggesting that the modified nucleotides in U2 snRNA might indeed play a role in splicing. Further analysis confirmed that U2 snRNA was not pseudouridylated in the extracts.
2.2. Definitive Evidence That Spliceosomal snRNA Modifications Are Required for snRNP Assembly and Pre-mRNA Splicing
In an effort to clarify the effect of modifications in pre-mRNA splicing, Yu et al. examined U2 snRNP biogenesis and spliceosome assembly in Xenopus oocytes (29). First, through the injection of an antisense U2 DNA oligonucleotide, endogenous U2 snRNA was depleted from Xenopus oocytes. The U2-depleted oocytes were then supplemented with in vitro synthesized U2 snRNA and incubated for a short period. Under these conditions, U2 modifications largely did not occur. Radiolabeled adenovirus pre-mRNA was then injected, and pre-mRNA splicing and spliceosome assembly were assayed using denaturing gel electrophoresis and native gel electrophoresis, respectively (29). Interestingly, unmodified or hypomodified U2 snRNA was unable to support splicing. Detailed analyses showed that such U2 snRNA failed to participate in the assembly of any of the higher-order splicing complexes (i.e., complexes A, B, and C). Anti-snRNP immunoprecipitation and glycerol gradient sedimentation were used to further examine U2 snRNP assembly. Experimental data indicated that U2 snRNA lacking modifications could only form a 12S snRNP; no significant 17S snRNP, which is a mature form of U2 snRNP, was observed. These results indicated that U2 snRNA modification is required for the transition from the 12S snRNP to a functional 17S snRNP. Interestingly, however, it was observed that upon longer reconstitution periods, in vitro synthesized U2 snRNA was assembled into primarily a 17S particle, thus regaining its ability to support splicing. Analysis of the modification status of U2 snRNA over time indicated that in vitro synthesized U2 snRNA was efficiently modified in the Xenopus oocyte reconstitution system following prolonged incubations. Thus, a nice correlation between the modification status of U2 snRNA and its ability to participate in pre-mRNA splicing was established. In addition, through the construction of chimeric U2 snRNAs in which various regions of U2 snRNA contained no modifications, Yu et al. were able to demonstrate that modifications within the 27.5′-most nucleotides were required for pre-mRNA splicing (29). The functional importance of these modified nucleotides has been later confirmed by the Luhrmann group (30). More recently, using 5FU-containing U2 snRNA, a U2-specific pseudouridylation inhibitor, Zhao and Yu further dissected the importance of U2 snRNA modifications. The experimental results revealed a requirement for pseudouridylation within the branch site recognition region for efficient pre-mRNA splicing and spliceosome assembly in Xenopus oocytes (31). The earlier studies failed to identify these pseudouridines (downstream from the 27.5′-most nucleotides) as important due to the fact that in the Xenopus reconstitution system these nucleotides are modified extremely fast, making it difficult to examine their importance under the particular conditions without specific pseudouridylation inhibitors.
2.3. Genetic Dissection of Spliceosomal snRNA Modification in Saccharomyces cerevisiae
The pre-mRNA splicing machinery of S. cerevisiae is one of the most extensively studied systems with regard to spliceosome assembly and pre-mRNA splicing catalysis. This fact coupled with the awesome power of yeast genetics makes S. cerevisiae an ideal organism for dissecting the function of posttranscriptionally modified nucleotides present in the spliceosomal snRNAs. Surprisingly, to date no 2′-O-methylations have been observed in the spliceosomal snRNAs of S. cerevisiae (our unpublished data). However, in 1999 Massenet et al. identified six pseudouridines, five of which are conserved in mammals (32). There are two within U1, three in the U2 branch site recognition region, and one within U5. The genes which encode the enzymes responsible for U2 snRNA pseudouridylation have all been identified (Pus1p, ψ44; Pus7p, ψ35; ψ42 is catalyzed by the box H/ACA RNP snR81) (32-34).
To investigate the role of ψ35 in U2 snRNA, Ma et al. constructed a deletion of pus7. Interestingly, while viable, the pus7-Δ displayed reduced fitness when grown in media containing high concentrations of NaCl, or when in competition with a wild-type strain (34). To further clarify the role of ψ35, Yang et al. screened the pus7-Δ strain against a collection of U2 snRNA mutants (35). Semi-quantitative PCR indicated an accumulation of pre-mRNA in the pus7-Δ strain when in combination with U40G or U40Δ mutations in U2 snRNA. Pre-mRNA accumulation varied depending on the transcript analyzed, with fold increases ranging from 0 to >10.
A detailed analysis of the other yeast spliceosomal snRNA pseudouridylations has yet to be described. However, the fact that they are conserved throughout evolution is suggestive of function. It will be interesting to see whether the pseudouridylations present within yeast spliceosomal snRNAs function synergistically.
3. Ribosomal RNA Modifications
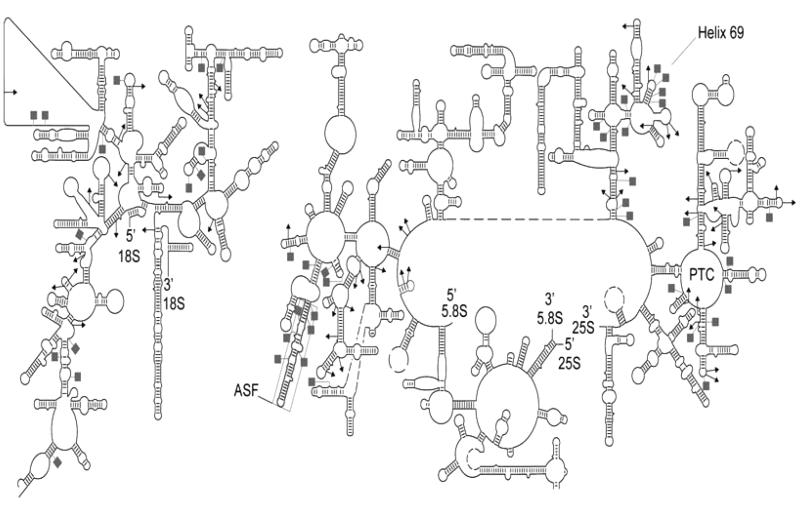
Ribosomal RNAs (rRNAs), integral components of the ribosome, have evolutionary conserved structures important for the process of translation despite some sequence variation among organisms. A frequent trait of rRNAs is the presence of modified nucleotides (Fig. 1.3). Similar to the spliceosomal snRNAs, the most common modifications in rRNAs are pseudouridylation and 2′-O-methylation.
Fig. 1.3.
Secondary structure depiction of S. cerevisiae cytoplasmic ribosome (modified from (37)). The 18S rRNA is depicted on the left, while the 25S–5.8S is shown on the right. The PTC, ASF, and Helix 69 are apparent in the structure. Modification sites in the rRNA species are illustrated as follows: pseudouridylation sites (ψ) – gray squares and 2′-O-methylation sites (Nm) – black triangles.
Several lines of evidence suggest that rRNA modifications contribute to ribosome function. For instance, while rRNAs do not exhibit a significant difference in size or number of ribosomal proteins from prokaryotic to lower and higher eukaryotic organisms, the number of modifications increases along with the evolutionary complexity of the organism (36). Furthermore, three-dimensional (3D) maps deduced for Escherichia coli and cytoplasmic S. cerevisiae ribosomes indicate the clustering of modifications in functionally important regions of the ribosome, such as the peptidyl transferase center (PTC), the A, P, and E sites of tRNA and mRNA binding, the polypeptide exit tunnel, and sites of subunit–subunit interaction (37). Together, these data strongly suggest that rRNA posttranscriptional modifications influence ribosome function.
3.1. Global Analysis of rRNA Modifications
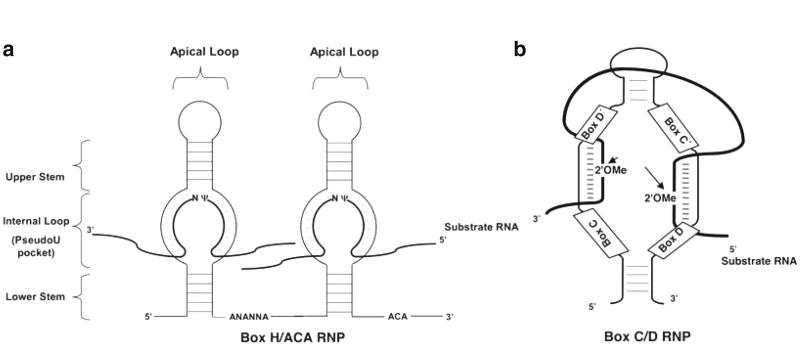
It has long been known that archaeal and eukaryotic rRNA modifications, including 2′-O-methylation and pseudouridylation, are catalyzed by box C/D RNPs and box H/ACA RNPs, respectively. Specifically, the box C/D and H/ACA snoRNAs (Fig. 1.4) are each associated with four evolutionary conserved core proteins of which only one is considered to have the catalytic activity required for the modification reaction. In yeast, Nop1p (fibrillarin in human) is the 2′-O-methyltransferase associated with box C/D RNPs, while Cbf5p (dyskerin in humans, NAP57 in rat, and Nop60B in Drosophila) is the pseudouridine synthase associated with box H/ACA RNPs. Using this information, it became possible to specifically disrupt the catalytic center of the enzyme, thus permitting an examination of the effect of a global depletion of either 2′-O-methylations (Nm) or pseudouridines (ψ) on ribosome biogenesis and function.
Fig. 1.4.
Schematic depiction of box H/ACA and C/D RNAs. (a) Minimal components of a eukaryotic pseudouridylation guide H/ACA. The RNA adapts a Hairpin-Hinge-Hairpin-Tail structure. Present within the hinge region is the box H, the ACA motif typically lies three nucleotides from the 3′-end of the RNA. Pseudouridylation is targeted to substrate RNAs by complementary basepairing interactions between the internal loop (pseudouridylation pocket) and nucleotides flanking the target uridine. (b) Secondary structure of a box C/D RNA. Boxes C, D, C′, and D′ are shown. The 2′-O-ME represents the target 2′-O-methylation site that is always the fifth nucleotide from box D or D′.
To assess the function of Nop1p in ribosome biogenesis, Tollervey et al. constructed a series of mutations within the coding sequence of NOP1 (38). S. cerevisiae expressing Nop1p carrying the double mutation A175V and P219S were defective both in growth and in rRNA methylation at high temperature (36°C). Surprisingly, although rRNA/pre-rRNA methylation was severely inhibited, there was only a slight impairment of 18S and 25S synthesis. However, the production of mature 60S ribosomal subunits was delayed when monitored by sucrose gradient centrifugation. Consistently, a similar mild processing defect was observed when yeast cells were exposed to the methylation inhibitor ethionine. These results suggested that rRNA methylation played only a minor role in the processing of pre-rRNA, but might contribute significantly to the assembly of ribosome subunits (or the combination of the two led to temperature-sensitive phenotype).
In 1999, point mutations within the pseudouridine synthase Cbf5p permitted an assessment of the role of rRNA pseudouridylation in ribosome biogenesis (39). Mutation of the universally conserved catalytic aspartate residue (D95 in S. cerevisiae) resulted in no detectable pseudouridine formation in 18S and 25S rRNAs. S. cerevisiae strains expressing catalytically deficient Cbf5p exhibited severe growth defects, and contained a reduction in the level of both 18S and 25S rRNAs. In addition, sucrose gradient analysis of ribosome assembly indicated a strong defect in both 40S and 60S subunit biogenesis. These data are strongly indicative of a role for rRNA pseudouridylation in rRNA processing and subunit assembly.
3.2. Functional Analysis of rRNA Modifications by Deleting Individual snoRNAs
The generation of 3D maps for the yeast ribosome made possible the realization that modifications cluster within 3D space (37). This information, coupled with the fact that the box H/ACA RNAs responsible for directing the pseudouridylation of virtually all rRNA pseudouridine residues have been identified, permitted the use of genetics in addressing the functional role pseudouridylation plays in specific regions of the ribosome.
In 2003, the Fournier group systematically analyzed the importance of pseudouridylation within the peptidyl transferase center (PTC) by deleting the snoRNA genes responsible for guiding pseudouridylation within the PTC (40). Located within the PTC are six ψs which are guided by five box H/ACA snoRNAs (snR34 for ψ2822. and ψ2876; snR46 for ψ2861; snR10 for ψ2919; snR37 for ψ2940; and snR42 for ψ2971). Interestingly, deletion of any snoRNA alone (with the exception of snR10) resulted in only minor if any phenotype; however, deletion of several or all six simultaneously resulted in a significant (20–45%) decrease in translation rate when measured by in vivo [35S] methionine incorporation. It should be noted that snR10 not only directs formation of ψ2919 within the A loop close to tRNA binding, but it is also involved in the processing of pre-rRNA. Surprisingly, expression of a mutant, in which a point mutation in snR10 disrupts its ability to guide modification while maintaining its ability to process pre-rRNA, displayed a 30% lower rate of translation as well as altered polysome profiles (40).
As none of the modifications are within 13 Å of the proposed site of peptide bond formation, it is unlikely that the chemistry of protein synthesis is affected. However, it is reasonable to expect that these modifications may modulate other critical events taking place during translation, such as tRNA binding or translocation through the ribosome. In line with this notion, G2918, the nucleotide adjacent to ψ2919, is known to form a Watson–Crick basepair with C75 in the CCA end of tRNA when in the A site (41). Given the stabilizing effect of ψ on neighboring nucleotides (see below), it is possible that ψ2919 modifies A site function by promoting stable interactions between nearby nucleotides. Indeed, interactions between nucleotides adjacent to ψ2919 have been shown to affect translation fidelity in E. coli (42).
The region directly above the A site is referred to as the A site finger (ASF) region and contains the richest concentration of ψ within rRNA (8–10 ψs depending on the organism; 10 in S. cerevisiae) (43). The ASF is believed to function during the translocation of tRNA through the ribosome (44). Interestingly, depletion of various combinations of ψ from within this region in S. cerevisiae significantly impacts ribosome function, including decreased amounts of LSU, impaired polysome formation, and reduced translation rate (43). Thus, similar to ψs near the PTC, it is probable that ψs within the ASF function synergistically to stabilize interactions between neighboring nucleotides as well as to optimize rRNA structure.
The large and small subunits of the ribosome are joined by a series of bridges, which are conserved throughout evolution. In addition to functioning as “molecular glue,” holding the 40S and 60S subunits together during protein synthesis, these bridges have been proposed to have additional functions including the transmission of signals from functional centers of the subunits to the modulation of ribosome–tRNA interactions during tRNA translocation (45-47). As with other regions of the ribosome which have well-characterized functions, the intersubunit bridges possess a wealth of posttranscriptionally modified nucleotides (37).
One such structure in yeast, helix 69 (H69), contains a rich cluster of pseudouridines, which are evolutionarily conserved from E. coli to human (three ψ in E. coli, four ψ and one Nm in yeast, and five ψ and one Nm in human). H69 interacts with both the A and P site tRNAs, both ribosomal subunits, and H44 (in the small subunit) which is part of the decoding center (48). Two of these modifications ψ2258 and ψ2260 are the most conserved pseudouridines from E. coli to human (37, 49). These modifications are catalyzed by RluD in E. coli (50, 51), hU19 in humans (52), and snr191 in yeast (53). Interestingly, a single deletion of snR91 led to a slight growth disadvantage in yeast (53). Liang et al. systemically addressed the function of modifications within H69 of S. cerevisiae by constructing deletion strains lacking snoRNAs that target this region for modification. As has been observed with the modifications in other regions, loss of any single or two modifications resulted in no apparent growth phenotype or translational impairment under standard conditions. However, loss of two pseudouridines at positions ψ2264 and ψ2266 results in increased susceptibility to the aminoglycoside neomycin. Neomycin resistance was further decreased by loss of the Am2256 modification. These data clearly suggest that these modifications influence ribosome structure. Furthermore, cells lacking three or more modifications from this region display numerous translation-related phenotypes including reduced translation rates (ranging from 25 to 60% depending on modifications disrupted), reduced translational fidelity, and reduced amounts of rRNA due to faster turnover (54). These data clearly demonstrate that posttranscriptional modifications can affect both ribosome synthesis and function, and add to the notion that posttranscriptionally modified nucleotides function in concert to manipulate ribosome performance.
While most research regarding rRNA modification has been focused on pseudouridylation, attempts have been made to address the role of individual rRNA 2′-O-methylations. However, single deletions of box C/D snoRNAs targeting upward of 51 2′-O-methylations have been made in yeast and have not shown an effect on either ribosome function or cell growth (55). More recently, Esguerra et al. screened 20 individual box C/D snoRNA deletion strains for phenotypic consequences in response to various environmental challenges (56). Surprisingly, all 20 individual deletion strains displayed phenotypes, primarily during the phase of adaptation to the environmental stress. Although this data is indicative of a function for individual modifications, further detailed molecular and biochemical dissection is required to understand the nature of these phenotypes. Overall, deletions of individual snoRNAs targeting rRNA within a specific 3D region of the ribosome, an approach that has been quite successful in eliciting the function of pseudouridines, has yet to be attempted for 2′-O-methylation.
3.3. rRNA Modification and Translational Recoding
During translation elongation, the mRNA is translated on the ribosome following strict rules of decoding. Although this process is thought to be very accurate, there has been an accumulation of data over the past 15 years, which indicate that the frequency of unconventional decoding, or “recoding,” can reach as high as 40% (57, 58). Such phenomena may include events such as readthrough of stop codons, frameshifting, and ribosomal hopping (59-61)
Recently, several assays have been developed to study the role of rRNA modifications in translational fidelity. For instance, in vivo bicistronic dual-luciferase reporters have been used to quantitatively measure changes in programmed −1 and programmed +1 ribosomal frameshifting (PRF), suppression of non-sense codons, and fidelity of aminoacylated-tRNA (aa-tRNA) selection (62, 63). Interestingly, yeast deficient in both the box C/D snoRNA snR52 and the 2′-O-methyltransferase Sbp1p displayed significant defects in –1 PRF, while all snoRNA deletion strains examined thus far have failed to allow the detection of any defects in +1 PRF. Remarkably, however, several snoRNA deletion strains were hyperaccurate in their ability to recognize translation termination codons. Specifically snR37Δ, snR10Δ, and a double mutant spb1DA/snR52Δ strains were all hyperaccurate at mediating translation termination (63).
More recently, we have characterized translation fidelity and recoding in the cbf5D95A strain where ribosomal RNAs are void of pseudouridine (see above) (Karijolich, Kantartzis, and Yu, unpublished data). Interestingly, the efficiency of programmed −1 and +1 frameshifting, as well as cap-independent translation is severely impaired. Rather surprisingly, yeasts lacking detectable pseudouridylation were several fold more efficient at translation termination than their wild-type counterpart. Along the same line, it has been demonstrated that cap-independent translation is severely impaired in hypomorphic dyskerin mice, which have a reduced level of rRNA pseudouridylation (64, 65).
Although a fairly large amount of functional data has been gathered regarding rRNA modifications and translation fidelity, a clear and concise picture of how these modifications function is still lacking. Nonetheless, it is clear that the majority of these modifications do not affect cell viability or ribosome performance when deleted alone, at least under “normal” laboratory conditions. Importantly, the data accumulated thus far suggests that these modifications function synergistically to modulate the structure and function of the ribosome.
4. Cytotoxicity Associated with 5FU Is a Result of Inhibition of Pseudouridylation
5FU is a chemotherapeutic agent that is prescribed for a variety of malignancies including bowel, breast, stomach, and esophagus. Although 5FU has been in use for nearly half a century, the mechanism behind its therapeutic effect has yet to be established (66-68). Initially it was hypothesized that 5FU affects DNA metabolism. 5FU, when converted into 5-fluorodeoxyuridine monophosphate (5FdUMP), inhibits the activity of thymidylate synthase, which is required for the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP). Lack of synthesis of dTMP, in turn, results in an inhibitory effect on the production of its downstream product dTTP and hence affecting DNA synthesis. Consequently, dUMP accumulates, resulting in elevated synthesis of its downstream product dUTP and the incorporation of dUTP into DNA, thus resulting in DNA damage. Paradoxically, however, when 5FU-exposed cells are treated with thymidine, which can be converted into dTMP through the action of thymidine kinase (a pathway that is independent of the thymidylate synthase pathway), 5FU-mediated cytotoxic and apoptotic effects remain, suggesting that DNA metabolism is not the primary target of 5FU.
Given that 5FU can be readily converted into 5-fluorouridine triphosphate (5FUTP), a ribonucleotide analog that can be incorporated into RNAs, it has been proposed that 5FU may directly affect RNA metabolism. Indeed, past research has shown that 5FU is readily incorporated into the long single pre-rRNA transcript that is processed to become the 18S, 5.8S, and 28S rRNAs. In this regard, it has been reported that 5FU-treated cells may have defects in rRNA processing (67, 69-72). However, whether this defect is a direct result from 5FU incorporation remains largely unclear.
More recently, data from other labs, as well as our own, now suggest that the cytotoxic effect of 5FU is mainly RNA based and is a result of the inhibition of pseudouridylation. For instance, 5FU-treated HeLa cells show a dramatic accumulation of pre-mRNA, which is accompanied by a significant reduction in the amount of pseudouridine present in U2 snRNA (73). Furthermore, in contrast to the fact that U2 snRNA isolated from uracil-treated HeLa cells efficiently reconstitute pre-mRNA splicing in Xenopus oocytes, U2 snRNA purified from 5FU-treated HeLa cells failed to reconstitute pre-mRNA splicing (73). Further demonstrating a role for the process of pseudouridylation in the cytotoxic effects of 5FU, Hoskins and Butler showed that a mutation in the catalytic subunit of Cbf5p, the pseudouridine synthase-associated box H/ACA RNPs (see above), suppresses the 5FU hypersensitivity of an yeast rrp6-Δ strain (74). Given that ribosomal RNAs are modified almost exclusively by box H/ACA RNPs in yeast, it would be of interest to see whether rRNA pseudouridylation is affected in the 5FU-treated rrp6-Δ strain. Moreover, the effects of 5FU on translation fidelity and recoding are unknown and warrant investigation.
5. Biophysical Contribution of Posttranscriptionally Modified Nucleotides
To date the data is clear, posttranscriptional modifications within the spliceosomal snRNAs and rRNA are important for pre-mRNA splicing and protein synthesis, respectively. However, the mechanism by which these modifications exert the effect are only beginning to be unraveled. It is well established that both 2′-O-methylation and pseudouridylation have distinct chemical properties from their unmodified counterparts. These distinct chemical properties have the potential to impact numerous aspects of the modified RNA, including structure, thermal stability, and biochemical interactions (75). In each case, the structural, thermodynamic, and biochemical contributions depend on the structural context and can extend beyond the site of modification. In this regard, 1H NMR, UV, and CD spectroscopy have demonstrated that short RNA fragments containing pseudouridine are significantly more stable than if the same RNA contained uridine (76). Conformation stabilization appears to be an intrinsic property of pseudouridine at the nucleotide level, and is mediated by both an increase in base stacking and the ability to coordinate a water molecule through the extra hydrogen bond present (76, 77). Similarly, data also suggest that 2′-O-methylation is capable of leading to increased stability in RNA conformations. For instance, 2′-O-methylation alters the hydration sphere around the oxygen resulting in the blockage of sugar edge interactions (78-80). In addition, the methyl group alters the ability of the ribose to engage in hydrogen bonding. Furthermore, it also plays a role in protecting the RNA from hydrolysis by alkali and nucleases. In addition, there is a direct correlation between an organism’s optimal growth temperature and the number of 2′-O-methylations present in rRNA (81), as was reported for tRNA (75, 82, 83), suggesting that 2′-O-methylation may indeed result in increased conformational stability.
In recent years, significant efforts have been made to understand the molecular mechanism by which U2 snRNA pseudouridylation contributes to pre-mRNA splicing. In this regard, the crystal structure of a self-complementary RNA designed to mimic the S. cerevisiae U2 snRNA–branchpoint interaction was determined in the absence of pseudouridine (84). Interestingly, the adenosines 5′ of the expected adenosine were bulged out. Subsequently, the Greebaum group determined solution structures of the U2 snRNA–branchpoint interaction either in the presence or absence of pseudouridine (85, 86). NMR data coupled with 2-aminopurine fluorescence titration data indicated that the presence of the pseudouridine was required for the extrahelical positioning of the expected branchpoint adenosine. This has recently been revalidated by Lin and Kielkopf who determined the crystal structure of the U2 snRNA–branchpoint interaction in the presence of pseudouridine (87). In support of the biophysical data, Valadkhan and Manley have shown that the change of a single uridine in the branch site recognition region of U2 snRNA to pseudouridine greatly enhances the production of X-RNA, a product generated by a splicing-related branching reaction in a cell- and protein-free system (88, 89).
6. Concluding Remarks
In the past decade or so, research into the functionality of posttranscriptional modifications, in particular 2′-O-methylation and pseudouridylation, has quickened. While the data is quite clear that modified nucleotides are important for fully functional spliceosomes and ribosomes, exactly how the modifications exert their effects is unclear. Furthermore, many fundamental questions remain unexplored. For instance, are posttranscriptional modifications reversible? Do modifications have a role during the catalytic phase of pre-mRNA splicing and/or protein synthesis? To answer this question, a detailed structural mechanistic analysis seems to be necessary. Furthermore, there are a number of modified residues within the spliceosomal U snRNAs and rRNAs that have never been tested for a function. With so many unanswered questions, the field of RNA modification is sure to see some exciting results in the near future.
Acknowledgments
We would like to thank the members of the Yu lab for insightful discussions. Our work was supported by grant GM62937 (to Yi-Tao Yu) from the National Institute of Health. John Karijolich was supported by a NIH Institutional Ruth L. Kirschstein National Research Service Award GM068411.
References
- 1.Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Yu YT, Scharl EC, Smith CM, Steitz JA. The RNA World. 2nd Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 487–524. [Google Scholar]
- 5.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Cohn WE, Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature. 1951;167:483–484. [Google Scholar]
- 7.Hodnett JL, Busch H. Isolation and characterization of uridylic acid-rich 7 S ribonucleic acid of rat liver nuclei. J Biol Chem. 1968;243:6334–6342. [PubMed] [Google Scholar]
- 8.Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968;38:289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, Busch H. Studies on the nuclear and nucleolar ribonucleic acid of regenerating rat liver. J Biol Chem. 1965;240:3960–3966. [PubMed] [Google Scholar]
- 10.Lerner MR, Steitz JA. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy R, Busch H. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag Press; Heidelberg: 1988. pp. 1–37. [Google Scholar]
- 12.Patton JR. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry. 1993;32:8939–8944. doi: 10.1021/bi00085a027. [DOI] [PubMed] [Google Scholar]
- 13.Patton JR. Multiple pseudouridine synthase activities for small nuclear RNAs. Biochem J. 1993;290:595–600. doi: 10.1042/bj2900595. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McPheeters DS, Fabrizio P, Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989;3:2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- 15.Will CL, Rumpler S, Klein Gunnewiek J, van Venrooij WJ, Luhrmann R. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucl Acids Res. 1996;24:4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wersig C, Bindereif A. Reconstitution of functional mammalian U4 small nuclear ribonucleoprotein: Sm protein binding is not essential for splicing in vitro. Mol Cell Biol. 1992;12:1460–1468. doi: 10.1128/mcb.12.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segault V, Will CL, Sproat BS, Luhrmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabrizio P, McPheeters DS, Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 19.Fabrizio P, Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- 20.Fabrizio P, Abelson J. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucl Acids Res. 1992;20:3659–3664. doi: 10.1093/nar/20.14.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolff T, Bindereif A. Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J. 1992;11:345–359. doi: 10.1002/j.1460-2075.1992.tb05057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff T, Menssen R, Hammel J, Bindereif A. Splicing function of mammalian U6 small nuclear RNA: conserved positions in central domain and helix I are essential during the first and second step of pre-mRNA splicing. Proc Natl Acad Sci USA. 1994;91:903–907. doi: 10.1073/pnas.91.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff T, Bindereif A. Mutational analysis of human U6 RNA: stabilizing the intramolecular helix blocks the spliceosomal assembly pathway. Biochim Biophys Acta. 1995;1263:39–44. doi: 10.1016/0167-4781(95)00085-u. [DOI] [PubMed] [Google Scholar]
- 24.Wolff T, Bindereif A. Conformational changes of U6 RNA during the spliceosome cycle: an intramolecular helix is essential both for initiating the U4–U6 interaction and for the first step of slicing. Genes Dev. 1993;7:1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- 25.Yu YT, Maroney PA, Nilsen TW. Functional reconstitution of U6 snRNA in nematode cis- and trans-splicing: U6 can serve as both a branch acceptor and a 5′ exon. Cell. 1993;75:1049–1059. doi: 10.1016/0092-8674(93)90315-h. [DOI] [PubMed] [Google Scholar]
- 26.Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- 27.Pan ZQ, Prives C. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 1989;3:1887–1898. doi: 10.1101/gad.3.12a.1887. [DOI] [PubMed] [Google Scholar]
- 28.McPheeters DS, Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- 29.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massenet S, Branlant C. A limited number of pseudouridine residues in the human atac spliceosomal UsnRNAs as compared to human major spliceosomal UsnR-NAs. RNA. 1999;5:1495–1503. doi: 10.1017/s1355838299991537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22:1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280:6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 36.Lapeyre B. Conserved Ribosomal RNA Modification and Their Putative Roles in Ribosome Biogenesis and Translation. Springer; Heidelberg/Berlin: 2005. [Google Scholar]
- 37.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 38.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 39.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 41.Kim DF, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 42.O’Connor M, Dahlberg AE. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc Natl Acad Sci USA. 1993;90:9214–9218. doi: 10.1073/pnas.90.19.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J Biol Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J Biol Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 45.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11:91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 46.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodnina MV, Daviter T, Gromadski K, Wintermeyer W. Structural dynamics of ribosomal RNA during decoding on the ribosome. Biochimie. 2002;84:745–754. doi: 10.1016/s0300-9084(02)01409-8. [DOI] [PubMed] [Google Scholar]
- 48.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 49.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 50.Raychaudhuri S, Conrad J, Hall BG, Ofengand J. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA. 1998;4:1407–1417. doi: 10.1017/s1355838298981146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry. 1998;37:15951–15957. doi: 10.1021/bi981002n. [DOI] [PubMed] [Google Scholar]
- 52.Bortolin ML, Kiss T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 53.Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang X-H, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 56.Esguerra J, Warringer J, Blomberg A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA. 2008;14:649–656. doi: 10.1261/rna.845808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 58.Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- 59.Baranov PV, Gurvich OL, Hammer AW, Gesteland RF, Atkins JF. Recode 2003. Nucl Acids Res. 2003;31:87–89. doi: 10.1093/nar/gkg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 61.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 62.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baxter-Roshek JLP, Alexey N, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2006;2:174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 65.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 66.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 67.Ghoshal K, Jacob ST. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem Pharmacol. 1997;53:1569–1575. doi: 10.1016/s0006-2952(97)00040-3. [DOI] [PubMed] [Google Scholar]
- 68.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 69.Greenhalgh DA, Parish JH. Effects of 5-fluorouracil on cytotoxicity and RNA metabolism in human colonic carcinoma cells. Cancer Chemother Pharmacol. 1989;25:37–44. doi: 10.1007/BF00694336. [DOI] [PubMed] [Google Scholar]
- 70.Herrick D, Kufe DW. Lethality associated with incorporation of 5-fluorouracil into preribosomal RNA. Mol Pharmacol. 1984;26:135–140. [PubMed] [Google Scholar]
- 71.Cory JG, Breland JC, Carter GL. Effect of 5-fluorouracil on RNA metabolism in Novikoff hepatoma cells. Cancer Res. 1979;39:4905–4913. [PubMed] [Google Scholar]
- 72.Greenhalgh DA, Parish JH. Effect of 5-fluorouracil combination therapy on RNA processing in human colonic carcinoma cells. Br J Cancer. 1990;61:415–419. doi: 10.1038/bjc.1990.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao X, Yu YT. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucl Acids Res. 2007;35:550–558. doi: 10.1093/nar/gkl1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoskins J, Butler JS. RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p. Genetics. 2008;179:323–330. doi: 10.1534/genetics.107.082727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucl Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 76.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucl Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudouridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 78.Auffinger P, Westhof E. Rules governing the orientation of the 2′-hydroxyl group in RNA. J Mol Biol. 1997;274:54–63. doi: 10.1006/jmbi.1997.1370. [DOI] [PubMed] [Google Scholar]
- 79.Auffinger P, Westhof E. Hydration of RNA base pairs. J Biomol Struct Dyn. 1998;16:693–707. doi: 10.1080/07391102.1998.10508281. [DOI] [PubMed] [Google Scholar]
- 80.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucl Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agris PF, Koh H, Soll D. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys. 1973;154:277–282. doi: 10.1016/0003-9861(73)90058-1. [DOI] [PubMed] [Google Scholar]
- 83.Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. The role of post-transcriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry. 1994;33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 84.Berglund JA, Rosbash M, Schultz SC. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA. 2001;7:682–691. doi: 10.1017/s1355838201002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–845. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9:958–965. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 87.Lin Y, Kielkopf CL. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry. 2008;47:5503–5514. doi: 10.1021/bi7022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–707. doi: 10.1038/35099500. [DOI] [PubMed] [Google Scholar]
- 89.Valadkhan S, Manley JL. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9:892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]