Abstract
The human neutrophil-specific adhesion molecule CD177 (also known as the NB1 alloantigen) becomes upregulated on the cell surface in a number of inflammatory settings. We recently showed that CD177 functions as a novel heterophilic counterreceptor for the endothelial junctional protein PECAM-1 (CD31), an interaction that is mediated by membrane-proximal PECAM-1 IgD 6, which is known to harbor an S536N single nucleotide polymorphism of two major isoforms V98N536G643 and L98S536R643 and a yet-to-be-determined region on CD177. In vitro transendothelial migration experiments revealed that CD177+ neutrophils migrated significantly faster through HUVECs expressing the LSR, compared with the VNG, allelic variant of PECAM-1 and that this correlated with the decreased ability of anti-PECAM-1 Ab of ITIM tyrosine phosphorylation in HUVECs expressing the LSR allelic variant relative to the VNG allelic variant. Moreover, engagement of PECAM-1 with rCD177-Fc (to mimic heterophilic CD177 binding) suppressed Ab-induced tyrosine phosphorylation to a greater extent in cells expressing the LSR isoform compared with the VNG isoform, with a corresponding increased higher level of β-catenin phosphorylation. These data suggest that heterophilic PECAM-1/CD177 interactions affect the phosphorylation state of PECAM-1 and endothelial cell junctional integrity in such a way as to facilitate neutrophil transmigration in a previously unrecognized allele-specific manner.
Neutrophils are the most abundant leukocytes in blood and function as the first line of defense in the innate immune response. Neutrophils detect bacterial components, such as LPS and fMLP, via Toll-like or G-protein coupled receptors (1, 2), resulting in upregulation of migratory activities and neutrophil accumulation at sites of acute inflammation, vascular injury, or infection.
Neutrophil recruitment is a tightly regulated process and involves a multistep cascade of adhesive and migratory events that are mediated by three classes of adhesion receptors: selectins, integrins, and adhesion receptors of the Ig superfamily (3–5). One important Ig superfamily member is PECAM-1, a cell adhesion and signaling receptor that is expressed on platelets, monocytes, neutrophils and some T cells, as well as abundantly at endothelial cell–cell junctions (5, 6). PECAM-1 is composed of six extracellular IgDs (IgD1–IgD6), and IgD1 is known to mediate cation-independent homophilic interactions that play an important role during monocyte and neutrophil transendothelial migration (7, 8).
In addition to PECAM-1 IgD1-mediated homophilic interactions, we recently reported that CD177 (also known as NB1 Ag), a neutrophil-specific 58–64-kDa GPI-anchored member of cysteine-rich Ly-6 family, functions as a novel heterophilic binding partner that engages PECAM-1 in membrane-proximal IgD6 (9). A characteristic feature of CD177 is its variable expression on the surface of neutrophil subpopulations; CD177+ neutrophils in individuals can vary from 0 to 100% (10). However, the molecular basis of heterogeneous CD177 expression is not completely understood. In different clinical conditions, such as myeloproliferative disorder, essential thrombocythemia, and after G-CSF administration, CD177 becomes significantly upregulated on the neutrophil surface (10, 11).
Recently, three linked single nucleotide polymorphisms (SNPs) within the PECAM-1 gene have been identified that encode amino acid substitutions within IgD1 (exon 3; L98V), IgD6 (exon 8; S536N), and the cytoplasmic domain (exon 12; R643G), resulting in the expression of two major PECAM-1 isoforms within the human population, termed LSR and VNG (frequency 0.42 versus 0.58) (12). Because the S536N dimorphism is proximal to the CD177 binding site within IgD6 of PECAM-1, the purpose of the present investigation was to further examine the effect of the S536N dimorphism on CD177-dependent neutrophil migration.
Materials and Methods
Abs and reagents
mAbs PECAM-1.1 (specific for IgD5), PECAM-1.3 (against IgD1), and PECAM-1.2 (against D6) were produced and characterized as described (7). MAb Gi18 specific for IgD1 of PECAM-1 and Gi11 (against JAM-C) were generated in our laboratory (13). mAb CD62e for detection of E-selectin was purchased from Serotec (Dusseldorf, Germany); mAbs specific for phosphorylated epidermal growth factor receptor (Y1173), ERK (T202/Y204), and β-catenin (T41/S45) and mAb against ICAM-1 were purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Hybridoma cells producing mAb 7D8 specific for CD177 was a gift from Dr. D. Stroncek (National Institutes of Health, Bethesda, MD). A polyclonal anti-peptide Ab specific for the phosphorylated form of tyrosine 686 (anti-pY686) was produced in rabbits and used to detect PECAM-1 ITIM phosphorylation. Unlabeled and labeled secondary Abs were obtained from DakoCytomation (Glostrup, Denmark). Protease inhibitor mixture and chemoattractants fMLP, TNF-α, leukotriene B4 (LTB4), and IL-8 were from Sigma-Aldrich (Taufkirchen, Germany). FITC-labeled albumin, calcein, and 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxy- fluorescein acetoxymethyl ester were from Invitrogen (Karlsruhe, Germany). Pneumolysin (PLY) was a generous gift from Dr. T. Mitchell, Glasgow, U.K.
Genotyping of HUVECs and neutrophils for PECAM-1 polymorphisms
Total RNAwas extracted using the peqGOLD RNAPure kit, according to the manufacturer's instructions (peqLAB, Erlangen, Germany). RNA (1 μg) was reverse transcribed using a Ready-To-Go kit (GE Healthcare, Munich, Germany) with random hexamer primers, as recommended by the manufacturer (Invitrogen). Specific primer pairs encompassing nucleotides 189– 673 (5′-TGTGCCTGCAGTCTTCACTC-3′ and 5′-CAGAACAGTTGACCCTCACG-3′) and 1795–2287 (5′-GGATCTGGTCCCATCACCTA-3′ and 5′-CCGTGTACTGCACGTCTGAG-3′) were designed according to the database (accession number: NM_000442.3, www.ncbi.nlm.nih.gov/nuccore/110347450?report=fasta) to amplify PECAM-1 polymorphic regions containing the L98V, S536N, and R643G dimorphisms. An aliquot of 2 μl cDNA was amplified with 5 μl each primer (5 μM), 8 μl 2′ deoxynucleoside 5′ triphosphate (1.25 mM each 2′ deoxynucleoside 5′ triphosphate), and 1 μl AmpliTaq Gold Polymerase (5 U/μl; Applied Biosystems, Weiterstadt, Germany) in a total volume of 50 μl for 30 cycles in a Thermal Cycler (Thermo Electron, Dreieich, Germany) under the following conditions: denaturation 95°C, 1 min; annealing 56°C, 1 min; and extension 72°C, 1 min. PCR products were sequenced with PCR primers (see above) using a Dye Terminator Cycle sequencing kit on an ABI 3100 DNA sequencer (Applied Biosystems).
Maintenance of HUVECs
Primary HUVECs derived from umbilical cords of single individuals were purchased from Lonza (Basel, Switzerland) and were cultured in endothelial cell basal medium (Lonza) in T-75 flasks, as recommended by the manufacturer. Cells were grown (∼2 d) up to 70–80% confluence and were subsequently used for experiments or were split by treatment with Accutase (PAA Laboratories, Cölbe, Germany). Only HUVECs maintained for less than four passages were used in this study. Aliquots of cells were frozen in liquid nitrogen.
Flow cytometric analysis of neutrophils and endothelial cells
Neutrophils were isolated from EDTA-anticoagulated whole blood from healthy donors by dextran sedimentation and gradient centrifugation. Aliquots of 103 washed fresh neutrophils were incubated with 20 μg FITC-labeled Gi18 or 7D8 against PECAM-1 or CD177, respectively, for 30 min at 4°C. After washing with PBS, fluorescence-labeled neutrophils were analyzed on a FACSCalibur flow cytometer (BD Biosciences, Heidelberg, Germany). In some cases, neutrophils were stimulated with fMLP (10−8 M) or PLY (25 ng/ml) for 30 min at room temperature prior to labeling with mAbs. HUVECs were isolated as described above and incubated with 20 μg FITC-labeled mAbs Gi18, Gi11, ICAM-1, or CD62e for 30 min at 4°C. Cells were washed with PBS and analyzed by flow cytometry, as above. For ICAM-1 and CD62e expression analysis, cells were stimulated with TNF-α (20 ng/ml; Immuno Tools, Friesoythe, Germany) for 2 h at 37°C prior to incubation with mAbs.
Permeability assays
The permeability of HUVECs was assessed by the passage of FITC-albumin (Mr, 56,000; Sigma-Aldrich), as previously described (14). Briefly, 3 × 105 HUVECs with known PECAM-1 phenotypes (LSR or VNG) were plated onto 48 Transwells fibronectin-coated 3-μm-pore-size polycarbonate membrane inserts (Costar, Cambridge, MA) and left for 2 d to form confluent monolayers. Confluency of cells was monitored by microscopy. Aliquots of 200 μl RPMI 1640 media (Life Technologies, Carlsbad, CA) containing FITC-albumin (40 μg/ml) were applied in the upper chamber. Every 15 min for 1 h, a sample from the bottom chamber was read in triplicate in a fluorescent microtiter plate reader (BioTek, Bad Friedrichshall, Germany). After incubation with 200 μl RPMI 1640 media containing 0.2 U/ml thrombin (Siemens Healthcare Diagnostic, Marburg, Germany) or histamine (3.2 × 10−5 mol/l; Sigma-Aldrich) at 37°C for 15 min, the passage of FITC-albumin was measured as described above. Additional experiments were conducted with HUVECs treated with 2.5 μg/ml rCD177/Fc or rJAM-C/Fc for 30 min. In some experiments, cells were pretreated with 2.5 μg/ml PECAM-1 mAb against domain 5 (PECAM-1.1) at 37°C for 15 min prior to performing permeability assays. All data were from at least four independent experiments, and statistical analysis was performed using SPSS software (IBM, Munich, Germany).
Neutrophil transmigration assay
Neutrophil transmigration through HUVECs was performed as previously described (15). In brief, PECAM-1–phenotyped HUVECs (1 × 105) were cultured on 6.5-mm Transwells with fibronectin-coated 3-μm-pore-size polycarbonate membrane inserts (Costar), as described above. Human neutrophils were obtained from CD177-phenotyped blood donors and were isolated by Ficoll (Sigma-Aldrich) gradient centrifugation followed by hypotonic red cell lysis. Phenotyped neutrophils (7.5 × 107 cells/ml) in RPMI 1640 medium (Life Technologies) were labeled with 30 μl 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxy-fluorescein acetoxymethyl ester (2.5 μg/ml). Aliquots of 200 μl labeled neutrophils (5 × 106) were placed in the upper chamber, and cells were allowed to migrate at 37°C to the lower chamber containing fMLP (10−8 M), IL-8 (50 ng/ml), or LTB4 (10−4 M) as chemoattractants. After 90 min, neutrophils found in the lower chamber were measured in triplicate in a fluorescent microtiterplate reader. For inhibition experiments, HUVECs were treated with PECAM-1.3 or -1.2 at a concentration of 2.5 μg/ml for 20 min at room temperature prior to migration assay. In some experiments, neutrophils were labeled with calcein (10 μg/ml) in RPMI 1640 at room temperature for 30 min prior stimulation with fMLP (10−8 M) or PLY (25 ng/ml).
Stimulation of HUVECs with rCD177/Fc, PECAM-1 mAbs, and H2O2
HUVECs were cultured on six-well plates (Greiner, Frickenhausen, Germany) for 48 h until confluent. The media were changed to endothelial basal medium serum-free medium 18 h before the experiment. Cells were stimulated with rCD177/Fc (2.5 μg/ml), or mAb PECAM-1.1, PECAM-1.2 (2.5 μg/ml), PECAM-1.3 (2.5 μg/ml) for 60 min at 37°C, or with H2O2 (0.9 mM, Merck, Darmstadt, Germany) for 10 min at 37°C. Stimulation was followed by further incubation (30 min) with a secondary Ab to achieve cross-linking (final concentration 2.5 μg/ml). In some experiments, stimulation with PECAM-1.1 or H2O2 was followed by treatment with rCD177/Fc for 30 min at 37°C. Cells were then lysed with 100 μl 20 mM TBS, 1% Triton X-100, 7.5 μl Protease Inhibitor Cocktail, and 10 ml 5% EDTA for 30 min at 4°C. After centrifugation at 10,000 × g for 30 min at 4°C, supernatants were collected, and the protein concentration was measured using a bicinchoninic acid assay (Pierce, Rockford, IL). Cell lysates (10 μg) were analyzed by SDS-PAGE/Western blot analysis using mAb Gi18 against PECAM-1 (2.5 μg/ml), a rabbit polyclonal Ab specific for phosphorylated PECAM-1 (1 μg/ml), or phosphorylated β-catenin (1:1000 dilution) or GAPDH (1:1000 dilution). Proteins were visualized using HRP-labeled donkey anti-mouse IgG or goat anti-rabbit IgG followed by chemiluminescence detection (ECL plus; GE Healthcare, Piscataway, NJ).
Production of L98S536 and V98N536 rPECAM-1 proteins
A construct encoding the extracellular domain of L98S536 PECAM-1 (residues 1–574) in the expression vector pcDNA3 was used as a template for the preparation of the V98N536 PECAM-1 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The vector was transfected into CHO cells, and high-expression clones were selected. The clones were grown in CELLine factory flasks as per the manufacturer's instructions (Wilson Wolf Manufacturing, New Brighton, MN), and culture supernatant was collected. The PECAM-1 isoforms were purified using PECAM-1.3 affinity columns prepared with the Amino Link Plus Immobilization Kit (Pierce, Rockford, IL) and analyzed on SDS-PAGE gels for purity.
Production of CD177 protein
Full-length CD177 cDNA containing the human Ig Fc domain in the plg plus vector were cloned into pIB/V5-His by T/A cloning, as recommended by the manufacturer (Invitrogen, Karlsruhe, Germany), and then transformed into TOP 10 competent Escherichia coli (Invitrogen). Positive clones were screened by PCR. Sequences from positive clones were validated by nucleotide sequence analysis on ABI 3100 DNA sequencer before transfection. High five cells were transfected with 1 μg plasmid and subcloned in the presence of blasticidin (20 mg/ml; Invitrogen, Karlsruhe, Germany) for stable expression. Soluble rCD177 was collected from cell culture supernatants and purified using mAb 7D8 affinity column, as previously described (9). Native CD177 protein was purified from human granulocytes, as described previously (16).
Surface plasmon resonance analysis
Surface plasmon resonance (SPR) analysis was performed on a ProteOn XPR36 system (Bio-Rad, Hercules, CA), as previously described (9). The purified soluble PECAM-1 proteins (VN or LS allelic form), at a concentration of 100 μg/ml (in 10 mM sodium acetate buffer [pH 4.5]), were immobilized on a general layer medium sensor chip by amine coupling using standard procedures. CD177 (1 μM in PBS) or mAb PECAM-1.2 (5– 80 nM in PBS) were injected as analyte over the chip at a flow rate of 20 and 100 μl/min, respectively, in a total volume of 250 μl at 25°C. The sensorgrams were evaluated using the ProteOn evaluation software package.
Results
Genotyping HUVECs and neutrophils
To identify the allelic isoforms of PECAM-1 present in neutrophils and HUVECs, mRNA was isolated from HUVECs (n = 15) and neutrophils (n = 10) derived from different donors and amplified by RT-PCR. Two regions encompassing the three SNPs (L98V, S536N, and R643G) were analyzed by direct DNA sequencing (Fig. 1). Because these three SNPs are transmitted as a block with strong linkage disequilibrium, three phenotypes of PECAM-1 homozygous for VNG or LSR or heterozygous for VNG/LSR were found in our cohort. Of these, two homozygous VNG, two heterozygous VNG/LSR, and two homozygous LSR HUVEC lines were established and used in subsequent studies; they were designated NN, NS, and SS to denote the IgD6 phenotype. Neutrophils were also typed for the S536N polymorphism; only neutrophils heterozygous for the S536N polymorphism were used to exclude the possible influence of this dimorphism on neutrophil transmigration.
Figure 1.
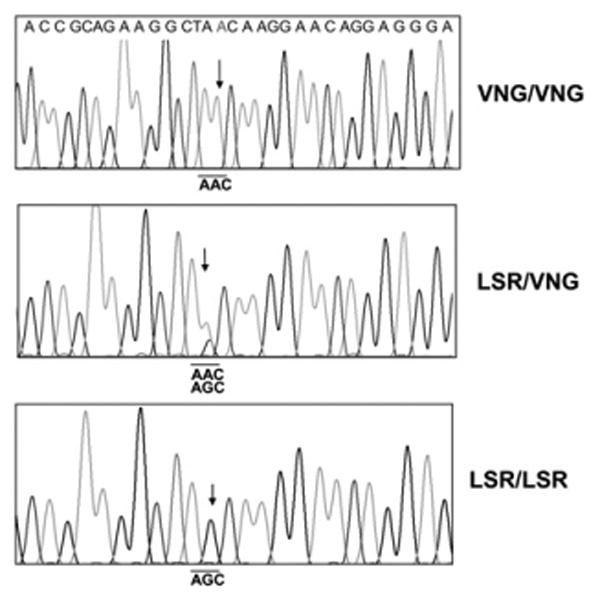
Nucleotide sequencing analysis of polymorphic PECAM-1 from three different HUVECs. mRNA extracted from HUVECs were reverse transcribed into cDNA and amplified with PECAM-1–specific primers and sequenced. Arrows indicate the position of SNP of PECAM-1 encoding asparagine/serine dimorphism (underline) at position 536 located in IgD6.
Analysis of PECAM-1 expression on different phenotyped HUVECs
Because the barrier integrity of HUVECs can be affected by variable surface density of PECAM-1 and other adhesive molecules, we compared the expression of these molecules on our endothelial cells bearing NN or SS allelic variants of PECAM-1 by flow cytometry. As shown in Fig. 2A, no significant difference in PECAM-1 expression was detected. Similar observations were noted with other adhesion molecules, such as JAM-C, E-selectin, and ICAM-1. These results indicated that HUVECs used in this study expressed equal levels of adhesion molecules on their surface.
Figure 2.
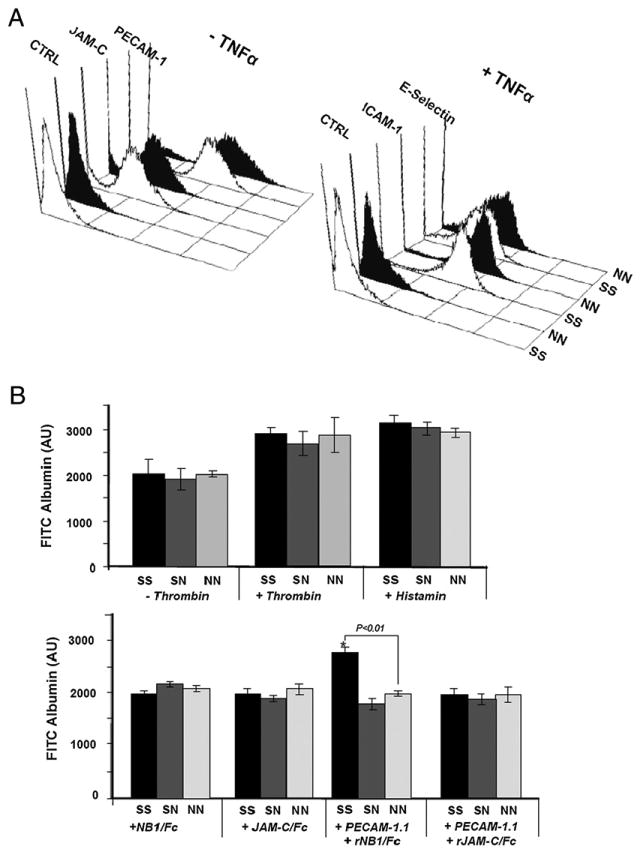
A, FACS analysis of adhesion molecules on SS (white) and NN (black) phenotyped HUVECs. HUVECs in resting (−TNF-α) or stimulated conditions (+TNF-α) were labeled with mAbs against PECAM-1, JAM-C, ICAM-1, E-Selectin, and mIgG (as control). Bound Abs were detected by FITC-labeled secondary Ab and were analyzed by flow cytometry. B, Permeability of three phenotyped HUVEC lines. HUVECs were cultured on fibronectin-coated polycarbonate filter chambers for 48 h. The passage of FITC-albumin through a confluent monolayer of cells at different time periods (5–60 min) was measured in the presence and absence of 0.2 U/ml thrombin or 3.2 × 10−5 mol/l histamine (upper panel). The lower panel shows permeability after treatment with mAb PECAM-1.1 in the presence and absence of rCD177/Fc or rJAM-C/Fc (as control). The intensity of migrated FITC-albumin in the lower chamber was measured by fluorescence microtiter plate reader. *p < 0.01; FITC-albumin passage between homozygous SS and NN (n = 4). AU, arbitrary unit.
Effect of PECAM-1 polymorphisms on vascular integrity
To examine the basal permeability of HUVECs expressing the SS, SN, and NN allelic isoforms of PECAM-1, we compared the flux of FITC-albumin through HUVEC monolayers. As shown in Fig. 2B, there was no significant difference in the permeability in any of the HUVEC lines 60 min after the addition of FITC-albumin. Similar results were obtained when HUVECs were treated with low-dose thrombin or histamine prior to permeability analysis (Fig. 2B). These data demonstrate that the LSR and VNG allelic isoforms of PECAM-1 are able to maintain vascular integrity to a similar extent.
To address the question of whether endothelial-basal permeability can be affected by CD177, HUVECs were treated with rCD177/Fc protein or rJAM-C/Fc as a control. After treatment with both proteins, no significant change and no allelic-dependent difference in cell permeability were observed. Interestingly, treatment of HUVECs with rCD177/Fc in the presence of anti–PECAM-1 Ab led to a significant increase in permeability in the SS, but not the NN, isoform of PECAM-1 (Fig. 2B), indicating that rCD177/Fc may influence endothelial permeability in allelic-dependent manner under stimulated, but not resting, conditions.
Effect of PECAM-1 polymorphisms on neutrophil transendothelial migration
The cell surface expression of CD177 on neutrophils is heterogeneous and varies widely among different individuals (17). Similarly, a 2–3-fold difference in the surface expression of PECAM-1 on neutrophils has been observed (M. Novinska and P.J. Newman, unpublished observations). Because differences in the expression of either of these cell surface receptors might influence the rate and/or extent of transendothelial migration, we determined their expression level prior to performing the transmigration assay. Neutrophils derived from six blood donors were surface labeled with FITC-conjugated anti-CD177 and anti–PECAM-1 and analyzed by flow cytometry. Neutrophils expressing high or low levels of CD177, but having similar PECAM-1 expression, were selected for further studies (Fig. 3). As shown in Fig. 4A, CD177High neutrophils transmigrated to a significantly greater extent through the homozygous SS, compared with heterozygous SN or homozygous NN, HUVEC monolayer toward fMLP. These differences were not detectable when CD177Null neutrophils were applied (Fig. 4A). These results indicate that the PECAM-1 S536N dimorphism within IgD6 has functional consequences for neutrophil transendothelial migration. To exclude the possibility that the genetically linked V98L dimorphism within IgD1, which mediates homophilic adhesion, might contribute to the observed difference in neutrophil transmigration, these experiments were repeated in the presence of mAbs PECAM-1.3 (specific for IgD1) or PECAM-1.2 (specific for IgD6). As shown in Fig. 4B, the addition of mAb PECAM-1.2 eliminated the phenotype-specific quantitative difference in transmigration efficiency, while suppressing the total degree of transmigration of CD177+ neutrophils across HUVEC monolayers. In contrast, the addition of PECAM-1.3 (specific for IgD1), which blocks homophilic interactions (7) and loosens endothelial junctions, overshadows the impact of heterophilic interaction and, thereby, eliminates the PECAM-1 allele-dependent effects on neutrophil migration. Thus, IgD6 plays a more significant role in the CD177-facilitated component of this process than does IgD1.
Figure 3.
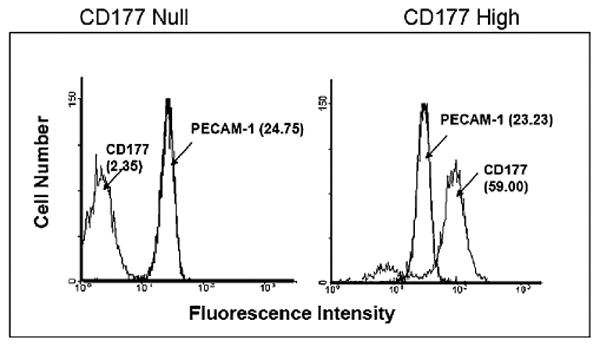
Analysis of CD177 and PECAM-1 surface expression in neutrophils by FACS. Neutrophils of CD177− (left panel) and CD177+ (right panel) individuals were labeled with mAb 7D8 or mAb Gi18. After washings, bound Abs were detected by FITC-labeled secondary Ab and were analyzed by flow cytometry.
Figure 4.
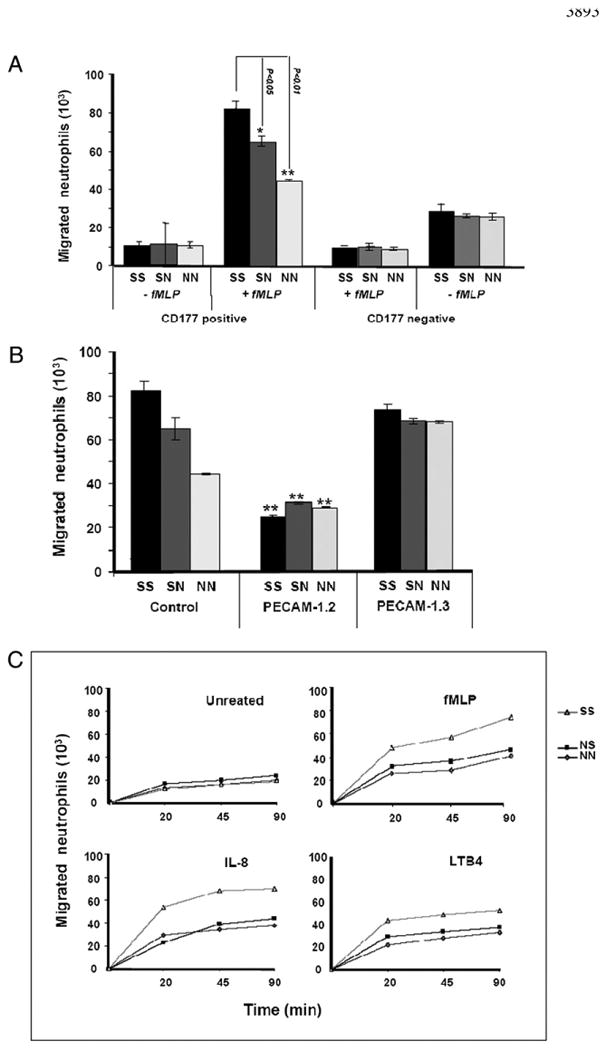
A, Analysis of neutrophil transmigration through PECAM-1–phenotyped HUVECs. HUVECs were cultured on fibronectin-coated polycarbonate membranes in transmigration chambers. After 48 h, fluorescence-labeled neutrophils (CD177+ or CD177−; see Fig. 3) were allowed to transmigrate through HUVECs for 90 min toward fMLP (10−8 M) or without fMLP (as control). Fluorescence intensity of migrated neutrophils in the lower chamber was measured by fluorescent microtiter plate reader (n = 4). B, Inhibition of neutrophil migration through PECAM-1–phenotyped HUVEC mAb PECAM-1 against IgD6 (PECAM-1.2) and IgD1 (PECAM-1.3). Fluorescence-labeled CD177+ neutrophils were allowed to pass through untreated and treated HUVECs toward fMLP and were measured as above (n = 4). C, Analysis of neutrophil transmigration through PECAM-1–phenotyped HUVECs with different chemoattractants. Fluorescence-labeled neutrophils (CD177+) were allowed to transmigrate through HUVECs for 90 min toward fMLP (10−8M), IL-8 (50 ng/ml), or LTB-4 (10−4 M) or without chemoattractant (as control) and were measured as above. *p < 0.05; **p < 0.01; neutrophil migration between homozygous SS and two NS and NN phenotypes (n = 4).
Effect of chemoattractants on kinetics of neutrophil transmigration in PECAM-1 allelic-dependent manner
To analyze the impact of different chemoattractants on PECAM-1 allelic-dependent migration of neutrophils, neutrophil migration through phenotyped HUVECs toward fMLP, IL-8, and LTB4 were compared in a time-dependent manner (Fig. 4C). Neutrophils migrated faster through HUVECs phenotyped for SS in comparison with NS and NN in response to all three chemoattractants, indicating that PECAM-1 allelic-dependent neutrophil migration is not chemoattractant specific.
To further characterize the influence of the S536N polymorphism on neutrophil transendothelial migration, we activated neutrophils with the bacterial toxin PLY prior to reperforming the transendothelial cell migration experiments. As shown in Fig. 5A, PLY treatment resulted in a significant upregulation of CD177 in the CD177− but not CD177+ neutrophil subpopulation, while depressing PECAM-1 expression. Despite the lower levels of PECAM-1, PLY-treated neutrophils exhibited slightly increased transendothelial migration across all HUVEC phenotypes (Fig. 5B), suggesting that neutrophil CD177, and not neutrophil PECAM-1, plays a dominant role in this process.
Figure 5.
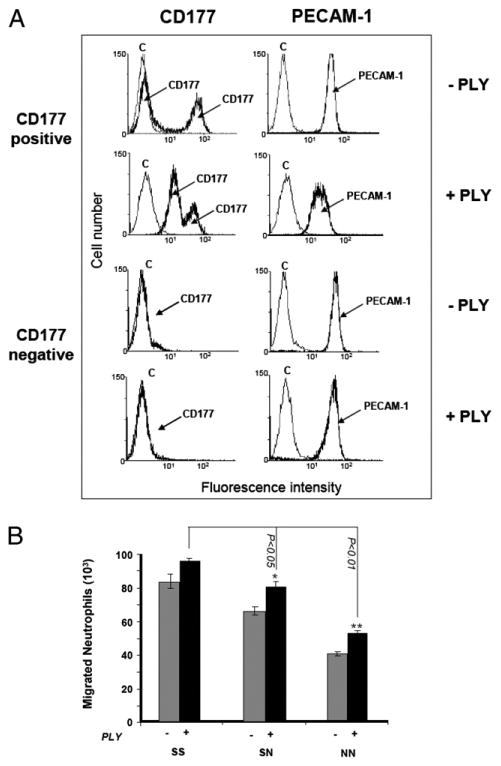
A, Analysis of CD177 and PECAM-1 surface expression on phenotyped neutrophils (CD177+ or CD177− neutrophils) before and after treatment with PLY. Neutrophils were treated with 25 ng/ml PLY for 30 min and labeled with mAbs 7D8 (anti-CD177) or Gi18 (anti–PECAM-1). After washing, bound mAb was detected with FITC-labeled secondary Ab and analyzed by FACS. B, PLY-untreated (−) or -treated (+) fluorescence-labeled neutrophils were allowed to migrate through HUVECs toward fMLP and were measured as above. *p < 0.05; **p < 0.01; neutrophil migration between homozygous SS and two NS and NN phenotypes (n = 4).
Allele-specific differences in PECAM-1 ITIM phosphorylation
Previous studies showed that the level of PECAM-1 ITIM phosphorylation correlates with endothelial cell barrier integrity (18, 19). To determine whether binding of domain-specific anti–PECAM-1 mAbs or CD177 might differentially stimulate phosphorylation of PECAM-1 expressed in SS versus NN homozygous HUVEC lines, these reagents were added to the cells, and coimmunoprecipitation/phosphotyrosine immunoblot analysis was performed. As shown in Fig. 6A, although CD177 by itself was unable to induce significant tyrosine phosphorylation of PECAM-1 ITIMs, engagement with IgD5-specific mAb PECAM-1.1 or IgD6-specific mAb PECAM-1.2, but not with IgD1-specific mAb PECAM-1.3, induced significant PECAM-1 tyrosine phosphorylation. Of note, engagement by mAb PECAM-1.2 induced less tyrosine phosphorylation in HUVECs expressing the SS allelic isoform of PECAM-1 than it did in NN-bearing endothelial cells. Because mAb PECAM-1.1 caused equal PECAM-1 phosphorylation in both isoforms, we sought to investigate the influence of CD177 on PECAM-1.1–pretreated HUVECs. Interestingly, rCD177/Fc suppressed PECAM-1 phosphorylation significantly in SS HUVECs compared with NN HUVECs. To further demonstrate the specificity of this phenomenon, we examined the phosphorylation of epidermal growth factor receptor and ERK, but we did not observe any PECAM-1 allele-specific differences (Supplemental Fig. 1). These effects were also not due to the differential affinity of mAb PECAM-1.2 or CD177 for the SS versus NN form of PECAM-1. SPR analysis showed that mAb PECAM-1.2 and CD177 bound equally to SS and NN allelic forms of PECAM-1 protein with similar binding kinetics (Fig. 7). Interestingly, postengagement of IgD6 by CD177 selectively diminished the degree of PECAM-1 tyrosine phosphorylation induced by the addition of H2O2 (Fig. 6B) in SS- but not NN-bearing HUVECs.
Figure 6.
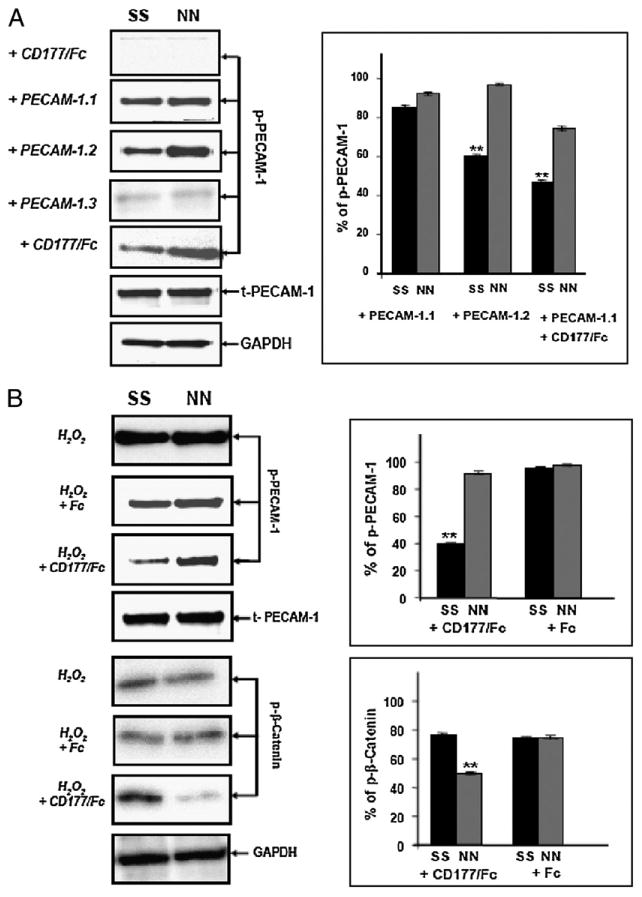
Immunoblot analysis of PECAM-1 phosphorylation in NN- or SS-phenotyped HUVECs after different stimulation. A, HUVECs were stimulated with rCD177/Fc, mAb PECAM-1.1 or PECAM-1.2 or PECAM-1.3 and PECAM-1.1 followed by rCD177/Fc (2.5 μg/ml for 60 min). After lysis, aliquots of protein (5 μg) were run on 10% SDS-PAGE. After blotting, proteins were stained with anti–PECAM-1 pY686 Ab (1 μg/ml), mAb Gi18 against PECAM-1 (2.5 μg/ml), or mAb against GAPDH (2.5 μg/ml) and were visualized with HRP-labeled secondary Abs using a chemiluminescence system. B, HUVECs were stimulated with H2O2 in the presence of rCD177/Fc or Fc alone (2.5 μg/ml for 60 min). After blotting, proteins were visualized with anti–PECAM-1 pY686 Ab (1 μg/ml), mAb Gi18 against PECAM-1 (2.5 μg/ml), anti–β-catenin pT41/S45 (2.5 μg/ml), or GAPDH (2.5 μg/ml), as described above. Bar graphs (right panels) represent the ratio of phosphorylated PECAM-1 (p– PECAM-1) or phosphorylated β-catenin (p–β-catenin) to total PECAM-1. *p < 0.01; PECAM-1 phosphorylation level between homozygous SS and NN.
Figure 7.
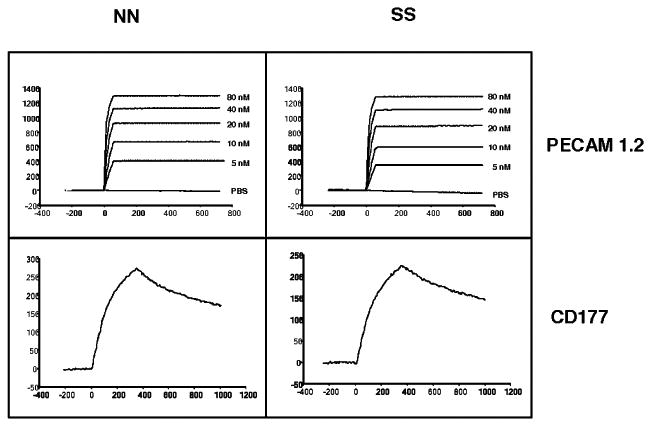
Effect of the PECAM-1 allelic form on ligand binding. Binding of mAb PECAM-1.2 and CD177 with the SS and NN allelic isoforms of PECAM-1 protein was determined in real time using SPR. Purified PECAM-1 isoforms were immobilized on a sensor chip. MAb PECAM-1.2 (upper panels) or purified CD177 (lower panels) was injected with a flow rate of 100 or 20 μl/ml, respectively, at 25°C. Note that CD177 bound with similar affinity to either PECAM-1 isoform as did mAb PECAM-1.2.
Several lines of evidence suggest that PECAM-1 ITIM phosphorylation might regulate endothelial permeability by phosphorylation/degradation of the adherent junction protein β-catenin (18, 19). Our permeability experiments in which we add the IgD5-specific mAb PECAM-1.1 to prestimulated HUVECs treated with rCD177/FC showed increased permeability in HUVECs expressing the SS versus NN isoform of PECAM-1 (Fig. 3B), and this was correlated with suppression of PECAM-1 phosphorylation (Fig. 6B). Thus, the ability of CD177 to suppress PECAM-1 phosphorylation in SS homozygous HUVECs might account for the different degree of β-catenin phosphorylation. To prove this hypothesis, we analyzed β-catenin phosphorylation in NN- and SS-bearing HUVECs after stimulation with H2O2 in the presence or absence of rCD177/Fc. Indeed, treatment of these cells with H2O2 and rCD177/Fc induced significantly greater β-catenin phosphorylation in SS- versus NN-bearing HUVECs (Fig. 6B). In contrast, equal β-catenin phosphorylation was observed in both cells treated with H2O2 alone. Taken together, our results suggest that PECAM-1 allele-dependent phosphorylation of β-catenin following CD177 engagement may be involved in the difference in endothelial cell permeability that we observed.
Discussion
Previous studies showed that the neutrophil-specific Ag CD177 supports transendothelial migration by interacting with membrane-proximal IgD6 of endothelial PECAM-1 (9). In the present investigation, we found that the S536N dimorphism located in IgD6 of endothelial PECAM-1 has a significant influence on the transendothelial migration of neutrophils (Fig. 4A). The rate of neutrophil transmigration through SS homozygous HUVECs was faster than through NN HUVECs, an effect that was dependent on the expression of neutrophil CD177 but independent of neutrophil PECAM-1 (Fig. 3A). Moreover, engagement of PECAM-1 by CD177 selectively suppressed PECAM-1 ITIM phosphorylation in SS but not NN homozygous HUVECs (Fig. 6B). Taken together, these data suggest that heterophilic PECAM-1/CD177 interactions that occur during neutrophil diapedesis reduce the overall PECAM-1 phosphorylation state, leading to weakened junctional stability that is conducive to neutrophil transmigration in a previously unrecognized allele-specific manner.
The S536N dimorphism within PECAM-1 IgD6 is in strong linkage disequilibrium with two other SNPs that occur within the coding region of the molecule, resulting in two common PECAM-1 haplotypes that have been termed LSR and VNG (12). Goodman et al. (20) were the first to describe an influence of these isoforms on leukocyte/endothelial cell interaction: heterozygous (LSR/VNG) monocytes were reported to adhere better to endothelium under conditions of flow than were LSR/VNG homozygous cells. This influence was attributed to PECAM-1 expressed on monocytes, rather than to endothelial cell PECAM-1. These findings are difficult to compare with our study because monocytes do not express CD177 (10), and adhesion under flow requires different molecular interactions than transmigration through static endothelial cells.
It is unlikely that any of the three amino acid polymorphisms of PECAM-1 by itself is solely responsible for the allele-specific effects that we observed downstream of PECAM-1/CD177 interaction. The L98V dimorphism located in IgD1 is known to mediate cation-independent homophilic interaction between PECAM-1 molecules on adjacent cells (21). Inhibition of this interaction affects allele-specific neutrophil transmigration (Fig. 4B), indicating that homophilic interactions between neutrophil and endothelial cell PECAM-1 do not play a major role in the allele-specific differences that were observed. The S536N dimorphism within IgD6 did not affect its binding affinity for CD177 (Fig. 7), and blocking IgD6 eliminated the differences in neutrophil transmigration rates between the two haplotypes. Finally, the R643G dimorphism is located within the cytoplasmic domain; however, its physical relationship to the ITIM phosphorylation sites has not been determined.
PECAM-1 is an adhesion and signaling receptor that is expressed abundantly at endothelial intracellular borders (22, 23), and it becomes phosphorylated when endothelial cell junctions are perturbed (18, 19). Elrayess et al. (24) reported higher tyrosine phosphorylation levels in VNG compared with LSR HUVECs, a finding that is consistent with the results in our study (Fig. 6A). Interestingly, although engaging endothelial cell PECAM-1 with a soluble dimeric form of CD177 did not by itself induce PECAM-1 phosphorylation, it was able to suppress PECAM-1 phosphorylation induced by Ab-mediated cross-linking or H2O2 treatment. Biswas et al. (19) reported that phosphorylated PECAM-1 ITIMs serve as a molecular scaffold that simultaneously recruits tyrosine-phosphorylated β-catenin and the protein-tyrosine phosphatase Src homology region 2 domain-containing phosphatase 1 (SHP)-2. Dephosphorylation of phosphorylated β-catenin by PECAM-1– bound SHP-2, in turn, allows β-catenin to rebind VE-cadherin, thereby supporting reassembly of the adherens junctional complex. Our finding that the binding of CD177 selectively suppresses PECAM-1 ITIM phosphorylation in SS (but not NN) homozygous HUVECs is consistent with the observed increase in FITC-albumin leakage as well as neutrophil transendothelial migration across endothelial cell monolayers expressing this allelic isoform of PECAM-1. Because our results showed that CD177-mediated suppression of PECAM-1 ITIM tyrosine phosphorylation leads to greater β-catenin phosphorylation in the PECAM-1 SS phenotype, it would be expected to delay the recruitment of SHP-2 and β-catenin and postpone restoration of endothelial cell junctional integrity in these cells. The R643G dimorphism is situated closed to the ITIM phosphorylation sites, but it seems more likely that two or all three dimorphisms together induce molecular changes that allow PECAM-1 to respond differently toward CD177 binding.
CD177 becomes significantly upregulated on the surface of circulating neutrophils in response to bacterial infections (11). PLY is a potent bacterial toxin produced during infections by Streptococcus pneumoniae, and it plays a role in the recruitment of neutrophils to luminal surfaces of the lungs during pneumonia (25). In this study, we linked these two observations by finding that PLY treatment results in upregulation of CD177 and enhanced PECAM-1 polymorphism-dependent neutrophil transendothelial migration. These data suggest that neutrophil CD177 and endothelial PECAM-1 contribute to the innate immune response to pneumonia infection and that the magnitude of the response may be affected by the allelic form of PECAM-1 that is expressed. Therefore, these findings add to the growing body of genetic-association studies linking PECAM-1 polymorphisms to other inherited and infectious diseases (25–29).
Despite quantitative differences in CD177 expression among healthy individuals, the proportion of CD177+ neutrophils in any individual seems to be relatively constant over time, with some notable exceptions (17). Recent reports of a genetic association between G42C polymorphism in the CD177 gene and the size of the CD177+ neutrophil population (30, 31) suggest that genetic and external factors can influence the size of the CD177+ subpopulation. However, the molecular mechanisms underlying this association are not clear. Additionally, we found a reciprocal regulation between CD177 and PECAM-1 on neutrophils. Treatment of these cells with fMLP (9) and with PLY (this study) caused significant upregulation of CD177, while depressing PECAM-1 expression on neutrophils. However, the reason for the different cross-talk between CD177 and PECAM-1 on neutrophils caused by different stimuli is unknown. In some situations, downregulation of PECAM-1 on neutrophils may attenuate the role of homophilic PECAM-1 interaction, while augmenting heterophilic interactions between CD177 and PECAM-1 on endothelial cells.
In conclusion, this study highlights the importance of heterophilic CD177/PECAM-1 interactions in neutrophil transendothelial migration. This interaction depends on the expression level of CD177 on neutrophils as well as on the allelic form of PECAM-1 expressed at endothelial cell junctions that, during the inflammatory response, become phosphorylated to different extents. Although beyond the scope of the present investigation, future studies to examine this hypothesis may shed considerable light on the complex interplay between neutrophil cell surface molecules and endothelial cell junctional proteins that takes place during acute and chronic inflammatory states.
Supplementary Material
Acknowledgments
We thank Olga Eva and Cathy Paddock for excellent technical assistance.
This work was supported by funding from Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (Klinische Forschergruppe Giessen), by Grant HL-40926 (to P.J.N. and D.K.N.) from the National Institutes of Health, and is part of the Ph.D. thesis of B.B.
Abbreviations used in this paper
- AU
arbitrary unit
- LTB4
leukotriene B4
- p–β-catenin
phosphorylated β-catenin
- p–PECAM-1
phosphorylated PECAM-1
- PLY
pneumolysin
- SHP
Src homology region 2 domain-containing phosphatase 1
- SNP
single nucleotide polymorphism
- SPR
surface plasmon resonance
Footnotes
The online version of this article contains supplemental material.
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 2.Springer TA. Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 4.Rao RM, Yang L, Garcia-Cardena G, Luscinskas FW. Endothelial-dependent mechanisms of leukocyte recruitment to the vascular wall. Circ Res. 2007;101:234–247. doi: 10.1161/CIRCRESAHA.107.151860b. [DOI] [PubMed] [Google Scholar]
- 5.Muller WA, Randolph GJ. Migration of leukocytes across endothelium and beyond: molecules involved in the transmigration and fate of monocytes. J Leukoc Biol. 1999;66:698–704. doi: 10.1002/jlb.66.5.698. [DOI] [PubMed] [Google Scholar]
- 6.Newman PJ. The biology of PECAM-1. J Clin Invest. 1997;100:25–29. [PubMed] [Google Scholar]
- 7.Sun QH, DeLisser HM, Zukowski MM, Paddock C, Albelda SM, Newman PJ. Individually distinct Ig homology domains in PECAM-1 regulate homophilic binding and modulate receptor affinity. J Biol Chem. 1996;271:11090–11098. doi: 10.1074/jbc.271.19.11090. [DOI] [PubMed] [Google Scholar]
- 8.Newton JP, Buckley CD, Jones EY, Simmons DL. Residues on both faces of the first immunoglobulin fold contribute to homophilic binding sites of PECAM-1/CD31. J Biol Chem. 1997;272:20555–20563. doi: 10.1074/jbc.272.33.20555. [DOI] [PubMed] [Google Scholar]
- 9.Sachs UJ, Andrei-Selmer CL, Maniar A, Weiss T, Paddock C, Orlova VV, Choi EY, Newman PJ, Preissner KT, Chavakis T, Santoso S. The neutrophil-specific antigen CD177 is a counter-receptor for platelet endothelial cell adhesion molecule-1 (CD31) J Biol Chem. 2007;282:23603–23612. doi: 10.1074/jbc.M701120200. [DOI] [PubMed] [Google Scholar]
- 10.Stroncek DF. Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr Opin Hematol. 2007;14:688–693. doi: 10.1097/MOH.0b013e3282efed9e. [DOI] [PubMed] [Google Scholar]
- 11.Göhring K, Wolff J, Doppl W, Schmidt KL, Fenchel K, Pralle H, Sibelius U, Bux J. Neutrophil CD177 (NB1 gp, HNA-2a) expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol. 2004;126:252–254. doi: 10.1111/j.1365-2141.2004.05027.x. [DOI] [PubMed] [Google Scholar]
- 12.Novinska MS, Pietz BC, Ellis TM, Newman DK, Newman PJ. The alleles of PECAM-1. Gene. 2006;376:95–101. doi: 10.1016/j.gene.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroll H, Sun QH, Santoso S. Platelet endothelial cell adhesion molecule-1 (PECAM-1) is a target glycoprotein in drug-induced thrombocytopenia. Blood. 2000;96:1409–1414. [PubMed] [Google Scholar]
- 14.Biosciences BD. In vitro study of cytokine-mediated activation of endothelial cell permeability using BD Falcon Cell Culture Inserts. Technical Bulletin. 2008;413 [Google Scholar]
- 15.Smith WB, Gamble JR, Clark-Lewis I, Vadas MA. Interleukin-8 induces neutrophil transendothelial migration. Immunology. 1991;72:65–72. [PMC free article] [PubMed] [Google Scholar]
- 16.Kissel K, Santoso S, Hofmann C, Stroncek D, Bux J. Molecular basis of the neutrophil glycoprotein NB1 (CD177) involved in the pathogenesis of immune neutropenias and transfusion reactions. Eur J Immunol. 2001;31:1301–1309. doi: 10.1002/1521-4141(200105)31:5<1301::AID-IMMU1301>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 17.Goldschmeding R, van Dalen CM, Faber N, Calafat J, Huizinga TW, van der Schoot CE, Clement LT, von dem Borne AE. Further characterization of the NB 1 antigen as a variably expressed 56-62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br J Haematol. 1992;81:336–345. doi: 10.1111/j.1365-2141.1992.tb08237.x. [DOI] [PubMed] [Google Scholar]
- 18.Esser S, Lampugnani MG, Corada M, Dejana E, Risau W. Vascular endothelial growth factor induces VE-cadherin tyrosine phosphorylation in endothelial cells. J Cell Sci. 1998;111:1853–1865. doi: 10.1242/jcs.111.13.1853. [DOI] [PubMed] [Google Scholar]
- 19.Biswas P, Canosa S, Schoenfeld D, Schoenfeld J, Li P, Cheas LC, Zhang J, Cordova A, Sumpio B, Madri JA. PECAM-1 affects GSK-3beta-mediated beta-catenin phosphorylation and degradation. Am J Pathol. 2006;169:314–324. doi: 10.2353/ajpath.2006.051112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman RS, Kirton CM, Oostingh GJ, Schön MP, Clark MR, Bradley JA, Taylor CJ. PECAM-1 polymorphism affects monocyte adhesion to endothelial cells. Transplantation. 2008;85:471–477. doi: 10.1097/TP.0b013e3181622d65. [DOI] [PubMed] [Google Scholar]
- 21.Fawcett J, Buckley C, Holness CL, Bird IN, Spragg JH, Saunders J, Harris A, Simmons DL. Mapping the homotypic binding sites in CD31 and the role of CD31 adhesion in the formation of interendothelial cell contacts. J Cell Biol. 1995;128:1229–1241. doi: 10.1083/jcb.128.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayalon O, Sabanai H, Lampugnani MG, Dejana E, Geiger B. Spatial and temporal relationships between cadherins and PECAM-1 in cell-cell junctions of human endothelial cells. J Cell Biol. 1994;126:247–258. doi: 10.1083/jcb.126.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Maschio A, Zanetti A, Corada M, Rival Y, Ruco L, Lampugnani MG, Dejana E. Polymorphonuclear leukocyte adhesion triggers the disorganization of endothelial cell-to-cell adherens junctions. J Cell Biol. 1996;135:497–510. doi: 10.1083/jcb.135.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elrayess MA, Webb KE, Whittall RA, Kabir J, Hawe E, Syvänne M, Taskinen MR, Frick MH, Nieminen MS, Kesäniemi YA, et al. A novel functional polymorphism in the PECAM-1 gene (53G>A) is associated with progression of atherosclerosis in the LOCAT and REGRESS studies. Atherosclerosis. 2003;168:131–138. doi: 10.1016/s0021-9150(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 25.Moreland JG, Bailey G. Neutrophil transendothelial migration in vitro to Streptococcus pneumoniae is pneumolysin dependent. Am J Physiol Lung Cell Mol Physiol. 2006;290:L833–840. doi: 10.1152/ajplung.00333.2005. [DOI] [PubMed] [Google Scholar]
- 26.Wenzel K, Baumann G, Felix SB. The homozygous combination of Leu125Val and Ser563Asn polymorphisms in the PECAM1 (CD31) gene is associated with early severe coronary heart disease. Hum Mutat. 1999;14:545. doi: 10.1002/(SICI)1098-1004(199912)14:6<545::AID-HUMU20>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi M, Looareesuwan S, Ubalee R, Tasanor O, Suzuki F, Wattanagoon Y, Na-Bangchang K, Kimura A, Aikawa M, Hirayama K. Association of adhesion molecule PECAM-1/CD31 polymorphism with susceptibility to cerebral malaria in Thais. Parasitol Int. 2001;50:235–239. doi: 10.1016/s1383-5769(01)00082-4. [DOI] [PubMed] [Google Scholar]
- 28.Listì F, Caruso C, Balistreri CR, Grimaldi MP, Caruso M, Caimi G, Hoffmann E, Lio D, Candore G. PECAM-1/CD31 in infarction and longevity. Ann N Y Acad Sci. 2007;1100:132–139. doi: 10.1196/annals.1395.011. [DOI] [PubMed] [Google Scholar]
- 29.Wei YS, Lan Y, Liu YG, Meng LQ, Xu QQ, Xie HY. Platelet-endothelial cell adhesion molecule-1 gene polymorphism and its soluble level are associated with ischemic stroke. DNA Cell Biol. 2009;28:151–158. doi: 10.1089/dna.2008.0817. [DOI] [PubMed] [Google Scholar]
- 30.Wolff J, Brendel C, Fink L, Bohle RM, Kissel K, Bux J. Lack of NB1 GP (CD177/HNA-2a) gene transcription in NB1 GP- neutrophils from NB1 GP-expressing individuals and association of low expression with NB1 gene polymorphisms. Blood. 2003;102:731–733. doi: 10.1182/blood-2002-09-2831. [DOI] [PubMed] [Google Scholar]
- 31.Caruccio L, Walkovich K, Bettinotti M, Schuller R, Stroncek D. CD177 polymorphisms: correlation between high-frequency single nucleotide polymorphisms and neutrophil surface protein expression. Transfusion. 2004;44:78–82. doi: 10.1046/j.0041-1132.2004.00606.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


