Abstract
Background
Subphenotypes have been identified within heterogeneous syndromes such as asthma and breast cancer, with important therapeutic implications. Whether subphenotypes exist within the acute respiratory distress syndrome (ARDS), another heterogeneous syndrome, is unknown.
Methods
We applied latent class modeling to identify subphenotypes using clinical and biological data from two NHLBI ARDS randomized controlled trials; modeling was conducted independently in each cohort. We then tested the association of subphenotypes with clinical outcomes in both cohorts and with the response to positive end-expiratory pressure (PEEP) in the second cohort.
Findings
Independent latent class models indicated that a two-class (i.e. two subphenotype) model was optimal for both cohorts. In both cohorts, we identified a hyperinflammatory subphenotype (Phenotype 2) that was characterized by higher plasma levels of inflammatory biomarkers, a higher prevalence of vasopressor use, lower serum bicarbonate, and a higher prevalence of sepsis, compared to Phenotype 1. Subjects in Phenotype 2 had higher mortality and fewer ventilator-free and organ failure-free days in both cohorts. In the second cohort, the effects of ventilation strategy on mortality, ventilator and organ failure-free days differed significantly by phenotype (p=0.003–0.049 for interactions).
Interpretation
Latent class models identify two subphenotypes within ARDS, one of which is characterized by more severe inflammation, shock, and metabolic acidosis and by significantly worse clinical outcomes. Response to treatment in a randomized trial of PEEP strategies differed based on subphenotype. Identification of ARDS subphenotypes may be useful in selecting patients for clinical trials.
Funding
National Institutes of Health
INTRODUCTION
The acute respiratory distress syndrome (ARDS) is a heterogeneous syndrome first identified in 1967 and defined by the clinical criteria of bilateral pulmonary opacities on chest radiograph, arterial hypoxemia (PaO2/FiO2 ratio < 300), and exclusion of cardiac failure as the primary etiology of the syndrome.(1–3) This definition was derived empirically based on clinical experience, with the hypothesis that it would identify patients with non-cardiogenic pulmonary edema, characterized by increased protein permeability of the alveolar-capillary membrane. Since the time of the original identification of ARDS and increasingly over the past two decades, there has been recognition of the clinical and biological heterogeneity within the syndrome(4, 5); this heterogeneity may reflect our incomplete understanding of the biology of ARDS and likely contributes to the poor track record of Phase II/III trials of novel therapies in patients with ARDS.(6) As a result, some investigators have proposed subdividing ARDS based on clinical risk factor, or by direct vs. indirect etiology of lung injury; however, at present there is no consensus in the field on the appropriate approach to reducing ARDS heterogeneity.
In contrast to ARDS, research in airways disease and cancer has made substantial progress towards identifying subphenotypes of disease, with important therapeutic implications. For example, subphenotypes based on the presence or absence of Th2-dependent inflammation have recently been identified within asthma, with important mechanistic and therapeutic implications.(7) This insight has led to new targeted treatments, such as a monoclonal antibody to IL-13 that is particularly effective in individuals with Th2-predominant inflammation.(8) Despite widespread recognition of the heterogeneity within common critical illness syndromes such as sepsis and ARDS, and some evidence suggesting that subphenotypes may exist within severe sepsis,(6, 9, 10) there is little data on whether such subphenotypes exist within ARDS.
Latent class analysis (LCA) is a well-validated statistical technique that uses mixture modeling to find the best fitting model for a set of data, based on the hypothesis that the data contains a number of unobserved groups or classes. The statistical approaches underlying this method were originally developed over a century ago by investigators analyzing whether a population of crabs in fact consisted of two subspecies.(11) In contrast to traditional regression analyses, in which the goal is to understand the relationship of pre-specified independent variables to a known outcome, LCA models ask whether there are subgroups of patients defined by a combination of the baseline variables, without mandating consideration of the outcome. Latent class-based methods have been extensively used in the social sciences and in other medical disciplines (12, 13) for instance in identification of asthma subphenotypes(14) but have not been highly utilized in critical care. We sought to capitalize on the wealth of clinical and biological data available from two NHLBI-sponsored ARDS Network randomized controlled trials by using LCA methods to attempt to identify and validate novel subphenotypes of ARDS and test their association with clinical outcomes and response to treatment.
MATERIALS AND METHODS
Study Design
Clinical and biological data were obtained from patients enrolled in the NHLBI ARDS Network’s randomized controlled trials of lower tidal volume ventilation (referred to here as ARMA)(15–17) and higher vs. lower positive end-expiratory pressure (PEEP) (trial referred to here as ALVEOLI).(18) Details of the original trials have been previously published in full. Patients randomized to higher tidal volume ventilation in the ARMA trial (n=429) were excluded a priori from the analysis due to the effect of higher tidal volumes on mortality, which would have precluded analysis of the association between latent class and clinical outcomes (Figure 1). Analyses were conducted first in the ARMA trial lower tidal volume patients (n=473, Figure 1). Analyses were then repeated independently in the ALVEOLI trial (n=549, no patients excluded), to test whether the findings would generalize to an independent sample. All clinical data (other than outcomes) and biological data used for this analysis were collected at study baseline (pre-randomization). Patients were enrolled in the ARMA study between 1996 and 1999; patients were enrolled in the ALVEOLI study between 1999 and 2002. Additional details on the original trials are available in the online supplement.
Figure 1.
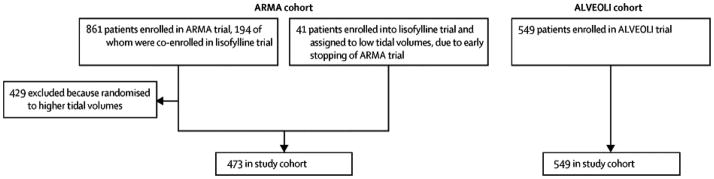
Flow diagram of patient inclusion in the first (ARMA) cohort and second (ALVEOLI) cohort.
Assay Procedures
Plasma samples used for this analysis were drawn at the time of randomization, which was < 36 hours from the time of meeting ARDS criteria in both studies. Plasma biomarkers were measured in duplicate using enzyme-linked immunoassay techniques (ELISA). Details of the methods used to perform the assays have been previously published.(4, 19–25)
Statistical Analysis
Baseline clinical data and biomarker levels were considered as class-defining variables in the LCA model; classification was conducted without consideration of clinical outcomes. Details on clinical variable selection, data cleaning and a complete list of the clinical variables included in the LCA models (Table S1) are in the online supplement. In addition to the clinical data, we included as inputs in the LCA model the eight plasma biomarkers previously associated with poor clinical outcomes in ARDS and previously measured in both samples: surfactant protein D (SP-D),(23) von Willebrand Factor antigen (vWF),(19) soluble intercellular adhesion molecule-1 (sICAM-1),(24) interleukins 6 and 8 (IL-6, IL-8),(21) soluble tumor necrosis factor receptor-1 (sTNFr-1),(22) plasminogen activator inhibitor-1 (PAI-1),(20) and protein C.(20) The datasets used, by virtue of being derived from randomized controlled trials with intensive on-site auditing and data quality checks, were largely complete; however, there were some variables with some missing data (Table S1).
Statistical analyses were conducted using Mplus v7.11. Basic two group comparisons between the two cohorts were conducted using the t-test, Wilcoxon rank sum, or chi squared test as appropriate. Next, we fitted a series of latent class models, first using the ARMA cohort and then repeated independently using the ALVEOLI cohort. Criteria for model selection were based on the Bayesian Information Criteria, the Vuong-Lo-Mendell-Rubin (VLMR) likelihood ratio test, and the size of the smallest class. Latent class model estimation was based on full-information maximum likelihood methods as implemented in Mplus. This approach allows for the use of all data from all patients, including those missing some data, in estimating the latent class models. Additional details on the latent class modeling procedures are available in the online supplement.
Once the number of classes was determined, the associations between class and clinical outcomes (90 day mortality, ventilator-free and organ failure-free days) were tested using the approach developed by Lanza (26). This method incorporates the degree of uncertainty of class membership. Finally, for the second cohort, in which patients were randomized to lower or higher PEEP, we tested models of each outcome using class, treatment assignment and their interaction as covariates to determine whether there was a differential treatment effect based on latent class. This analysis was conducted using Poisson regression for ventilator-free and organ failure-free days and using logistic regression for mortality.
RESULTS
Clinical Features of Cohorts
Baseline clinical data on patients in both cohorts are presented in Table 1. Several significant differences between the two cohorts were noted, including primary ARDS risk factor, severity of illness as measured by APACHE III scores, and prevalence of vasopressor use at enrollment. In addition, several ventilator parameters at randomization differed considerably between the two cohorts, likely reflecting changes in practice resulting from the publication of the results of the first trial (of lower tidal volume ventilation) that had been adopted at the time of the start of the second trial. Baseline biomarker data are presented in Table 2. As with the clinical data, there were substantial differences in the baseline biomarker levels between cohorts, likely reflecting differences in severity of illness and/or in pre-randomization ventilation parameters.
Table 1.
Comparison of Key Clinical Data Points (Pre-Randomization) Between ARMA (n=473) and ALVEOLI (n=549) Cohorts
| Clinical Variable* | ARMA Cohort | ALVEOLI Cohort | p-value |
|---|---|---|---|
| Age, years | 51 ± 17 | 51 ± 17 | 0.96 |
| Female gender, n (%) | 284 (60%) | 302 (55%) | 0.09 |
| Caucasian race, n (%) | 355 (75%) | 412 (75%) | 0.93 |
| ARDS Risk Factor, n (%)† | |||
| Trauma | 59 (13%) | 45 (9%) | |
| Sepsis | 125 (27%) | 120 (23%) | < 0.001 |
| Aspiration | 72 (16%) | 84 (16%) | |
| Pneumonia | 145 (32%) | 221 (42%) | |
| Other | 60 (13%) | 52 (10%) | |
| APACHE III score | 82 ± 29 | 94 ± 32 | < 0.001 |
| On vasopressors at enrollment, n (%) | 134/338** (40%) | 144/549 (26%) | < 0.001 |
| PaO2/FiO2 ratio | 132 ± 60 | 128 ± 58 | 0.25 |
| Maximum temperature, °C | 38.5 ± 0.9 | 38.5 ± 1.0 | 0.09 |
| Lowest systolic blood pressure, mm Hg | 88 ± 19 | 88 ± 17 | 0.90 |
| Maximum heart rate, beats per minute | 127 ± 22 | 125 ± 24 | 0.31 |
| Maximum respiratory rate, breaths per minute | 30 ± 10 | 33 ± 10 | < 0.001 |
| Urine output, prior 24 hrs, L | 2.02 (1.24–2.98) | 1.85 (1.13–2.93) | 0.06 |
| Lowest hematocrit, % | 30 ± 6 | 30 ± 6 | 0.64 |
| Peak white blood cell count, thousands | 12.3 (8.9–17.9) | 13.0 (8.7–18.2) | 0.68 |
| Platelets, thousands | 164 ± 121 | 177 ± 124 | 0.10 |
| Lowest sodium | 137±5 | 137 ± 5 | 0.86 |
| Highest creatinine, mg/dL | 1.10 (0.80–1.70) | 1.10 (0.8–1.9) | 0.70 |
| Lowest glucose | 135 ± 57 | 133 ± 64 | 0.68 |
| Lowest albumin | 2.2 ± 0.6 | 2.1 ± 0.6 | < 0.01 |
| Total bilirubin | 1.0 (0.60–2.10) | .80 (0.5–1.5) | < .001 |
| Bicarbonate | 21 ± 5 | 22 ± 6 | 0.24 |
| Tidal volume, mL | 671 ± 126 | 511 ± 119 | < 0.001 |
| Total minute ventilation, L/min | 13 ± 4 | 12 ± 4 | < 0.001 |
| PEEP, cm H20 | 8.6 ± 3.8 | 9.5 ± 4.3 | < 0.001 |
| Plateau pressure, cm H20 | 30 ± 8 | 27 ± 7 | < 0.001 |
| Mean airway pressure, cm H20 | 16 ± 5 | 16 ± 5 | 0.38 |
| PaCO2, mm Hg | 37 ± 8 | 39 ± 9 | < 0.001 |
| Body Mass Index | 27 ± 7 | 27 ± 7 | 0.53 |
Data shown as mean ± standard deviation, n (%), or median (interquartile range) as appropriate.
Demoninator = 338 due to missing data
Denominator = 461 for ARMA, 522 for ALVEOLI due to missing data
Table 2.
Comparison of Key Biomarker Values (Pre-Randomization) Between ARMA and ALVEOLI Cohorts
| Biomarker* | ARMA Cohort | ALVEOLI Cohort | p-value |
|---|---|---|---|
| Protein C (% control) | 47 (32–66) | 78 (45–122) | < .001 |
| Plasminogen activator inhibitor-1 (ng/ml) | 70 (40–138) | 61 (30–144) | .002 |
| Interleukin-6 (pg/ml) | 264 (109–766) | 238 (93–741) | .26 |
| Interleukin-8 (pg/ml) | 43 (20–93) | 40 (16–98) | 0.001 |
| Soluble tumor necrosis factor receptor-I (pg/ml) | 3255 (2128–5600) | 4265 (2599–8448) | < .001 |
| Soluble intercellular adhesion molecule-1 (ng/ml) | 627 (345–1038) | 924 (605–1385) | < .001 |
| Surfactant Protein D (ng/ml) | 84 (40–162) | 101 (50–218) | .004 |
| Von Willebrand Factor antigen (% control) | 284 (173–436) | 398 (247–624) | < .001 |
Data shown as median (interquartile range)
Latent-Class Modeling: Identification of Number of Phenotypes
In each cohort, latent-class models suggested that a two-class model provided the optimal fit. Specifically, Table 3 displays a summary of the model fits for 2 through 5 classes for both cohorts. In both cohorts, the p-value testing the number of classes indicated that a 2-class model was a significant improvement over a 1-class model, but that the 3-class model did not significantly increase the explanatory power. At the same time, the value of the Bayesian Information Criteria continued to decrease as the number of classes increased; this decrease suggests that the addition of more classes is worth the added model complexity. This decrease was also seen in both the Akakie Information Criteria and sample-sized adjusted-Bayesian Information Criteria (data not shown). To ensure that a two-class model provided the optimal fit, we also explored a three class model, which produced one class with an N of only 46 in the ARMA cohort. In the ALVEOLI cohort, the third class consisted of only four cases. While the decrease in the BIC would suggest adding additional classes to the model, upon consideration of the p-value (favoring a two-class model) and the small number of subjects in the third class, the two class model was retained. For simplicity, we will henceforth refer to the two classes as Phenotypes 1 and 2, respectively. In the 2-class model, the average latent class probabilities for the most likely class in the first cohort were .95 for Phenotype 1 and .92 for Phenotype 2; in the second cohort, the analogous probabilities were .97 and .94, indicating good model fit and very strong probabilities of class assignment (Figure S1).
Table 3.
Fit statistics for latent class models from two to five classes
| ARMA Cohort:
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Number of Individuals Per Class/Subphenotype
| ||||||||
| Number of classes | BIC | Entropy* | N1 | N2 | N3 | N4 | N5 | p-value** |
| 2 | 39947.9 | .78 | 318 | 155 | .036 | |||
| 3 | 39760.2 | .88 | 308 | 119 | 46 | .59 | ||
| 4 | 39656.7 | .86 | 212 | 126 | 43 | 92 | .28 | |
| 5 | 39583.8 | .86 | 150 | 120 | 36 | 36 | 131 | .64 |
| ALVEOLI Cohort:
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Number of Individuals Per Class/Subphenotype
| ||||||||
| Number of classes | BIC | Entropy* | N1 | N2 | N3 | N4 | N5 | p-value** |
| 2 | 49709.5 | .87 | 404 | 145 | .016 | |||
| 3 | 49383.7 | .92 | 400 | 145 | 4 | .58 | ||
| 4 | 49098.8 | .94 | 386 | 129 | 4 | 30 | .35 | |
| 5 | 48955.1 | .87 | 242 | 154 | 4 | 30 | 119 | .80 |
Abbreviations: BIC = Bayesian Information Criterion
Entropy is an index of how well the classes are separated. It ranges from zero to one and values around .8 and up are generally considered a sign of a useful model.
By Vuong-Lo-Mendell-Rubin test, testing whether the number of classes provides improved model fit compared to a model using one fewer class.
Clinical and Biological Characteristics of Each Phenotype
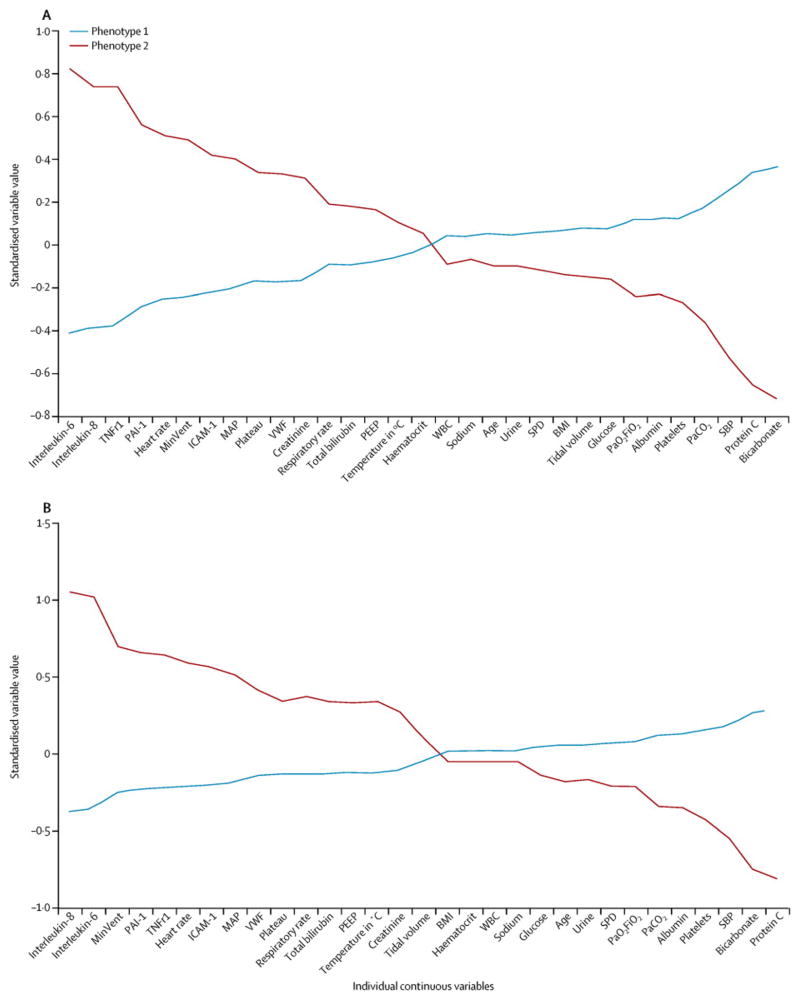
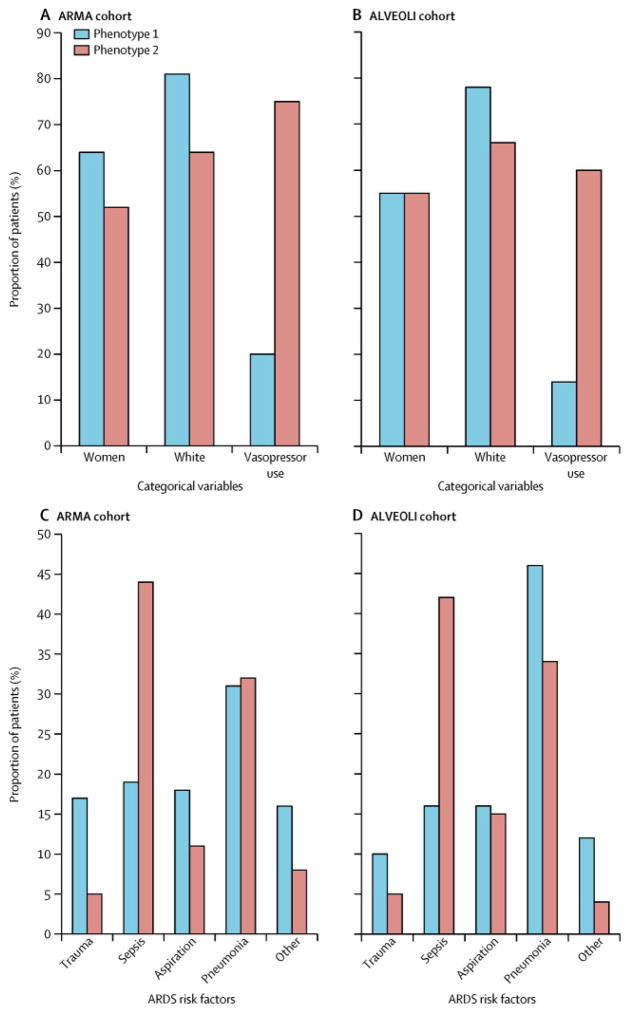
We next sought to understand the clinical and biological features that distinguished each phenotype. To accomplish this, given the high probabilities of class membership, we assigned study participants to their most likely phenotype and examined the mean values of the variables used in the model for each phenotype. Figure 2A shows the continuous variables for the two phenotypes in the ARMA cohort, sorted by the degree of separation between the phenotypes. Compared with Phenotype 1, Phenotype 2 was defined by considerably higher plasma levels of IL-6, IL-8, sTNFr-1, and PAI-1; higher heart rate and total minute ventilation; and lower systolic blood pressure, bicarbonate, and Protein C. Figures 3A and 3B show differences in the categorical variables between the phenotypes in the ARMA cohort. While gender and race differed significantly but not markedly between the phenotypes, vasopressor use at baseline was more than three times as common in Phenotype 2 compared to Phenotype 1. Furthermore, as shown in Figure 3B, subjects in Phenotype 1 were more likely to have trauma-associated ARDS and less likely to have sepsis-associated ARDS.
Figure 2.
Differences in the standardized values of each variable by phenotype on the y-axis, with the individual continuous variables along the x-axis, for the ARMA cohort (Figure 2A) and the ALVEOLI cohort (Figure 2B). The variables are sorted based on the degree of separation between the classes from maximum positive separation on the left (i.e. Phenotype 2 higher than Phenotype 1) to maximum negative separation on the right (i.e. Phenotype 2 lower than Phenotype 1). Variable standardization, in which all means are scaled to zero and standard deviations to one, is described in the Supplementary Methods; a value of +1 for the standardized variable signifies that the mean value for a given phenotype was one standard deviation higher than the mean value in the cohort as a whole.
Abbreviations: IL = interleukin; TNFr1 = tumor necrosis factor receptor-1; PAI-1 = plasminogen activator inhibitor-1; MinVent = total minute ventilation; ICAM-1 = intercellular adhesion molecule-1; MAP = mean airway pressure; VWF = von Willebrand Factor; Creat = creatinine; Resp Rate = respiratory rate; Tbili = total bilirubin; PEEP = positive end expiratory pressure; Temp = temperature in Celsius; Hct = hematocrit; Urine = urine output over prior 24h; SP-D = surfactant protein D; BMI = body mass index; TV = tidal volume; Gluc = glucose; SBP = systolic blood pressure; ProtC = Protein C; Bicarb = bicarbonate; Aspir = aspiration; Pneum = pneumonia.
Figure 3.
Figure 3A. Differences in categorical variables based on phenotype assignment in the ARMA cohort; p=0.007 for comparison of gender, p<0.0001 for other comparisons.
Figure 3B. Differences in ARDS risk factor by phenotype in the ARMA cohort; p<0.0001.
Figure 3C. Differences in categorical variables based on phenotype assignment in the ALVEOLI cohort; p=0.96 for comparison of gender, p=0.004 for race, p<0.0001 for vasopressors.
Figure 3D. Differences in ARDS risk factor by phenotype in the ALVEOLI cohort; p<0.0001.
As described in the methods, the latent class models were derived again independently in the ALVEOLI cohort, and the contribution of the key variables is presented in Figures 2B, 3C and 3D. The characteristics of the two subphenotypes in this cohort were remarkably similar to those in the ARMA cohort, with one phenotype (Phenotype 2) characterized by more profound inflammation, acidosis, and shock compared to the other phenotype (Phenotype 1). Specifically, as in the ARMA cohort, Phenotype 2 was characterized by higher plasma levels of inflammatory biomarkers, higher heart rate and minute ventilation, and by lower systolic blood pressure, bicarbonate, and Protein C, compared with Phenotype 2 (Figure 2B). As in the ARMA cohort, there were marked and significant differences in vasopressor use and in ARDS risk factor between the two phenotypes (Figures 3C and 3D).
Phenotype Prediction with Reduced Number of Variables
In order to determine whether phenotype prediction would be potentially feasible using a reduced number of variables, we used the measures with the greatest difference in mean absolute values between phenotypes in the ARMA cohort as predictive markers in receiver-operator characteristic curve analysis. Using three variables (IL-6, sTNFr-1, and vasopressor use [Yes/No]), the area under the curve for phenotype prediction was 0.937 in the ARMA cohort and 0.929 in the ALVEOLI cohort, suggesting that phenotype can be accurately predicted with a modest number of variables (Table S2). The addition of 1–2 additional variables further increased the area under the curve slightly in both cohorts (Table S2).
Association between Phenotype and Clinical Outcomes
In order to determine whether the two phenotypes had different natural histories, we tested the association between probable phenotype assignment and clinical outcomes, incorporating the degree of uncertainty regarding phenotype assignment as described in the methods. In the ARMA cohort, subjects in Phenotype 2 had significantly fewer organ failure-free and ventilator-free days, compared with subjects in Phenotype 1 (Table 4). Furthermore, subjects in Phenotype 2 had significantly higher mortality compared to Phenotype 1 (44% vs 23%, p=0.006; Table 4). Likewise, in the ALVEOLI cohort, subjects in Phenotype 2 had dramatically worse clinical outcomes than those in Phenotype 1, including a markedly higher mortality rate (51% vs 19%, p<0.001; Table 4). Clinical outcomes analyzed without adjustment for uncertainty regarding phenotype assignment showed a similar pattern (Table S3).
Table 4.
Association between Phenotype Assignment and Clinical Outcomes, Adjusted for Degree of Uncertainty Regarding Phenotype Assignment
| ARMA Cohort | ALVEOLI Cohort | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Clinical Outcome | Phenotype 1 (n=318) | Phenotype 2 (n=155) | p-value | Phenotype 1 (n=404) | Phenotype 2 (n=145) | p-value |
| Ventilator Free Days | 17.8 | 7.7 | <0.001 | 18.4 | 8.3 | <0.001 |
| Organ Failure Free Days | 14.5 | 8.0 | <0.001 | 16.5 | 8.4 | <0.001 |
| Mortality (90-day) | 23% | 44% | 0.006 | 19% | 51% | <0.001 |
Values are estimated means that take into account the uncertainty of class membership.
Effect of Treatment Strategy on Outcomes, Stratified By Phenotype
Finally, we used data from the ALVEOLI trial to determine whether there were differences in response to randomly assigned treatment (mechanical ventilation with higher vs. lower positive-end expiratory pressure, PEEP) based on phenotype. We found that PEEP strategy had significantly different effects on mortality in the two different phenotypes (p=0.049 for interaction). Specifically, within Phenotype 1, patients randomized to the higher PEEP strategy had a mortality of 48/202 (24%), vs. 33/202 (16%) in patients randomized to the low PEEP strategy. In Phenotype 2, patients randomized to the low PEEP strategy had a mortality of 36/71 (51%), vs. 31/74 (42%) in patients randomized to high PEEP. Likewise, there were even stronger interactions between phenotype and PEEP strategy for the outcomes of ventilator-free and organ failure-free days, reflecting significantly differential effects of high vs. low PEEP on these clinical outcomes in the two different phenotypes (Table 5).
Table 5.
Differences in response to PEEP strategy by probable latent class assignment, in ALVEOLI Cohort
| Phenotype 1 (n=404) | Phenotype 2 (n=145) | ||||
|---|---|---|---|---|---|
|
| |||||
| Low PEEP (n=202) | High PEEP (n=202) | Low PEEP (n=71) | High PEEP (n=74) | p-value* | |
| Dead at 90 days, n (%) | 33 (16%) | 48 (24%) | 36 (51%) | 31 (42%) | 0.049 |
| Ventilator Free Days, median (IQR) | 20 (10, 25) | 21 (3, 24) | 2 (0, 21) | 4.5 (0, 20) | 0.018 |
| Organ Failure Free Days, median (IQR) | 22 (11, 26) | 22 (9, 26) | 4 (0, 18) | 6.5 (0, 21) | 0.003 |
p-value for interaction between PEEP assignment and latent class
As a sensitivity analysis to determine whether general severity of illness scores could supplant phenotype identification, we tested for interactions between APACHE III score and PEEP. In contrast to the analyses using phenotype, there were no significant interactions between APACHE score and PEEP strategy for the outcomes of mortality, organ failure free days or ventilator-free days (p=0.58–0.99 for interactions).
DISCUSSION
Clinicians caring for patients with ARDS and researchers studying ARDS have long appreciated the heterogeneity within this complex syndrome, yet the critical care community has lacked empirical data on whether or how to refine our definitions and further subdivide ARDS. This study provides evidence that there are two subphenotypes within the broader phenotype of ARDS, with different natural histories, clinical characteristics, biomarker profiles, and clinical outcomes. These subphenotypes were evident in independent analyses of two clinical trial samples, despite considerable differences in the baseline clinical and biological profiles of these two cohorts. Further, subphenotype was strongly and consistently associated with clinical outcomes in both cohorts, with marked differences in ventilator-free and organ failure free days and in mortality. Perhaps most significantly, the two subphenotypes had different responses to treatment (lower vs. higher PEEP) in the ALVEOLI trial, suggesting that identification of subphenotypes may be critically important for future clinical trials in ARDS.
Taken together, the variables that characterize Phenotype 2 (high plasma levels of inflammatory biomarkers, severe shock and metabolic acidosis) paint a portrait of a hyper-inflammatory ARDS subphenotype that may afflict patients across the demographic spectrum of age, gender, race-ethnicity, and etiology of ARDS (though there are some differences in the latter factors by subphenotype). In contrast, the portrait of Phenotype 1 painted by this data is of a clinical syndrome characterized by less severe inflammation and shock. Interestingly, neither the severity of ARDS (PaO2/FiO2 ratio), nor the severity of renal or hepatic failure, nor the extent of leukocytosis distinguished the two phenotypes from each other, since the specified variables had relatively similar values in the two phenotypes (Figure 2). In concert with the results of our sensitivity analysis that incorporated APACHE scores, these data suggest that phenotype membership is not merely a reflection of severity of illness as measured by traditional prognostic indices.
Importantly, no single clinical or biological variable was sufficient to identify subphenotype; put differently, none of the clinical features typically used to subdivide ARDS - such as ARDS risk factor, presence or absence of sepsis, direct vs. indirect lung injury, or the use of vasopressors - were associated purely with one or the other subphenotype (Figure 3). When considered as a group, however, the clinical variables that characterize phenotype assignment form a coherent and plausible cluster that has face validity from a clinical and research perspective. For instance, Phenotype 2 is characterized by a high prevalence of vasopressor use, more severe acidosis, and a high minute ventilation – a constellation of clinical data points that forms a recognizable pattern of more severe ARDS and systemic injury to the practicing intensivist. Hypothetically, if Phenotype 2 had been characterized by a low prevalence of vasopressor use, severe acidosis, and a low minute ventilation, then it would not seem recognizable from a clinical perspective, in contrast to the combination of variables that resulted from the Phenotype 2 model.
Among the continuous variables, it is notable that in general the plasma protein biomarkers contributed more prominently to the phenotype definitions than did most of the clinical variables (including clinically utilized biomarkers such as serum creatinine and white blood cell count; Figure 2). This finding suggests that the plasma protein biomarkers may be capturing aspects of pathophysiology that are not otherwise well-captured in our clinical data, and also that development of the capability to measure these biomarkers in an expedient point-of-care method may be necessary in order to incorporate subphenotype determination into clinical trials.
Two important branch points in the analytic strategy deserve further mention. First, we deliberated carefully at the inception of the analyses over whether to include the high tidal volume patients in the analyses of the ARMA cohort, which would have enabled us to test for an interaction between tidal volume and subphenotype in that cohort. We ultimately decided not to include these patients, because to do so would have precluded analyses of the association between subphenotype and mortality in that cohort. We thought this aspect of the analysis was too important to discard, and therefore decided to exclude the high tidal volume patients. Second, the ultimate decision as to the optimal number of classes identified by the LCA models requires consideration of a number of different factors. Latent class models seek to find the best model fit, assuming that there are “X” latent classes in the data. If there are only 2 classes but we fit a 3 class model, the third class will be forced in by selecting a small number of cases with a more extreme or unique set of values. The p-value for the VLMR test strongly suggests that a two-class model is preferable to a three class model, in both cohorts. That data, combined with the small size of the 3rd class, led us to focus on a two-class model. A class that is very small, relatively, offers little information and may represent more of an anomaly than a useful finding.
While latent class modeling has not to our knowledge been previously applied in classical ARDS cohorts, Shah and colleagues recently used latent class-based models to identify subphenotypes within primary graft dysfunction (PGD), a type of acute lung injury that occurs after lung transplantation.(27) These models focused exclusively on the timing of onset and resolution of lung dysfunction, using only grades of PGD at various timepoints to generate latent classes, and found that patients with severe persistent dysfunction (one of three identified classes) had the worst clinical outcomes. This approach differs from ours in its focus on repeated measures of one clinical input (grade of PGD) to generate latent class and lack of inclusion of biological markers; whether consideration of additional clinical data points and/or biomarkers as class-defining variables would lead to identification of different PGD subphenotypes remains unknown.
The finding of differential response to PEEP by ARDS subphenotype has face validity in light of other studies that have noted interactions between PEEP response and ARDS severity. A recent meta-analysis of 2299 ARDS patients (including the 549 patients used in our derivation cohort) reported that those with a PaO2/FiO2 less than 200 had a 5% lower hospital mortality with higher PEEP strategies compared with lower PEEP strategies (p=0.049).(28, 29) Two important contrasts with our approach deserve mention. First, while PaO2/FiO2 ratio was considered in subphenotype identification, it was not one of the variables that contributed most prominently to the classification (Figure 2). Second, the interaction between PEEP and subphenotype that we identified is quantitatively larger and was statistically significant in a much smaller sample size than the meta-analysis, suggesting that the interaction between subphenotype and PEEP response is substantially stronger than that between PaO2/FiO2 ratio and PEEP response. It is important to emphasize that while the interactions identified between PEEP and clinical outcomes are potentially provocative, we are reticent to make any recommendations for clinical care on the basis of a subgroup analysis. Rather, we view these results as hypothesis-generating, and think that they support the need for more rapid and/or bedside assays of the molecular phenotype of critically ill patients to validate these findings in future trials.
Our study has several strengths. First, it should be emphasized that the latent class models were generated independently in each of the two cohorts. Specifically, findings from the first cohort were not considered in the modeling strategy in the second cohort. Given this approach, the similarity of the findings in the two cohorts is noteworthy. Likewise, since clinical outcomes were not considered as class-defining variables, the strengths and consistency of the associations between subphenotype and clinical outcomes are striking. Second, because we studied patients within the framework of a randomized controlled trial, we are able to draw stronger conclusions about causal associations between treatment (with PEEP) and clinical outcomes, with the usual caveats regarding subgroup analyses. Third, by virtue of leveraging patients enrolled in multicenter trials, the samples studied reflect demographically diverse cohorts of ARDS patients. Fourth, the two cohorts differed substantially on many clinical and biological measures (Tables 1 and 2), strengthening the generalizability of our findings and making the similarity of the subphenotypes identified in the two cohorts more significant.
This study has some limitations. First, the patients included in these analyses were drawn from randomized controlled trials of ARDS; different subphenotypes may be present in less carefully selected ARDS patient populations. Second, the biomarkers included in these analyses were limited to those that had been measured already in both cohorts. While these biomarkers have value for prognosis and pathogenesis, other informative biomarkers have emerged in ARDS research over the past several years, including angiopoietin-2,(30–32) the receptor for advanced glycation endproducts,(33) club (formerly known as Clara) cell 16,(34) brain natriuretic peptide,(35) interleukin-1 receptor antagonist(36) and others. Consideration of these biomarkers, and/or of alternative genomic or metabolomic markers, may result in more comprehensive subphenotypes being identified or may lead to the recognition of these biomarkers as important classifiers. Third, analyses of possible classifying variables was limited to the data collected in the original studies; clinical variables such as alcohol use,(37) cigarette smoking,(38) or other comorbidities may potentially contribute to subphenotype identification but were not available for this analysis. Likewise, considerable histopathologic variability has been demonstrated within ARDS in autopsy series; whether or how consideration of pathology findings would influence subphenotype identification remains unknown.(39, 40)
In summary, our analysis has identified two different subphenotypes within two independent cohorts of patients with ARDS. These two subphenotypes have markedly different natural histories, clinical and biological characteristics, clinical outcomes, and response to treatment, fulfilling the criteria necessary to define an subphenotype.(7) We suggest that these findings provide proof-of-concept that the clinical syndrome of ARDS contains distinct subphenotypes and should prompt future studies aimed at further elucidating these subphenotypes with comprehensive clinical and biological data. Given the differential response to treatment by subphenotype identified herein, this area of research has the potential to directly inform future randomized controlled trials of novel treatments for ARDS.
Supplementary Material
Research in Context.
Systematic Review
We did not carry out a systematic review prior to the inception of these analyses. Some prior analyses have attempted to identify ARDS subgroups using traditional regression-based methods; however, to our knowledge, this manuscript represents the first report of the use of latent class models to identify subphenotypes of ARDS within two separate heterogeneous samples of ARDS patients.
Interpretation
Subphenotypes have been identified within heterogeneous syndromes such as asthma and breast cancer, with important therapeutic implications. Whether subphenotypes exist within ARDS, another clinically heterogeneous syndrome, is unknown. These analyses identify two subphenotypes within ARDS, one of which is characterized by more severe inflammation, shock, and metabolic acidosis, significantly worse clinical outcomes, and a differential response to treatment with positive end-expiratory pressure. These findings provide proof-of-concept that the clinical syndrome of ARDS contains distinct subphenotypes and should prompt future studies aimed at further elucidating these subphenotypes with comprehensive clinical and biological data, a novel approach that could inform the design of future randomized controlled trials of new treatments for ARDS.
Acknowledgments
Support: This work was supported by contracts (NO1-HR 46054-46064) with the National Heart, Lung, and Blood Institute (NHLBI). Dr. Calfee was supported by HL090833 and 110969. Dr. Matthay was supported by HL 51856. Dr. Ware was supported by HL103836 and HL112656.
Role of the funding source: The funding sources had no role in the study design, analysis or interpretation of the data, or writing of the report for these analyses. The corresponding author (CC) had full access to all of the data and the final responsibility to submit for publication.
Footnotes
Author contributions: CC designed the study, participated in data cleaning and analysis, interpreted the data, and drafted and revised the manuscript. KD performed the data cleaning and analysis, contributed to data interpretation, and critically revised the manuscript. LW, TT, and PP provided the clinical and biomarker data, contributed to data interpretation, and critically revised the manuscript. MM contributed to study design, data analysis and interpretation and critically revised the manuscript. All authors had access to the raw data. All authors provided final approval of the version to be published. Drs. Delucchi, Thompson, Parsons and Matthay are Full Professors.
Conflicts of Interest: Dr. Calfee reports grants from NIH/NHLBI during the conduct of the study; grants from Glaxo Smith Kline, personal fees from Cerus Corp, outside the submitted work. Dr. Delucchi has nothing to disclose. Dr. Thompson reports grants from National Institutes of Health during the conduct of the study. Dr. Parsons has nothing to disclose. Dr. Ware has nothing to disclose. Dr. Matthay reports grants from NHLBI during the conduct of the study; grants from GlaxoSmithKline, other from RocheGenetec, personal fees from Cerus Inc, other from French Society of Intensive Care, outside the submitted work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet. 1967 Aug 12;2(7511):319–23. doi: 10.1016/s0140-6736(67)90168-7. [DOI] [PubMed] [Google Scholar]
- 2.Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994 Mar;149(3 Pt 1):818–24. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 3.Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012 Jun 20;307(23):2526–33. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 4.Calfee CS, Eisner MD, Ware LB, et al. Trauma-associated lung injury differs clinically and biologically from acute lung injury due to other clinical disorders. Crit Care Med. 2007 Oct;35(10):2243–50. doi: 10.1097/01.ccm.0000280434.33451.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tejera P, Meyer NJ, Chen F, et al. Distinct and replicable genetic risk factors for acute respiratory distress syndrome of pulmonary or extrapulmonary origin. Journal of medical genetics. 2012 Nov;49(11):671–80. doi: 10.1136/jmedgenet-2012-100972. Epub 2012/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank AJ, Thompson BT. Pharmacological treatments for acute respiratory distress syndrome. Current opinion in critical care. 2010 Feb;16(1):62–8. doi: 10.1097/MCC.0b013e328334b151. Epub 2009/12/03. eng. [DOI] [PubMed] [Google Scholar]
- 7.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012 May;18(5):716–25. doi: 10.1038/nm.2678. Epub 2012/05/09. [DOI] [PubMed] [Google Scholar]
- 8.Corren J, Lemanske RF, Hanania NA, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011 Sep 22;365(12):1088–98. doi: 10.1056/NEJMoa1106469. Epub 2011/08/05. [DOI] [PubMed] [Google Scholar]
- 9.Wong HR, Cvijanovich NZ, Allen GL, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011 Nov;39(11):2511–7. doi: 10.1097/CCM.0b013e3182257675. Epub 2011/06/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iskander KN, Osuchowski MF, Stearns-Kurosawa DJ, et al. Sepsis: multiple abnormalities, heterogeneous responses, and evolving understanding. Physiol Rev. 2013 Jul;93(3):1247–88. doi: 10.1152/physrev.00037.2012. Epub 2013/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLachlan G, Peel D. Finite Mixture Models. New York: John Wiley & Sons; 2000. [Google Scholar]
- 12.Ezzedine K, Le Thuaut A, Jouary T, Ballanger F, Taieb A, Bastuji-Garin S. Latent Class Analysis of a series of 717 patients with vitiligo allows the identification of two clinical subtypes. Pigment cell & melanoma research. 2013 Oct 16; doi: 10.1111/pcmr.12186. Epub 2013/10/17. [DOI] [PubMed] [Google Scholar]
- 13.Rindskopf R, Rindskopf W. The value of latent class analysis in medical diagnosis. Stat Med. 1986;5:21–7. doi: 10.1002/sim.4780050105. [DOI] [PubMed] [Google Scholar]
- 14.Depner M, Fuchs O, Genuneit J, et al. Clinical and epidemiologic phenotypes of childhood asthma. Am J Respir Crit Care Med. 2014 Jan 15;189(2):129–38. doi: 10.1164/rccm.201307-1198OC. Epub 2013/11/29. [DOI] [PubMed] [Google Scholar]
- 15.The Acute Respiratory Distress Syndrome Network. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med. 2000 May 4;342(18):1301–8. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 16.The Acute Respiratory Distress Syndrome Network. Ketoconazole for early treatment of acute lung injury and acute respiratory distress syndrome: a randomized controlled trial. JAMA. 2000 Apr 19;283(15):1995–2002. doi: 10.1001/jama.283.15.1995. [DOI] [PubMed] [Google Scholar]
- 17.The Acute Respiratory Distress Syndrome Network. Randomized, placebo-controlled trial of lisofylline for early treatment of acute lung injury and acute respiratory distress syndrome. Crit Care Med. 2002 Jan;30(1):1–6. doi: 10.1097/00003246-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 18.The Acute Respiratory Distress Syndrome Network. Higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N Engl J Med. 2004 Jul 22;351(4):327–36. doi: 10.1056/NEJMoa032193. [DOI] [PubMed] [Google Scholar]
- 19.Ware LB, Eisner MD, Thompson BT, Parsons PE, Matthay MA. Significance of von Willebrand factor in septic and nonseptic patients with acute lung injury. Am J Respir Crit Care Med. 2004 Oct 1;170(7):766–72. doi: 10.1164/rccm.200310-1434OC. [DOI] [PubMed] [Google Scholar]
- 20.Ware LB, Matthay MA, Parsons PE, Thompson BT, Januzzi JL, Eisner MD. Pathogenetic and prognostic significance of altered coagulation and fibrinolysis in acute lung injury/acute respiratory distress syndrome. Crit Care Med. 2007 Aug;35(8):1821–8. doi: 10.1097/01.CCM.0000221922.08878.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons PE, Eisner MD, Thompson BT, et al. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med. 2005 Jan;33(1):1–6. doi: 10.1097/01.ccm.0000149854.61192.dc. discussion 230–2. [DOI] [PubMed] [Google Scholar]
- 22.Parsons PE, Matthay MA, Ware LB, Eisner MD. Elevated plasma levels of soluble TNF receptors are associated with morbidity and mortality in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2005 Mar;288(3):L426–31. doi: 10.1152/ajplung.00302.2004. [DOI] [PubMed] [Google Scholar]
- 23.Eisner MD, Parsons P, Matthay MA, Ware L, Greene K. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax. 2003 Nov;58(11):983–8. doi: 10.1136/thorax.58.11.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calfee CS, Eisner MD, Parsons PE, et al. Soluble intercellular adhesion molecule-1 (sICAM-1) and clinical outcomes in patients with acute lung injury. Intensive Care Med. 2008;35(2):248–57. doi: 10.1007/s00134-008-1235-0. Epub 2008 Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ware LB, Koyama T, Billheimer DD, et al. Prognostic and Pathogenetic Value of Combining Clinical and Biochemical Indices in Patients with Acute Lung Injury. Chest. 2010 Oct 26;137(2):288–96. doi: 10.1378/chest.09-1484. Epub 2009 Oct 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanza ST, Tan X, Bray BC. Latent Class Analysis With Distal Outcomes: A Flexible Model-Based Approach. Struct Equ Modeling. 2013 Jan 1;20(1):1–26. doi: 10.1080/10705511.2013.742377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah RJ, Diamond JM, Cantu E, et al. Latent class analysis identifies distinct phenotypes of primary graft dysfunction after lung transplantation. Chest. 2013 Aug;144(2):616–22. doi: 10.1378/chest.12-1480. Epub 2013/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briel M, Meade M, Mercat A, et al. Higher vs lower positive end-expiratory pressure in patients with acute lung injury and acute respiratory distress syndrome: Systematic review and meta-analysis. JAMA. 2010;303(9):865–73. doi: 10.1001/jama.2010.218. [DOI] [PubMed] [Google Scholar]
- 29.Rubenfeld GD. How much peep in acute lung injury. JAMA. 2010;303(9):883–4. doi: 10.1001/jama.2010.226. [DOI] [PubMed] [Google Scholar]
- 30.Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med. 2012 Jun;40(6):1731–7. doi: 10.1097/CCM.0b013e3182451c87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal A, Matthay MA, Kangelaris KN, et al. Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am J Respir Crit Care Med. 2013;187(7):736–42. doi: 10.1164/rccm.201208-1460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer NJ, Li M, Feng R, et al. ANGPT2 Genetic Variant Is Associated with Trauma-associated Acute Lung Injury and Altered Plasma Angiopoietin-2 Isoform Ratio. Am J Respir Crit Care Med 2011. 2011 May 15;183(10):1344–53. doi: 10.1164/rccm.201005-0701OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calfee CS, Ware LB, Eisner MD, et al. Plasma Receptor for Advanced End-Products and Clinical Outcomes in Acute Lung Injury. Thorax. 2008;63(12):1083–9. doi: 10.1136/thx.2008.095588. Epub 2008 Jun 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kropski JA, Fremont RD, Calfee CS, Ware LB. Clara cell protein (CC16), a marker of lung epithelial injury, is decreased in plasma and pulmonary edema fluid from patients with acute lung injury. Chest. 2009 Jun;135(6):1440–7. doi: 10.1378/chest.08-2465. Epub 2009/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fremont RD, Koyama T, Calfee CS, et al. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma. 2010 May;68(5):1121–7. doi: 10.1097/TA.0b013e3181c40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer NJ, Feng R, Li M, et al. IL1RN coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL-1 receptor antagonist. Am J Respir Crit Care Med. 2013 May 1;187(9):950–9. doi: 10.1164/rccm.201208-1501OC. Epub 2013/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss M, Bucher B, Moore FA, Moore EE, Parsons PE. The role of chronic alcohol abuse in the development of acute respiratory distress syndrome in adults. JAMA. 1996 Jan 3;275(1):50–4. [PubMed] [Google Scholar]
- 38.Calfee CS, Matthay MA, Eisner MD, et al. Active and passive cigarette smoking and acute lung injury after severe blunt trauma. American Journal Of Respiratory And Critical Care Medicine. 2011;183(12):1660–5. doi: 10.1164/rccm.201011-1802OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thille AW, Esteban A, Fernandez-Segoviano P, et al. Comparison of the Berlin definition for acute respiratory distress syndrome with autopsy. Am J Respir Crit Care Med. 2013 Apr 1;187(7):761–7. doi: 10.1164/rccm.201211-1981OC. Epub 2013/02/02. [DOI] [PubMed] [Google Scholar]
- 40.Thille AW, Esteban A, Fernandez-Segoviano P, et al. Chronology of histological lesions in acute respiratory distress syndrome with diffuse alveolar damage: a prospective cohort study of clinical autopsies. The lancet Respiratory medicine. 2013 Jul;1(5):395–401. doi: 10.1016/S2213-2600(13)70053-5. Epub 2014/01/17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.