Abstract
Proinflammatory cytokine TWEAK has now emerged as a key mediator of skeletal muscle-wasting in many catabolic conditions. However, the mechanisms by which TWEAK induces muscle proteolysis remain poorly understood. Here, we have investigated the role of ubiquitin-proteasome system, autophagy, and caspases in TWEAK-induced muscle wasting. Addition of TWEAK to C2C12 myotubes stimulated the ubiquitination of myosin heavy chain (MyHC) and augmented the expression of E3 ubiquitin ligase MuRF1. Pretreatment of myotubes with proteasome inhibitors MG132 or lactacystin or knockdown of MuRF1 by RNAi blocked the TWEAK-induced degradation of MyHC and myotube atrophy. TWEAK increased the expression of several autophagy-related molecules. Moreover, the inhibitors of autophagy improved the levels of MyHC in TWEAK-treated myotubes. TWEAK also increased activity of caspases in C2C12 myotubes. Pan-caspase or caspase 3 inhibitory peptide inhibited the TWEAK-induced loss of MyHC and myotube diameter. Our study demonstrates that nuclear factor-kappa B (NF-κB) transcription factor is essential for TWEAK-induced expression of MuRF1 and Beclin1. Furthermore, our results suggest that caspases contribute, at least in part, to the activation of NF-κB in response to TWEAK treatment. Collectively, the present study provides novel insight into the mechanisms of action of TWEAK in skeletal muscle.
Keywords: Skeletal muscle atrophy, TWEAK, Ubiquitin-proteasome system, NF-kappa B, MuRF1, Capsases, Autophagy
INTRODUCTION
Skeletal muscle-wasting or atrophy is a debilitating consequence of many catabolic conditions such as diabetes, cancer, sepsis, AIDS, heart failure, denervation, immobilization, and sarcopenia and is associated with reduced quality of life and increased mortality (Acharyya and Guttridge, 2007; Jackman and Kandarian, 2004; Kandarian and Stevenson, 2002). Earlier reports have suggested that inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α) and interleukin-1β (IL-1β) play a pivotal role in loss of skeletal muscle mass especially in chronic diseases (Argiles et al., 2005; Argiles et al., 2009; Späte and Schulze, 2004). We have recently reported that proinflammatory cytokine TNF-like weak inducer of apoptosis (TWEAK) is a major inducer of skeletal muscle-wasting both in vitro and in vivo (Dogra et al., 2007a). At equimolar concentrations, TWEAK causes more pronounced degradation of thick filament protein myosin heavy chain (MyHC) in cultured myotubes compared to TNF-α, a well-known muscle-wasting cytokine and structural homologue of TWEAK (Dogra et al., 2007a). Furthermore, we have shown that TWEAK is a major mediator of loss of skeletal muscle mass and function in response to denervation suggesting its role in physiological atrophy (Mittal et al., 2010). However, the mechanisms by which TWEAK induces loss of skeletal muscle mass remain poorly understood.
It has been consistently observed that skeletal muscle wasting involves the activation of the ATP-dependent ubiquitin-proteasome system which accelerates myofibril proteolysis (Solomon and Goldberg, 1996). Moreover, in almost all muscle-wasting conditions, the expression of two E3 ubiquitin ligases MuRF1 and MAFbx has been found to be highly up-regulated which label the target proteins for degradation by 26S proteasome (Bodine et al., 2001; Cao et al., 2005; Gomes et al., 2001). Their catabolic role in skeletal muscle has been supported by the findings that targeted deletion of MuRF1 or MAFbx rescues muscle atrophy in response to distinct stimuli including cancer cachexia, denervation, hind limb suspension, and inflammatory cytokines (Bodine et al., 2001; Glass, 2010; Gomes et al., 2001). Moreover, a few substrates which MuRF1 and MAFbx target for ubiquitination in atrophying skeletal muscle have now been identified (Clarke et al., 2007; Cohen et al., 2009; Kedar et al., 2004; Tintignac et al., 2005).
In addition to ubiquitin-proteasome system, the autophagy-lysosomal system has also been suggested to play important role in muscle proteolysis in atrophy conditions (Attaix and Bechet, 2007; Mammucari et al., 2007; Mammucari et al., 2008). In autophagy, a double membrane vesicle structure known as autophagosome sequesters a portion of cytoplasm including insoluble ubiquitinated proteins and fuses with lysosome where the contents of the vesicles are degraded (Levine and Kroemer, 2008; Mizushima et al., 2008). Autophagosome formation and elevated expression of several genes of this pathway have been observed in skeletal muscle of mice in response to starvation and denervation (Mammucari et al., 2007; Romanello et al., 2010; Zhao et al., 2007). Although the ubiquitin-proteasome and autophagy were initially considered as independent pathways serving distinct functions, accumulating evidence suggests that these two proteolytic systems can function in a cooperative manner to stimulate muscle wasting (Mammucari et al., 2007; Zhao et al., 2007).
Published reports suggest that skeletal muscle wasting may also require the activation of other proteolytic systems such as caspases and calpains upstream of ubiquitin-proteasome and autophagy systems (Lee et al., 2004; Tidball and Spencer, 2002). Caspase3 has been shown to cleave actinomyosin complexes in vitro, generating monomeric actin and actin fragments, including a characteristic 14-kDa fragment, which is subsequently degraded by the 26S proteasome (Du et al., 2004). In animal models of systemic catabolic conditions such as diabetes, caspase-3 activation and subsequent actin cleavage have been found to be initial critical steps in the loss of skeletal muscle mass (Du et al., 2004). Furthermore, Caspase3-null mice have been found to be somewhat resistant to denervation-induced skeletal muscle loss (Plant et al., 2009). However, it remains unknown whether TWEAK affects the activation of specific caspases and whether caspases are involved in TWEAK-induced muscle-wasting.
Nuclear factor-kappa B (NF-κB) is a proinflammatory transcription factor that regulates the expression of a plethora of genes including those involved in muscle proteolysis and fibrosis (Kumar et al., 2004). Increased activation of NF-κB has been consistently observed in skeletal muscle in different types of atrophy (Li et al., 2008). The role of NF-κB in muscle wasting is well-supported by the observations that constitutive activation of NF-κB causes severe muscle-wasting in mice (Cai et al., 2004) and specific inhibition of NF-κB has been found to block muscle atrophy in response to tumor growth, inflammatory cytokines, denervation, and unloading (Cai et al., 2004; Dogra et al., 2007a; Hunter and Kandarian, 2004; Mourkioti et al., 2006). NF-κB has been found to induce the expression of several components of ubiquitin-proteasome system including ubiquitin conjugating E2 enzyme UBcH2/E220k (Li et al., 2003) , MuRF1 (Cai et al., 2004) and the proteasome C3 subunit (Du et al., 2000). The activation of NF-κB may also promote muscle proteolysis through enhancing the activity of autophagy-lysosomal system (Criollo et al., 2010).
While the TWEAK has been found to activate NF-κB and ubiquitin-proteasome system (Dogra et al., 2007a; Mittal et al., 2010), it remains unknown whether TWEAK also affects the activity of autophagy-lysosomal system and caspases in skeletal muscle. Furthermore, a direct role for any of these proteolytic systems in TWEAK-induced muscle atrophy has not been yet determined. It also remains unknown whether there is a cross-talk between ubiquitin-proteasome, autophagy, and caspases to induce the degradation of muscle protein and whether NF-κB regulates the expression of some of the components of these proteolytic systems in TWEAK-treated muscle cells.
In the present study, we have investigated the role of ubiquitin-proteasome, autophagy, caspases, and NF-κB signaling system in TWEAK-induced muscle-wasting. Our results show that TWEAK stimulates the activation of ubiquitin-proteasome system, autophagy, and caspases in cultured myotubes. Specific inhibition of either of these proteolytic systems is sufficient to considerably reduce TWEAK-induced muscle wasting. Furthermore, our study demonstrates that NF-κB is required for the expression of MuRF1 and Beclin1, the critical components of ubiquitin-proteasome and autophagy systems, respectively.
MATERIALS AND METHODS
Materials
Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS), were obtained from Invitrogen (Carlsbad, CA). Recombinant TWEAK protein, caspase-3 inhibitor (i.e. Ac-DMQD-CHO), pan-caspase inhibitor (i.e. Z-VAD-FMK), and a control peptide (i.e. Z-FA-FMK) were purchased from R&D Systems. Protease inhibitor mixture, horse serum, β-actin antibody, and 3-methyladenine (3-MA) were from Sigma Chemical Co. (St. Louis, MO). Effectene transfection reagent was obtained from Qiagen (Valencia, CA). Antibodies against Beclin1, LC3B, MuRF1, caspase 3, and tubulin were from Cell Signaling Technology, Inc. (Beverly, MA). MF-20 antibody was obtained from the Developmental Studies Hybridoma Bank of the University of Iowa. Anti-ubiquitin, MG132, and lactacystin were purchased from Calbiochem (San Diego, CA). 32P-γ-ATP was from MP Biomedicals (Solon, OH). Consensus NF-κB oligonucleotides were purchased from Promega.
Cell Culture
C2C12 (a mouse myoblastic cell line) were obtained from American Type Culture Collection (Rockville, MD). The cells were grown in DMEM containing 10% FBS. Differentiation in C2C12 myoblast cultures was induced by replacing the medium with differentiation medium (2% horse serum in DMEM) (Dogra et al., 2006; Dogra et al., 2007b).
Short hairpin RNA (shRNA)
Validated plasmids encoding short hairpin RNA (shRNA) for murine IKKβ and Beclin1 and negative control were purchased from Sigma Chemical Company (Saint Louis, MO). All shRNA plasmids were amplified by transforming in E. Coli and purified using endotoxin-free maxi plasmid kit (Qiagen). The plasmids were introduced in C2C12 myoblasts using Effectene transfection reagents. Since shRNA plasmids contained puromycin resistant gene, the transfected cells were selected in the presence of 1.6 µg/ml puromycin (Sigma) for 72h. The knockdown of IKKβ and Beclin1 was confirmed by performing Western blot.
Adenoviral Vectors
MuRF1 shRNA adenoviral construct and a control shRNA viral construct targeting no known RNA sequence used for this study have been previously described (Clarke et al., 2007). Construction of control adenoviral vector (Ad.Control) and IκBαΔN-expressing adenoviral vector (Ad.IκBαΔN) has been reported in a previously published article (Dogra et al., 2007a). The purified viruses were used at a final concentration of 2 × 107 pfu/ml.
Transient Transfection and MuRF1 Reporter Gene Activity
Cells plated in 24-well tissue culture plates were transfected with MuRF1-Luc reporter plasmid (kindly provided by Dr. Stewart Lecker) using Effectene transfection reagent according to the protocol suggested by the manufacturer (Qiagen). Transfection efficiency was controlled by cotransfection of myoblasts with Renilla luciferase encoding plasmid pRL-TK (Promega). Specimens were processed for luciferase expression using a Dual luciferase assay system with reporter lysis buffer per the manufacturer’s instructions (Promega). Luciferase measurements were made using a luminometer (Berthold Detection Systems).
Stable Transfection
For stable transfection, C2C12 myoblasts were plated in 6-well tissue culture plate at a density of 2 × 105 cells/well. Next day, the cells were transfected with 0.4 µg/well control or specific shRNA plasmid along with pBabe-puro plasmid (in 1:10 ratio) using Effectene transfection reagent following a protocol suggested by the manufacturer (Qiagen). After 36h, the transfected cells were selected in the presence of 1.6 µg/ml puromycin (Sigma) for 6 days followed by addition of differentiation medium and incubation for additional 96h. The knockdown of specific gene was confirmed by performing Western blot.
Immunocytochemistry and Myotube Diameter Analyses
C2C12 myotubes were treated with TWEAK for 72h followed by immunostaining for MyHC protein using MF20 antibody as described (Dogra et al., 2007a). The pictures of the myotubes were taken using a digital camera and diameter of the myotubes was measured using NIS Elements BR 3.00 software (Nikon). Myotube diameter was quantified as follows: 10 fields were chosen randomly and 10 myotubes were measured per field. The average diameter per myotube was calculated as the mean of the five measurements taken along the length of the myotube.
Immunoprecipitation and Western Blotting
Immunoprecipitation and Western blot were performed following a standard method as described (Dogra et al., 2006; Dogra et al., 2007b). All antibodies were used at a dilution of 1:1000 for immunoblotting.
Electrophoretic Mobility Shift Assay (EMSA)
The activation of NF-κB transcription factor was measured by EMSA. A detailed procedure for the preparation of nuclear and cytoplasmic extracts and EMSA has been described previously (Dogra et al., 2006).
Quantitative Real-Time PCR (QRT-PCR)
Quantitative real time PCR was performed to measure the mRNA levels of different genes following a method as described (Dogra et al., 2006). The sequences of the primers used are as follows: Caspase 3: 5’-GGA GGC TGA CTT CCT GTA TGC TTA-3’ (forward), 5’-AAT TCC GTT GCC ACC TTC CT-3’ (reverse); Caspase 8: 5’-TGC TAT TGC TGA AGA ACT GGG C-3’ (forward), 5’-TTT CCC GCA GCC TCA GAA AT-3’(reverse). Sequence of all other primers has been described in our previously published article (Mittal et al., 2010). Data normalization was done using the endogenous control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin, and the normalized values were subjected to a 2−ΔΔCt formula to calculate the fold change between the control and experimental groups. The formula and its derivations were obtained from the ABI Prism 7900 sequence detection system user guide.
Caspase Activity Assay
Pan-caspase activity was determined using a homogeneous caspase assay kit (Roche) according to supplier’s instructions.
Statistical Analyses
All the experiments were repeated at least three times unless otherwise stated. Results are expressed as mean ± SD. The Student t test was used to compare quantitative data populations with normal distributions and equal variance. A value of p<0.01 was considered statistically significant unless otherwise specified.
RESULTS
Ubiquitin-proteasome system is involved in TWEAK-induced degradation of myosin heavy chain (MyHC) and atrophy in C2C12 myotubes
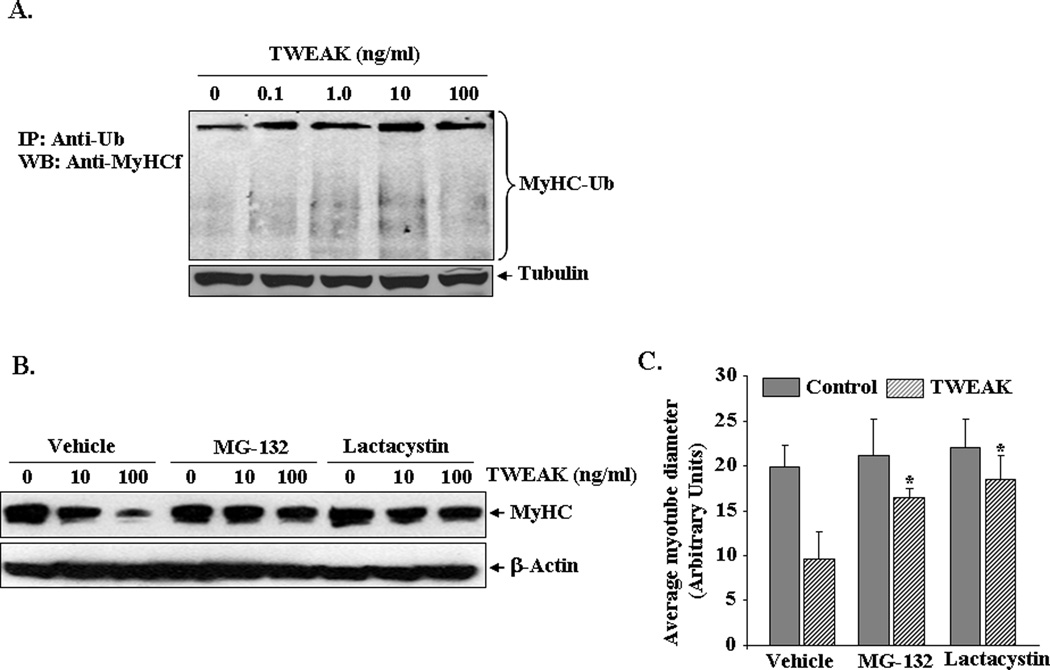
We have previously reported that TWEAK reduces the protein levels of MyHC and induces atrophy in cultured C2C12 myotubes (Dogra et al., 2007a). We first investigated whether the loss of MyHC in response to TWEAK occurs through the activation of ubiquitin-proteasome system. C2C12 myotubes were treated with increasing concentrations of TWEAK for 12h and cell extracts made were immunoprecipitated with anti-ubiquitin (Ub) followed by Western blot using anti-MyHC. As shown in Figure 1A, the mature form of MyHC was readily immunoprecipitated with ubiquitin antibody suggesting the potential role of ubiquitin-proteasome pathway in the turnover of MyHC. We also detected that levels associated with the mature form of MyHC are increased in response to TWEAK treatment (Figure 1A). Moreover, smaller MyHC-reactive fragments, which generally represent proteolyzed intermediates of this protein, were visible in TWEAK-treated samples (Acharyya et al., 2004). These results prompted us to investigate whether the inhibition of proteasome system would block the TWEAK-induced degradation of MyHC in C2C12 myotubes. C2C12 myotubes were pre-incubated with proteasome inhibitor MG132 or lactacystin for 3h followed by addition of TWEAK. After 72h, the cell extracts made were analyzed by Western blot using MF-20 (specific for MyHC-fast type) antibody. Interestingly, TWEAK-induced loss of MyHC was considerably reduced upon treatment of myotubes with MG132 or lactacystin (Figure 1B). As a measure of myotube atrophy, in a parallel experiment, we also studied the effects of MG132 and lactacystin on myotube diameter. Average myotube diameter was significantly higher in TWEAK-treated cultures containing MG132 or lactacystin compared to corresponding myotubes treated with vehicle (Figure 1C). These results indicate that TWEAK induced MyHC degradation and myotube atrophy involves ubiquitin-proteasome system.
FIGURE 1. Role of the ubiquitin-proteasome system in TWEAK-induced proteolysis and atrophy.
A) C2C12 myotubes were incubated with increasing amounts of TWEAK protein for 12h. Protein extracts were made and an equal amount of protein (400 µg/sample) was immunoprecipitated (IP) with ubiquitin antibody (4 µg/sample) overnight followed by Western blot (WB) for MyHC. Data presented here show that treatment of myotubes with TWEAK increases the ubiquitination of MyHC. The immunoblot for MyHC was intentionally overexposed to highlight differences in MyHC protein products coupled to ubiquitin (bracketed). Levels of an unrelated protein tubulin remained unchanged after treatment with TWEAK (lower panel). B). C2C12 myotubes were incubated with proteasome inhibitor MG132 (2µg/ml) or lactacystin (2µg/ml) for 3h followed by addition of TWEAK for 72h. The cell extracts made were analyzed by Western blot using MyHC antibody. Representative immunoblots from two independent experiments presented here show that MG132 and lactacystin reduces the TWEAK-induced degradation of MyHC in myotubes (upper panel). These treatments did not affect the levels of an unrelated protein β-actin (lower panel). C). C2C12 myotubes were preincubated with MG132 (2µg/ml) or lactacystin (2µg/ml) for 3h followed by treatment with TWEAK for 72h. The cultures were then immunostained using MF-20 antibody and average myotube diameter was calculated. Data from two independents experiments each done in triplicate presented here show that MG132 and lactacystin significantly prevented the loss of myotube diameter in TWEAK-treated myotubes. *p<0.01, values significantly different from corresponding myotubes treated with vehicle alone.
E3 ubiquitin ligase MuRF1 is essential for TWEAK-induced MyHC degradation and atrophy in cultured myotubes
E3 ubiquitin ligase MuRF1, an important component of ubiquitin-proteasome system in skeletal muscle, has been reported to catalyze the conjugation of ubiquitin to MyHC in dexamethasone-treated muscle cells (Clarke et al., 2007). We sought to determine whether TWEAK induces the degradation of MyHC through augmenting the expression of MuRF1. Consistent with our previously published report (Dogra et al., 2007a), treatment with TWEAK significantly increased the MuRF1 transcript levels in C2C12 myotubes (Figure 2A). Furthermore, Western blot analysis showed that TWEAK also augments the protein levels of MuRF1 in C2C12 myotubes (Figure 2A). Using MuRF1 promoter reporter construct, we also found that TWEAK enhanced the activity of MuRF1 promoter in myotubes (Figure 2B). To understand the role of MuRF1 in TWEAK-induced degradation of MyHC, C2C12 myotubes were transduced with adenoviral vector expressing either a scrambled short hairpin RNA (shRNA) or MuRF1 shRNA (Clarke et al., 2007). Suppression of MuRF1 levels upon transduction with MuRF1 shRNA adenoviral vector (Ad.MuRF1 shRNA) was confirmed by performing Western blot (Figure 2C). Interestingly, depletion of MuRF1 considerably increased the levels of MyHC in TWEAK-treated C2C12 myotubes (Figure 2C). Furthermore, myotube diameter was also found to be significantly improved in MuRF1 shRNA-expressing cultures compared to control shRNA-expressing cultures after 72h of addition of TWEAK (Figure 2D). These results suggest that TWEAK induces the degradation of MyHC and myotube atrophy through augmenting the expression of MuRF1.
FIGURE 2. Involvement of MuRF1 in TWEAK-induced myotube atrophy.
A). C2C12 myotubes were treated with 100 ng/ml TWEAK for 6h. The mRNA and protein levels of MuRF1 were measured by QRT-PCR and Western blot, respectively. Data presented here show that TWEAK increases transcript and protein levels of MuRF1 in myotubes. The levels of unrelated protein tubulin remained unaffected upon treatment with TWEAK. B). C2C12 myoblasts were transiently transfected with MuRF1-Luc reporter construct and Renilla plasmid (1:50 ratio) followed incubation in DM for 96h. The cells were then treated with indicated concentrations of TWEAK for 18h and luciferase activity in cell extracts was measured using Dual luciferase activity assay kit (Promega). Data presented here demonstrate that TWEAK significantly (*p<0.05) increases MuRF1 promoter activity in C2C12 myotubes. C). C2C12 myotubes were transduced with Ad.Control (Ad.Con) or Ad.MuRF1 shRNA expressing vectors (MOI, 1:200) for 48h. The cells were then treated with indicated concentrations of TWEAK. Representative immunoblots from two independent experiments presented here show that overexpression of MuRF1 shRNA drastically reduces the levels of MuRF1 (upper panel) and inhibited TWEAK-induced loss of MyHC (middle panel) without affecting the levels of tubulin (lower panel). Black line indicates that intervening lanes have been spliced out. D). Measurement of myotube diameter in control or MuRF1-expressing shRNA cultures 72h after TWEAK (100 ng/ml) treatment showed that knockdown of MuRF1 preserves myotube size. *p<0.05, values significantly different from TWEAK-treated control shRNA-expressing cultures.
TWEAK augments the expression of autophagy-related genes in myotubes
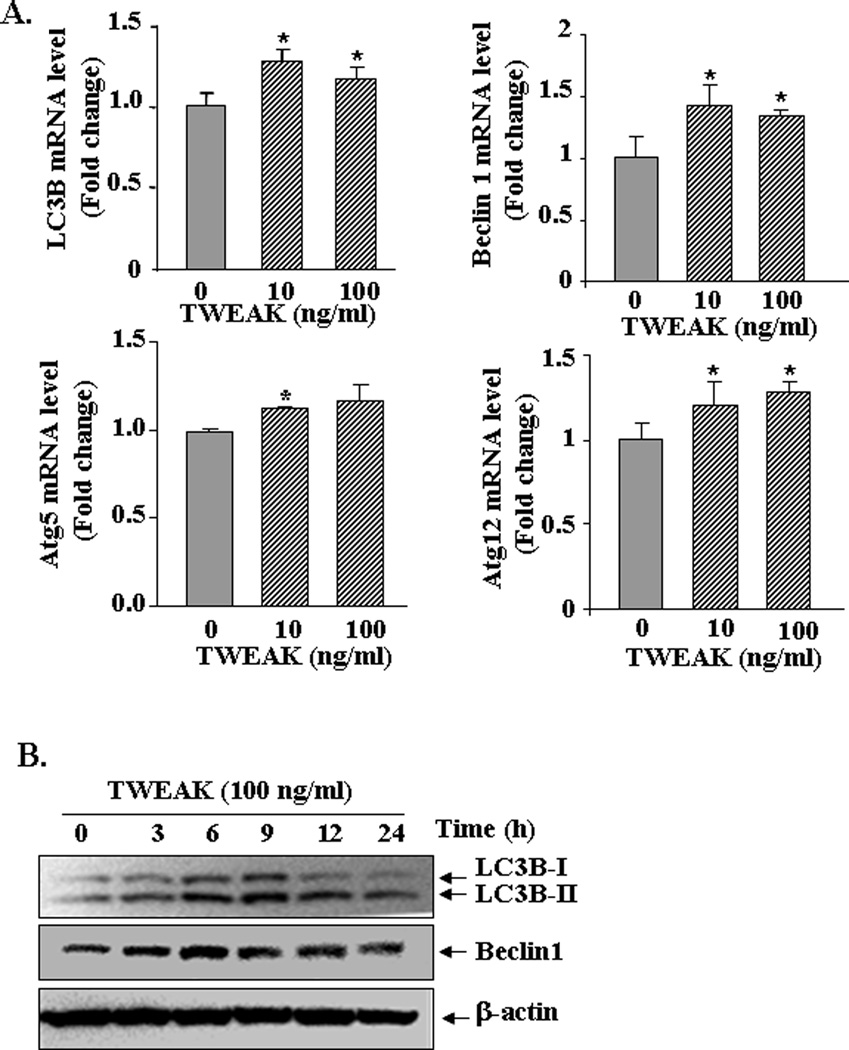
Published reports suggest that muscle-wasting stimuli such as starvation and denervation induce the expression of several autophagy related genes in skeletal muscle cells (Mammucari et al., 2007; Sandri, 2010; Zhao et al., 2007). We next investigated whether TWEAK affects the activity of autophagy-lysosomal system in cultured myotubes. C2C12 myotubes were treated with TWEAK protein and the transcript levels of major autophagy-related genes were measured by QRT-PCR assay. As shown in Figure 3A, TWEAK significantly increased the transcript levels of Beclin1, LC3B, Atg5 and Atg12 in C2C12 myotubes. Furthermore, by performing Western blot, we also studied whether TWEAK induces the conversion of LC3B-I protein into LC3B-II, a critical step in the activation of autophagy system (Sandri, 2010). We observed that the protein levels of both LC3B-I and LC3B-II were increased in C2C12 myotubes in a time-dependent manner in response to TWEAK (Figure 3B). TWEAK also enhanced the protein level of Beclin 1 (~ 2.3 fold at 6h) in C2C12 myotubes (Figure 3B) suggesting that TWEAK augments the activity of autophagy-lysosomal pathway.
FIGURE 3. Effects of TWEAK on expression of autophagy-related genes in myotubes.
A). C2C12 myotubes were treated with indicated amounts of TWEAK. After 6h, the transcript levels of Beclin1, LC3B, Atg5, and Atg12 were measured by QRT-PCR technique. Data from three independent experiments each done in triplicate presented here demonstrate that TWEAK up-regulates the expression of Beclin1, LC3B, Atg5, and Atg12 in C2C12 myotubes. *p<0.05, values significantly different from TWEAK-untreated myotubes. B). C2C12 myotubes were incubated with 100 ng/ml TWEAK for indicated time intervals and protein extracts made were analyzed by Western blot. Representative immunoblots from two independent experiments presented here show that TWEAK increases the protein levels of LC3B (upper panel) and Beclin1 (middle panel) without affecting the levels of an unrelated protein β-actin (lower panel) in C2C12 myotubes. TWEAK also increased the conversion of LC3B-I into LC3B-II protein (upper panel).
Autophagy inhibitors rescue TWEAK-induced atrophy in myotubes
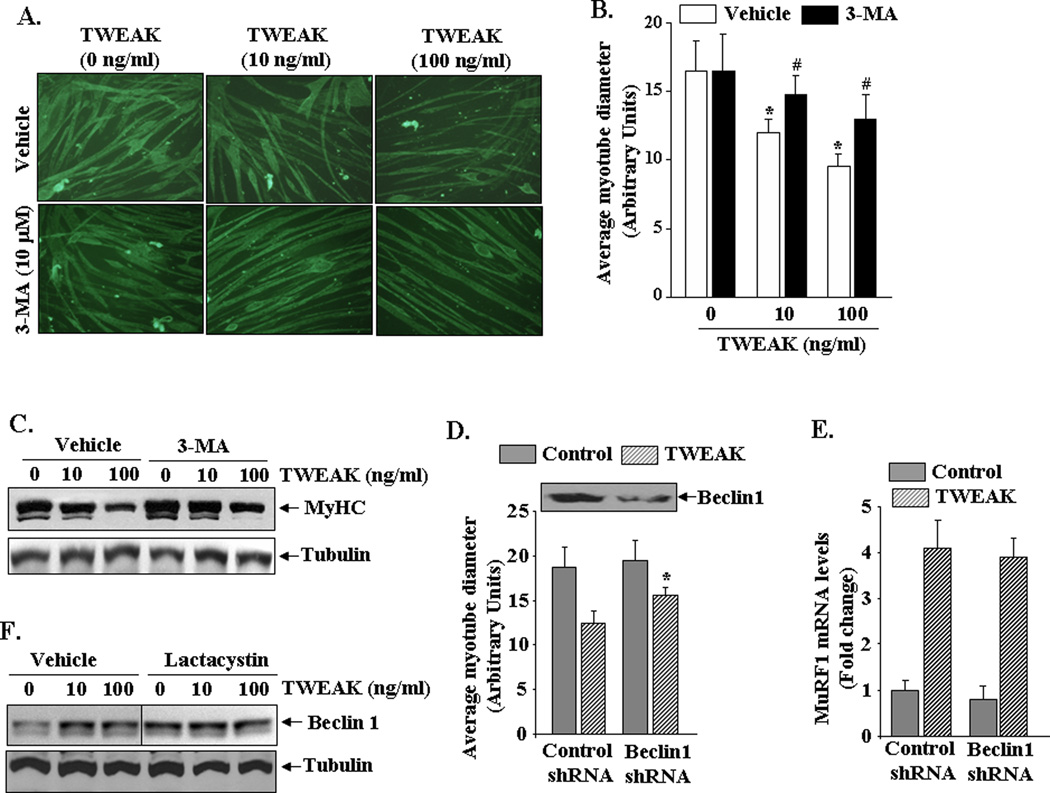
To further understand the role of autophagy in TWEAK-induced muscle atrophy, we used 3-methyladenine (3-MA), a well-known pharmacological inhibitor of macroautophagy (Seglen and Gordon, 1982). C2C12 myotubes were pretreated with 3-MA for 3h followed by incubation with TWEAK for additional 72h and staining of myotube cultures with MF20 antibody. As shown in Figure 4A, Figure 3-MA prevented TWEAK-induced atrophy in C2C12 cultures. Quantitative estimation of myotube diameter confirmed that 3-MA preserved myotube size in response to TWEAK treatment (Figure 4B). We next studied the effects of 3-MA on levels of MyHC in TWEAK-treated C2C12 myotubes. A moderate increase in MyHC levels was noticeable in TWEAK-treated myotubes incubated with 3-MA (Figure 4C). Interestingly, 3-MA was less effective in preventing the loss of MyHC compared to proteasome inhibitors (Figure 1C) or knockdown of MuRF1 (Figure 2C).
FIGURE 4. Role of autophagy in TWEAK-induced myotube atrophy.
A). C2C12 myotubes were pre incubated with 10 µM 3-methyladenine (3-MA) followed by addition of indicated amounts of TWEAK. After 72h, the cells were immunostained with MyHC antibody. Representative photomicrographs from two independent experiments presented here demonstrate that 3-MA preserves myotube size in TWEAK-treated cultures. B). Quantification of average myotube diameter in control and TWEAK-treated cultures incubated with vehicle or 3-MA. *p<0.01, values significantly different from TWEAK-untreated myotubes incubated with vehicle alone. #p<0.01, values significantly different from TWEAK-treated myotubes incubated with vehicle alone. C). C2C12 myotubes were pre-incubated with autophagy inhibitor 3-MA for 3h followed by addition of TWEAK for 72h and measurement of MyHC levels by Western blot. Representative immunoblot presented here shows that 3-MA attenuates TWEAK-induced degradation of MyHC in myotubes (upper panel). The levels of tubulin remained unchanged after treatment with 3-MA and/or TWEAK. D). C2C12 myoblasts were stably transfected with control shRNA or Beclin1 shRNA-expressing plasmid followed by addition of DM for 96h. Immunoblot (inset) shows protein level of Beclin1 in control and Beclin1 shRNA-transfected untreated myotubes. Cells were treated with 100 ng/ml of TWEAK for 72h followed by staining with MF-20 antibody and measurement of myotube diameter. Data presented here demonstrate that knockdown of Beclin1 significantly improved mean myotube diameter in TWEAK-treated cultures. *p<0.05, values significantly different from TWEAK-treated control shRNA expressing cultures. E). Control or Beclin1 shRNA-expressing myotubes were treated with 100 ng/ml TWEAK for 6h and relative mRNA levels of MuRF1 were measured by QRT-PCR. No significant difference in MuRF1 mRNA level was noticeable after knockdown of Beclin1 in TWEAK-treated cultures. F). C2C12 myotubes were incubated with lactacystin (2µg/ml) for 3h followed by addition of TWEAK for 72h. The cell extracts made were analyzed by Western blot using Beclin1 and tubulin antibody. Black line indicates that intervening lanes have been spliced out
We also used siRNA technique to investigate the role of autophagy in TWEAK-induced muscle-wasting. Beclin1 is a critical component of autophagy system and is required for autophagosome formation in mammalian systems (Liang et al., 1999). C2C12 myoblasts were stably transfected with plasmid vector expressing a control shRNA or Beclin1 shRNA followed by their differentiation into myotubes. Knockdown of Beclin1 did not affect the myotube formation in C2C12 cultures (data not shown). However, we found that knockdown of Beclin1 significantly improved myotube diameter in TWEAK-treated cultures (Figure 4D) providing additional evidence regarding potential role of autophagy in TWEAK-induced atrophy. We also investigated whether autophagy affects the ubiquitin-proteasome system and vice versa, in response to TWEAK. As a measure of the activation of ubiquitin-proteasome system, we quantified the transcript levels of MuRF1 in C2C12 myotubes transfected with control or Beclin1 shRNA. While TWEAK increased the expression of MuRF1 in both control and Beclin1 shRNA-expressing C2C12 myotubes, the levels of MuRF1 were comparable between them (Figure 4E). Furthermore, treatment with proteasome inhibitor lactacystin increased the basal level of Beclin1 in myotubes though no further increase was observed after TWEAK treatment (Figure 4F). These results indicate that when ubiquitin-proteasome system is not functional, TWEAK can still induce muscle proteolysis through the activation of autophagy-lysosomal system.
Caspases are involved in TWEAK-induced myotube atrophy
Since caspases have been previously found to be involved in some types of muscle atrophy (Du et al., 2004; Plant et al., 2009; Supinski et al., 2010), we investigated whether TWEAK affects the expression and/or activation of caspases in cultured myotubes. C2C12 myotubes were treated with TWEAK for 24h and the cell extracts made were analyzed for pan-caspase activity using a commercially available kit (Roche). As shown in Figure 5A, TWEAK significantly increased the activation of caspases in cultured myotubes. Published reports suggest that caspase 8 (an initiator caspase) and caspase 3 (an effector caspase) are some of the important caspases involved in muscle proteolysis (Du et al., 2004; Supinski et al., 2009; Supinski et al., 2010). Interestingly, we found that treatment with TWEAK significantly increased the mRNA levels of both caspase 8 and caspase 3 in myotubes (Figure 5B). Furthermore, Western blot analysis showed that TWEAK augments the protein levels of pro-caspase 3 and its conversion into active caspase 3 (Figure 5C).
FIGURE 5. Caspases are involved in TWEAK-induced atrophy in cultured myotubes.
A). C2C12 myotubes were treated with indicated concentration of TWEAK for 24h followed by preparation of cell extracts and measurement of homogenous caspase activity using a kit. Data presented here demonstrate that TWEAK significantly (*p<0.05) increases the activation of caspases in C2C12 myotubes. B). C2C12 myotubes were treated with indicated amounts of TWEAK. After 9h, the mRNA levels of caspase 3 and caspase 8 were measured by QRT-PCR. Data presented here demonstrate that TWEAK up-regulates the transcript levels of both Caspase 3 and caspase 8 in myotubes. *p<0.05, values significantly different from TWEAK-untreated myotubes. C). Western blot analysis showed that TWEAK increases the amount of pro-caspase 3 protein and its cleavage to active caspase 3 without affecting the levels of unrelated protein β-actin in myotubes. D). C2C12 myotubes were incubated with 10 µM of Z-FA-FMK, Z-VAD-FMK or Ac-DMQD-CHO peptide for 3h followed by addition of indicated amounts of TWEAK for 72. The cell extracts made were analyzed by Western blot using MyHC antibody. The representative immunoblot presented here shows that Z-VAD-FMK or Ac-DMQD-CHO reduces the TWEAK-induced degradation of MyHC (upper panel) without affecting the levels of β-actin (lower panel) in myotubes. E). C2C12 myotubes were incubated with 10 µM of Z-FA-FMK, Z-VAD-FMK or Ac-DMQD-CHO peptide for 3h followed by addition of 100 ng/ml TWEAK for 72h. The cultures were then immunostained with MyHC antibody and myotube diameter was measured. Data from three independent experiments presented here show that Z-VAD-FMK or Ac-DMQD-CHO improves average myotube diameter in TWEAK-treated cultures. *p<0.01, values significantly different from TWEAK-treated myotubes incubated with Z-FA-FMK.
To investigate the role of caspases in TWEAK-induced myotube atrophy, we employed specific cell permeable peptide inhibitors of pan-caspases and caspase-3. C2C12 myotubes were preincubated with control peptide Z-FA-FMK, pan-caspase inhibitor Z-VAD-FMK, or caspase-3 inhibitor Ac-DMQD-CHO for 3h followed by incubation with TWEAK for 72h and measurement of MyHC levels by Western blot. Although both Z-VAD-FMK and Ac-DMQD-CHO were found to be effective in blocking TWEAK-induced loss of MyHC in C2C12 myotubes, Z-VAD-FMK was found to be more potent compared to Ac-DMQD-CHO (Fig 5D). Consistent with MyHC levels, incubation with Z-VAD-FMK or Ac-DMQD-CHO significantly improved myotube diameter in TWEAK-treated C2C12 cultures (Figure 5E). These results suggest that caspase-3 is one of the caspases involved in TWEAK-induced muscle-wasting.
NF-κB is involved in TWEAK-induced expression of MuRF1 and Beclin1 in cultured myotubes
NF-κB is a major transcription factor involved in different types of muscle atrophy (Li et al., 2008). We have previously reported that the inhibition of NF-κB prevents the TWEAK-induced degradation of MyHC in cultured myotubes (Dogra et al., 2007a). Recent studies have also indicated that NF-κB controls the expression of MuRF1 in skeletal muscle in atrophic conditions (Cai et al., 2004; Mourkioti et al., 2006). We investigated whether NF-κB plays a role in TWEAK-induced expression of MuRF1 and Beclin1, the major components of ubiquitin-proteasome and autophagy systems, respectively. C2C12 myoblasts were stably transfected with plasmid vectors expressing either control or IKKβ (an upstream activator of classical NF-κB pathway (Li et al., 2008)) shRNA followed by their differentiation into myotubes. The cells were then treated with TWEAK for 6h and relative mRNA levels of MuRF1 and Beclin1 were measured by QRT-PCR assay. Interestingly, knockdown of IKKβ significantly reduced the transcript levels of both MuRF1 (Figure 6A) and Beclin1 (Figure 6B) in TWEAK-treated myotubes. To further validate the role of NF-κB in TWEAK-induced expression of MuRF1 and Beclin1, we studied the effects of adenoviral-mediated overexpression of a degradation-resistant mutant of IκBα protein (i.e. IκBαΔN), an inhibitor of NF-κB (Dogra et al., 2007a; Li et al., 2008) in C2C12 myotubes. Our results showed that TWEAK-induced expression of MuRF1 or Beclin1 was significantly reduced in IκBαΔN-expressing myotubes (Figure 6C and Figure 6D). Taken together, these findings suggest that TWEAK augments the expression of both MuRF1 and Beclin1 through activation of NF-κB transcription factor.
FIGURE 6. NF-κB activation is essential for TWEAK-induced expression of MuRF1 and Beclin1 in myotubes.
C2C12 myoblasts were stably transfected with control shRNA or IKKβ shRNA-expressing plasmid followed by incubation in DM for additional 96h. The cells were then treated with TWEAK (100 ng/ml) for 6h and the mRNA levels of MuRF1 and Beclin1 were measured by QRT-PCR. Data from two independent experiments each done in triplicate presented here demonstrate that knockdown of IKKβ inhibited the TWEAK-induced expression of A) MuRF1 and B) Beclin1 in myotubes. *p<0.05, values significantly different from TWEAK-treated myotubes transfected with control shRNA. C2C12 myotubes were transduced with control (Ad.Con) or IκBαΔN-expressing adenovirus (Ad. IκBαΔN), the cells were treated with 100 ng/ml TWEAK for 6h, and the mRNA levels of MuRF1 and Beclin1 were measured by QRT-PCR. Data presented here show that overexpression of IκBαΔN protein significantly inhibits the TWEAK-induced expression of C) MuRF1 and D) Beclin1 in myotubes. *p<0.05, values significantly different from TWEAK-treated myotubes transduced with Ad.Con.
Role of caspases in the activation of NF-κB and expression of MuRF1 in myotubes in response to TWEAK
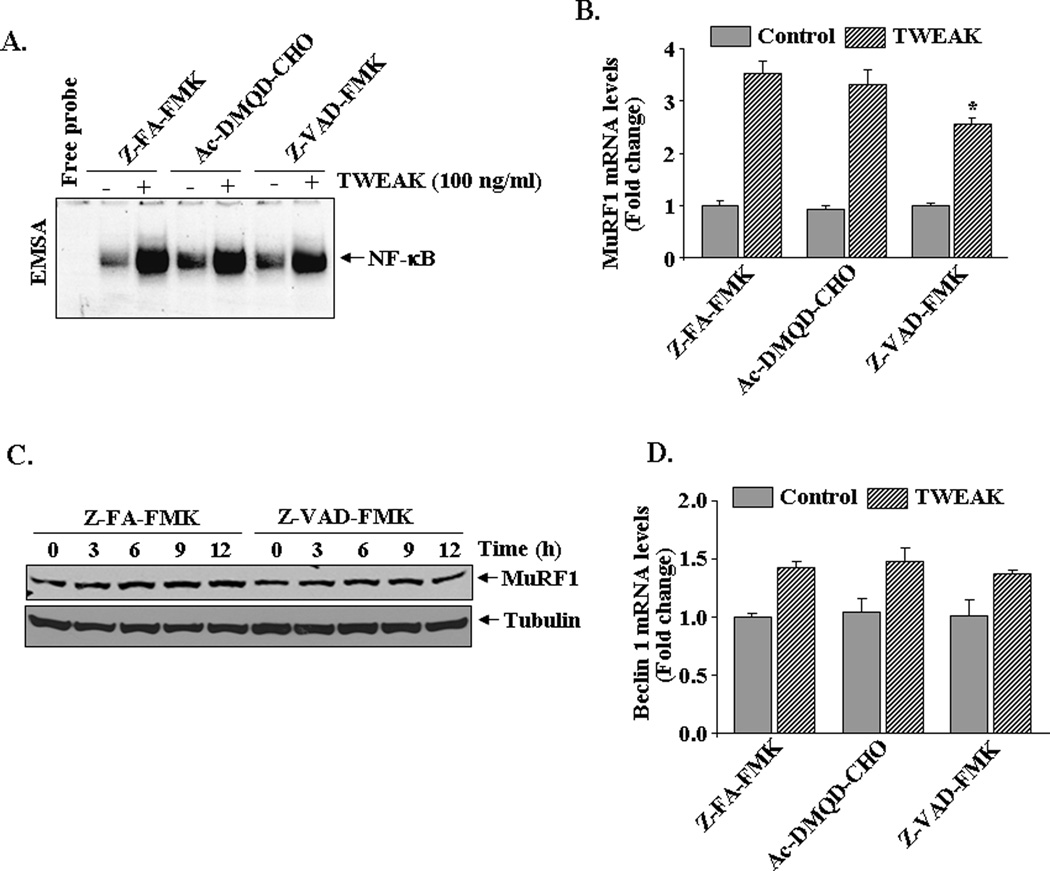
Recent reports also indicate that caspases may also be involved in the activation of NF-κB in response to specific stimuli (Lamkanfi et al., 2007). We sought to investigate the role of caspases in TWEAK-induced activation of NF-κB in cultured myotubes. C2C12 myotubes were preincubated with Z-FA-FMK, Ac-DMQD-CHO, or Z-VAD-FMK for 3h followed by treatment with TWEAK for 1h. The activation of NF-κB was measured by EMSA. As shown in Figure 7A, treatment with Caspase 3 inhibitor Ac-DMQD-CHO did not affect the activation of NF-κB in TWEAK-treated myotubes. Though moderately, pan-caspase inhibitor Z-VAD-FMK reduced the levels of NF-κB (~45%) in TWEAK-treated myotubes (Figure 7A).
FIGURE 7. Role of caspases in TWEAK-induced activation of NF-κB and MuRF1 expression.
A). C2C12 myotubes were incubated with 10 µM of Z-FA-FMK, Z-VAD-FMK or Ac-DMQD-CHO peptide for 3h followed by addition of 100 ng/ml TWEAK for 1h and measurement of NF-κB DNA-binding activity by EMSA. Representative EMSA gel presented here from three independent experiments presented here demonstrates that treatment with Z-VAD-FMK partially inhibits the TWEAK-induced activation of NF-κB in myotubes. B). C2C12 myotubes were incubated with 10 µM of Z-FA-FMK, Z-VAD-FMK or Ac-DMQD-CHO peptide for 3h followed by treatment with TWEAK (100 ng/ml) for 6h and measurement of MuRF1 mRNA levels by QRT-PCR. Data presented here show that Z-VAD-FMK significantly inhibits the TWEAK-induced expression of MuRF1 in myotubes. *p<0.05, values significantly different from TWEAK-treated myotubes incubated with Z-FA-FMK. C). Representative immunoblots presented here demonstrate that Z-VAD-FMK reduces the protein levels of MuRF1 in C2C12 myotubes in response to TWEAK. The levels of tubulin were not affected by either of these treatments. D). C2C12 myotubes were incubated with 10 µM of Z-FA-FMK, Z-VAD-FMK, or Ac-DMQD-CHO peptide for 3h, treated with TWEAK (100 ng/ml) for 6h, and mRNA levels of Beclin1 were measured by QRT-PCR. Data from three independent experiments presented here show that neither Z-VAD-FMK nor Ac-DMQD-CHO had any significantly effect on TWEAK-induced expression of Beclin1 in myotubes.
We also investigated whether caspases mediate muscle atrophy through up-regulation of the components of ubiquitin-proteasome or autophagy system in TWEAK-treated myotubes. C2C12 myotubes were preincubated with Z-FA-FMK (control peptide), Ac-DMQD-CHO (caspase 3 inhibitor), or Z-VAD-FMK (pan-caspase inhibitor) for 3h followed by addition of TWEAK. After 6h, mRNA level of MuRF1 was measured by QRT-PCR technique. As shown in Figure 7B, inhibitor of pan caspases but not caspase 3 significantly reduced the TWEAK-induced expression of MuRF1 in C2C12 myotubes. Consistent with mRNA levels, we also found that pan caspases inhibitor was effective in reducing the protein levels of MuRF1 in TWEAK-treated myotubes (Figure 7C). However, treatment of myotubes with pan-caspase or caspase 3 peptide inhibitor did not have any significant effect on TWEAK-induced expression of Beclin1 in C2C12 myotubes (Figure 7D). These results indicate that though not major, caspases may have a role in the activation of NF-κB and induction of MuRF1 in TWEAK-treated myotubes.
DISCUSSION
TWEAK is a multifunctional cytokine involved in regulation of inflammatory and immune responses (Maecker et al., 2005; Winkles, 2008). In our search for novel mediators of muscle-wasting, we previously identified TWEAK as a potent muscle-wasting cytokine (Dogra et al., 2007a). The present study was undertaken to understand the role of various proteolytic systems in TWEAK-induced muscle atrophy. Our results demonstrate that TWEAK stimulates the activity of ubiquitin-proteasome system, autophagy and caspases and inhibition of any one of these proteolytic systems is sufficient to significantly rescue muscle atrophy in response to TWEAK. A schematic representation of the roles that various proteolytic systems play in TWEAK-induced muscle atrophy, supported by our experiments in this study, is depicted in Figure 8.
FIGURE 8. Schematic representation of the mechanisms of action of TWEAK in skeletal muscle.
The role of NF-κB and various proteolytic systems and a potential cross-talk between them in TWEAK-treated myotubes leading to myotube atrophy is depicted in this figure. Broken line indicates relatively weaker connection.
The ubiquitin-proteasome system has been suggested to degrade many contractile proteins and plays a major role in skeletal muscle-wasting (Solomon and Goldberg, 1996). Different and multiple events in the ubiquitination, deubiquitination, and proteolytic machineries are responsible for the activation of the system and subsequent muscle-wasting (Cao et al., 2005). Though skeletal muscle-wasting involves the degradation of several muscle proteins, MyHC is one of the most important proteins rapidly degraded in response to different catabolic stimuli including cancer cachexia, inflammatory cytokine cocktail, and dexamethasone (Acharyya et al., 2004; Clarke et al., 2007; Ladner et al., 2003). Our previously published report (Dogra et al., 2007a) and experiments in this study suggest that TWEAK alone is sufficient to stimulate proteolysis leading to significant loss of MyHC with concomitant myotube atrophy. The present study also suggests that the activation of ubiquitin-proteasome system is a prominent mechanism by which TWEAK causes muscle-wasting in cultured myotubes. The ubiquitylation of MyHC was rapidly increased and proteasome inhibitors considerably preserved levels of MyHC and spared myotube size in response to TWEAK (Figure 1).
In catabolic states where proteolysis is increased, two genes specific to muscle atrophy, MuRF1 and MAFbx, are highly upregulated (Cao et al., 2005; Glass, 2010). Recently, it was reported that MuRF1 associates and catalyzes the ubiquitylation of thick filament protein MyHC in skeletal muscle in response to dexamethasone (Clarke et al., 2007). The results of the present study suggest that MuRF1 contributes to the TWEAK-induced loss of MyHC and myotube atrophy (Figure 2). Our experiments also demonstrate that TWEAK induces the expression of several genes involved in autophagy-lysosomal pathway and macroautophagy inhibitor 3-MA prevents TWEAK-induced loss of myotube diameter (Figs. 3 and 4). Although it is still unclear whether autophagy promotes or prevents cell damage and leads to improvement or worsening of disease outcome, the presence of autophagic vacuoles is a prominent and characteristic structural feature in different types of atrophying muscle (Sandri, 2010). For example, autophagy is highly up-regulated in response to fasting and denervation. Previous studies have shown that overexpression of constitutively active Foxo3 stimulates proteolysis and initially both proteasomal and lysosomal systems contribute equally to the proteolysis of myofibrillar proteins but at later time points lysosomal pathway account for ~70% of the total proteolysis (Zhao et al., 2007). It has also been shown that the phosphatidylinositol 3-kinase (PI3K)-Akt signaling inhibits atrophy by suppressing the activity of FOXO family transcription factors which regulates the expression of several genes involved in ubiquitin-proteasome and autophagy systems (Mammucari et al., 2007). Interestingly, we have previously reported that TWEAK down-regulates the activity of Akt and reduces the phosphorylation of FOXO resulting in their increased activation and nuclear localization (Dogra et al., 2007a). Therefore, one of the mechanisms by which TWEAK augments the activity of ubiquitin-proteasome system and autophagy in myotubes could be through the inhibition of Akt-mTOR pathway. Until recently, autophagy was considered mostly as proteolytic mechanism responsible for degradation of bulk of muscle protein in atrophying conditions (Sandri, 2010). However, a recent study has shown that muscle-specific inhibition of autophagy induces myopathy and exaggerates muscle loss in response to denervation and starvation raising an interesting possibility that basal level of autophagy may be essential for clearance of already damaged and/or ubiquitinated proteins in atrophying muscle (Masiero et al., 2009). This notion is also supported by autophagy-deficient Pompe mice where a larger accumulation of ubiquitinated protein was noticeable (Raben et al., 2008). Although their precise mechanisms of action remain unclear, our experiments indicate that both proteasomal and autophagy systems are involved in TWEAK-induced atrophy. Our study also suggests a possible link between these two proteolytic systems because inhibitors of proteasome increased the expression of Beclin1 in myotubes (Figure 4F).
Accumulating evidence suggests that other proteolytic enzymes such a caspases may be required for initial cleavage of the myofibrillar components, thereby accelerating disassembly and degradation by the ubiquitin-proteasome system (Cohen et al., 2009; Du et al., 2004). Activation of caspase 3 during atrophy is at least one of the mechanisms involved in the initial steps of myofibrillar degradation. Although the ubiquitin proteasome system can degrade monomeric actin or myosin, it does not break down actomyosin complexes or myofibrils (Cohen et al., 2009; Solomon and Goldberg, 1996). The role of caspase 3 in actin fragmentation during atrophy is supported by the observation that recombinant caspase-3 could increase actin fragmentation of purified actomyosin, and protein extracts prepared from whole muscle or L6 myotubes (Du et al., 2004). More recently, it has been found that activation of caspase 8, an immediate upstream activator of caspase 3, is increased in skeletal muscle in response to many catabolic stimuli such as cytokine cocktail, endotoxin, and during sarcopenia further supporting the role of caspases in muscle proteolysis (Baker and Hepple, 2006; Supinski et al., 2009; Supinski et al., 2010).
Our results showed that while TWEAK does not cause any noticeable cell death in myotubes (data not shown), it augments the expression and activation of caspases in C2C12 myotubes (Figure 5A). Our results also provide initial evidence that caspases are involved in TWEAK-induced loss of MyHC. Since pan caspase inhibitor was more effective compared to caspase 3 inhibitor in preventing the loss of MyHC and myotube diameter, it appears that in addition to caspase 3, some other caspases are also involved in TWEAK-induced muscle atrophy. While MuRF1 E3 ligase has been found to ubiquitinate MyHC leading to its degradation by 26S proteasome, knockdown of MuRF1 did not completely prevent loss of MyHC in response to TWEAK (Figure 2C) or dexamethasone (Clarke et al., 2007) indicating that other proteolytic systems especially caspases may also contribute to loss of MyHC in atrophy conditions. Furthermore, it is also possible that the activation of non-mitochondrial caspase pathways is an initial event leading to the disassembly of myofilamnents and subsequent degradation of soluble MyHC by MuRF1-dependent mechanisms (Cohen et al., 2009).
It has been consistently observed that the activation of NF-κB in skeletal muscle is increased in many catabolic conditions (Li et al., 2008). Inhibition of NF-κB has been found to prevent loss of MyHC in TWEAK-treated myotubes (Li et al., 2008). Our results demonstrate that one of the mechanisms by which NF-κB mediates TWEAK-induced muscle atrophy is through augmenting the expression of MuRF1 (Figure 6) which is consistent with a previously published report (Mourkioti et al., 2006) and data in this study (Figure 6) that NF-κB is essential for MuRF1 expression in atrophying skeletal muscle. We also found that NF-κB induces the expression of Beclin1 in myotubes in response to TWEAK. Although the exact mechanisms by which TWEAK up-regulates Beclin1 expression remain unknown, a recently published report has suggested that NF-κB promotes autophagy in some other cell-types (Criollo et al., 2010). Furthermore, activation of caspases seems to contribute, at least in part, to the activation of NF-κB in response to TWEAK supported by our results that pan-caspase inhibitor diminished the activation of NF-κB in response to TWEAK treatment (Figure 7A). Since pan-caspase inhibitor was also able to reduce the expression of MuRF1, these results indicate a possible cross-talk between caspases and ubiquitin-proteasome system through NF-κB.
Based on the findings in this report, a putative sequence of events that is initiated in response to TWEAK and leads to muscle atrophy is presented in Figure 8. While more investigations are required to understanding the mechanisms of action of TWEAK in skeletal muscle, the present study provides initial evidence that TWEAK-induced muscle atrophy involves coordinated activation of multiple proteolytic systems and NF-κB. Similar mechanisms might be involved in skeletal muscle-wasting in response to other stimuli.
ACKNOWLEDGEMENTS
This work was supported by a National Institute of Health grant (AG029623) to AK. We sincerely thank Dr. Stewart H. Lecker for providing MuRF1-Luc construct and Dr. David J. Glass for adenoviral vector for MuRF1 shRNA used in this study.
Abbreviations
- DM
Differentiation medium
- 3-MA
3-methyladenine
- MAFBx
muscle atrophy F-box
- MuRF1
muscle RING-finger 1
- MyHC
myosin heavy chain
- NF-κB
nuclear factor-kappa B
- QRT-PCR
quantitative real-time PCR
- RNAi
RNA interference
- shRNA
short hairpin RNA
- TWEAK
TNF like weak inducer of apoptosis
REFERENCES
- Acharyya S, Guttridge DC. Cancer cachexia signaling pathways continue to emerge yet much still points to the proteasome. Clin Cancer Res. 2007;13(5):1356–1361. doi: 10.1158/1078-0432.CCR-06-2307. [DOI] [PubMed] [Google Scholar]
- Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest. 2004;114(3):370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiles JM, Busquets S, Lopez-Soriano FJ. The pivotal role of cytokines in muscle wasting during cancer. Int J Biochem Cell Biol. 2005;37(10):2036–2046. doi: 10.1016/j.biocel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Argiles JM, Busquets S, Toledo M, Lopez-Soriano FJ. The role of cytokines in cancer cachexia. Curr Opin Support Palliat Care. 2009;3(4):263–268. doi: 10.1097/SPC.0b013e3283311d09. [DOI] [PubMed] [Google Scholar]
- Attaix D, Bechet D. FoxO3 controls dangerous proteolytic liaisons. Cell Metab. 2007;6(6):425–427. doi: 10.1016/j.cmet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Hepple RT. Elevated caspase and AIF gene expression correlate with progression of sarcopenia during aging in male F344BN rats. Exp Gerontol. 2006;41(11):1149–1156. doi: 10.1016/j.exger.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119(2):285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Cao PR, Kim HJ, Lecker SH. Ubiquitin-protein ligases in muscle wasting. Int J Biochem Cell Biol. 2005;37(10):2088–2097. doi: 10.1016/j.biocel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, Tailler M, Delahaye N, Tesniere A, De Stefano D, Younes AB, Harper F, Pierron G, Lavandero S, Zitvogel L, Israel A, Baud V, Kroemer G. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29(3):619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra C, Changotra H, Mohan S, Kumar A. Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-kappaB and degradation of MyoD protein. J Biol Chem. 2006;281(15):10327–10336. doi: 10.1074/jbc.M511131200. [DOI] [PubMed] [Google Scholar]
- Dogra C, Changotra H, Wedhas N, Qin X, Wergedal JE, Kumar A. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 2007a;21(8):1857–1869. doi: 10.1096/fj.06-7537com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra C, Hall SL, Wedhas N, Linkhart TA, Kumar A. Fibroblast growth factor inducible 14 (Fn14) is required for the expression of myogenic regulatory factors and differentiation of myoblasts into myotubes. Evidence for TWEAK-independent functions of Fn14 during myogenesis. J Biol Chem. 2007b;282(20):15000–15010. doi: 10.1074/jbc.M608668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Mitch WE, Wang X, Price SR. Glucocorticoids induce proteasome C3 subunit expression in L6 muscle cells by opposing the suppression of its transcription by NF-kappa B. J Biol Chem. 2000;275(26):19661–19666. doi: 10.1074/jbc.M907258199. [DOI] [PubMed] [Google Scholar]
- Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE. Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest. 2004;113(1):115–123. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care. 2010;13(3):225–229. doi: 10.1097/mco.0b013e32833862df. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RB, Kandarian SC. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J Clin Invest. 2004;114(10):1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287(4):C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Kandarian SC, Stevenson EJ. Molecular events in skeletal muscle during disuse atrophy. Exerc Sport Sci Rev. 2002;30(3):111–116. doi: 10.1097/00003677-200207000-00004. [DOI] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci U S A. 2004;101(52):18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear factor-kappaB: its role in health and disease. J Mol Med. 2004;82(7):434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- Ladner KJ, Caligiuri MA, Guttridge DC. Tumor necrosis factor-regulated biphasic activation of NF-kappa B is required for cytokine-induced loss of skeletal muscle gene products. J Biol Chem. 2003;278(4):2294–2303. doi: 10.1074/jbc.M207129200. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Vanden Berghe T, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14(1):44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol. 2004;15(6):1537–1545. doi: 10.1097/01.asn.0000127211.86206.e1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Malhotra S, Kumar A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J Mol Med. 2008;86(10):1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Lecker SH, Chen Y, Waddell ID, Goldberg AL, Reid MB. TNF-alpha increases ubiquitin-conjugating activity in skeletal muscle by up-regulating UbcH2/E220k. FASEB J. 2003;17:1048–1057. doi: 10.1096/fj.02-0759com. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Maecker H, Varfolomeev E, Kischkel F, Lawrence D, LeBlanc H, Lee W, Hurst S, Danilenko D, Li J, Filvaroff E, Yang B, Daniel D, Ashkenazi A. TWEAK attenuates the transition from innate to adaptive immunity. Cell. 2005;123(5):931–944. doi: 10.1016/j.cell.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4(4):524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Mittal A, Bhatnagar S, Kumar A, Lach-Trifilieff E, Wauters S, Li H, Makonchuk DY, Glass DJ, Kumar A. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol. 2010;188(6):833–849. doi: 10.1083/jcb.200909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti F, Kratsios P, Luedde T, Song YH, Delafontaine P, Adami R, Parente V, Bottinelli R, Pasparakis M, Rosenthal N. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116(11):2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant PJ, Bain JR, Correa JE, Woo M, Batt J. Absence of caspase-3 protects against denervation-induced skeletal muscle atrophy. J Appl Physiol. 2009;107(1):224–234. doi: 10.1152/japplphysiol.90932.2008. [DOI] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17(24):3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29(10):1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584(7):1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon V, Goldberg AL. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J Biol Chem. 1996;271(43):26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- Späte U, Schulze PC. Proinflammatory cytokines and skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7(3):265–269. doi: 10.1097/00075197-200405000-00005. [DOI] [PubMed] [Google Scholar]
- Supinski GS, Ji X, Callahan LA. The JNK MAP kinase pathway contributes to the development of endotoxin-induced diaphragm caspase activation. Am J Physiol Regul Integr Comp Physiol. 2009;297(3):R825–R834. doi: 10.1152/ajpregu.90849.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supinski GS, Ji XY, Callahan LA. p38 Mitogen-activated protein kinase modulates endotoxin-induced diaphragm caspase activation. Am J Respir Cell Mol Biol. 2010;43(1):121–127. doi: 10.1165/rcmb.2008-0395OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J Physiol. 2002;545(Pt 3):819–828. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem. 2005;280(4):2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7(5):411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]