Abstract
Networks in biology can appear complex and difficult to decipher. We illustrate how to interpret biological networks with the help of frequently used visualization and analysis patterns.
Networks represent relationships. In a biological context, many different types of relationships can be measured, such as physical interactions between proteins or genetic interactions revealed by combinations of mutations. When large collections of diverse relationships are generated from several different high-throughput experimental analyses of a single biological system, network visualization and analysis can prove particularly useful1–3.
To illustrate how data visualized as a network can be easier to interpret than long lists of proteins, interactions and correlations, we analyze an example network representing the yeast chromosome maintenance and duplication machinery (Fig. 1). Networks are often analyzed using methods—which we term ‘visualization and analysis patterns’— to infer new hypotheses about protein function, pathway components and links between known processes. We apply these patterns to our example network and provide references for further reading to tutorials that describe specialized network analysis software.
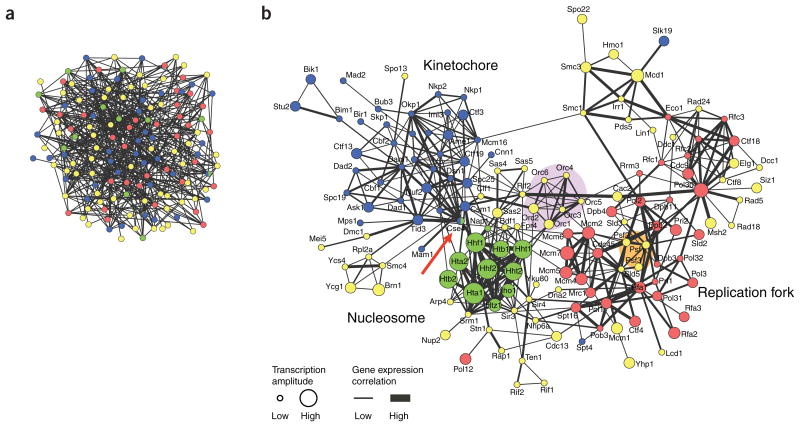
Figure 1.
Network visualization of chromosome maintenance and duplication machinery in baker's yeast, Saccharomyces cerevisiae. Nodes represent proteins that are annotated as being located on the chromosome by the Gene Ontology project7 (for clarity, the suffix ‘p’ has been removed from yeast protein names). Node colors specify chromosomal location subcategories: red, replication fork; green, nucleosome; blue, kinetochore; yellow, other chromosome components. Edges represent protein-protein interactions that were manually extracted from publications by BioGRID database curators4 (which could include small- and large-scale experiments). (a) Without specific layout, the network looks like a ‘jumbled mess’ and cannot be interpreted. (b) The same network after applying the force-directed layout and adding gene expression data of cells monitored during one round of the cell cycle are visually annotated on the network (data are from ref. 8). Edges are drawn thicker when the Pearson correlation between transcript profiles is higher. Node size corresponds to the transcriptional amplitude (root mean square of the time-course expression values), which is a measure of how much expression changes over the cell cycle. The network was visualized using Cytoscape software6. Interesting regions were manually emphasized (shading and red arrow) and node labels placed for clarity using Adobe Illustrator.
Mapping biological data to a network
In Figure 1, yeast proteins involved in chromosome maintenance and duplication are shown as nodes in a network. Nodes are connected by links, called edges. Edges represent physical protein interactions that are experimentally measured using techniques such as yeast two-hybrid screens or protein pull-down followed by mass spectrometry. We retrieved the protein interaction data from the BioGRID database4. Data about protein function and gene expression will be used to help interpret the network using the visualization and analysis patterns described below.
Visualization pattern one: layout
The first step to make a network more intelligible is to organize the nodes. With no organization, the nodes are a jumbled mess (Fig. 1a). Fortunately, many automatic methods for laying out networks are available in easy-to-use software tools5,6. Most interaction networks can be reasonably well organized using automated layout methods that place connected nodes near each other and untangle the lines (Fig. 1b and Box 1). This makes it easier to apply the analysis patterns we describe later in the text.
Box 1. How to lay out a network.
Methods to automatically organize networks (that is, layout algorithms) enable interesting relationships within data to be seen more easily. Most networks can be visualized by using a ‘spring-embedded’ or ‘force-directed’ layout algorithm, based on the idea of edges ‘pulling together’ nodes that ‘repel’ each other. Other, more specific, layout algorithms are available, such as ‘hierarchical’ algorithms, which are useful for displaying taxonomy trees or regulatory cascades. Edge length is determined by the layout algorithm only for visualization purposes and does not convey biological information. Typical network visualization software contains many layout options. A practical approach to choose among these is to try a force-directed layout first, or hierarchical if the network is tree-like, and then try others to see which one best arranges a given network.
Automatic network layout works well for many small- and medium-sized networks (e.g., 50–500 nodes). It is rarely perfect, however, and most networks are more easily interpreted after subsequent manual node rearrangement that can be performed using network visualization software5,6. Larger networks, especially those with many edges, are often too tangled to be effectively visualized and interpreted, resulting in the ‘hairball’ network phenomenon (Fig. 1a). In these cases, it can be useful to break down the network into smaller parts, such as specific pathways or interesting sets of proteins, and explore them separately. Exceedingly tangled networks, lacking apparent structure, can also result from the presence of too many false positives or weak interactions. One way to address this problem is to reduce the number of edges, such as by increasing stringency to keep only the edges with the highest confidence.
Visualization pattern two: visual features
Networks offer a way of seeing relationships between data gathered using different experimental techniques. These complementary pieces of information can be conveyed by drawing nodes and edges with different ‘visual features’—such as shapes, sizes, colors and line thicknesses. Here, we use visual features to display protein function annotation and gene expression data.
In Figure 1b, node color represents the subcellular localization of a protein. A protein is colored according to whether it localizes to the replication fork (red), nucleosome (green), kinetochore (blue) or other chromosome components (yellow). We obtained these localization data from the Gene Ontology (GO) database7, but the same information could be gathered from other sources, such as experiments or computational prediction.
The size of a node and the thickness of an edge convey gene expression data8. Larger nodes are proteins whose corresponding mRNA changes substantially over the course of the cell cycle. Edge thickness represents gene expression correlation between interacting proteins: the thicker the edge, the more strongly correlated the gene expression profiles during the cell cycle.
Simultaneously visualizing all of these attributes—localization (color), expression level (size) and expression correlation (edge thickness)—reveals that many green nodes are large and highly connected with thick edges, suggesting that the nucleosome (green color) is dynamically (large size) and coordinately (thick edges) regulated at the mRNA level.
Analysis pattern one: ‘guilt by association’ protein function prediction
A network may be used to infer protein function based on interactions. One common way of doing this is to infer that the function of an unannotated protein may be similar to that of its neighbors—the proteins it is connected to in the network—if many of those neighbors are annotated with the same function. This principle is called ‘guilt by association’.
In Figure 1b, the proteins Psf1, Psf2 and Psf3 (shaded in orange) are not specifically assigned to the replication fork (red nodes) but are localized to chromosomes, according to Gene Ontology annotation. However, their interactions with many replication fork proteins suggest that they are involved in DNA replication. In fact, they are known members of the GINS complex, responsible for the assembly of the DNA replication machinery. Similarly, the interaction partners of Cse4 (red arrow) belong to the kinetochore and nucleosome, suggesting a multifunctional role for Cse4 at the interface between these two systems. This inference is consistent with the known capability of Cse4 to assemble a specialized nucleosome on centromeric DNA, which is required for kinetochore assembly.
Analysis pattern two: highly interconnected nodes (clusters)
Dense interconnections in protein interaction networks are characteristic of protein complexes or pathways. In Figure 1b, this is exemplified by the proteins Orc1, Orc2, Orc3, Orc4, Orc5 and Orc6 (shaded in violet), which display more connections with each other than with other proteins. In fact, they are known members of the yeast origin recognition complex (ORC), responsible for the loading of the replication machinery onto DNA.
The ORC is an example of a known complex, but this analysis pattern can also be used to identify novel complexes of unnanotated proteins and new components of known systems. For instance, in an application of the guilt-by-association pattern, we might predict that uncharacterized proteins that cluster with a known complex are unidentified members of that complex9.
Analysis pattern three: global system relationships
Once known or new systems (pathways or complexes) have been identified using protein function annotation or clustering, a broad overview of the network reveals global system-level relationships. In Figure 1b, the nucleosome and replication fork are characterized by high correlation within group members (thick edges) and consistent transcriptional modulation over the cell cycle (large node sizes). They are not directly physically connected, however, and there is no evidence of transcriptional correlation between their members, which indicates that they play roles at different points in the cell cycle.
Pros, cons and challenges of network representation
We've described above the basics of network representation. The approach is valuable for data integration and may increase data coverage and confidence. Coverage of a biological system is increased by combining complementary perspectives from different types of experiments, each able to reveal different aspects of the system. Data confidence may be assessed by identifying regions of the network where independent experimental techniques agree and are therefore more likely to be correct. This is particularly valuable when studying high-throughput or other data sets affected by noise and incompleteness.
Networks are well-defined mathematical objects (Fig. 2). Thus, analysis patterns can be implemented computationally, enabling automated and unbiased hypothesis generation2,6. Such approaches are powerful, in that they can efficiently find and calculate statistical significance for specific patterns in very large data sets. Even so, as automated methods take time to develop and may not always be accurate, experts are still needed to interpret the results and ensure biological relevance.
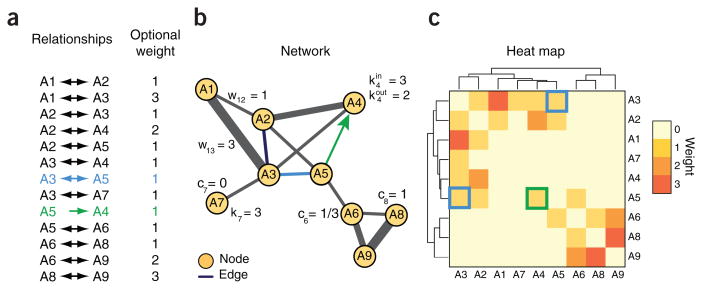
Figure 2.
Mathematical representation of networks and three alternate visualizations of the same data. (a) List of relationships with optional ‘weight’ (often denoted with the letter w), which represent attributes such as relationship significance or stength. Relationships can be undirected (e.g., A3 ↔ A5, shown in blue) or directed (e.g., A5 → A4, shown in green). (b) Network view. Networks are mathematically grounded in the field of graph theory, in which they are commonly denoted G = (V, E), (G, graph; V, a set of vertices or nodes; E, a set of edges). Some commonly encountered mathematical concepts include the node degree (ki), which is the number of edges attached to a node, and the clustering coefficient (ci), which counts the number of edges among the neighbors of a node, divided by the maximal possible number of such edges. If edges have directions, it is useful to distinguish between the in-degree (kini) and the out-degree (kouti). The node degree distribution and average clustering coefficient have been used to characterize different types of networks13. (c) Heat map view. Nodes are represented along the sides of the heat map and elements of the map (small squares) are colored according to edge weight, with higher weights having a darker color. Similar rows and columns are placed adjacent to each other, as shown by the similarity tree on each map axis. This view is useful for finding nodes with similar neighbors14.
Networks may also be used to represent many types of biological data, not just physical interactions. For instance, protein sequence similarity can be mapped to edges and protein families can be defined as clusters. Box 2 reviews interaction types commonly encountered in molecular biology and genetics.
Box 2. Examples of node relationships in biology.
Numerous types of node relationships occur in biological networks. The most common can be organized into several categories.
Physical interactions. These occur between biomolecules in direct contact. For instance, protein-protein interactions are important in processes such as protein-complex formation, signal transduction and transport15.
Regulatory interactions. These are directed activation or inhibition events. For instance, in gene-expression regulation, a transcription factor is connected to its targets by directed edges16.
Genetic interactions. These connect genes whose concurrent genetic perturbation leads to a phenotypic result different than expected from the combination of single effects. For instance, synthetic lethal interactions connect genes that weakly affect organism viability when deleted individually, but are lethal when deleted in combination. Genetic interactions are useful to study gene function, and to identify complexes and pathways that work together to control essential functions17.
Similarity relationships. These link biological objects that are similar according to a common attribute. Many different similarity measures can be used, such as protein sequence similarity or gene coexpression based on correlated transcriptional profiles. Similarity relationships are useful to identify groups of functionally related genes or proteins18.
Not all aspects of biological systems are easily represented using a network approach, so information can be lost in the mapping. The dynamic nature of a physical system—such as a biological pathway with many molecular components and states that vary in concentration and location over time—is not easily mapped to a static two-dimensional network representation. Relationships involving more than two objects are also difficult to represent in a network of pairwise edges. For example, biochemical reactions typically involve at least three participants (substrate, enzyme and product). Also, hierarchical structure in networks, such as in a pathway with subprocesses or a complex with subunits, is not easily represented.
Alternative network representations that more faithfully represent biological systems have been proposed10–12 (http://www.biopax. org/), but no general and standard solution has yet emerged. Nonetheless, in many situations commonly encountered in molecular and cell biology, the use of simple networks combined with the patterns described here can be and have been effectively applied to arrive at novel biological insights. Being aware of these patterns should make it easier to see how they have been used and refined in network-based studies.
Acknowledgments
D.G. is financially supported by the Swiss National Science Foundation (Grant PBELA—120936).
Footnotes
Daniele Merico and David Gfeller contributed equally to this work.
References
- 1.Pujana MA, et al. Nat Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- 2.Mummery-Widmer JL, et al. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser AG, Marcotte EM. Nat Genet. 2004;36:559–564. doi: 10.1038/ng1370. [DOI] [PubMed] [Google Scholar]
- 4.Stark C, et al. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, et al. Nucleic Acids Res. 2007;35:W625–632. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cline MS, et al. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spellman PT, et al. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunsalus KC, et al. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 10.Hu Z, et al. Nat Biotechnol. 2007;25:547–554. doi: 10.1038/nbt1304. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda K, Takagi T. Bioinformatics. 2001;17:829–837. doi: 10.1093/bioinformatics/17.9.829. [DOI] [PubMed] [Google Scholar]
- 12.Le Novère N, et al. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 13.Strogatz SH. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 14.Collins SR, et al. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 15.Reguly T, et al. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson EH, et al. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 17.Boone C, Bussey H, Andrews BJ. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 18.Stuart JM, Segal E, Koller D, Kim SK. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]