Abstract
Aicardi-Goutières syndrome is a mendelian mimic of congenital infection and also shows overlap with systemic lupus erythematosus at both a clinical and biochemical level. The recent identification of mutations in TREX1 and genes encoding the RNASEH2 complex and studies of the function of TREX1 in DNA metabolism have defined a previously unknown mechanism for the initiation of autoimmunity by interferon-stimulatory nucleic acid. Here we describe mutations in SAMHD1 as the cause of AGS at the AGS5 locus and present data to show that SAMHD1 may act as a negative regulator of the cell-intrinsic antiviral response.
Aicardi-Goutières syndrome (AGS, MIM225750) is a genetically determined encephalopathy whose clinical importance is magnified because it closely mimics (and hence is often misdiagnosed as) the sequelae of congenital infection1. AGS and congenital virus infection are both associated with an increased production of interferon alpha (IFN-α)2. Furthermore, a disturbance of IFN-α homeostasis is considered central to the pathogenesis of the autoimmune disorder systemic lupus erythematosus (SLE)3. In keeping with this, some children with AGS also develop an early-onset form of SLE4–7. These related clinical observations led us to predict in 2003 that elucidation of the genetic basis of AGS would identify cellular components with key roles in the pathogenesis of acquired autoimmune disease8.
In 2006 we reported that recessive mutations in any of the genes encoding the 3′→5′ exonuclease TREX1 (TREX1, previously known as AGS1)9 or the three nonallelic components of the RNASEH2 endonuclease complex (RNASEH2B, RNASEH2C and RNASEH2A, also known as AGS2, AGS3 and AGS4, respectively)10 result in AGS. We further showed that heterozygous TREX1 mutations can cause both a dominant form of AGS and a cutaneous subtype of SLE, called familial chilblain lupus (CHBL, MIM610448)11. Subsequently, others have demonstrated heterozygous TREX1 mutations in a cohort of individuals with SLE12.
Because TREX1 and RNASEH2 are nucleases, we hypothesized that these proteins are involved in clearing cellular nucleic acid debris, and that a failure of waste removal may result in immune activation, specifically triggering the innate immune response that is more normally induced by viral nucleic acid9. Subsequently, Yang et al.13 demonstrated that human TREX1 deficiency does indeed result in the intracellular accumulation of abnormal single-stranded DNA species. Furthermore, Stetson et al.14 have shown that in Trex1-null mice, a type I interferon response accrues from activation of a TLR-independent pathway involving IRF3. Collectively, these studies have allowed the definition of a novel cell-intrinsic mechanism for the initiation of autoimmunity by interferon-stimulatory nucleic acid, and they offer a mechanistic explanation for the phenotypic overlap of AGS with congenital infection and SLE. That is, in the absence of TREX1 or RNASEH2 activity, endogenous nucleic acids accumulate and are sensed as viral or ‘nonself’, leading to the induction of an IFN-α–mediated immune response. The identification of additional cellular components involved in this important aspect of innate immunity is therefore of great interest.
In a genotype–phenotype analysis, we previously showed that 17% of families with AGS do not have identifiable mutations in TREX1, RNASEH2B, RNASEH2C or RNASEH2A15. In four consanguineous families (AGS73, AGS84, AGS109, AGS128) screening negative for mutations in these genes, we undertook extended genetic analysis. First, on the basis of genotypic discordance between siblings or an absence of homozygosity, we showed that these pedigrees were incompatible with linkage to TREX1, RNASEH2B, RNASEH2C or RNASHN2A (data not shown), thus indicating further genetic heterogeneity. In order to identify other genes involved in AGS, we conducted a SNP array genome-wide scan using affected individuals from these four families, and identified a 2-cM region of shared homozygosity on chromosome 20q11 (Supplementary Fig. 1 online). High-density microsatellite genotyping in this interval was then carried out using the four mapping families plus a further two consanguineous AGS pedigrees (AGS76 and AGS126) and two non-consanguineous AGS sibships of Maltese origin (AGS79, AGS104). Again, none of these families had identifiable mutations in TREX1, RNASEH2A, RNASEH2B or RNASEH2C. Microsatellite genotyping confirmed a region of homozygosity, between the markers D20S195 and D20S478, common to all six consanguineous pedigrees (Supplementary Fig. 2 online). Additionally, within this interval, the two Maltese sibships had a shared 1.4-Mb region of homozygosity with identical alleles at seven polymorphic markers. Such an observation was highly suggestive of an ancestral haplotype, especially as founder mutations in the Maltese population have been previously identified in two other disorders16. On the basis of this smaller region, we considered the AGS5 critical interval to contain 20 RefSeqannotated genes. An overlapping autozygous region in an Arab family (AGS165) with three affected children indicated a further possible reduction of the AGS5 critical interval to a <1-Mb region between D20SG1 and D20S834.
Sequencing of the genes BLCAP, CTNNBL1 and TGIF2 within this interval did not identify pathogenic mutations. However, in eight of the nine pedigrees used to define the AGS5 locus, and in one further family (AGS145) not included in the initial mapping panel, we identified homozygous mutations in the SAMHD1 gene (Table 1, Fig. 1 and Supplementary Table 1 online). Both Maltese families carried the identical C>T transition in exon 4. Three additional homozygous mutations (one nonsense, one missense and one 12-nt deletion producing an in-frame loss of four amino acids) were identified in this same exon. Homozygous mutations were also found in exons 5 and 10 (both missense), intron 14 (acceptor splice site) and exon 15 (nonsense). A further three probands were identified as compound heterozygotes, each carrying a missense mutation in exon 4 and a second missense (AGS82, AGS92) or nonsense (AGS91) mutation in exon 7, 5 and 12, respectively. In one additional proband (AGS156) we could identify only a single missense SAMHD1 mutation, which was maternally inherited.
Table 1.
Details of ancestry, pedigree structure and sequence alterations, with corresponding amino-acid changes, in families with mutations in SAMHD1
| Family | Ancestry | Tested | Nucleotide alteration |
Exon | Amino-acid alteration |
Parental consanguinity |
|---|---|---|---|---|---|---|
| AGS76 | Hungarian | 2A, F | Hom 625G>A | 5 | G209S | Third cousins |
| AGS79 | Maltese | 2A | Hom 433C>T | 4 | R145X | No |
| AGS82 | French | 2A, 1U, M, F | Het 368A>C | 4 | H123P | No |
| Het 760A>G | 7 | M254V | ||||
| AGS84 | Pakistani | 1A | Hom 1609-1G>C | (Intron 14) | Splice acceptor | First cousins |
| AGS91 | Fijian | 1A, M, F | Het 434G>A | 4 | R145Q | No |
| Het 1324C>T | 12 | R442X | ||||
| AGS92 | Canadian | 1A, M, F | Het 427C>T | 4 | R143C | No |
| Het 602T>A | 5 | I201N | ||||
| AGS104 | Maltese | 2A | Hom 433C>T | 4 | R145X | No |
| AGS109 | Moroccan | 1A | Hom 1153A>G | 10 | M385V | First cousins |
| AGS126 | Moroccan | 1A | Hom 428G>A | 4 | R143H | First cousins |
| AGS128 | Indian | 1A, M, F | Hom 445C>T | 4 | Q149X | First cousins |
| AGS145 | French | 1A | Hom 1642C>T | 15 | Q548X | No |
| AGS156 | Ashkenazi | 1A, M, F | Het 1106T>C | 10 | L369S | No |
| AGS165 | Arab | 3A, M, F | Hom 359_370del | 4 | 120_123del | First cousins |
A, affected; U, unaffected; F, father; M, mother; Hom, homozygous; Het, heterozygous.
Figure 1.
SAMHD1 is the AGS5 gene. Schematic of the AGS5 critical region and the SAMHD1 gene and protein. (a) Genetic map of chromosome 20q. The AGS5 critical interval is defined by the overlapping homozygous segment in AGS165 and the ancestral haplotype shared by two Maltese famillies (marker distances derived from the UCSC Browser, March 2006 assembly). (b) SAMHD1 spans 59,532 bp of genomic sequence (chromosome 20:34,954,059– 35,013,590) in 16 exons and encodes a 626 amino-acid (AA) protein with a molecular weight of 72.2 kDa. (c) Schematic of the SAMHD1 protein indicating position of identified substitutions and conserved domains.
All amino-acid alterations involved highly conserved residues (Supplementary Figs. 3 and 4 online). All mutations segregated with the disease in the families investigated and all parents tested were heterozygous for a relevant familial mutation. To investigate the effect of the intron 14 1609-1G>C mutation on splicing, we used RNA extracted from lymphoblastoid cells of the affected individual (AGS84) for RT-PCR. This showed that the predominant transcript resulted, as expected, from skipping of exon 15; full-length transcripts, and transcripts missing both exons 14 and 15 were also seen, albeit at lower levels (Supplementary Fig. 5 online). We did not identify SAMHD1 sequence changes in probands from a further 26 families fulfilling clinical criteria for a diagnosis of AGS and screening negative for mutations in TREX1, RNASEH2B, RNASEH2C and RNASEH2A.
SAMHD1 spans 59,532 bp of genomic sequence in 16 exons and encodes a 626 amino-acid protein. Using an antibody raised against recombinant SAMHD1, we determined wild-type SAMHD1 to be exclusively localized to the nucleus in control fibroblasts (Fig. 2a). In cells of an affected individual (AGS128) carrying the Q149X substitution, SAMHD1 was also localized to the nucleus, confirming that the derived mRNA is not subject to nonsense-mediated decay, and indicating that the first 149 amino acids of the protein are sufficient for nuclear localization (Supplementary Fig. 6 online). However, expression was diminished and localization to internal nuclear structures was less clear. We corroborated these data by showing that both full-length SAMHD1 and the Q149X N-terminal green fluorescent protein (GFP)–tagged fusion proteins were able to confer nuclear localization (Fig. 2b,c), and that the Q149X substitution resulted in reduced protein expression and an altered distribution within the nucleus.
Figure 2.
Localization of SAMHD1. (a) Localization of SAMHD1 in MRC-5 control and Q149X fibroblast cells (AGS128). Cells immunolabeled with DAPI (DNA), α-tubulin (to show microtubules) and antibody to SAMHD1. Scale bar, 10 µm. Top, specific nuclear localization of SAMHD1 in MRC-5 control cells. Bottom, nuclear localization is maintained in subject fibroblasts expressing the Q149X nonsense substitution (nonsignificant difference from MRC-5 cells), although expression is reduced and localization to nuclear structures is less well defined. (b) Localization of wild-type (WT) GFP-SAMHD1 and GFP-SAMHD1 carrying a Q149X substitution in HeLa cells. Cells immunolabeled with DAPI (DNA), α-tubulin and GFP. Scale bar, 10 µm. Top, GFP is expressed throughout the cell. Middle, specific nuclear localization of SAMHD1-GFP was seen in >98% of cells. Bottom, nuclear localization but with reduced expression and an altered intranuclear distribution is maintained in cells expressing a SAMHD1-GFP carrying the Q149X nonsense substitution. (c) Immunoblot of cell lysates containing expressed GFP constructs. Lane 1, molecular weight marker; lane 2, Optimem; lane 3, GFP control; lane 4, wild-type SAMHD1-GFP; lane 5, Q149X SAMHD1-GFP. In lane 4, full-length wild-type SAMHD1-GFP is indicated by the band between 75 and 100 kDa, whereas the smaller bands suggest protein degradation. A truncated Q149X-GFP gene product is seen in lane 5. FL, full-length; TP, truncated protein; PD, protein degradation; GFP, GFP construct.
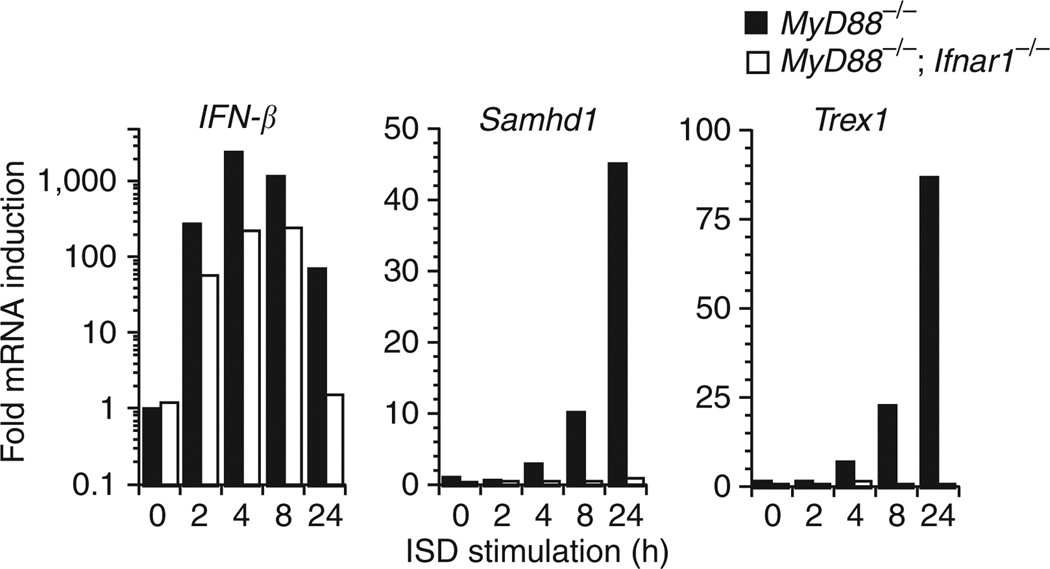
SAMHD1 was originally identified in a human dendritic cell cDNA library17 as an ortholog of the mouse IFN-γ–induced gene Mg11, hence the alternative name dendritic cell–derived IFN-γ–induced protein (DCIP). Other evidence implicates this protein in immune function, as it is upregulated in response to viral infections18–20 and may have a role in mediating TNF-α proinflammatory responses21. The AGS1 protein TREX1 is induced as part of the IFN-stimulatory DNA (ISD) response, a cytosolic TLR-independent antiviral pathway that detects DNA and triggers immune activation through IRF3 (ref. 14). To determine whether SAMHD1 is similarly induced, we transfected macrophages from MyD88 knockout mice with immunostimulatory DNA and observed an upregulation of Samhd1 expression (Fig. 3). Performing the identical experiment in macrophages from MyD88/Ifnar1 double knockout mice showed this expression to be interferon dependent. The latter result is consistent with reported data demonstrating induction of SAMHD1 by type 1 interferons (both IFN-α and IFN-β)22. The use of MyD88-null cells rules out the possibility of a TLR contribution to the above expression data and indicates that like TREX1, SAMHD1 may act as a negative regulator of the ISD response.
Figure 3.
Fold induction of IFN-β, Samhd1 and Trex1 following transfection with interferon-stimulatory DNA. We transfected bone marrow–derived macrophages from MyD88−/− and MyD88/Ifnar1 double knockout mice with calf-thymus interferon-stimulatory DNA and recorded fold induction of IFN-β, Samhd1 and Trex1 at the specified time points.
SAMHD1 is the only identified human protein in which a sterile alpha motif (SAM) and an HD domain occur in tandem. SAMs are 65–70 residues in length and can serve as protein-interaction modules mediating interactions with other SAM domain and non-SAM-domain– containing proteins23. Additionally, the SAM domains of Saccharomyces cerevisiae Vts1p and its Drosophila melanogaster homolog Smaug bind an RNA stem-pentaloop hairpin24. Modeling of the available structure of the N-terminal SAM domain of SAMHD1 (PDB 2E8O) with a similar RNA hairpin suggested differences in charge, which would preclude RNA hairpin binding (Supplementary Fig. 7 online). Additionally, using surface plasmon resonance (SPR), we were unable to identify an interaction with a variety of nucleic acid ligands (Supplementary Table 2 online). The HD domain, characterized by a motif with a doublet of divalent-cation-coordinating histidine and aspartic acid residues, is found in a diverse superfamily of enzymes with a predicted or known phosphohydrolase activity25. It is noteworthy that nucleotides are the substrates of five HD-domain enzymes characterized to date26, and a sixth, YhaM27, has a known exonuclease activity. Further work will be necessary to determine whether SAMHD1 also acts as a nuclease.
The elucidation of the genetic basis and cellular pathology of AGS is providing insights into key pathways of the innate immune response28–30. Our present findings suggest that like TREX1, SAMHD1 has a protective role in preventing self-activation of innate immunity by cell-intrinsic components. Further characterization of these cellular components is likely to influence our understanding of common autoimmune pathologies and general principles of antiviral defense.
ONLINE METHODS
Affected individuals and families
All affected individuals included in this study fulfilled diagnostic criteria for Aicardi-Goutières syndrome, with neurological features of an early-onset encephalopathy, negative investigations for common prenatal infections, intracranial calcification in a typical distribution, a cerebrospinal fluid lymphocytosis >5 cells/mm3 and/or >2 IU/ml of IFN-α in the cerebrospinal fluid and/or chilblains. With consent, blood samples were obtained from affected children, their parents and unaffected siblings. Genomic DNA was extracted from peripheral blood leukocytes by standard methods. The study was approved by the Leeds (East) Research Ethics Committee (reference number 07/Q1206/7).
Genotyping
Genome-wide scans were conducted in four families (AGS73, AGS84, AGS109, AGS128) using the Affymetrix GeneChip SNP Human Mapping 50,000 array (50K Xba240). We collated and standardized these data to a single recent build using the SNPsetter program and analyzed the combined data using IBDFinder. We conducted high-density genotyping using established microsatellite markers and novel markers (D20SG1, D20SG2, D20SG5, D20SG6, D20SG7; Supplementary Table 3 online) derived from the UCSC Human Genome Browser sequence (March 2006 freeze). Microsatellite genotyping was done by standard PCR amplification using fluorescent primers and analysis on an ABI 3130.
Mutation detection
We designed primers to amplify the coding exons of SAMHD1 (Supplementary Table 3). Purified PCR amplification products were sequenced using BigDye terminator chemistry and an ABI 3130 DNA sequencer. Mutation description is based on the reference cDNA sequence NM_015474; nucleotide numbering begins at the first A of the ATG initiator codon.
RT-PCR
We extracted RNA from subject lymphoblastoid cell pellets and HEK293 cells using a standard Trizol extraction. Reverse-transcription PCR of SAMHD1 was conducted using the Qiagen one-step rtPCR kit with primers SAMHD1GFPF and SAMHD1GFPR (Supplementary Table 3) to give a product of 1,893 bp. A PCR of the region around exon 15 was done using primers cDNA5F (in exon 11) and SAMHD1GFPR (Ex16/3′UTR) and the cDNA as template to give an expected product size of 648 bp. Products were visualized by agarose gel electrophoresis, gel-purified and sequenced.
Immunofluorescence microscopy
To determine the cellular localization of SAMHD1, we cultured MRC-5 control cells and subject fibroblasts carrying the Q149X substitution on glass coverslips for 24 h. Cells were fixed for 5 min in ice-cold methanol and labeled with 4,6-diamidino-2-phenylindole (DAPI, Calbiochem) for DNA, α-tubulin to show the microtubules (rat α-tubulin MCAP77, Serotec), and SAMHD1 (rabbit SAMHD1 12586-1-AP, Proteintech Group) using rabbit or rat secondary antibodies (Invitrogen Molecular Probes highly cross-absorbed IgG-specific Alexa conjugates). Immunofluorescence analysis was conducted using a Nikon TE2000 motorized microscope with a Hamamatsu Orca ER monochrome camera and Nikon Plan Apo VC (×100 or ×60) lens with a numerical aperture of 1.40, using IP lab software. Greater than 630 cells over three experimental series were scored for nuclear and cytoplasmic staining (Supplementary Table 4 online).
Production and expression of SAMHD1-GFP constructs
We generated a SAMHD1 cDNA expression cassette by reverse transcription and PCR of human glioma RNA. This was cloned into a holding vector and the mutation resulting in Q149X was introduced by site-directed mutagenesis (QuickChange: Stratagene) using primers Q149XF and Q149XR (Supplementary Table 3). These constructs were then subcloned into the pcDNA 6.2/N-EmGFP-GW/ TOPO vector (Invitrogen), sequenced to verify that they were correct, and transiently transfected into HeLa cells using Lipofectamine 2000. We determined localization of the constructs using immunofluorescence microscopy with a mouse monoclonal GFP antibody (Invitrogen). Cells were scored as above. HeLa cells transiently expressing the GFP constructs (as above) were harvested by scraping into 1× PBS. Cell lysates in RIPA buffer were run on 7.5% SDS-PAGE gels and immunoblotted using a rabbit polyclonal GFP antibody (Invitrogen).
Macrophage transfection and ISD response
Bone marrow–derived macrophages (700,000 per well of 12-well plates) from MyD88 null and MyD88/Ifnar1 double knockout mice were transfected with 3 µg of calf thymus DNA complexed with Lipofectamine 2000. Cells were harvested into RNA-Bee at the indicated time points, and quantitative RT-PCR was done with primers specific for mouse IFN-β, SAMHD1, and HPRT. The expression of each gene was calculated relative to untreated samples, by dividing each dCT value by the untreated value, and normalized for HPRT within each sample.
URLs
SNPsetter and IBDFinder programs, http://dna.leeds.ac.uk/; CDART (Conserved Domain Architecture Retrieval Tool), http://www.ncbi.nlm.nih. gov/Structure/lexington/lexington.cgi; ClustalW2, www.ebi.ac.uk.
Supplementary Material
ACKNOWLEDGMENTS
We thank the participating families with Aicardi-Goutières syndrome for the use of genetic samples and clinical information. We thank all clinicians for contributing samples not included in the current manuscript. We thank C. Ponting and E. Morrison for helpful discussions and R. Smith for technical support in preparing images. This work was supported by BDF Newlife, the Royal Society, a Wellcome Trust VIP award to G.I.R., the National Institutes for Health Research Manchester Biomedical Research Centre, and the International Aicardi-Goutières syndrome Association (IAGSA).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
G.I.R. performed genotyping and sequencing with contributions from T.A.B., T.L., H.G. and M.A. G.I.R. and J.B. undertook localization studies. A.A. and I.W.M. performed SPR experiments. I.M.C. carried out the SNP analysis. D.B.S. and R.L.B. performed the ISD studies. R.M.J. and J.C.F. undertook protein modeling. All other co-authors identified subjects with AGS and performed related clinical and laboratory studies. D.T.B. provided critical input into project direction and manuscript preparation. Y.J.C. designed and supervised the project and wrote the manuscript.
References
- 1.Crow YJ, Livingston JH. Aicardi-Goutieres syndrome: an important Mendelian mimic of congenital infection. Dev. Med. Child Neurol. 2008;50:410–416. doi: 10.1111/j.1469-8749.2008.02062.x. [DOI] [PubMed] [Google Scholar]
- 2.Dussaix E, Lebon P, Ponsot G, Huault G, Tardieu M. Intrathecal synthesis of different alpha-interferons in patients with various neurological diseases. Acta Neurol. Scand. 1985;71:504–509. doi: 10.1111/j.1600-0404.1985.tb03235.x. [DOI] [PubMed] [Google Scholar]
- 3.Crow MK. Type I interferon in systemic lupus erythematosus. Curr. Top. Microbiol. Immunol. 2007;316:359–386. doi: 10.1007/978-3-540-71329-6_17. [DOI] [PubMed] [Google Scholar]
- 4.Dale RC, Tang SP, Heckmatt JZ, Tatnall FM. Familial systemic lupus erythematosus and congenital infection-like syndrome. Neuropediatrics. 2000;31:155–158. doi: 10.1055/s-2000-7492. [DOI] [PubMed] [Google Scholar]
- 5.Aicardi J, Goutieres F. Systemic lupus erythematosus or Aicardi-Goutieres syndrome? Neuropediatrics. 2000;31:113. doi: 10.1055/s-2000-7533. [DOI] [PubMed] [Google Scholar]
- 6.De Laet C, et al. Phenotypic overlap between infantile systemic lupus erythematosus and Aicardi-Goutieres syndrome. Neuropediatrics. 2005;36:399–402. doi: 10.1055/s-2005-873058. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen M, Skullerud K, Bakke SJ, Lebon P, Jahnsen FL. Cerebral thrombotic microangiopathy and antiphospholipid antibodies in Aicardi-Goutieres syndrome–report of two sisters. Neuropediatrics. 2005;36:40–44. doi: 10.1055/s-2004-830532. [DOI] [PubMed] [Google Scholar]
- 8.Crow YJ, et al. Cree encephalitis is allelic with Aicardi-Goutieres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J. Med. Genet. 2003;40:183–187. doi: 10.1136/jmg.40.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crow YJ, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 10.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 11.Rice G, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 13.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice G, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrugia R, et al. Molecular genetics of tetrahydrobiopterin (BH4) deficiency in the Maltese population. Mol. Genet. Metab. 2007;90:277–283. doi: 10.1016/j.ymgme.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol. Lett. 2000;74:221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 18.Hartman ZC, et al. Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J. Virol. 2007;81:1796–1812. doi: 10.1128/JVI.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prehaud C, Megret F, Lafage M, Lafon M. Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J. Virol. 2005;79:12893–12904. doi: 10.1128/JVI.79.20.12893-12904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao D, Peng D, Li L, Zhang Q, Zhang C. Inhibition of G1P3 expression found in the differential display study on respiratory syncytial virus infection. Virol. J. 2008;5:114. doi: 10.1186/1743-422X-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao W, Bao Z, Cheng C, Mok YK, Wong WS. Dendritic cell-derived interferon-gamma-induced protein mediates tumor necrosis factor-alpha stimulation of human lung fibroblasts. Proteomics. 2008;8:2640–2650. doi: 10.1002/pmic.200700954. [DOI] [PubMed] [Google Scholar]
- 22.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 23.Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 24.Oberstrass FC, et al. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat. Struct. Mol. Biol. 2006;13:160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 25.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 26.Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 2008;378:215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oussenko IA, Sanchez R, Bechhofer DH. Bacillus subtilis YhaM, a member of a new family of 3′-to-5′ exonucleases in gram-positive bacteria. J. Bacteriol. 2002;184:6250–6259. doi: 10.1128/JB.184.22.6250-6259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcón-Riquelme ME. Nucleic acid by-products and chronic inflammation. Nat. Genet. 2006;38:866–867. doi: 10.1038/ng0806-866. [DOI] [PubMed] [Google Scholar]
- 29.Coscoy L, Raulet DH. DNA mismanagement leads to immune system oversight. Cell. 2007;131:836–838. doi: 10.1016/j.cell.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhoj VG, Chen ZJ. Linking retroelements to autoimmunity. Cell. 2008;134:569–571. doi: 10.1016/j.cell.2008.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.