Abstract
Altered brain connectivity has been widely considered as a genetic risk mechanism for schizophrenia. Of the many susceptibility genes identified so far, ZNF804A (rs1344706) is the first common genetic variant associated with schizophrenia on a genome-wide level. Previous fMRI studies have found that carriers of rs1344706 exhibit altered functional connectivity. However, the relationship between ZNF804A and white matter structural connectivity in patients of schizophrenia remains unknown. In this study, 100 patients with schizophrenia and 69 healthy controls were genotyped at the single nucleotide polymorphism rs1344706. Diffusion tensor imaging (DTI) was conducted and analyzed with tract-based spatial statistics. Systematic statistical analysis was conducted on multiple diffusion indices, including fractional anisotropy, axial diffusivity, radial diffusivity, and mean diffusivity. Unpaired two-sample t-test revealed significant differences in fractional anisotropy and diffusivity between schizophrenia and control groups. A two-way ANOVA analysis was conducted to assess the main effects of and the interaction between schizophrenia and ZNF804A. Although significant main effects of the diagnosis of schizophrenia were found on radial diffusivity, no association between the ZNF804A (rs1344706) and white matter connectivity was found in the entire group of subjects or in a selected subgroup of age-matched subjects (n = 72).
Keywords: ZNF804A, Schizophrenia, White matter, Diffusion tensor imaging (DTI), Tract-based spatial statistics (TBSS)
1. Introduction
Schizophrenia (SZ) has consistently showed high heritability, with heritability estimated at 73–90% [24]. In a recent genome-wide association study, a single nucleotide polymorphism (SNP) rs1344706 in ZNF804A was identified to be associated with schizophrenia [16]. This association has been replicated in multiple independent samples [4], including the Han Chinese population [31]. ZNF804A is known to be expressed in the brain and is associated with white matter development. In particular, studies on zfp804a, the mouse homologue of ZNF804A, suggested that ZNF804A may be involved in the regulation of early neurodevelopment [2]. Furthermore, bioinformatic analyses of the conserved mammalian sequence around rs1344706 suggested that the variant was associated with myelin transcription factor 1 and octamer-binding factor 6 [4]. Both transcription factors are involved in oligodendrocyte differentiation and proliferation [3,15], two critical processes contributing to abnormal white matter connectivity that has been widely implicated in schizophrenia. Despite this confirmed association between ZNF804A and schizophrenia, the relationship between rs1344706 and white matter connectivity remains to be elucidated [4].
Imaging genetics provides a unique tool for exploring and evaluating the functional impact of genetic polymorphisms [1]. Using functional MRI (fMRI), Esslinger et al. found that healthy carriers of rs1344706 exhibit no changes in regional brain activity but pronounced gene dosage-dependent reduced connectivity of dorsolateral prefrontal cortex across hemispheres and increased connectivity with hippocampus [5,6]. Using T1-weighted MRI, Wei et al. found that white matter (WM) intensity is increased in the left prefrontal lobe of patients carried with the risk-allele compared to non-risk allele carriers [28]. Although these findings are consistent with the “dysconnectivity” hypothesis of schizophrenia [19], neither fMRI nor T1-weighted MRI provides measure of white matter structural connectivity. In an attempt to address this issue, Voineskos et al. applied diffusion tensor imaging (DTI) to measure connectivity of major frontotemporal and interhemispheric white matter tracts in healthy carriers of the risk variant rs1344706 [26]. While cortical thickness was found to be reduced, no effect of the risk variant on microstructural integrity of white matter tracts was found. The effect of the risk variant on the white matter connectivity in patients with schizophrenia, however, remains to be unknown. Schizophrenia is a complex brain disorder involving multiple risk genes and environmental factors. It is possible that ZNF804A may interact with other risk factors in patients with schizophrenia. Such interaction may result in different neurobiological phenotypes that maybe detectable by MRI [25,14].
The goal of the current study was to examine the effects of ZNF804A gene polymorphism rs1344706 on the WM connectivity in schizophrenia using the methods of DTI and tract-based spatial statistics (TBSS), an unbiased and hypothesis-free whole-brain approach. The effects of rs1344706 on five indices of diffusion were assessed using the general linear model.
2. Methods
2.1. Subjects
100 patients with schizophrenia (n = 77) or schizophreniform disorder (after follow-up, a diagnosis of schizophrenia was established; n = 23) were recruited in the Third Affiliated Hospital of Sun Yat-sen University. All participants were Han Chinese. The inclusion criteria were as follows: (a) age being between 18 and 45 years; (b) years of education being greater than 9 and (c) had to be right-handed (assessed by a 10-item questionnaire, the Edinburgh Handedness Inventory [17]). The exclusion criteria were: (a) presented with chronic neurological disorders; (b) had a history of alcohol or substance abuse; (c) had a history of electroconvulsive therapy. Diagnosis based on the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) was made by an experienced psychiatrist using patient version of the Structured Clinical Interview for DSM-IV (SCID) [7]. Family psychiatric history was obtained by interviewing the patients and their relatives when possible, who provided information on family history in details during the clinical interview. This study adopted the definition of family history as described by Xu et al. [29]. Patients with positive family history (PFH) were defined as having at least one relative with schizophrenia in their first-degree or second-degree relatives; otherwise, they were defined as patients with negative family history (NFH). The Positive and Negative Syndrome Scale (PANSS) [11] was used to measure psychopathologic symptoms at the time of imaging.
69 healthy volunteers (all Han Chinese, unpaid) from the local community were recruited by advertisement. To be included in the study, these volunteers and their first-degree or second degree relatives [29] should not have history of mental disorder based on DSM-IV. Interview and assessment were conducted by the same experienced psychiatrist using SCID as for the patient group. Other inclusion and exclusion criteria were the same as those of the patients.
2.2. Genotyping
Genotyping was performed as described previously [28]. Briefly, genomic DNA from leukocytes in blood was amplified by polymerase chain reaction (PCR) to generate a 443 bp product spanning rs1344706. Primers were as follows: upper GAATCTAGA GTCAT-GCAGG, and lower CAAGTTATTC TCTAGAGTCC.
2.3. Image acquisition and analysis
Images were acquired on a 1.5-T GE Signa Twinspeed MRI scanner (General Electric Medical System, Milwaukee, WI, USA) equipped with a quadrature birdcage head coil. Diffusion-weighted images were acquired with a single-shot echo planar imaging (EPI) sequence. The diffusion gradients were applied along fifteen non-collinear directions (b = 1000 s/mm2), together with an acquisition of “b0 image” (b = 0). 35 contiguous axial slices were acquired with a slice thickness of 4 mm and no gap. The acquisition parameters were as follows: repetition time = 11,000 ms; echo time = 74.7 ms; matrix = 128 × 128; field of view = 240 mm × 240 mm.
Diffusion data were analyzed using the Diffusion Toolbox distributed with the FSL's software package (University of Oxford, UK) [22]. All diffusion-weighted images were first linearly registered to the corresponding non-diffusion weighted images. Diffusion tensors were obtained based on DTI and diagonalized to derive the fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (L1) and the radial diffusivity (L2 and L3).
Voxelwise statistical analysis of the FA, MD, L1, L2 and L3 values was carried out using TBSS [23], a program implemented in FSL [22]. The steps of this analysis are as follows. First, all subjects' FA images were skull-stripped and aligned into the MNI152 standard space using the nonlinear registration tool (FNIRT) within FSL. Next, the mean FA volume was created and thinned to create a mean FA skeleton, which represents the centers of all tracts common to the entire group of subjects. Each subject's aligned FA data were then projected onto this skeleton and the resulting data were fed into voxelwise cross-subject statistical testing.
An unpaired two-sample t-test was first performed to compare patients and controls without accounting for genetic differences. Then, a full-factorial two-way analysis of variance (ANOVA) was implemented within the framework of the general linear model using the FEAT algorithm in FSL [22]. Within this model, the first factor characterized if the subject had schizophrenia, and the second factor accounted for the presence of the risk-allele rs1344706 (T). Maps of t-statistics for main effects and interaction were generated by permutation testing (n = 500) using the randomize tool in FSL for each of the aforementioned tensor characteristics. To correct for multiple comparisons, statistical significance was inferred using the Threshold-Free Cluster Enhancement method [21]. After family-wise error correction, only those clusters with acceptable p-values were retained (p < 0.05).
To address potential confounds that may be caused by age-related white matter structural differences and unequal sample sizes, the analysis was carried out both in the entire group of subjects (ALL; n = 169) and in a selected subgroup (SEL; n = 72) of age-matched subjects. The selected subgroup included 36 healthy subjects (GG = 12, GT = 12, TT = 12, mean age = 27.11 y.o., standard deviation of age = 4.44 y.o., minimum age = 20 y.o., maximum age = 38 y.o.) and 36 patients (GG = 12, GT=12, TT = 12, mean age = 27.55 y.o., standard deviation of age = 4.43 y.o., minimum age = 20 y.o., maximum age = 37 y.o.). Furthermore, a series of 2-sample t-tests was also conducted for the entire group (ALL) and for the selected group (SEL) to determine whether white matter differences are merely due to ZNF804A.
The white matter tracts were identified based on a DTI white matter atlas (JHU ICBM-DTI-81, Johns Hopkins University).
2.4. Statistical analysis of demographics
Analysis of variance tests (for numeric variables) and χ2 tests (for categorical variables) were conducted with the Statistical Package for Social Sciences, version 15.0 (SPSS Inc., Chicago, IL), to detect demographic differences in relation to diagnosis and genotype.
3. Results
3.1. Clinical data
The patient group (GG = 24, GT = 58, TT = 18, p = 0.10) and the healthy control group (GG = 12, GT = 35, TT = 22, p = 0.76) were both in Hardy–Weinberg equilibrium as calculated with GENEPOP [20].
As shown in Table 1, there were no significant differences (p > 0.05) between genotype subgroups in sex, age, or years of education within each diagnostic group or within the total sample. There were no significant differences (p > 0.05) between the patients and the healthy controls in all aforementioned demographic variables. Within the patient group, the genotype subgroups did not differ significantly (p > 0.05) in the subtypes of the disease, the duration of psychosis, total scores of the PANSS, positive scores, negatives scores, general scores, age at onset of psychosis or family psychotic history. In addition, no significant differences were found in the types (first or second generation) of or in the maximal total dosage (chlorpromazine equivalents [10]) of antipsychotic medication during the 2 months before the MRI scanning.
Table 1.
Demographic and clinical characteristics for schizophrenic patients and healthy subjects.
| Means (SD) | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Main effect of diagnosis | Main effect of ZNF804A | Diagnosis × ZNF804A interaction | ||||||
|
|
|
|
||||||
| Controls (n = 69) | Patients (n= 100) | G homozygous (n = 36) | T carriers (n = 133) | HC | SP | |||
|
|
|
|||||||
| G homozygous (n = 12) | T carriers (n = 57) | G homozygous (n = 24) | T carriers (n = 76) | |||||
| Age (years) | 25.4 (5.7) | 26.5 (6.9) | 27.3 (5.9) | 26.0 (6.4) | 27.3 (4.4) | 25.0 (5.9) | 27.3 (6.7) | 26.2 (7.0) |
| Sex (F/M) | 30/39 | 47/53 | 16/20 | 61/72 | 5/7 | 25/32 | 11/13 | 36/40 |
| Years of education | 12.9(2.9) | 12.2(2.9) | 12.8(3.0) | 12.4(2.8) | 13.7 (2.9) | 12.7(2.9) | 12.3(3.1) | 12.2(2.8) |
| Subtypes of SZ (Para/Dis/Un) | NA | NA | NA | NA | NA | NA | 13/1/10 | 47/7/22 |
| Age of onset (years) | NA | NA | NA | NA | NA | NA | 24.7 (6.2) | 23.9 (6.3) |
| Duration of psychosis (weeks) | NA | NA | NA | NA | NA | NA | 124.2(146.2) | 126.7(182.5) |
| Family psychotic history (PFH/NFH) | NA | NA | NA | NA | NA | NA | 3/21 | 13/63 |
| Positive scores of PANSS | NA | NA | NA | NA | NA | NA | 21.8 (6.3) | 24.4 (5.0) |
| Negative scores of PANSS | NA | NA | NA | NA | NA | NA | 17.5(7.3) | 14.4(5.6) |
| General scores of PANSS | NA | NA | NA | NA | NA | NA | 36.3 (5.5) | 14.4(5.6) |
| Total scores of PANSS | NA | NA | NA | NA | NA | NA | 75.9(13.1) | 75.2(11.1) |
| Antipsychotic type (none/lst/2nd generation) | NA | NA | NA | NA | NA | NA | 19/1/4 | 56/4/16 |
| Antipsychotic dose, CPZ equivalents | NA | NA | NA | NA | NA | NA | 420.0(152.5) | 356.8 (200.4) |
Abbreviations: CPZ, chlorpromazine hydrochloride; Dis, disorganized type; HC, healthy controls; NFH, negative family history; Para, paranoid type; PANSS, Positive and Negative Syndrome Scale; PFH, positive family history; SP, schizophrenic patients; Un, undifferentiated type.
3.2. Schizophrenia versus controls without considering genetic effects
In the unpaired two-sample t-test without considering the effect of ZNF804A, TBSS revealed significant differences between the patient and control groups. FA broadly decreased in near all major fiber tracts in the patients (p < 0.05) (Fig. 1a), consistent with previous reports [9]. The spatial extent of significant regions was generally reduced in the SEL group compared to the ALL group. However the locations of significant differences were similar (Fig. 1a). In regions of decreased FA, a congruent increase in the minimum radial diffusivity L3 was also observed (p < 0.05).
Fig. 1.
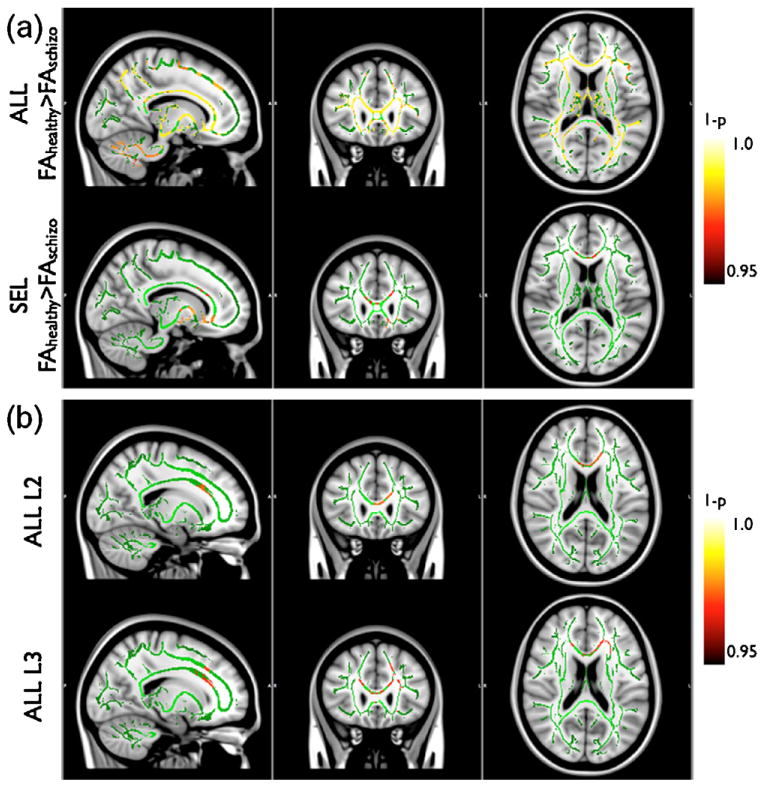
TBSS analysis of the effect of schizophrenia and ZNF804A rs1344706 on structural brain connectivity. (a) Unpaired two-sample t-test between schizophrenia and controls based on FA. The top row illustrates the tract-specific statistical significance of the hypothesis that FA of healthy controls is higher than that of schizophrenia in the ALL group. The bottom row shows the results in the SEL group. (b) 2 × 2 ANOVA analysis of the main effect of schizophrenia on the radial diffusivity in white matter fiber tracts in the ALL group. The top row shows the statistical significance maps analyzed based on the second eigenvalue (i.e. the larger radial diffusivity denoted by L2) of the diffusion tensor. The bottom row shows the statistical significance maps based on the third eigenvalue (i.e. the smaller radial diffusivity denoted by L3). Note that the most significant foci found in the SEL group coincide with the unpaired t-test in (a). No significant main effect or interaction was found for ZNF804A rs1344706. The background gray-scale T1-weighted images provide anatomical references. The green color highlights the skeletons of white matter tracts constructed by FSL The “hot” color-scale bar on the right represents the statistical significance (1 − p) with color intensity. These “hot” colors are overlaid on the track skeletons such that brighter voxels represent higher significance (p < 0.05).
3.3. Association between ZNF804A and the integrity of white matter
3.3.1. Main effect of Schizophrenia
In the ALL group, significant clusters were found in the radial diffusivities L2 and L3 (p < 0.05) based on 2 × 2 ANOVA (Fig. 1b). The results were noticeably more significant for the smaller radial diffusivity (L3). As shown in Fig. 1 b, significant regions in the L3 map included bilateral parts of the genu of the corpus callosum (cc; MNI 14, 29,15 and MNI −12, 26,19), bilateral anterior branches of the corona radiata (acr; MNI ±16, 25, 25) as well as left prefrontal projections of the corona radiata (lpfcr; MNI −16, 28, 37), left parts of the anterior thalamic radiation (latr; MNI −26, 22, 20) and discrete anterior portions of the left uncinate fasciculus (luf; MNI −23, 27, −8). With family-wise error correction, no significant cluster was found in FA, MD and L1 maps (p > 0.05).
With a threshold of p = 0.05, all of the above results were consistent in the SEL group although the spatial extent of significant regions was slightly smaller than that of the ALL group.
3.3.2. Main effect of ZNF804A
The results of the 2 × 2 ANOVA showed no significant cluster (p > 0.05) in any of the tensor derived characteristics (FA, MD, L1, L2, L3). This result was consistent between the ALL group and the SEL group.
3.3.3. Interaction between ZNF804A and schizophrenia
The tract-based statistics did not establish any interaction (p > 0.05) between the risk-allele and the disease for any of the reported tensor characteristics. This result again was consistent between the ALL group and the SEL group.
3.3.4. Unpaired 2-sample t-test
To compare risk-allele carriers versus non-risk-allele carriers, two null hypotheses were tested: (1) DTI indices in GT or TT are greater than GG and (2) DTI indices in GT or TT are smaller than GG. None of these comparisons revealed any significant group difference within the control or patient groups even at a relatively relaxed threshold of p < 0.1.
4. Discussion
In this study, no association was found between rs1344706 of ZNF804A and the integrity of white matter (WM) fiber tract in patients with schizophrenia. While the main effect of schizophrenia was significant, the main effect of the risk variant and their interaction with the fiber tract integrity were not significant in any of the five major DTI indices. Our study indicates that, while ZNF804A is associated with schizophrenia, the risk gene is not associated with abnormalities in white matter structural connectivity based on DTI and TBSS.
Many studies have suggested that abnormal WM may be the primary neuromechanism of schizophrenia and these abnormalities may be heritable [12,30,19,8]. In this context, our finding is surprising given that previous imaging studies of ZNF804A have suggested potential abnormalities in white matter connectivity. For instance, a number of fMRI studies have found that the risk variant was associated with functional connections [5,6,18,27]. In addition, our previous study based on structural MRI has found that rs1344706 was associated with WM T1-weighted image intensity in the prefrontal lobes and hippocampi of both patients with schizophrenia and healthy controls [28]. Our study based on state-of-the-art DTI techniques thus provided one of the first direct evidence of the lack of association between ZNF804A and abnormal DTI structural connectivity in schizophrenia. This null finding expands the complex role of genetics may play in schizophrenia. More cautions are thus required in interpreting the effect of susceptibility genes in schizophrenia.
Our DTI analysis using tract-based spatial statistics was comprehensive and robust. This technique inherently trades off spatial resolution against increased statistical power and analyze the whole brain simultaneously [13,23]. To offer a more complete assessment of WM integrity, an integrated analysis of multiple diffusion indices was analyzed. Each index offers its own physiological sensitivity. For example, the axial diffusivity refers to rate of diffusion along the direction of fiber tracts, while the radial diffusivity measures the rate of diffusion in directions perpendicular to the tracts. Additional unpaired bidirectional 2-sample t-tests were carried out to compare risk-allele carriers versus non-risk-allele carriers among healthy subjects and patients with schizophrenia, respectively. These comparisons also did not show association between the risk variant and WM fiber tract integrity based on DTI and TBSS.
5. Study limitations
Although the DTI methods for imaging acquisition and processing were standard, the number of diffusion gradient directions at 15, though sufficient, is relatively small. Slice thickness at 4 mm, for the purpose of improving signal-to-noise ratio, was thicker than most DTI studies. Future studies would benefit from higher spatial resolution and more diffusion directions. A second limitation is that the effect of medication was excluded. Although the medication was balanced between different groups, this factor might obscure potentially smaller genetic effects. To further investigate the potential effect of medication, first-episode schizophrenia without medication should be considered in future studies. A third limitation is the effect of age. Although we conducted an analysis using a selected age-matched group, our study did not fully evaluate the effect of age. A comprehensive analysis considering all variables may require a much larger sample size.
6. Conclusion
In conclusion, based on DTI methods, our study did not reveal association between the SNP rs1344706 in ZNF804A and white matter fiber tract integrity in patients with schizophrenia.
Highlights.
-
▸
Imaging genetics of ZNF804A (rs1344706) in schizophrenia.
-
▸
Whole-brain track-based DTI analysis of the effect of rs1344706.
-
▸
rs1344706 is not associated with brain fiber tract integrity in schizophrenia.
Acknowledgments
The authors thank Vivian Bancroft-Wu for her editorial assistance. The research was funded by grants from the R&D Special Fund for Health Profession (grant no. 201002003), the Natural Science Foundation of China (grant nos. 81071093; 81101028 and 30900485) and the Technology Project of the Guangdong Province China (grant nos. 2011B031800073 and 2011B031800101). Data were analyzed at the Brain Imaging Analysis Center of Duke University Medical Center using a Linux cluster partially supported by the National Institutes of Health (1S10RR025561).
Abbreviations
- FA
fractional anisotropy
- NFH
negative family history
- PFH
positive family history
- ROI
regions of interesting
- SEL
selected group
- SZ
schizophrenia
- WM
white matter
Footnotes
Conflict of interest: No conflict of interest declared.
Contributors: Dr. Zhao J designed the study along with Drs. Wei Q and Diao F. Drs. Wei Q, Kang Z, Diao F, Xiaoli Wu, Li L and Zheng L collected the original imaging data. Drs. Liu C, Guidon A, Wei Q managed and analyzed the imaging data. Dr. Wei Q wrote the first draft of the manuscript, Drs. Wei Q, Zhao J, Liu C, Zhang J, Hu M and Guo X revised the paper. All authors contributed to and have approved the final manuscript.
Contributor Information
Chunlei Liu, Email: chunlei.liu@duke.edu.
Jingping Zhao, Email: zhaojingpingcsu@163.com, weiql@mail.sysu.edu.cn.
References
- 1.Bigos KL, Weinberger DR. Imaging genetics – days of future past. Neuroimage. 2010;53:804–809. doi: 10.1016/j.neuroimage.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 2.Chung HJ, Lee JY, Deocaris CC, Min H, Kim SH, Kim MH. Mouse homologue of the schizophrenia susceptibility gene ZNF804A as a target of Hoxc8. Journal of Biomedicine and Biotechnology 2010. 2010:231708. doi: 10.1155/2010/231708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collarini EJ, Kuhn R, Marshall CJ, Monuki ES, Lemke G, Richardson WD. Down-regulation of the POU transcription factor SCIP is an early event in oligodendrocyte differentiation in vitro. Development. 1992;116:193–200. doi: 10.1242/dev.116.1.193. [DOI] [PubMed] [Google Scholar]
- 4.Donohoe G, Morris DW, Corvin A. The psychosis susceptibility gene ZNF804A: associations, functions, and phenotypes. Schizophrenia Bulletin. 2010;36:904–909. doi: 10.1093/schbul/sbq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, Haddad L, Mier D, Opitz Von Boberfeld C, Raab K, Witt SH, Rietschel M, Cichon S, Meyer-Lindenberg A. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 6.Esslinger C, Kirsch P, Haddad L, Mier D, Sauer C, Erk S, Schnell K, Arnold C, Witt SH, Rietschel M, Cichon S, Walter H, Meyer-Lindenberg A. Cognitive state and connectivity effects of the genome-wide significant psychosis variant in ZNF804A. Neuroimage. 2011;54:2514–2523. doi: 10.1016/j.neuroimage.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 7.S R, G M, First MB. Structured Clinical Interview for DSM-IV Axis I Disorders, Patient Edition (SCID-P) Version. Biometrics Research; New York: 1995. [Google Scholar]
- 8.Hao Y, Yan Q, Liu H, Xu L, Xue Z, Song X, Kaneko Y, Jiang T, Liu Z, Shan B. Schizophrenia patients and their healthy siblings share disruption of white matter integrity in the left prefrontal cortex and the hippocampus but not the anterior cingulate cortex. Schizophrenia Research. 2009;114:128–135. doi: 10.1016/j.schres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Jeong B, Wible CG, Hashimoto R, Kubicki M. Functional and anatomical connectivity abnormalities in left inferior frontal gyrus in schizophrenia. Human Brain Mapping. 2009;30:4138–4151. doi: 10.1002/hbm.20835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kane JM, Leucht S, Carpenter D, Docherty JP. The expert consensus guideline series. Optimizing pharmacologic treatment of psychotic disorders. Introduction: methods, commentary, and summary. Journal of Clinical Psychiatry. 2003;64(Suppl. 12):5–19. [PubMed] [Google Scholar]
- 11.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophrenia Bulletin. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 12.Konrad A, Winterer G. Disturbed structural connectivity in schizophrenia primary factor in pathology or epiphenomenon? Schizophrenia Bulletin. 2008;34:72–92. doi: 10.1093/schbul/sbm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melonakos ED, Shenton ME, Rathi Y, Terry DP, Bouix S, Kubicki M. Voxel-based morphometry (VBM) studies in schizophrenia – can white matter changes be reliably detected with VBM? Psychiatry Research. 2011;193:65–70. doi: 10.1016/j.pscychresns.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicodemus KK, Law AJ, Radulescu E, Luna A, Kolachana B, Vakkalanka R, Rujescu D, Giegling I, Straub RE, McGee K, Gold B, Dean M, Muglia P, Callicott JH, Tan HY, Weinberger DR. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Archives of General Psychiatry. 2010;67:991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen JA, Berndt JA, Hudson LD, Armstrong RC. Myelin transcription factor 1 (Myt1) modulates the proliferation and differentiation of oligodendrocyte lineage cells. Molecular and Cellular Neurosciences. 2004;25:111–123. doi: 10.1016/j.mcn.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 16.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nature Genetics. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 17.Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- 18.Paulus FM, Krach S, Bedenbender J, Pyka M, Sommer J, Krug A, Knake S, Nothen MM, Witt SH, Rietschel M, Kircher T, Jansen A. Partial support for ZNF804A genotype-dependent alterations in prefrontal connectivity. Human Brain Mapping. 2011 doi: 10.1002/hbm.21434. http://dx.doi.org/10.1002/hbm.21434. [DOI] [PMC free article] [PubMed]
- 19.Pettersson-Yeo W, Allen P, Benetti S, McGuire P, Mechelli A. Dysconnectivity in schizophrenia: where are we now? Neuroscience and Biobehavioral Reviews. 2011;35:1110–1124. doi: 10.1016/j.neubiorev.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 21.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 22.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl. 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 23.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 25.Tost H, Meyer-Lindenberg A. Puzzling over schizophrenia: schizophrenia, social environment and the brain. Nature Medicine. 2012;18:211–213. doi: 10.1038/nm.2671. [DOI] [PubMed] [Google Scholar]
- 26.Voineskos AN, Lerch JP, Felsky D, Tiwari A, Rajji TK, Miranda D, Lobaugh NJ, Pollock BG, Mulsant BH, Kennedy JL. The ZNF804Agene: characterization of a novel neural risk mechanism for the major psychoses. Neuropsychopharmacol. 2011;36:1871–1878. doi: 10.1038/npp.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter H, Schnell K, Erk S, Arnold C, Kirsch P, Esslinger C, Mier D, Schmitgen MM, Rietschel M, Witt SH, Nothen MM, Cichon S, Meyer-Lindenberg A. Effects of a genome-wide supported psychosis risk variant on neural activation during a theory-of-mind task. Molecular Psychiatry. 2010;18 doi: 10.1038/mp.2010.18. http://dx.doi.org/10.1038/mp.2010. [DOI] [PubMed] [Google Scholar]
- 28.Wei Q, Kang Z, Diao F, Shan B, Li L, Zheng L, Guo X, Liu C, Zhang J, Zhao J. Association of the ZNF804A gene polymorphism rs1344706 with white matter density changes in Chinese schizophrenia. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2012;36:122–127. doi: 10.1016/j.pnpbp.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature Genetics. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 30.Zalesky A, Fornito A, Seal ML, Cocchi L, Westin CF, Bullmore ET, Egan GF, Pantelis C. Disrupted axonal fiber connectivity in schizophrenia. Biological Psychiatry. 2011;69:80–89. doi: 10.1016/j.biopsych.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R, Lu SM, Qiu C, Liu XG, Gao CG, Guo TW, Valenzuela RK, Deng HW, Ma J. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Molecular Psychiatry. 2010;16:360–361. doi: 10.1038/mp.2010.55. [DOI] [PubMed] [Google Scholar]


