Abstract
The metabolic adaptations that support oncogenic growth can also render cancer cells dependent on certain nutrients. Along with the Warburg effect, increased utilization of glutamine is one of the metabolic hallmarks of the transformed state. Glutamine catabolism is positively regulated by multiple oncogenic signals, including those transmitted by the Rho family of GTPases and by c-Myc. The recent identification of mechanistically distinct inhibitors of glutaminase, which can selectively block cellular transformation, has revived interest in the possibility of targeting glutamine metabolism in cancer therapy. Here, we outline the regulation and roles of glutamine metabolism within cancer cells and discuss possible strategies for, and the consequences of, impacting these processes therapeutically.
Cancer cell metabolism & glutamine addiction
Interest in the metabolic changes characteristic of malignant transformation has undergone a renaissance of sorts in the cancer biology and pharmaceutical communities. However, the recognition that an important connection exists between cellular metabolism and cancer began nearly a century ago with the work of Otto Warburg [1–3]. Warburg found that rapidly proliferating tumor cells exhibit elevated glucose uptake and glycolytic flux, and furthermore that much of the pyruvate generated by glycolysis is reduced to lactate rather than undergoing mitochondrial oxidation via the tricarboxylic acid (TCA) cycle (Figure 1). This phenomenon persists even under aerobic conditions (‘aerobic glycolysis’), and is known as the Warburg effect [4]. Warburg proposed that aerobic glycolysis was caused by defective mitochondria in cancer cells, but it is now known that mitochondrial dysfunction is relatively rare and that most tumors have an unimpaired capacity for oxidative phosphorylation [5]. In fact, the most important selective advantages provided by the Warburg effect are still debated. Although aerobic glycolysis is an inefficient way to produce ATP (2 ATP/glucose vs ~36 ATP/glucose by complete oxidation), a high glycolytic flux can generate ATP rapidly and furthermore can provide a biosynthetic advantage by supplying precursors and reducing equivalents for the synthesis of macromolecules [4]. The mechanisms underlying the Warburg effect are also not yet fully resolved, although it is increasingly clear that a number of oncogenes and tumor suppressors contribute to the phenomenon. The PI3K/Akt/mTORC1 signaling axis, for example, is a key regulator of aerobic glycolysis and biosynthesis, driving the surface expression of nutrient transporters and the upregulation of glycolytic enzymes [6]. The HIF transcription factor also upregulates expression of glucose transporters and glycolytic enzymes in response to hypoxia and growth factors (or loss of the von Hippel–Landau [VHL] tumor suppressor), and the oncogenic transcription factor c-Myc similarly induces expression of proteins important for glycolysis [6].
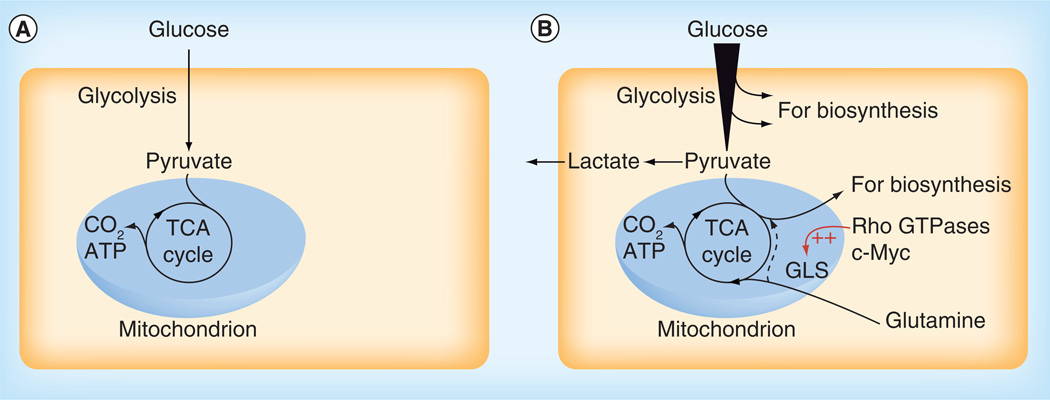
Figure 1. Cell proliferation requires metabolic reprogramming.
(A) In non-proliferating cells under aerobic conditions, metabolic fuels such as glucose typically undergo complete oxidation to CO2 in mitochondria via the TCA cycle. Energy released during this series of reactions is used to generate a proton electrochemical gradient across the inner mitochondrial membrane, which in turn drives ATP synthesis. (B) In proliferating cells there is an increased demand for precursors for protein, nucleotide and lipid production, in addition to ATP. Nutrient uptake is consequently enhanced and metabolic intermediates are diverted from glycolysis and the TCA cycle into biosynthetic pathways. For example, citrate from the TCA cycle can be exported from the mitochondrion to support lipogenesis in the cytosol. Reduction of pyruvate to lactate, catalyzed by lactate dehydrogenase, regenerates NAD+ to sustain glycolytic flux. Glutamine often serves as an anaplerotic substrate to maintain TCA cycle function, through its conversion by GLS and glutamate dehydrogenase to the TCA cycle intermediate α-ketoglutarate. Anaplerotic α-ketoglutarate can undergo oxidative metabolism in the TCA cycle or, during hypoxia or in cells with mitochondrial defects, reductive metabolism to citrate to support biosynthesis (dashed line).
TCA: Tricarboxylic acid.
A second major change in the metabolic program of many cancer cells, and the primary focus of this review, is the alteration of glutamine metabolism. Glutamine is the major carrier of nitrogen between organs, and the most abundant amino acid in plasma [7]. It is also a key nutrient for numerous intracellular processes including oxidative metabolism and ATP generation, biosynthesis of proteins, lipids and nucleic acids, and also redox homeostasis and the regulation of signal transduction pathways [8–10]. Although most mammalian cells are capable of synthesizing glutamine, the demand for this amino acid can become so great during rapid proliferation that an additional extracellular supply is required; hence glutamine is considered conditionally essential [11]. Indeed, many cancer cells are ‘glutamine addicted’, and cannot survive in the absence of an exogenous glutamine supply [12,13].
An important step in the elevation of glutamine catabolism is the activation of the mitochondrial enzyme glutaminase, which catalyzes the hydrolysis of glutamine to generate glutamate and ammonium. The subsequent deamination of glutamate releases a second ammonium to yield the TCA cycle intermediate α-ketoglutarate (α-KG), a reaction catalyzed by glutamate dehydrogenase (GLUD1). This series of reactions is particularly important in rapidly proliferating cells, in which a considerable proportion of the TCA cycle metabolite citrate is exported from mitochondria in order to generate cytosolic acetyl-CoA for lipid biosynthesis [14]. Replenishment of TCA cycle intermediates (anaplerosis) is therefore required, and glutamine often serves as the key anaplerotic substrate through its conversion via glutamate to α-KG (Figure 1).
Mammals express two genes for glutaminase enzymes [15–17]. The GLS gene encodes a protein initially characterized in kidney and thus called kidney-type glutaminase (KGA), although this enzyme and its shorter splice variant glutaminase C (GAC), collectively referred to as GLS, are now known to be widely distributed [18–20]. The KGA and GAC isoforms share identical N-terminal and catalytic domains, encoded by exons 1–14 of the GLS gene, but have distinct C-termini derived from exon 15 in the case of GAC and exons 16–19 in the case of KGA [21]. Upregulation of GLS, in particular the GAC iso-form, is common in cancer cells and the degree of GLS overexpression correlates with both the degree of malignancy and the tumor grade in human breast cancer samples [22,23]. The GLS2 gene encodes a protein originally discovered and characterized in liver, which has thus been referred to as liver-type glutaminase and, more recently, as glutaminase 2 (GLS2) [15].
Both KGA and GAC can be activated by inorganic phosphate (Pi), and this activation correlates closely with a dimer-to-tetramer transition for each enzyme [7, 22]. As the concentration of Pi is raised the apparent catalytic constant, kcatapp, increases and simultaneously the apparent Michaelis constant, Kmapp, decreases; consequently the catalytic efficiency rises dramatically, especially in the case of GAC [22]. x-ray crystal structures of GAC and KGA in different states indicate that the positioning of a key loop within each monomer (Glu312 to Pro329), located between the active site and the dimer–dimer interface, is critical for mediating tetramerization-induced activation [22,24]. Given the ability of Pi to promote tetramerization and activation of GAC and KGA, it has been proposed that the elevated mitochondrial Pi levels found under hypoxic conditions, which are commonly encountered in the tumor microenvironment, could be one trigger for GLS activation [22].
Oncogenic alterations affecting glutamine metabolism
At least two classes of cellular signals regulate glutamine metabolism, influencing both the expression level and the enzymatic activity of GLS. The transcription factor c-Myc can suppress the expression of microRNAs miR-23a and miR-23b and, in doing so, upregulates GLS (specifically GAC) expression [13,25]. Independent of changes in GAC expression, oncogenic diffuse B-cell lymphoma protein (Dbl), a GEF for Rho GTPases and oncogenic variants of downstream Rho GTPases are able to signal to activate GAC in a manner that is dependent on NF-κB [23]. Mitochondria isolated from Dbl- or Rho GTPase-transformed NIH-3T3 fibroblasts demonstrate significantly higher basal glutaminase activity than mitochondria isolated from non-transformed cells [23]. Furthermore, the enzymatic activity of GAC immunoprecipitated from Dbl-transformed cells is elevated relative to GAC from non-transformed cells, indicating the presence of activating post-translational modification(s) [23]. Indeed, when GAC isolated from Dbl-transformed cells is treated with alkaline phosphatase, basal enzymatic activity is dramatically reduced [23]. Collectively, these findings point to phosphorylation events underlying the activation of GAC in transformed cells. Similarly, phosphorylation-dependent regulation of KGA activity downstream of the Raf-Mek-Erk signaling axis occurs in response to EGF stimulation [24].
It is becoming clear that, in addition to c-Myc and Dbl, many other oncogenic signals and environmental conditions can impact cellular glutamine metabolism. Loss of the retinoblastoma tumor suppressor, for example, leads to a marked increase in glutamine uptake and catabolism, and renders mouse embryonic fibroblasts dependent on exogenous glutamine [26]. Cells transformed by KRAS also illustrate increased expression of genes associated with glutamine metabolism and a corresponding increased utilization of glutamine for anabolic synthesis [27]. In fact, KRAS signaling appears to induce glutamine dependence, since the deleterious effects of glutamine withdrawal in KRAS-driven cells can be rescued by expression of a dominant-negative GEF for Ras [28]. Downstream of Ras, the Raf-MEK-ERK signaling pathway has been implicated in the upregulation of glutamine uptake and metabolism [24,29]. A recent study using human pancreatic ductal adenocarcinoma cells identified a novel KRAS-regulated metabolic pathway, through which glutamine supports cell growth [30]. Proliferation of KRAS-mutant pancreatic ductal adenocarcinoma cells depends on GLS-catalyzed production of glutamate, but not on downstream deamination of glutamate to α-KG; instead, transaminase-mediated glutamate metabolism is essential for growth. Glutamine-derived aspartate is subsequently transported into the cytoplasm where it is converted by aspartate transaminase into oxaloacetate, which can be used to generate malate and pyruvate. The series of reactions maintains NADPH levels and thus the cellular redox state [30].
Other recent studies have revealed that another pathway for glutamine metabolism can be essential under hypoxic conditions, and also in cancer cells with mitochondrial defects or loss of the VHL tumor suppressor [31–35]. In these situations, glutamine-derived α-KG undergoes reductive carboxylation by IDH1 or IDH2 to generate citrate, which can be exported from mitochondria to support lipogenesis (Figure 1). Activation of HIF is both necessary and sufficient for driving the reductive carboxylation phenotype in renal cell carcinoma, and suppression of HIF activity can induce a switch from glutamine-mediated lipogenesis back to glucose-mediated lipogenesis [32,35]. Furthermore, loss of VHL and consequent downstream activation of HIF renders renal cell carcinoma cells sensitive to inhibitors of GLS [35]. Evidently, the metabolic routes through which glutamine supports cancer cell proliferation vary with genetic background and with microenvironmental conditions. Nevertheless, it is increasingly clear that diverse oncogenic signals promote glutamine utilization and furthermore that hypoxia, a common condition within poorly vascularized tumors, increases glutamine dependence.
Strategies impacting glutamine metabolism
The reprogrammed metabolism that supports the proliferation and survival of cancer cells also leaves them vulnerable to therapeutic strategies that disrupt metabolic hallmarks of the transformed state. Indeed, several agents targeting cancer cell metabolism are already approved or in clinical trials [36,37]. Because many oncogenic alterations promote a converging metabolic phenotype, characterized by enhanced biosynthesis and cell-autonomous nutrient uptake, targeting metabolism has the potential to affect a broad spectrum of cancers [37]. The importance of glutamine for many critical processes in cancer cells, and the fact that glutamine metabolism is regulated by both oncogenes and tumor suppressors [13,23,25–27,38], makes this branch of cancer metabolism an attractive target for therapeutic strategies. In the sections below, possible approaches for impacting cancer cell glutamine metabolism are outlined, with a focus on the selective inhibition of GLS isoforms.
Depleting cancer cell glutamine supply
A key component of current therapeutic protocols for acute lymphoblastic leukemia (ALL) is the enzyme l-asparaginase, which catalyzes deamination of asparagine to aspartic acid. In addition to its activity towards asparagine, l-asparaginase catalyzes removal of the amide nitrogen from glutamine to form glutamic acid [39]. Studies of ALL patients treated with l-asparaginase demonstrate that serum l-asparaginase activity correlates strongly with asparagine and glutamine depletion in the blood (each greater than 90% in some cases), and that depletion of these amino acids correlates with improved treatment outcome [40,41]. The orphan drug phenylbutyrate has also been demonstrated to lower plasma glutamine concentrations,[42] and both glycerol- and sodium-phenylbutyrate are currently approved by the US FDA for treatment of hyperammonemia in patients with urea cycle disorders. In humans, phenylbutyrate is metabolized to phenylacetate, which is conjugated with glutamine to form phenylacetylglutamine and subsequently excreted [42]. Clinical trials indicate that phenylbutyrate treatment can be safely administered, and results in clinical improvement in some patients with hormone-refractory prostatic carcinoma and glioblastoma multiforme [43]. However, the extent to which plasma glutamine depletion contributes to this outcome is an open question, as at least some of the therapeutic effects of phenylbutyrate are likely to arise from its activity as a histone deacetylase inhibitor [44].
Inhibiting cancer cell glutamine uptake
The c-Myc-regulated transporter SLC1A5 (also called ASCT2) has been implicated in mediating net glutamine uptake in cancer cells, and its expression level is upregulated across a broad spectrum of primary human cancers [13,45]. High levels of SLC1A5 correlate with aggressive biological behavior and decreased patient survival, and proliferation of glutamine-dependent human colon carcinoma cell lines was slowed by inhibition of SLC1A5 [45,46]. The l-glutamine analog l-γ-glutamyl-p-nitroanilide is an inhibitor of SLC1A5-mediated transport, and antagonizes mTORC1 signaling by limiting cellular uptake of glutamine (see below) [47,48].
Recently, pharmacologic and genetic targeting of SLC1A5 was reported to decrease lung cancer cell growth and viability, in part through the effects on mTORC1 signaling [46,49]. It has also been proposed that one mechanism of action of the estrogen receptor modulators tamoxifen and raloxifene is the inhibition of cellular glutamine uptake through SLC1A5 [50].
Use of glutamine mimetics
The antitumor activity of l-glutamine mimetic anti-metabolites was first observed in the 1950s [51], and subsequent research focused on the molecules 6-diazo-5-oxo-l-norleucine (l-DON), acivicin and azaserine, all of which were isolated from Streptomyces species [51]. The primary effect of all three molecules is believed to be the inhibition of purine biosynthesis, although l-DON is also an inhibitor of glutaminase [52]. In spite of promising preclinical data, early clinical trials of each molecule revealed excessive toxicity, including neurotoxicity, myelosuppression, nausea and vomiting [51]. A more recent clinical study of l-DON in combination with PEGylated glutaminase demonstrated therapeutic activity and tolerable toxicity in patients with advanced refractory solid tumors [53]. Nevertheless, a key goal of current research efforts is to develop therapies that target aspects of tumor glutamine metabolism selectively in order to minimize toxic side effects in patients.
Selective inhibition of GLS
A renewed interest in the concept of targeting glutamine catabolism as a therapeutic approach in the treatment of cancer stems from the recent identification of two novel glutaminase inhibitors: 968 and BPTES [bis-2-(5-phenylacetamido-1,2,4-thiadiazoyl-2-yl)ethyl sulfide] (Figure 2). Neither 968 nor BPTES are glutamine mimetics [23,54], suggesting that it may be possible to inhibit GLS specifically without disrupting other aspects of glutamine metabolism. Given the high degree of toxicity observed with glutamine mimetics [51], the identification of allosteric inhibitors of glutaminase potentially represents a significant advance in the development of anticancer therapeutics targeting glutamine metabolism. Both 968 and BPTES are selective for the GLS isoenzyme and can inhibit both of the splice variants, KGA and GAC. However, despite these similarities the two molecules are structurally and functionally distinct, and thus BPTES and 968 can be considered as prototypes for two distinct classes of allosteric GLS inhibitors.
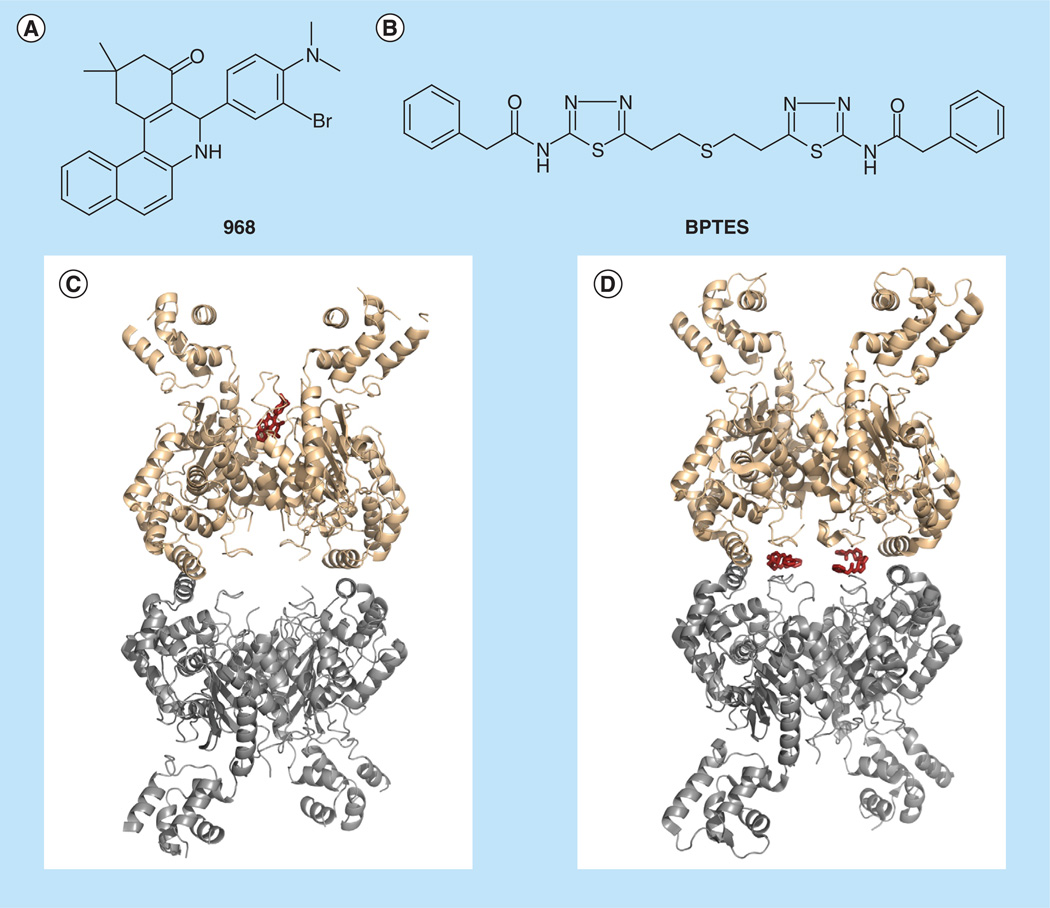
Figure 2. Allosteric inhibitors of GLS.
(A) Chemical structure of 968. (B) Chemical structure of BPTES. (C) Molecular docking model of 968 complexed with GAC. 968 is modeled to bind a hydrophobic pocket at the monomer-monomer interface of the GAC dimer. (D) x-ray crystallographic structure of human GAC in complex with BPTES (PDB 3UO9). Two BPTES molecules bind at the dimer-dimer interface of the GAC tetramer [56].
(C) adapted from [58].
BPTES can bind to the GLS tetramer and effectively inhibit its enzymatic activity. Biochemical studies determined that BPTES stabilizes the tetramer in an inactive conformation [54,55]. Structural studies of both KGA and GAC complexed with BPTES confirm this mechanism of action and reveal the BPTES binding site to be at the dimer–dimer interface (Figure 2) [24,56]. While BPTES is reasonably potent in vitro (IC50 ~60 nM), higher concentrations are needed to produce an effect on GLS in cells (IC50 ~20 µM)[24,5 6]. Moreover, the hydrophobic nature of BPTES (aqueous solubility ~1 µ g/ml) poses some challenges to the physiological delivery of the molecule [57]. Recent efforts to utilize the available structural information to rationally improve on the drug-like properties of BPTES resulted with an analog that retained potency while improving on the aqueous solubility [57].
Structural information regarding the nature of the 968 binding interaction with GLS is not available at this time, but molecular docking experiments of 968 on the GAC isoform place the molecule in a hydrophobic pocket between the N- and C-termini, at the monomer– monomer interface (Figure 2) [58]. The idea that BPTES and 968 may bind at two distinct allosteric sites is congruent with observations that BPTES and 968 achieve GLS inhibition by different mechanisms. In contrast to BPTES, 968 is largely ineffective at inhibiting GLS once the enzyme has been activated, creating a degree of confusion in the field as to the validity of 968 as a GLS inhibitor. A recent in vitro study demonstrated that although 968 is unable to inhibit GAC once it has been activated by phosphate (an allosteric activator of GAC), it is able to block the ability of phosphate to induce the activation of GAC (i.e., treatment of GAC with 968 prior to the addition of phosphate retains GAC in an inactive conformation) [58]. Thus, 968 is not a conventional inhibitor of an active enzyme, but functions by binding inactive GAC (presumably the monomeric and/or dimeric species) and retaining the inactive state.
The different mechanisms by which BPTES and 968 inhibit GAC are also reflected in their respective effects on cells. BPTES rapidly inhibits GAC and cell proliferation, whereas a lag occurs before the effects of 968 are observed [59]. As mentioned earlier, post-translational modification of GAC appears to be necessary for its activation downstream of the Rho GTPases. 968 would be ineffective at inhibiting the activated (and modified) enzyme and protein turnover would be required to allow 968 to bind the inactive (and unmodified) enzyme to bring a complete cessation of GAC activity. It has also been suggested that 968 may only inhibit the activation of GAC under specific signaling contexts [59], which would be in contrast to the general inhibition of GLS by BPTES. Like BPTES, 968 displays a low micromolar potency and poor aqueous solubility and there is a need for analogs with improved drug-like properties. Studies are also being undertaken to characterize potential off-target activities of 968 and BPTES, an important issue about which little is currently known. While both of these molecules can inhibit tumor growth in xenograph studies, it will ultimately be interesting to see which of these allosteric classes of glutaminase inhibitors, if either, yields the greater clinical efficacy in the treatment of cancer, given their distinct modes of action.
Metabolic fates of glutamine & consequences of targeting cancer cell glutamine metabolism
Due to the diverse roles of glutamine as a nutrient, the consequences of therapeutically targeting glutamine metabolism depend on the exact process being impacted. In proliferating cells, glutamine is utilized both for its γ-nitrogen (in nucleotide and hexosamine biosynthesis) and for its carbon skeleton and α-nitrogen (Figure 3). Therapies involving the use of glutamine mimetic antimetabolites or inhibitors of glutamine uptake will therefore have different outcomes to therapies that selectively target a single metabolic reaction.
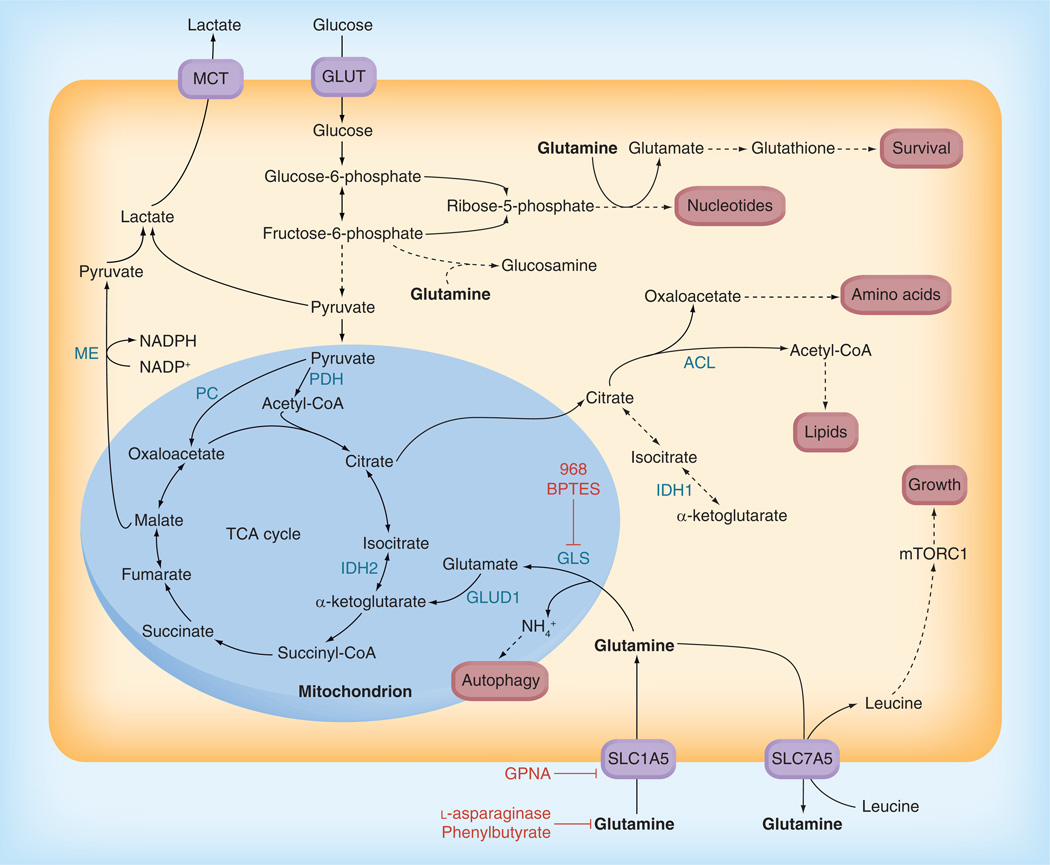
Figure 3. Core pathways of cellular glutamine metabolism.
Core pathways of glutamine metabolism, and their intersections with glucose metabolism, are depicted. 968 BPTES, GPNA, l-asparaginase and phenylbutyrate are inhibitors that target aspects of glutamine metabolism, and are shown in red. Glutamine is an essential nutrient for several processes that are important for cancer cell proliferation and survival. It serves as a nitrogen donor in nucleotide and glucosamine synthesis, a precursor for glutathione synthesis and a key anaplerotic substrate for maintenance of TCA cycle flux. Extracellular glutamine supplies can be depleted by l-asparaginase and by phenylbutyrate. Cellular uptake of glutamine is mediated by the transporter SLC1A5, which can be inhibited by the glutamine analog l-γ-glutamyl-p-nitroanilide. 968 and BPTES disrupt glutamine catabolism by allosterically inhibiting GLS.
ACL: ATP-citrate lyase; GLUD: Glutamate dehydrogenase; GLUT: Glucose transporter; IDH: Isocitrate dehydrogenase;
MCT: Monocarboxylate transporter; ME: Malic enzyme; PC: Pyruvate carboxylase; PDH: Pyruvate dehydrogenase; TCA: Tricarboxylic acid.
Effects on signaling pathways & gene expression
mTORC1 signaling
Glutamine metabolism can lead to the stimulation of multiple signaling pathways that promote cell growth and proliferation. A key signaling node influenced by glutamine is mTORC1, which regulates translation, cell cycle progression and autophagy [48]. mTORC1 integrates growth factor and nutrient signals, and, in the absence of amino acids, is unresponsive to growth factor stimulation [60,61]. Uptake of glutamine through SLC1A5, followed by its rapid efflux through the bidirectional transporter SLC7A5/SLC3A2 in exchange for essential amino acids, is the rate-limiting step for mTORC1 activation in both transformed and non-transformed cell lines [48]. Silencing of SLC1A5 inhibits signaling by mTORC1 to the translational machinery, and abolishes the growth and survival of human hepatoma cells [49,62]. It was recently demonstrated that glutamine catabolism itself, and the consequent generation of α-KG, are also critical for the lysosomal translocation and activation of mTORC1 in glutamine-dependent cervical carcinoma and osteosarcoma cell lines [63].
One important outcome of targeting glutamine uptake and/or catabolism, potentially in a broad range of cancers, will thus be the suppression of mTORC1 signaling, even in the presence of growth factor stimulation. Mutations that promote mTORC1 signaling (e.g., activation of PI3K and Akt, and loss of PTEN, among others) are among the most frequent genetic lesions in cancer, and a range of familial cancer syndromes also involve hyperactivated mTORC1 [64]. It is expected that targeting of mTOR signaling could have therapeutic efficacy in a range of cancers, but rational drug design of mTOR inhibitors has been hampered by the lack of high-resolution structural information [64]. As an alternative to classical ATP-competitive inhibition, attenuation of mTOR signaling through plasma glutamine and asparagine depletion using FDA-approved asparaginase has been proposed [64,65].
The hexosamine biosynthetic pathway
A distinct route through which glutamine influences cellular signaling is the hexosamine biosynthetic pathway (HBP), which requires both glucose and glutamine for production of UDP–GlcNAc, the substrate for protein glycosylation. The signaling activity of numerous proteins, including growth factor receptors for insulin, EGF and interleukins, requires N-linked or O-linked glycosylation [66]. The rate-limiting step in the HBP is the transfer of the amido-group of glutamine to fructose-6-phosphate by the enzyme glutamine:fructose-6-phosphate amido-transferase [67], and glutamine deprivation would therefore be expected to lower flux through this pathway. Intriguingly, in IL-3-dependent hematopoietic cells, inhibition of glutamine metabolism by l-DON blocked IL-3-dependent signaling to Stat5, even when the HBP was rescued by addition of GlcNAc. Supplementation with α-KG restored Stat5 phosphorylation, indicating that mitochondrial metabolism of glutamine, in addition to metabolism through the HBP, is required for robust signaling [68,69]. These studies support the emerging concept that the post-translational modifications that are important for driving cellular growth and proliferation can be impacted at the level of the metabolic pathways that generate the necessary precursors [66].
MAPK signaling
Under aerobic conditions, glutamine catabolism is essential for KRAS-mediated oncogenic growth [70]. Specifically, reactive oxygen species, generated during mitochondrial glutamine metabolism, are required for KRAS-induced transformation through their regulation of the ERKs [70]. Earlier studies similarly indicated an important role for glutamine in MAPK regulation, with both ERK and JNK stimulated by glutamine in enterocytes [71,72]. These signaling effects correlate with increased proliferation and decreased apoptosis, although the underlying mechanisms have not been elucidated.
Given the importance of glutamine supply and catabolism for signaling through various mitogenic pathways, it is expected that the therapeutic targeting of glutamine metabolism would impact the expression of proteins important for cell-cycle progression. Suppression of mTORC1 signaling following inhibition of glutamine uptake or catabolism, for example, would lower the cap-dependent translation of a subset of transcripts that are particularly important for oncogenesis [73,74]. Suppression of ERK and JNK signaling would likewise decrease AP-1-dependent gene transcription [71]. A recent study reported that inhibition of GLS by 968 led to decreased expression of a set of genes important for oncogenic growth, perhaps due to global changes in histone methylation [75]. Furthermore, work using HepG2 hepatoma cells revealed that the presence of glutamine facilitates the gene expression of SREBP targets, both by inducing SREBP-1a transcription and by stimulating SREBP processing [76].
Glutamate & ammonia signaling
In rapidly proliferating cells, including many cancer cells, a major metabolic fate of glutamine is the glutaminase-catalyzed conversion to glutamate and ammonia. Much of the glutamate is subsequently converted to α-KG for TCA cycle anaplerosis. However in certain cancers, including gliomas and melanoma, glutamate secretion stimulates tumor growth and survival through the activation of cell-surface glutamate receptors and their downstream effectors including MAPKs and PI3K [77]. The FDA-approved drug riluzole suppresses secretion of glutamate into the extracellular milieu, and reduces the proliferation, colony formation and invasive activity of melanoma cells [78]. It will be interesting to determine whether cancer cells that are driven by glutamatergic signaling are especially sensitive to inhibitors of GLS. A product of both the glutaminase- and GLUD1-catalyzed reactions, ammonia, also has roles as a signaling molecule, supporting basal autophagy in both transformed and non-transformed cells [79,80]. Since autophagy represents a cytoprotective response that limits tumor cell killing by cytotoxic chemotherapy, blockade of ammonia-driven autophagy via GLS inhibition could sensitize some cancers to treatment [79].
Effects on energy generation & biosynthesis
The metabolic fates of glutamine, and thus the consequences of targeting different aspects of glutamine metabolism, depend on the genetic background of the cancer and on micro-environmental conditions [81,82]. Glutamine starvation of some cancer cells, for example, can be rescued by supplementation with α-KG, indicating that the primary role of glutamine is as a carbon source for TCA cycle anaplerosis. In other cells; however, glutamine starvation can be rescued by addition of ammonia, implicating nitrogen donation as the critical function [81,82]. In c-Myc-driven fibroblasts, addition of TCA cycle intermediates is sufficient to prevent apoptosis on glutamine withdrawal but not sufficient to rescue proliferation, indicating that in this system glutamine is required both for TCA cycle anaplerosis and for other functions [83].
Nucleotide biosynthesis
The rate-limiting step in purine biosynthesis is catalyzed by glutamine 5-phosphoribosyl-1-pyrophosphate amidotransferase, which has a Michaelis constant for glutamine of 0.5–2.5 mM [84,85]. Under normal conditions, glutamine would not be rate-limiting for this reaction; however, glutamine depletion, for example during l-asparaginase treatment, is expected to impact this rate-limiting step [86]. Cultured leukemia cells treated with l-asparaginase are unable to progress into the S-phase of the cell cycle, possibly because low glutamine levels prevent sufficient nucleotide biosynthesis [87]. Indeed, a glutamine supply is critical for progression into S-phase in various cells [88–90], and it was recently demonstrated that whereas both glucose and glutamine are required for progression through the restriction point in mid-to-late G1 in HeLa cells, glutamine is the only substrate essential for progression through S-phase [91]. The G1-to-S transition can be rescued in KRAS-transformed fibroblasts by addition of all four deoxyribonucleotides, indicating that the arrest of proliferation accompanying glutamine withdrawal is due to depleted nucleotide levels [28]. Further illustrating the importance of glutamine as a nitrogen donor in proliferating cells, Meng et al. found that rescue of glutamine withdrawal by α-KG in Hep3B cells is dependent on an ability to generate glutamate from α-KG [82].
TCA cycle anaplerosis
For some proliferating cells, supplementation with α-KG alone is sufficient to rescue the proliferative arrest induced by glutamine withdrawal, indicating that TCA cycle anaplerosis rather than nitrogen donation is the primary metabolic role [81]. The major route for conversion of glutamine to α-KG is via GLUD1 (Figure 3). Some glutamate can also be formed as a result of amido-group transfer from glutamine during hexosamine and nucleotide biosynthesis, and some α-KG can be generated by transaminases. Once glutamine-derived carbon enters the TCA cycle in the form of α-KG, it has several possible fates (Figure 3). In SF188 glioblastoma cells, up to 90% of anaplerotic oxaloacetate is derived from glutamine, and furthermore, the conversion of glutamine to lactate generates sufficient NADPH to support fatty acid synthesis [92]. Secretion of alanine and ammonia accompanies this rapid glutamine catabolism, such that most of the amino groups from glutamine are lost from the cell rather than incorporated into biomolecules [92].
Consistent with the critical role of TCA cycle anaplerosis in cancer cell proliferation, a range of glutamine-dependent cancer cell lines are sensitive to silencing or inhibition of GLS [23,93]. Although loss of GLS suppresses proliferation, in some cases the induction of a compensatory anaplerotic mechanism mediated by pyruvate carboxylase (PC) allows the use of glucose- rather than glutamine-derived carbon for anaplerosis [93]. Low glutamine conditions render glioblastoma cells completely dependent on PC for proliferation; reciprocally, glucose deprivation causes them to become dependent on GLUD1, presumably as a mediator of glutamine-dependent anaplerosis [94]. These studies provide insight into the possibility of inhibiting glutamine-dependent TCA cycle anaplerosis (e.g., with 968 or BPTES) and indicate that high expression of PC could represent a means of resistance to GLS inhibitors.
In c-Myc-induced human Burkitt lymphoma P493 cells, entry of glucose-derived carbon into the TCA cycle is attenuated under hypoxia, whereas glutamine oxidation via the TCA cycle persists [95]. Upon complete withdrawal of glucose, the TCA cycle continues to function and is driven by glutamine. The proportions of viable and proliferating cell populations are almost identical in glucose-replete and -deplete conditions so long as glutamine is present. Inhibition of GLS by BPTES causes a decrease in ATP and glutathione levels, with a simultaneous increase in reactive oxygen species production. Strikingly, whereas BPTES treatment under aerobic conditions suppresses proliferation, under hypoxic conditions it results in cell death, an effect ascribed to glutamine’s critical roles in alleviating oxidative stress in addition to supporting bioenergetics.
In addition to deamidation, glutamine-derived carbon can also reach the TCA cycle through transamination [96], and recent studies indicate that inhibition of this process could be a promising strategy for cancer treatment [30,97,98]. The transaminase inhibitor amino-oxyacetate selectively suppresses proliferation of the aggressive breast cancer cell line MDA-MB-231 relative to normal human mammary epithelial cells, and similar effects were observed with siRNA knockdown of aspartate transaminase [97]. Treatment with amino-oxyacetate killed glutamine-dependent glioblastoma cells, in a manner that could be rescued by α-KG and was dependent on c-Myc expression [13]. Transaminase inhibitors have also been found to suppress both anchorage-dependent and anchorage-independent growth of lung carcinoma cells [98].
Reductive carboxylation
The central metabolic precursor for fatty acid biosynthesis is acetyl-CoA, which can be generated from pyruvate in the mitochondria by pyruvate dehydrogenase. Since acetyl-CoA cannot cross the inner mitochondrial membrane, it is exported to the cytosol via the citrate shuttle following its condensation with oxaloacetate in the TCA cycle (Figure 3). In the cytosol, citrate is converted back to acetyl-CoA and oxaloacetate in a reaction catalyzed by ATP citrate lyase. In addition to its synthesis from glycolytic pyruvate, citrate can also be generated by reductive carboxylation of α-KG [99]. Across a range of cancer cell lines, 10–25% of lipogenic acetyl-CoA is generated from glutamine via this reductive pathway; indeed, reductive metabolism is the primary route for incorporation of glutamine, glutamate and α-KG carbon into lipids [32]. Some of the reductive carboxylation of α-KG is catalyzed by cytosolic IDH1, as well as by mitochondrial IDH2 and/or IDH3.
In A549 lung carcinoma cells, glutamine dependence and reductive carboxylation flux increases under hypoxic conditions [32,34], such that glutamine-derived α-KG accounts for approximately 80% of the carbon used for de novo lipogenesis. Similarly, in melanoma cells, the major source of carbon for acetyl-CoA, citrate and fatty acids switches from glucose under normoxia to glutamine (via reductive carboxylation) under hypoxia [31]. The hypoxic switch to reductive glutamine metabolism is dependent on HIF, and constitutive activation of HIF is sufficient to induce the preferential reductive metabolism of α-KG even under normoxic conditions [32]. Tumor cells with mitochondrial defects, such as electron-transport chain mutations/inhibition, also use glutamine-dependent reductive carboxylation as the major pathway for citrate generation, and loss of electron-transport chain activity is sufficient to induce a switch from glucose to glutamine as the primary source of lipogenic carbon [33].
Together these studies indicate that mitochondrial defects/inhibition, and/or hypoxia, might sensitize cancer cells to inhibition of GLS. The fact that P493 cells are more sensitive to BPTES under hypoxic conditions could in part be explained by an increased reliance on glutamine-dependent reductive carboxylation for lipogenesis [95]. Intriguingly, cancer cells harboring neoenzymatic mutations in IDH1, which results in production of the oncometabolite 2-hydroxyglutarate, are also sensitized to GLS inhibition [100]. 2-hydroxyglutarate is generated primarily from glutamine-derived α-KG [100,101], and therefore tumors expressing mutant IDH might be especially susceptible to alterations in α-KG levels.
Effects on redox homeostasis
One of the key endogenous antioxidants is glutathione (GSH), a tripeptide of glutamate, cysteine and glycine. Not only is glutamine-derived glutamate directly incorporated into GSH molecules, but it is also required for uptake of cystine through the xc− cystine/glutamate antiporter to generate intracellular cysteine [90,102]. In addition to contributing to GSH synthesis, in some contexts glutamine could be important for the reduction of oxidized glutathione to GSH by NADPH, since oxidation of glutamine-derived malate by the malic enzyme is an important means of NADPH generation (Figure 3) [92]. A robust supply of GSH protects tumor cells from the oxidative stress associated with rapid metabolism and is critical for tumor cell survival [103,104]. Within cancer cells, higher GSH levels have been associated with increased resistance to chemotherapy and selective GSH depletion has been proposed as a potential cancer therapy [103].
Potential adverse consequences of targeting glutamine metabolism
As with all therapies, the potential side effects of strategies impacting glutamine metabolism must be seriously considered. The widespread use of l-asparaginase to lower plasma asparagine and glutamine concentrations in ALL patients demonstrates the potential for glutamine metabolism to be safely targeted, and also sheds light on potential toxicological consequences. For example, glutamine is known to be essential for the proliferation of lymphocytes, macrophages and neutrophils, and immunosuppression is a known side effect of l-asparaginase treatment, requiring close monitoring [11,105]. Evidence from early trials using glutamine-mimetic anti-metabolites, such as l-DON, indicates that these unselective molecules can cause excessive gastrointestinal toxicity and neurotoxicity. Within the brain, GLS converts glutamine into the neurotransmitter glutamate in neurons; astrocytes then take up synaptically released glutamate and convert it back to glutamine, which is subsequently transported back to neurons [106,107].
Studies of Gls-null and Gls-heterozygous mice highlight the importance of GLS within the CNS, and indicate that an inability to cross the blood–brain barrier is likely to be an advantageous property of drugs that target glutamine catabolism [107–111]. Although no gross or microscopic defects are detected in peripheral organs or in the CNS of Gls-null mice, the animals die during the first postnatal day [108]. Glutamate levels in the brains of neonatal Gls-null mice are approximately 50% lower than in wild type mice, and glutamatergic synaptic transmission is rapidly exhausted in cultured Gls-null neurons [108]. A possible cause of the observed neonatal demise is a hypoventilation phenotype, consistent with a glutamate-release deficit in neurons regulating respiration [108]. Additionally, renal GLS expression normally increases dramatically during acidosis, leading to excretion of NH4+ [112]; without this compensatory mechanism, Gls-null mice could develop respiratory acidosis [108]. Heterozygous mice, in contrast, survive and are remarkably normal in a wide-ranging battery of behavioral tests, in spite of a 50% global reduction of glutaminase activity [107].
Conclusion
It has become clear during the past decade that altered metabolism plays a critical, in some cases even causal, role in the development and maintenance of cancers. It is now accepted that virtually all oncogenes and tumor suppressors impact metabolic pathways [5]. Furthermore, mutations in certain metabolic enzymes (e.g., isocitrate dehydrogenase, succinate dehydrogenase and fumarate hydratase) are associated with both familial and sporadic human cancers [113]. With this realization has come a renewed interest in the possibility of selectively targeting the metabolism of cancer cells as a therapeutic strategy. The use of l-asparaginase to treat ALL by depleting plasma asparagine and glutamine levels and the promising outcome of the first use of dichloroacetate (which acts, at least in part, through its inhibition of the metabolic enzyme pyruvate dehydrogenase kinase) in glioblastoma patients [114,115], support the notion that cancer metabolism can be safely and effectively targeted in the clinic. The metabolic adaptations of cancer cells must balance the requirements for modestly increased ATP synthesis, dramatically upregulated macromolecular biosynthesis and maintenance of redox balance. By serving as a carbon source for energy generation, a carbon and nitrogen source for biosynthesis and a precursor of the cellular antioxidant glutathione, glutamine is able to contribute to each of these requirements.
The countless combinations of genetic alterations that are found in human neo-plasias mean that there is not a single rigid metabolic program that is characteristic of all transformed cells. This perhaps explains why some current anti-metabolite chemotherapies (e.g., those targeting nucleotide synthesis) are effective only for certain malignancies. A deeper understanding of the metabolic alterations within specific genetic contexts will allow for better-targeted therapeutic interventions. Furthermore, it seems highly likely that combination therapies based on drug synergisms will be especially important for exploiting therapeutic windows within which cancer cells, but not normal cells, are impacted [37]. Glucose and glutamine metabolic pathways, for example, might be able to compensate for one another under some circumstances. When glucose metabolism is impaired in glioblastoma cells, glutamine catabolism becomes essential for survival [94]; reciprocally, suppression of GLS expression causes cells to become fully dependent on glucose-driven TCA cycle anaplerosis via PC [93]. The implication is that PC inhibition could synergize with GLS inhibition.
A topic warranting further investigation is the role that GLS2 plays in cellular metabolism. GLS, in particular the GAC isoform, is upregulated downstream of oncogenes and downregulated by tumor suppressors, and is essential for growth of many cancer cells. In contrast, GLS2 is activated by the ‘universal’ tumor suppressor p53, and furthermore is significantly downregulated in liver tumors and can block transformed characteristics of some cancer cells when overexpressed [116–118]. Emphasizing the importance of genetic context, it was recently reported that GLS2 is significantly upregulated in neuroblastomas overexpressing N-Myc [119]. There are various possible explanations for the apparently different roles of two enzymes that catalyze the same reaction. Because the regulation of GLS and GLS2 is distinct, they will be called up under different conditions. The two enzymes have different kinetic characteristics, and therefore might influence energy metabolism and antioxidant defense in different manners [20]. There is also evidence that GLS2 may act, directly or indirectly, as a transcription factor [118]. Finally, it is possible that the different interactions of GLS and GLS2 with other proteins are responsible for their apparently different roles.
As our understanding deepens of the potential benefits and risks of targeting cancer cell metabolism as a therapeutic strategy, the development of new technologies to study tumor metabolism in vivo will be of great value. 18F-fluorodeoxyglucose positron emission tomography is routinely used to image tumors and relies on an enhanced rate of glucose uptake by cancer cells relative to their non-transformed neighbors. Analogs of amino acids (including glutamine) labeled with 18F or 11C have also been successfully used for positron emission tomography studies [120–124], and the ability to image glutamine uptake and metabolism could indicate specific tumor characteristics, and be used to monitor response to targeted therapeutic agents. Certain cancers, including lymphomas and prostatic carcinomas, lack the high rates of glucose uptake necessary for 18F-fluorodeoxyglucose positron emission tomography, and glutamine-based positron emission tomography could be particularly useful for imaging these diseases [120].
Future perspective
The recent identification of novel inhibitors of GLS, which can selectively block characteristics of the transformed state, represents an exciting advance from the toxicity associated with anti-metabolite glutamine mimetics. As the field develops, it will become increasingly important to elucidate details of the regulation of glutamine metabolism within cancer cells and its integration with other metabolic adaptations. This will not only highlight the potential strengths and weaknesses of glutamine-targeted therapies, but will also reveal ‘loopholes’ for the development of drug resistance, along with opportunities for drug synergism.
Executive summary.
Cancer cell metabolism & glutamine addiction
-
▪
Along with the Warburg effect, upregulated glutamine metabolism is a metabolic hallmark of cell proliferation and the transformed state.
Oncogenic alterations affecting glutamine metabolism
-
▪
Many oncogenic signals induce cellular glutamine dependence and regulate multiple metabolic pathways for glutamine.
Strategies impacting glutamine metabolism
-
▪
Early strategies for targeting cancer cell glutamine metabolism involved the use of glutamine mimetic anti-metabolites, such as l-DON. These molecules yielded promising preclinical data, but failed in clinical trials due to excessive toxicity, especially in the GI tract and the nervous system.
-
▪
More recently, inhibitors have been identified that target specific aspects of glutamine metabolism. The small molecules 968 and BPTES represent two classes of allosteric inhibitors of the glutaminase isoenzyme GLS.
-
▪
Inhibition of GLS can selectively suppress characteristics of cellular transformation, and 968 and BPTES both slow tumor growth in xenograft models.
Metabolic fates of glutamine & consequences of targeting cancer cell glutamine metabolism
-
▪
Important roles played by glutamine in cancer cells include TCA cycle anaplerosis, nitrogen donation for nucleotide and hexosamine biosynthesis, precursor provision for glutathione production, and stimulation of mitogenic signaling.
-
▪
Inhibiting aspects of cellular glutamine metabolism has the potential to impact numerous processes important for oncogenic growth. Possible side effects of targeting cancer cell glutamine metabolism include immunosuppression and neurotoxicity. Inability to cross the blood–brain barrier could be an advantageous property of any drug molecules that are developed.
Acknowledgements
The authors would like to thank C Westmiller for her expert secretarial assistance.
MJ Lukey acknowledges support from the Fleming Fellowship (Cornell University) and RA Cerione acknowledges support from NIH (GM040654, GM061762 and CA163255).
Key Terms
- Warburg effect
Phenomenon of enhanced glucose uptake and glycolytic flux, coupled to high rates of lactate secretion (rather than complete oxidation of glucose-derived carbon via the tricarboxylic acid cycle), which persists even under aerobic conditions.
- Glutamine catabolism
Series of metabolic reactions whereby glutamine is degraded. An important pathway in many proliferating cells is the conversion of glutamine to α-ketoglutarate (α-KG), for entry into the tricarboxylic acid cycle. The enzyme glutaminase catalyzes the hydrolysis of glutamine to glutamate and ammonium, and glutamate dehydrogenase subsequently catalyzes the deamination of glutamate to generate α-KG along with a second ammonium. The conversion of glutamate to α-KG can also be catalyzed by alanine transaminase and by aspartate transaminase.
- Anaplerosis
Replenishment of metabolic intermediates within a given pathway. For example, when the tricarboxylic acid (TCA) cycle intermediate citrate is exported from the mitochondrion to support lipid biosynthesis in the cytosol, anaplerosis must occur to sustain TCA cycle flux. Glutamine can serve as an anaplerotic substrate in this case, following its conversion via glutamate to the TCA cycle intermediate α-ketoglutarate.
- Rho GTPases
Family of signaling proteins that influences many cellular processes, including actin cytoskeleton rearrangements and cell-cycle progression. Rho GTPases play fundamental roles in several aspects of cancer progression, and were recently reported to upregulate the activity of the glutaminase isoenzyme GLS.
- Reductive carboxylation
Metabolic pathway through which α-ketoglutarate, often derived from glutamine, is reductively converted to citrate, as opposed to undergoing oxidative metabolism via the ‘forward’ reactions of the tricarboxylic acid cycle. Reductive carboxylation is particularly important under hypoxic conditions and in cells with mitochondrial defects. The citrate that is generated can be exported from the mitochondrion to support cytosolic lipogenesis.
- Cancer cell metabolism
Many cancer cells display metabolic phenotypes that are dramatically altered relative to their non-transformed counterparts. Widely conserved features of cancer cell metabolism include enhanced nutrient uptake, and rewiring of metabolic pathways to increase production of biosynthetic intermediates. Over recent years, it has become clear that many oncogenic proteins are involved in the reprogramming of cancer cell metabolism.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
- 1.Warburg O. Origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. Respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 3.Warburg O, Posener K, Negelein E. On the metabolism of carcinoma cells. Biochemische Zeitschrift. 1924;152:309–344. [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin. Cell Dev. Biol. 2012;23(4):362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Newsholme P. Why is l-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J. Nutr. 2001;131(9 Suppl.):2515S–2522S. doi: 10.1093/jn/131.9.2515S. discussion 2523S–2514S. [DOI] [PubMed] [Google Scholar]
- 12.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122(3168):501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 13.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl Acad. Sci. USA. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Icard P, Poulain L, Lincet H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim. Biophys. Acta. 2012;1825(1):111–116. doi: 10.1016/j.bbcan.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Aledo JC, Gomez-Fabre PM, Olalla L, Marquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome. 2000;11(12):1107–1110. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 16.Szeliga M, Obara-Michlewska M. Glutamine in neoplastic cells: focus on the expression and roles of glutaminases. Neurochem. Int. 2009;55(1–3):71–75. doi: 10.1016/j.neuint.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Marquez J, de la Oliva AR, Mates JM, Segura JA, Alonso FJ. Glutaminase: a multifaceted protein not only involved in generating glutamate. Neurochem. Int. 2006;48(6–7):465–471. doi: 10.1016/j.neuint.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Gomez-Fabre PM, Aledo JC, Del Castillo-Olivares A, et al. Molecular cloning, sequencing and expression studies of the human breast cancer cell glutaminase. Biochem. J. 2000;345(Pt 2):365–375. [PMC free article] [PubMed] [Google Scholar]
- 19.Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genomics. 1999;1(2):51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- 20.Mates JM, Segura JA, Martin-Rufian M, Campos-Sandoval JA, Alonso FJ, Marquez J. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr. Mol. Med. 2013;13(4):514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 21.Porter LD, Ibrahim H, Taylor L, Curthoys NP. Complexity and species variation of the kidney-type glutaminase gene. Physiol. Genomics. 2002;9(3):157–166. doi: 10.1152/physiolgenomics.00017.2002. [DOI] [PubMed] [Google Scholar]
- 22.Cassago A, Ferreira AP, Ferreira IM, et al. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl Acad. Sci. USA. 2012;109(4):1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. ▪ The discovery that GLS is essential for Rho GTPase-mediated transformation and the identification of 968 as an inhibitor of GLS.
- 24.Thangavelu K, Pan CQ, Karlberg T, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl Acad. Sci. USA. 2012;109(20):7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. ▪ The demonstration that expression levels of GLS can be regulated by the transcription factor c-Myc, via its suppression of the micro-RNAs miR-23a/b.
- 26.Reynolds MR, Lane AN, Robertson B, et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene. 2013 doi: 10.1038/onc.2012.635. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaglio D, Metallo CM, Gameiro PA, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS ONE. 2009;4(3):e4715. doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr EL, Kelman A, Wu GS, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185(2):1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. ▪ Identification of a novel KRAS-regulated pathway for glutamine metabolism, which requires activity of aspartate transaminase (GOT1).
- 31.Filipp FV, Scott DA, Ronai ZA, Osterman AL, Smith JW. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(3):375–383. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wise DR, Ward PS, Shay JE, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl Acad. Sci. USA. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. ▪ Demonstration that hypoxia or HIF activation promote the reductive carboxylation of glutamine-derived α-ketoglutarate, such that glutamine becomes a major source of the key metabolite citrate.
- 35.Gameiro PA, Yang J, Metelo AM, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat. Biotechnol. 2012;30(7):671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. ▪ A review of the therapeutic opportunities emerging from recent research into cancer cell metabolism.
- 38.van der Vos KE, Eliasson P, Proikas-Cezanne T, et al. Modulation of glutamine metabolism by the PI(3)K-PKB-FOXO network regulates autophagy. Nat. Cell. Biol. 2012;14(8):829–837. doi: 10.1038/ncb2536. [DOI] [PubMed] [Google Scholar]
- 39.Avramis VI, Panosyan EH. Pharmacokinetic/ pharmacodynamic relationships of asparaginase formulations: the past, the present and recommendations for the future. Clin. Pharmacokinet. 2005;44(4):367–393. doi: 10.2165/00003088-200544040-00003. [DOI] [PubMed] [Google Scholar]
- 40.Abshire TC, Pollock BH, Billett AL, Bradley P, Buchanan GR. Weekly polyethylene glycol conjugated l-asparaginase compared with biweekly dosing produces superior induction remission rates in childhood relapsed acute lymphoblastic leukemia: a Pediatric Oncology Group Study. Blood. 2000;96(5):1709–1715. [PubMed] [Google Scholar]
- 41.Grigoryan RS, Panosyan EH, Seibel NL, Gaynon PS, Avramis IA, Avramis VI. Changes of amino acid serum levels in pediatric patients with higher-risk acute lymphoblastic leukemia (CCG-1961) In Vivo. 2004;18(2):107–112. [PubMed] [Google Scholar]
- 42.Darmaun D, Welch S, Rini A, Sager BK, Altomare A, Haymond MW. Phenylbutyrate-induced glutamine depletion in humans: effect on leucine metabolism. Am. J. Physiol. 1998;274(5 Pt 1):E801–E807. doi: 10.1152/ajpendo.1998.274.5.E801. [DOI] [PubMed] [Google Scholar]
- 43.Thibault A, Cooper MR, Figg WD, et al. A Phase I and pharmacokinetic study of intravenous phenylacetate in patients with cancer. Cancer Res. 1994;54(7):1690–1694. [PubMed] [Google Scholar]
- 44.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 45.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 2005;15(4):254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Hassanein M, Hoeksema MD, Shiota M, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin. Cancer Res. 2013;19(3):560–570. doi: 10.1158/1078-0432.CCR-12-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esslinger CS, Cybulski KA, Rhoderick JF. Ngamma-aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg. Med. Chem. 2005;13(4):1111–1118. doi: 10.1016/j.bmc.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 48.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs BC, Finger RE, Onan MC, Bode BP. ASCT2 silencing regulates mammalian target-of-rapamycin growth and survival signaling in human hepatoma cells. Am. J. Physiol. Cell Physiol. 2007;293(1):C55–C63. doi: 10.1152/ajpcell.00330.2006. [DOI] [PubMed] [Google Scholar]
- 50.Todorova VK, Kaufmann Y, Luo S, Klimberg VS. Tamoxifen and raloxifene suppress the proliferation of estrogen receptor-negative cells through inhibition of glutamine uptake. Cancer Chemother. Pharmacol. 2011;67(2):285–291. doi: 10.1007/s00280-010-1316-y. [DOI] [PubMed] [Google Scholar]
- 51.Ahluwalia GS, Grem JL, Hao Z, Cooney DA. Metabolism and action of amino acid analog anti-cancer agents. Pharmacol. Ther. 1990;46(2):243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths M, Keast D, Patrick G, Crawford M, Palmer TN. The role of glutamine and glucose analogues in metabolic inhibition of human myeloid leukaemia in vitro . Int. J. Biochem. 1993;25(12):1749–1755. doi: 10.1016/0020-711x(88)90303-5. [DOI] [PubMed] [Google Scholar]
- 53.Mueller C, Al-Batran S, Jaeger E, et al. A Phase IIa study of PEGylated glutaminase (PEG-PGA) plus 6-diazo-5-oxo-l-norleucine (DON) in patients with advanced refractory solid tumors. J. Clin. Oncol. 2008;26(15) [Google Scholar]
- 54. Robinson MM, McBryant SJ, Tsukamoto T, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem. J. 2007;406(3):407–414. doi: 10.1042/BJ20070039. ▪ Identification of BPTES as a novel inhibitor of GLS.
- 55.Hartwick EW, Curthoys NP. BPTES inhibition of hGA(124–551), a truncated form of human kidney-type glutaminase. J. Enzyme Inhib. Med. Chem. 2012;27(6):861–867. doi: 10.3109/14756366.2011.622272. [DOI] [PubMed] [Google Scholar]
- 56. DeLaBarre B, Gross S, Fang C, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50(50):10764–10770. doi: 10.1021/bi201613d. ▪ The first full-length structure of GAC, in the presence and absence of the selective inhibitor BPTES.
- 57.Shukla K, Ferraris DV, Thomas AG, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 2012;55(23):10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katt WP, Ramachandran S, Erickson JW, Cerione RA. Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol. Cancer Ther. 2012;11(6):1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson KF, Erickson JW, Antonyak MA, Cerione RA. Rho GTPases and their roles in cancer metabolism. Trends Mol. Med. 2013;19(2):74–82. doi: 10.1016/j.molmed.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 61.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuchs BC, Perez JC, Suetterlin JE, Chaudhry SB, Bode BP. Inducible antisense RNA targeting amino acid transporter ATB0/ ASCT2 elicits apoptosis in human hepatoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286(3):G467–G478. doi: 10.1152/ajpgi.00344.2003. [DOI] [PubMed] [Google Scholar]
- 63.Duran RV, Oppliger W, Robitaille AM, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell. 2012;47(3):349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 64.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 65.Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent l-asparaginase. J. Biol. Chem. 2009;284(47):32742–32749. doi: 10.1074/jbc.M109.047910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Metallo CM, Vander Heiden MG. Metabolism strikes back: metabolic flux regulates cell signaling. Genes Dev. 2010;24(24):2717–2722. doi: 10.1101/gad.2010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ngoh GA, Facundo HT, Zafir A, Jones SP. O-GlcNAc signaling in the cardiovascular system. Circ. Res. 2010;107(2):171–185. doi: 10.1161/CIRCRESAHA.110.224675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wellen KE, Lu C, Mancuso A, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24(24):2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Donadio AC, Lobo C, Tosina M, et al. Antisense glutaminase inhibition modifies the O-GlcNAc pattern and flux through the hexosamine pathway in breast cancer cells. J. Cell Biochem. 2008;103(3):800–811. doi: 10.1002/jcb.21449. [DOI] [PubMed] [Google Scholar]
- 70.Weinberg F, Hamanaka R, Wheaton WW, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rhoads JM, Argenzio RA, Chen W, et al. l-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Physiol. 1997;272(5 Pt 1):G943–G953. doi: 10.1152/ajpgi.1997.272.5.G943. [DOI] [PubMed] [Google Scholar]
- 72.Larson SD, Li J, Chung DH, Evers BM. Molecular mechanisms contributing to glutamine-mediated intestinal cell survival. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293(6):G1262–G1271. doi: 10.1152/ajpgi.00254.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satheesha S, Cookson VJ, Coleman LJ, et al. Response to mTOR inhibition: activity of eIF4E predicts sensitivity in cell lines and acquired changes in eIF4E regulation in breast cancer. Mol. Cancer. 2011;10:19. doi: 10.1186/1476-4598-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simpson NE, Tryndyak VP, Pogribna M, Beland FA, Pogribny IP. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 2012;7(12):1413–1420. doi: 10.4161/epi.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inoue J, Ito Y, Shimada S, et al. Glutamine stimulates the gene expression and processing of sterol regulatory element binding proteins, thereby increasing the expression of their target genes. FEBS J. 2011;278(15):2739–2750. doi: 10.1111/j.1742-4658.2011.08204.x. [DOI] [PubMed] [Google Scholar]
- 77.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin. Cancer Res. 2012;18(16):4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le MN, Chan JL, Rosenberg SA, et al. The glutamate release inhibitor Riluzole decreases migration, invasion, and proliferation of melanoma cells. J. Invest. Dermatol. 2010;130(9):2240–2249. doi: 10.1038/jid.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci. Signal. 2010;3(119):ra31. doi: 10.1126/scisignal.2000911. ▪ Discovery that ammonia derived from glutamine catabolism can promote autophagy.
- 80.Cheong H, Lindsten T, Thompson CB. Autophagy and ammonia. Autophagy. 2012;8(1):122–123. doi: 10.4161/auto.8.1.18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9(19):3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 82.Meng M, Chen S, Lao T, Liang D, Sang N. Nitrogen anabolism underlies the importance of glutaminolysis in proliferating cells. Cell Cycle. 2010;9(19):3921–3932. doi: 10.4161/cc.9.19.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007;178(1):93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weber G, Prajda N, Lui MS, et al. Multi-enzyme-targeted chemotherapy by acivicin and actinomycin. Adv. Enzyme Regul. 1982;20:75–96. doi: 10.1016/0065-2571(82)90009-7. [DOI] [PubMed] [Google Scholar]
- 85.Wyngaarden JB. Regulation of purine biosynthesis and turnover. Adv. Enzyme Regul. 1976;14:25–42. doi: 10.1016/0065-2571(76)90006-6. [DOI] [PubMed] [Google Scholar]
- 86.Cory JG, Cory AH. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo. 2006;20(5):587–589. [PubMed] [Google Scholar]
- 87.Krejci O, Starkova J, Otova B, et al. Upregulation of asparagine synthetase fails to avert cell cycle arrest induced by l-asparaginase in TEL/AML1-positive leukaemic cells. Leukemia. 2004;18(3):434–441. doi: 10.1038/sj.leu.2403259. [DOI] [PubMed] [Google Scholar]
- 88.Chang WK, Yang KD, Shaio MF. Lymphocyte proliferation modulated by glutamine: involved in the endogenous redox reaction. Clin. Exp. Immunol. 1999;117(3):482–488. doi: 10.1046/j.1365-2249.1999.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horig H, Spagnoli GC, Filgueira L, et al. Exogenous glutamine requirement is confined to late events of T cell activation. J. Cell Biochem. 1993;53(4):343–351. doi: 10.1002/jcb.240530412. [DOI] [PubMed] [Google Scholar]
- 90.Roth E, Oehler R, Manhart N, et al. Regulative potential of glutamine - relation to glutathione metabolism. Nutrition. 2002;18(3):217–221. doi: 10.1016/s0899-9007(01)00797-3. [DOI] [PubMed] [Google Scholar]
- 91.Colombo SL, Palacios-Callender M, Frakich N, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl Acad. Sci. USA. 2011;108(52):21069–21074. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl Acad. Sci. USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cheng T, Sudderth J, Yang C, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl Acad. Sci. USA. 2011;108(21):8674–8679. doi: 10.1073/pnas.1016627108. ▪ The finding that an alternative anaplerotic pathway involving pyruvate carboxylase is crucial for survival of tumor cells during glutamine deprivation, and could be a means of resistance to drugs that target GLS.
- 94.Yang C, Sudderth J, Dang T, Bachoo RM, McDonald JG, DeBerardinis RJ. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69(20):7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Le A, Lane AN, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+−dependent malic enzyme. J. Biol. Chem. 1984;259(10):6215–6221. [PubMed] [Google Scholar]
- 97.Thornburg JM, Nelson KK, Clem BF, et al. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10(5):R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beuster G, Zarse K, Kaleta C, et al. Inhibition of alanine aminotransferase in silico and in vivo promotes mitochondrial metabolism to impair malignant growth. J. Biol. Chem. 2011;286(25):22323–22330. doi: 10.1074/jbc.M110.205229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holleran AL, Briscoe DA, Fiskum G, Kelleher JK. Glutamine metabolism in AS-30D hepatoma cells. Evidence for its conversion into lipids via reductive carboxylation. Mol. Cell Biochem. 1995;152(2):95–101. doi: 10.1007/BF01076071. [DOI] [PubMed] [Google Scholar]
- 100.Seltzer MJ, Bennett BD, Joshi AD, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lo M, Wang YZ, Gout PW. The x(c)-cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J. Cell Physiol. 2008;215(3):593–602. doi: 10.1002/jcp.21366. [DOI] [PubMed] [Google Scholar]
- 103.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006;43(2):143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 104.Mates JM, Segura JA, Alonso FJ, Marquez J. Oxidative stress in apoptosis and cancer: an update. Arch. Toxicol. 2012;86(11):1649–1665. doi: 10.1007/s00204-012-0906-3. [DOI] [PubMed] [Google Scholar]
- 105.Bunpo P, Murray B, Cundiff J, Brizius E, Aldrich CJ, Anthony TG. Alanyl-glutamine consumption modifies the suppressive effect of l-asparaginase on lymphocyte populations in mice. J. Nutr. 2008;138(2):338–343. doi: 10.1093/jn/138.2.338. [DOI] [PubMed] [Google Scholar]
- 106.Chaudhry FA, Reimer RJ, Edwards RH. The glutamine commute: take the N line and transfer to the A. J. Cell Biol. 2002;157(3):349–355. doi: 10.1083/jcb.200201070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gaisler-Salomon I, Miller GM, Chuhma N, et al. Glutaminase-deficient mice display hippocampal hypoactivity, insensitivity to pro-psychotic drugs and potentiated latent inhibition: relevance to schizophrenia. Neuropsychopharmacology. 2009;34(10):2305–2322. doi: 10.1038/npp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Masson J, Darmon M, Conjard A, et al. Mice lacking brain/kidney phosphate-activated glutaminase have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior and die shortly after birth. J. Neurosci. 2006;26(17):4660–4671. doi: 10.1523/JNEUROSCI.4241-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gaisler-Salomon I, Wang Y, Chuhma N, et al. Synaptic underpinnings of altered hippocampal function in glutaminase-deficient mice during maturation. Hippocampus. 2012;22(5):1027–1039. doi: 10.1002/hipo.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.El Hage M, Masson J, Conjard-Duplany A, Ferrier B, Baverel G, Martin G. Brain slices from glutaminase-deficient mice metabolize less glutamine: a cellular metabolomic study with carbon 13 NMR. J. Cereb. Blood Flow Metab. 2012;32(5):816–824. doi: 10.1038/jcbfm.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bae N, Wang Y, Li L, Rayport S, Lubec G. Network of brain protein level changes in glutaminase deficient fetal mice. J. Proteomics. 2013;80:236–249. doi: 10.1016/j.jprot.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Curthoys NP. Role of mitochondrial glutaminase in rat renal glutamine metabolism. J. Nutr. 2001;131(9 Suppl.):2491S–2495S. doi: 10.1093/jn/131.9.2491S. discussion 2496S–2497S. [DOI] [PubMed] [Google Scholar]
- 113.Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J. Mol. Med. (Berl.) 2011;89(3):213–220. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Michelakis ED, Sutendra G, Dromparis P, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci. Transl. Med. 2010;2(31):31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 115.Vander Heiden MG. Targeting cell metabolism in cancer patients. Sci. Transl. Med. 2010;2(31):31ed1. doi: 10.1126/scitranslmed.3001210. [DOI] [PubMed] [Google Scholar]
- 116.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl Acad. Sci. USA. 2010;107(16):7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl Acad. Sci. USA. 2010;107(16):7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Szeliga M, Obara-Michlewska M, Matyja E, et al. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57(9):1014–1023. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 119.Qing G, Li B, Vu A, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22(5):631–644. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rajagopalan KN, DeBerardinis RJ. Role of glutamine in cancer: therapeutic and imaging implications. J. Nucl. Med. 2011;52(7):1005–1008. doi: 10.2967/jnumed.110.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu A, Lee D, Shim H. Metabolic positron emission tomography imaging in cancer detection and therapy response. Semin. Oncol. 2011;38(1):55–69. doi: 10.1053/j.seminoncol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lieberman BP, Ploessl K, Wang L, et al. PET imaging of glutaminolysis in tumors by 18F-(2S,4R)4-fluoroglutamine. J. Nucl. Med. 2011;52(12):1947–1955. doi: 10.2967/jnumed.111.093815. [DOI] [PubMed] [Google Scholar]
- 123.Qu W, Oya S, Lieberman BP, et al. Preparation and characterization of l-[5–11C]-glutamine for metabolic imaging of tumors. J. Nucl. Med. 2012;53(1):98–105. doi: 10.2967/jnumed.111.093831. [DOI] [PubMed] [Google Scholar]
- 124.Qu W, Zha Z, Ploessl K, et al. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J. Am. Chem. Soc. 2011;133(4):1122–1133. doi: 10.1021/ja109203d. [DOI] [PubMed] [Google Scholar]