Abstract
End-to-end chromosome fusions that occur in the context of telomerase deficiency can trigger genomic duplications. For over 70 years these duplications have been attributed solely to Breakage-Fusion-Bridge cycles. To test this hypothesis, we examined end-to-end fusions isolated from C. elegans telomere replication mutants. Genome level rearrangements revealed fused chromosome ends possessing interrupted terminal duplications accompanied by template switching events. These features are very similar to disease-associated duplications of interstitial segments of the human genome. A model termed Fork Stalling and Template Switching has been proposed previously to explain such duplications, where promiscuous replication of large, non-contiguous segments of the genome occurs. Thus, a DNA synthesis-based process may create duplications that seal end-to-end fusions, in the absence of Breakage-Fusion-Bridge cycles.
Most human somatic cells are deficient for telomerase and experience progressive loss of telomeric DNA at chromosome ends. Critically shortened telomeres can elicit high levels of end-to-end chromosome fusion, resulting in genome rearrangements that commonly occur in developing tumors (1). In many organisms, the instability of dicentric chromosomes impedes elucidation of the initial structures of fusions events and therefore a mechanistic understanding of their genesis (Fig. 1A). Because Caenorhabditis elegans has holocentric chromosomes, end-to-end fusions derived from telomerase deficient backgrounds can be transmitted stably during mitosis and meiosis (Fig. 1A). In other organisms, subtelomeric duplications that occur at critically shortened telomeres have been attributed to chromosome fusion followed by breakage during mitosis: the breakage-fusion-bridge (BFB) model (2-3). To test the hypothesis, we genetically isolated C. elegans end-to-end chromosome fusions based on the meiotic non-disjunction phenotype that they cause when heterozygous (4-5). These fusions were isolated from mutants deficient for the trt-1 telomerase reverse transcriptase or for the DNA damage checkpoint gene mrt-2, which is required for telomerase-mediated telomere repeat addition (4-5). Homozygous fusions were then stably maintained, as assessed by genetic mapping and chromosome cytology, in strains possessing normal telomerase activity (6).
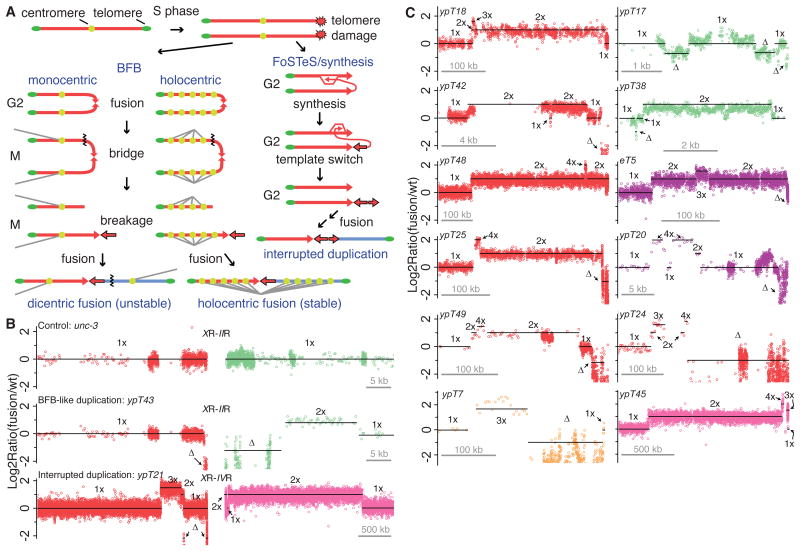
Figure 1. Fusion breakpoint architecture.
(A) Although BFB can explain terminal duplications (red arrows with black borders) at fusion breakpoints, a synthesis-based mechanism is plausible. Subsequent fusion to a second chromosome (blue) yields an unstable dicentric in most organisms, but a mitotically stable holocentric fusion in C. elegans. Black zigzag lines indicate impending breaks. (B) CGH plots of an unc-3 control with normal chromosome termini, and fusion strains with duplications at fused chromosome termini. Telomeres are in center of the panel. (C) Chromosome ends bearing interrupted duplications. Telomeres are on the right side of CGH plots. The Y-axes represent the ratio of signal intensity for fusion versus wildtype. The X-axes represent the distance from a chromosome end, where green, purple, pink, tan and red indicate chromosomes II, III, IV, V, and X, respectively. Symbols: Δ (terminal or internal deletion), 1× (wildtype copy number), 2× (duplication), 3× (triplication), 4× (quadruplication).
The molecular structures of 38 X-autosome end-to-end chromosome fusion events derived from C. elegans trt-1 mutant strains were investigated using a whole-genome oligonucleotide microarray by Comparative Genomic Hybridization (CGH) (7-8). Subtelomeric duplications were present at one (n=17), both (n=12), or neither (n=9) of the fused chromosome ends (Figs. S1-S2). Duplicated regions ranged in size from 100 bp to 2.1 Mb and were typically two, three or four times wildtype copy number (Tables S1 and S2). Subtelomeric deletions frequently occurred prior to duplication (6), presumably as a consequence of end-resection of critically shortened telomeres.
In the BFB model, breakage of a sister chromatid fusion during anaphase may yield daughter cells with broken chromosomes carrying either a terminal deletion or an inverted duplication (Fig. 1A) (9). BFB-type inverted duplications could occur in C. elegans if holocentric sister chromatids fuse in G2 and are then pulled towards opposite sides of the mitotic spindle and severed (Figs. 1A and S3). CGH and/or sequencing revealed uninterrupted duplications consistent with BFB for 18/41 fused chromosome termini (Figs. 2B, S5B, and S7). For example, the fusion ypT43 (XR-IIR) (italics = chromosome number; R = right end) is sealed by a 15.4 kb uninterrupted duplication of IIR that is inverted with respect to the parent chromosome II (Fig. S5B), suggesting sister chromatid fusion of IIR and breakage, prior to fusion with X. The remaining 23/41 fused chromosome termini possessed structures that ruled out BFB, such as duplications interrupted by non-duplicated or triplicated sequences (Figs. 1B-C, 2B-D, and 3).
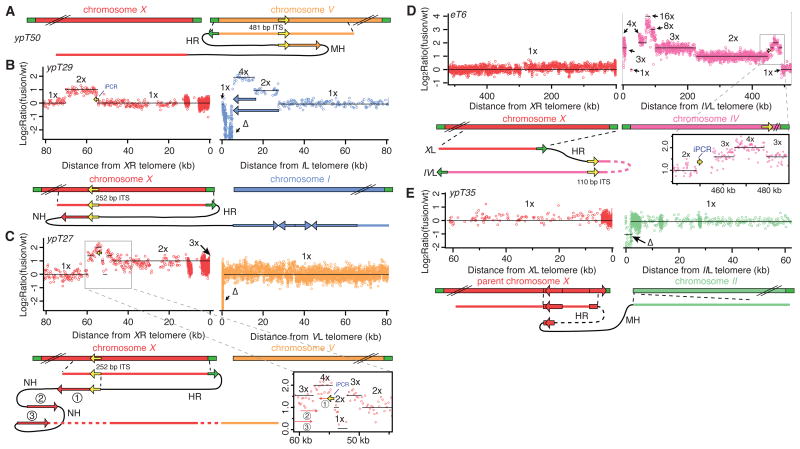
Figure 2. Homology-driven synthesis generates terminal duplications.
Recombination of an uncapped telomere (green arrows) and an interstitial telomeric sequence tract (ITS: yellow arrows) occurred for four fusions (A-D). (E) HR within a large inverted repeat for ypT35. Circled numbers show the predicted origins, order, and relative orientations of altered block arrow regions based on sequence analysis. Dashed lines indicate unknown structure of segments of chromosome. DNA sequence transitions exhibited NH (0 bp), MH (1 to 4 bp), or HR (>11 bp) of homology. Plots follow same scheme as Figure 1.
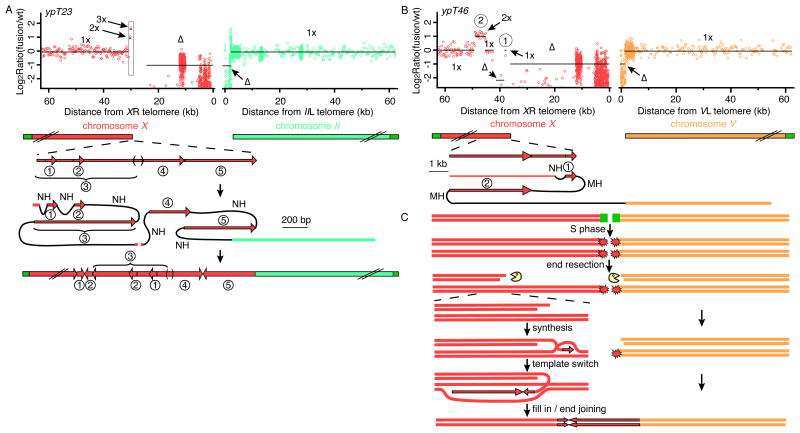
Figure 3. Genomic shards seal complex fusion breakpoints.
(A) ypT23 and (B) ypT46 fusion breakpoints. The gray box shows the position of a 1.8 kb locus from which the genomic shards originated. (C) Model of synthesis-based duplication for ypT46.
To characterize some of the chromosome rearrangements in detail, we used PCR analysis to amplify fusion junctions and thereby to reveal the orientation and connectivity of the chromosome segments. An inverse PCR approach employed primers targeted to a single chromosome end, which revealed recombination or end-joining with interstitial sites in the genome for complex fusions ypT21, ypT27, ypT29, ypT50 and eT6 (n= 26 tested) (Fig. S4 and Supporting Material). The fusion junctions coincided with borders of genomic duplications observed by CGH (Figs. 2B-D, S5A, and S7; iPCR in blue font on CGH plots). A second PCR approach employed robust, validated primers at borders of copy number changes to span rearrangement junctions predicted based on CGH data alone. Eighteen sequence transitions were recovered for 9 fusions (ypT27, ypT29, ypT50, ypT23, ypT46, ypT49, ypT43, ypT35, and ypT37; n=12 tested) (Table S3). Thus, PCR analysis demonstrated that the terminal duplications observed at fused chromosome ends were connected to the fused chromosome ends, suggesting that these duplications seal end-to-end fusions of critically shortened telomeres.
Most rearrangement junctions shared either no homology (NH) or 1 to 4 nucleotides of microhomology (MH), indicating that the fusions were products of non-homologous end-joining (NHEJ) events (Table S4). However, several fusion breakpoints contained longer stretches of homology (HR) corresponding to the telomeric repeat sequence TTAGGC. In the genome of C. elegans, many interstitial telomere sequence tracts (ITSs) appear within the terminal 5 Mb of each chromosome end, ranging in size from 12 bp to 486 bp and consisting of perfect telomere repeats interspersed among degenerate telomere repeats (Fig. S6). During creation of three fusions, ypT50, ypT29 and ypT27, an uncapped telomere recombined with the ITS located closest to its chromosome terminus, presumably the ITS of a sister chromatid (Figs. 2A-C, S7, and S8). For eT6, an interchromosomal telomere recombination event occurred where the XL telomere recombined with the fifth ITS from the IVL terminus (Figs. 2D and S6). This structure could have been formed if strand transfer occurred within or adjacent to the longest stretch of perfect telomere repeat sequence in each ITS. The identical 25-nucleotide sequence within the terminal XR ITS was targeted for ypT27 and ypT29 (Fig. S6). The amount of perfect telomere sequence remaining at the telomere recombination sites of ypT29, ypT27, ypT50, and eT6 was 29.5, 16.7, 18, and 24.7 repeats, respectively. Telomeres can adopt a highly conserved strand invasion structure termed the T-loop, whose minimal size in vitro is 148 bp or 24.7 repeats (10). Thus, telomere uncapping in the context of telomerase deficiency may result from an inability to form T-loops that may protect chromosome termini from aberrant recombination events (11-12).
Although telomere recombination events have been observed for genomic DNA containing end-to-end fusions from human cells and from C. elegans (13-14), their consequences have not been reported previously. The ypT27 and eT6 telomere recombination events each resulted in interrupted duplications (Fig. 2C-D). Recombination occurred at ITS tracts facing away from a chromosome end, yet elicited copy number changes on both sides of the targeted ITS, including much of the sequence between the ITS and its chromosome terminus (Fig. 2C-D). The ypT27 and eT6 structures are inconsistent with BFB cycles or break-induced replication (BIR), where a new replication fork is established at a site of homology at least 72 bp in length, followed by precise duplication of the chromosome terminus (15-16). Instead, multiple template switching and synthesis initiation events may create duplications at uncapped telomeres (Fig. S8). The ypT29 and ypT50 telomere recombination events each resulted in uninterrupted inverted duplications of 16 or 8 kb, respectively, structures that appear consistent with BFB (Figs. 2A-B and S7). However, the template-switching events observed for ypT27 and eT6 telomere recombination events suggest that the inverted duplications that sealed ypT29 and ypT50 may have been template-based and created by synthesis-dependent strand annealing (SDSA), where a simple break-primed synthesis reaction occurs and is resolved by an NHEJ reaction with a second uncapped telomere (17-19).
For two additional fusions, inverted duplications occurred at the left end of the X chromosome, which ends with a 5 kb terminal inverted repeat that is separated by ∼10 kb of intervening sequence. For ypT35 and ypT37, 0.8 and 1.9 kb tracts of DNA adjacent to the internal copy of a 5 kb inverted repeat were added to XL prior to end-joining with the left end of chromosome II (Figs. 2E and S9). Thus, strand invasion of the terminal XL repeat with the internal copy may have initiated an SDSA-like process, resulting in uninterrupted duplications analogous to those of ypT29 and ypT50 (Fig. 2A-B). Similar SDSA-like events may serve to initiate synthesis-mediated interrupted duplications (Figs. 1B-C, 2C-D). Seven additional fusions exhibit breakpoints at either 23 or 300 kb from XL, suggesting recurrent locus-driven repair events (Figs. S10-12).
Up to 32% of tumor genome fusion breakpoints show evidence of template switching, where segments of DNA near the breakpoint are duplicated and accompanied by deletion of intervening sequences (20-22). Such breakpoints are referred to as genomic shards or junctional sequences (20, 22). This molecular signature was apparent based on sequence analysis for one telomeric recombination event that was initiated by template switching events, ypT27 (Figs. 2C and S8), and for three additional fusion events, ypT23, ypT46, and ypT49 (Figs. 3 and S13).
Large-scale duplication of interstitial segments of the human genome can result in genomic disorders. These aberrations display hallmarks that we observed in 82% of end-to-end fusions (n=38): duplications interrupted by triplications and non-duplicated sequences, likely generated by template-switching, as well as breakpoints that are sealed by microhomology (23), suggesting a conserved process that may be relevant to metazoan genome evolution. Models where a stalled replication fork or DSB induces promiscuous DNA synthesis, termed Fork Stalling and Template Switching (FoSTeS) or microhomology-mediated BIR (mmBIR), have been proposed to explain the origin of such large spontaneous mitotic duplications (Figs. 2C-D, 3 and S13) (18-19). Transposon excision in C. elegans can lead to duplications that may result from template switching (24), consistent with a role for replication-based repair in non-terminal segments of nematode genomes.
While DNA bridges during mitosis are observed in cells with critically shortened telomeres, even in C. elegans (5), we propose that critically shortened telomeres commonly trigger synthesis events primed by microhomology or limited homology to create large-scale, interrupted subtelomeric duplications, in the absence of BFB cycles (Figs. 1, 2C-D, 3, S8 and S13). These interrupted duplications resemble interstitial mammalian genome aberrations attributed to FoSTeS/mmBIR, and they can be resolved by end-joining with a second dysfunctional chromosome end. BFB may function independently to promote duplication at uncapped telomeres prior to fusion.
Studies in E. coli have shown that genome duplications can occur in response to stress through a process similar to FoSTeS, and this has been hypothesized to be adaptive during evolution (18, 25-26). Telomere dysfunction drives amplification and deletion of genomic loci relevant to human cancer (27). Recurrent or non-recurrent recombination events similar to those described here could contribute to genome rearrangements that play critical roles in tumor development (2).
Supplementary Material
Acknowledgments
We thank J. Maydan for technical assistance, D. Reiner, J. Sekelsky, L. Shtessel, D. Shippen and T. Petes for comments on the manuscript, and D. Ramsden for discussion. SA and ML were supported by NIH Grants GM066228 and GM072150. DGM was supported by funds from Genome Canada and Genome British Columbia and the Michael Smith Research Foundation. DGM is a fellow of the Canadian Institute For Advanced Research.
Footnotes
References
- 1.Capper R, et al. Genes Dev. 2007;21:2495. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey SM, Murnane JP. Nucleic Acids Res. 2006;34:2408. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClintock B. Genetics. 1941;26:234. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed S, Hodgkin J. Nature. 2000;403:159. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 5.Meier B, et al. PLoS Genet. 2006;2:e18. doi: 10.1371/journal.pgen.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowden MR, Meier B, Lee TW, Hall J, Ahmed S. Genetics. 2008;180:741. doi: 10.1534/genetics.108.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallioniemi A, et al. Science. 1992;258:818. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 8.Mantripragada KK, et al. Int J Mol Med. 2004;13:273. [PubMed] [Google Scholar]
- 9.Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. Exp Cell Res. 2005;302:233. doi: 10.1016/j.yexcr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Stansel RM, de Lange T, Griffith JD. EMBO J. 2001;20:5532. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raices M, et al. Cell. 2008;132:745. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 12.Griffith JD, et al. Cell. 1999;97:503. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 13.Cheung I, Schertzer M, Rose A, Lansdorp PM. Nucleic Acids Res. 2006;34:96. doi: 10.1093/nar/gkj417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letsolo BT, Rowson J, Baird DM. Nucleic Acids Res. 2010;38:1841. doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosco G, Haber JE. Genetics. 1998;150:1037. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackett JA, Feldser DM, Greider CW. Cell. 2001;106:275. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 17.McVey M, Radut D, Sekelsky JJ. Genetics. 2004;168:2067. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Nat Rev Genet. 2009;10:551. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branzei D, Foiani M. Cell. 2007;131:1228. doi: 10.1016/j.cell.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Bignell GR, et al. Genome Res. 2007;17:1296. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell PJ, et al. Nat Genet. 2008;40:722. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens PJ, et al. Nature. 2009;462:1005. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JA, Carvalho CM, Lupski JR. Cell. 2007;131:1235. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Moerman DG, Kiff JE, Waterston RH. Nucleic Acids Res. 1991;19:5669. doi: 10.1093/nar/19.20.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stankiewicz P, Lupski JR. Annu Rev Med. 2010;61:437. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 26.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Hagan RC, et al. Cancer Cell. 2002;2:149. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.