Abstract
It has been suggested that bacterial resistance is selected within a mutation selection window of antibiotics. More recent studies showed that even extremely low concentration of antibiotic could select resistant bacteria in vitro. Yet little is known about the exact antibiotic concentration range that can effectively select for resistant organisms in animal gastrointestinal (GI) tract. In this study, the effect of different dosages of enrofloxacin on resistance and mutation development in rat GI tract E. coli was investigated by determining the number of resistant E. coli recoverable from rat fecal samples. Our data showed that high dose antibiotic treatment could effectively eliminate E. coli with single gyrA mutation in the early course of treatment, yet the eradication effects diminished upon prolonged treatment. Therapeutic and sub-therapeutic dose (1/10 and 1/100 of therapeutic doses) of enrofloxacin could effectively select for mutation in GI tract E. coli at the later course of enrofloxacin treatment and during the cessation periods. Surprisingly, very low dose of enrofloxacin (1/1000 therapeutic dose) could also select for mutation in GI tract E. coli at the later course of enrofloxacin treatment, only with slightly lower efficiency. No enrofloxacin-resistant E. coli could be selected at all test levels of enrofloxacin during long term treatment and the strength of antibiotic treatment does not alter the overall level of E. coli in rat GI tract. This study demonstrated that long term antibiotic treatment seems to be the major trigger for the development of target mutations in GI tract E. coli, which provided insight into the rational use of antibiotics in animal husbandry.
Keywords: target mutation, rat GI tract E. coli, enrofloxacin, resistance development
INTRODUCTION
The use of antibiotics in treatment of bacterial infections represents one of the most important inventions in human history. Since its discovery in the 1940s, antibiotics have saved millions of human lives and have also been widely used in the fields of veterinary medicine and agriculture in the past 70 years. However, due to the extensive use and misuse of antibiotics in various settings, most agents have lost their efficacy to bacteria as a result of emergence and spread of multiple-drug-resistant bacterial pathogens (Ferber, 2010). In the last decade, resistance to human first line drugs has increased significantly and the choices of treatment for serious bacterial infections have become extremely limited, threatening to take medicine back into the pre-antibiotic era (Hawkes et al., 2007; Brazier, 2008; Ferber, 2010; Srivastava et al., 2011; Ghafur, 2013).
The mechanisms of antimicrobial resistance in bacteria have been intensively investigated (Tenover, 2006). To date, the mechanisms of bacterial resistance to any antibiotic has been known to a certain extent. However, the driving forces mediating the selection and development of antibiotic resistance in bacteria as well as how resistance genes spread between different organisms and environment niches are complex and still not completely understood. Since resistance to antibiotics in bacteria has been evolving over a long period of time, a complex repertoire of resistance mechanisms have emerged. At this stage, it is still not clear whether mutation development or acquisition of antimicrobial resistance genes is the primary mechanism which is required to initiate the development of antimicrobial resistance in a bacterial population. Nevertheless, a wide range of transferable elements harboring antimicrobial resistance genes is known to be responsible for the rapid dissemination of resistance traits in bacteria (Zhao et al., 2010; Jiang et al., 2011; Zheng et al., 2012; Yang et al., 2014). Regardless of the nature of resistance mechanism, acquisition of antimicrobial resistance genes or genetic mutations responsible for bacterial antibiotic resistance, constant antibiotic pressure is commonly accepted to be the driving force of expansion of the resistant population. It has been suggested that resistant bacteria were selected at the concentration of antibiotics within the mutation selection window, a concentration between the minimal inhibitory concentration (MIC) and minimal mutation prevention concentration (MPC; Zhao and Drlica, 2002; Blondeau, 2009; Firsov et al., 2013). However, recent studies have shown that resistant bacteria can be selected at very low concentration of antibiotics in vitro. The concentration can be even lower than the concentration of antibiotics used for growth promotion purpose in animals, and the concentration of environmental antibiotic residues due to natural production by microorganisms and human contamination (Gullberg et al., 2011). However, these studies were conducted in in vitro model and could not explain how animal GI tract organisms respond to different concentrations of antibiotics, since growth promotional usage of antibiotics in animals is considered as the most important practice that selected for both antibiotic-resistant genes and bacterial pathogens (Kelly et al., 2004; Marshall and Levy, 2011; Looft et al., 2012). Animal GI tract is considered as a more complicated environment than any other ecosystems. Recent studies have demonstrated the role of bacterial stress responses in antibiotic resistance development (Srinivasan et al., 2012; Bernier et al., 2013; Duval and Lister, 2013). The environmental stresses that the bacteria encounter may lead to variation in physiological functions of the organisms, eliciting changes in the intrinsic mutation rate and abilities to survive drug action (Poole, 2012). Animal GI tract imposes a mixture of stresses to bacteria including low pH, oxidative stress, starvation stress, antimicrobial compounds, and interactions between microbiota, which may influence the development of antibiotic resistance in GI tract flora (Nguyen et al., 2011; Bernier et al., 2013). However, in contrast to in vitro condition, the physiological changes and fitness costs arisen during the response to these stresses and the development of mutations may in turn put the GI tract bacteria under adverse conditions which may affect the survival fitness of such organisms in the GI tract environment. The effect of these factors on the development of antimicrobial resistance in GI tract bacteria is not fully understood. In this study, a rat model was used to test the effect of different concentrations of antibiotics on the resistance development in E. coli in animal GI tract.
MATERIALS AND METHODS
ANTIBIOTIC AND ANIMALS
Enrofloxacin was purchased from Sangon Biotech company (Shanghai, China). The antibiotic was dissolved in 0.1 M NaOH at 10 mg/mL stock solution on the day of use. 11–15 weeks old, specified pathogen-free (SPF) male SD rats with body weight 250–350 g were used in all experiments. Animals were purchased from Guangdong Medical Laboratory Animal Center and were housed individually and allowed free access to food and water. They were examined twice every day for any clinical signs such as behavior, gastrointestinal (GI) function, respiratory distress, food, and water intake etc. The experimental protocol was approved by the Research Animal Care and Use Committee of the Hong Kong Polytechnic University.
ANTIMICROBIAL TREATMENT
Since fluoroquinolones are concentration-dependent antibiotic (Firsov et al., 2004), we checked the effect of different dosages of fluoroquinolone on the development of resistance to enrofloxacin in E. coli in animal GI tracts. Thirty male rats were equally divided into six groups: one group was treated with therapeutic dose of enrofloxacin (10 mg/kg body weight), one was treated with saline as control and other groups were treated with doses of 10-fold, 1/10, 1/100, and 1/1000 of the therapeutic dose.
The oral antibiotic treatment regimen lasted about 1 month including three antibiotic treatments and three cessation gaps between treatments. All the rats were subjected to different doses of enrofloxacin treatment for 5 days, then cessation of antibiotic treatment for 4 days, another enrofloxacin retreatment for 5 days, followed by another cessation of antibiotic treatment for 8 days, then enrofloxacin retreatment for another 5 days, and tracing for 3 more days without antibiotic treatment.
At the indicated time intervals, fresh feces (250–500 mg) were collected and re-suspended in 1 ml of saline. The suspension was mixed and diluted by 10-fold in saline. 100 μl of suspension was plated on MacConkey agar containing 0 mg/L, 0.125 mg/L, 0.5 mg/L, and 2 mg/L of enrofloxacin. The plates were incubated at 37°C for 12 h and the total colony counts were recorded. The colony forming units (CFUs) per gram of feces were determined. Colonies that showed typical morphology of E. coli on MacConkey plate (pink to rose-red, large regular colonies) were considered as E. coli and some of which were confirmed by 16SrRNA sequencing using primers (F: CCAGACTCCTACGGGAGGCAG, R:CGTATTACCGCGGCTGCTG).
ANTIMICROBIAL SUSCEPTIBILITY
Antimicrobial susceptibilities to enrofloxacin were determined by the agar dilution method in accordance with Clinical and Laboratory Standards Institute guidelines (CLSI, 2013) using E. coli strain ATCC 25922 as quality control. The MICs of enrofloxacin were determined following CLSI guidelines (CLSI, 2013).
DETERMINATION OF THE TARGET MUTATION IN E. coli
Presence of target mutations of the Quinolone-resistance determining regions (QRDR) of the gyrA and parC genes in E. coli were determined by PCR as previously described (Everett et al., 1996).
RESULTS
Thirty SPF male SD rats were selected for this experiment. Resistance background of GI tract E. coli of these rats were checked by plating 200~300 mg of fecal samples of the rats on MacConkey plates with 0, 0.125, 0.5, and 2 mg/L of enrofloxacin. All rats were found to contain a similar amount of E. coli in their GI tract (data not shown). Five rats exhibited a background of less susceptible E. coli which can grow on MacConkey plate containing 0.5 mg/L of enrofloxacin. No fecal E. coli was able to grow on MacConkey plate with 2 mg/L concentration of enrofloxacin (data not shown). Twenty colonies with typical morphology of E. coli from MacConkey plates were randomly selected and confirmed to be E. coli by 16SrRNA sequencing. Similar E. coli confirmation was performed for the following each group of experiment by randomly selecting 20~40 E. coli for 16SrRNA sequencing. All the checked colonies were confirmed to be E. coli (data not shown).
Upon background check, these rats were separated into six experimental groups. All five rats with a low background of E. coli with reduced susceptibility to enrofloxacin were grouped into one category and treated with a high dose of enrofloxacin (10-fold therapeutic dose). The rest of the 25 rats were randomly separated into 5 groups with one control group and 4 different treatment groups including therapeutic dose, 1/10, 1/100, and 1/1000 of the therapeutic dose treatments. The background of the high dose group animals is different from the others. The purpose of the high dose group study is different from other groups and intended to check the efficiency of higher dose of enrofloxacin on the clearance of less susceptible E. coli in animal GI tract.
EFFECT OF HIGH DOSE OF ANTIBIOTIC TREATMENT ON RAT GI TRACT E. coli
For the 10-fold therapeutic dose treatment group, the application of enrofloxacin caused gradual eradication of rat GI tract E. coli. The numbers of GI tract E. coli decreased gradually during the first three days of treatment; the reduction became dramatic at the fourth day of treatment and the number of E coli recoverable remained at a low level for the first treatment period. After withdrawing the antibiotics, E. coli appeared again at the second day after the cessation of antibiotic treatment and increased on the third and fourth day. At the beginning of the second course of treatment period, the numbers of GI tract E. coli almost returned back to the normal level. During the whole 5-days antibiotic treatment, the numbers of GI tract E. coli were not affected, and remained at the normal level, which lasted throughout the second antibiotic cessation period. The GI tract E. coli gradually decreased again upon the start of the third antibiotic treatment. The number of E. coli became dramatically reduced again at the third day of the third course of the treatment and then gradually recovered at the fourth and fifth days of the antibiotic treatment. The number of E. coli kept recovering upon cessation of antibiotic treatment (Figure 1Ai).
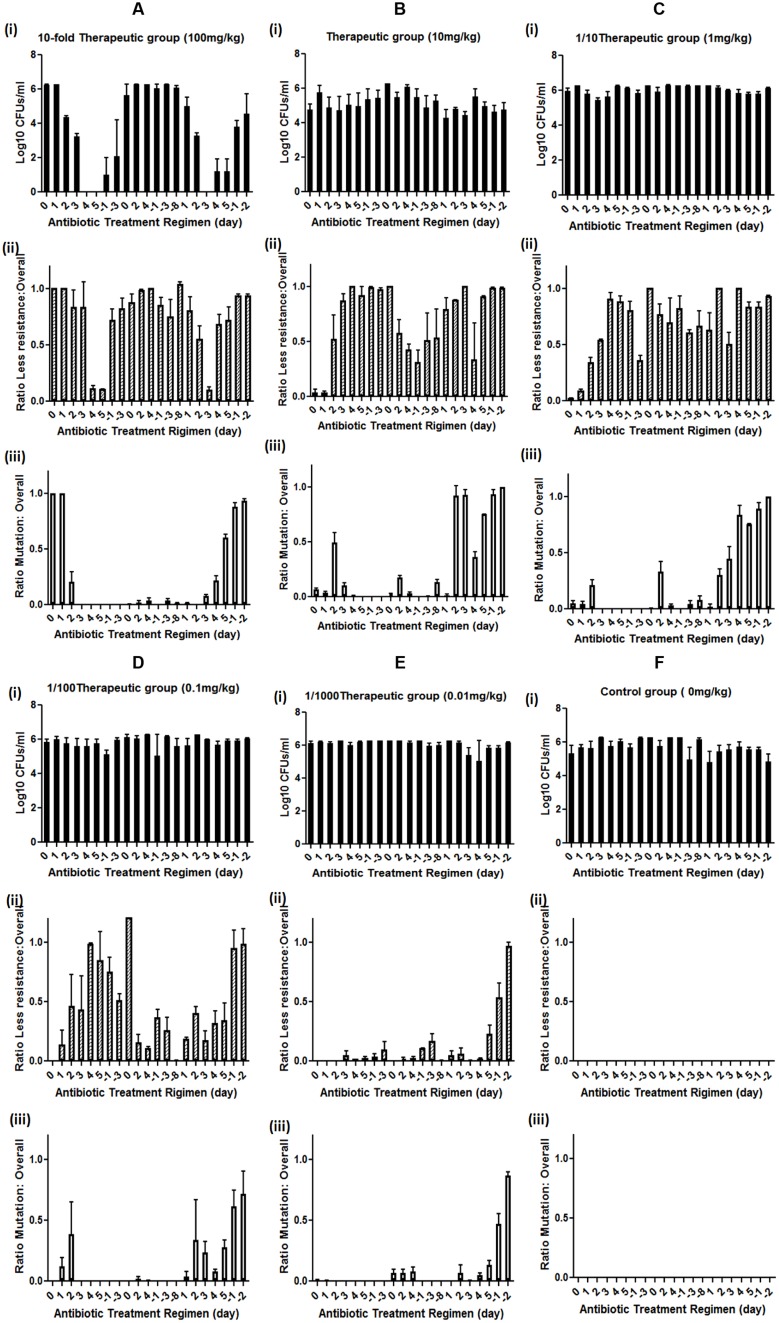
FIGURE 1.
Effect of different doses of enrofloxacin treatment on mutation development in rat GI tract E. coli. (A) high dose (10-fold therapeutic dose) enrofloxacin treatment, (B) therapeutic dose of enrofloxacin treatment, (C) sub-therapeutic dose (1/10 of therapeutic dose) of enrofloxacin treatment, (D) Low dose (1/100 of therapeutic dose) enrofloxacin treatment, (E) very low dose (1/1000 of therapeutic dose) of enrofloxacin treatment, (F) Control group; (i) Levels of rat GI tract E. coli during enrofloxacin treatment; (ii) levels of less susceptible (grown on MacConkey supplemented with 0.125 mg/L of enrofloxacin) GI tract E. coli during enrofloxacin treatment; (iii) Levels of rat GI tract E. coli with target mutation (grown on MacConkey supplemented with 0.5 mg/L of enrofloxacin) during enrofloxacin treatment. Y-axis represents the ratio between less susceptible E. coli or E. coli with target mutation versus overall GI tract E. coli. Positive day number represents the antibiotic treatment duration, negative day number represents the duration since the cessation of antibiotic, 0 represents the day of the application of antibiotic and the fecal E. coli were isolated before the antibiotic treatment at day 0. The number is the average of data from five rats in the group.
The effect of the antibiotic treatment to the development of fluoroquinolone-resistant E. coli was recorded. The number of less susceptible E. coli, which can grow at MacConkey plate with 0.125 mg/L was high before antibiotic treatment and slightly decreased on the second and third days of antibiotic treatment. The number of less susceptible E. coli became dramatically decreased on the fourth and fifth days of treatment. After withdrawing the antibiotic, the number of less susceptible E. coli reverted back to the normal level and remained so until the beginning the third course of the antibiotic treatment. The less susceptible E. coli slightly decreased at the first day of the third course of the antibiotic treatment and became undetectable at the third day of the treatment, then recovered to normal level throughout the rest of the treatment and non-treatment period (Figure 1Aii).
The E. coli that can grow on MacConkey with 0.5 mg/L enrofloxacin exhibited different response to high dose enrofloxacin treatment. The numbers of E. coli that can grow on MacConkey with 0.5 mg/L enrofloxacin reduced on the seconf day of treatment and became almost undetectable throughout the rest of the first, second and third treatments and non-antibiotic treatment period except for a period of brief appearance during the later course of the third treatment and antibiotic cessation period (Figure 1Aiii). Throughout the whole course of treatment, no E. coli isolates could be recovered on MacConkey plate with 2 mg/L of enrofloxacin, suggesting that high dose antibiotic could not select for fluoroquinolone-resistant GI tract E. coli.
Each of 20 E. coli isolates that grew on MacConkey with 0.125 mg/L and 0.5 mg/L enrofloxacin, respectively, were selected to check their MIC and target mutation profiles. E. coli that grew on MacConkey with 0.125 mg/L enrofloxacin exhibited a MIC range of enrofloxacin of 0.006~0.03 with no mutation in any of the four target genes gyrA, gyrB, parC, and parE; yet all E. coli strains that grew on MacConkey with 0.5 mg/L enrofloxacin exhibited MIC of enrofloxacin of 0.25~1 mg/L with single mutation in gyrA (S83L; Table 1). Therefore, the MacConkey plate with 0.5 mg/L enrofloxacin could be used to check for the mutation rate of GI tract E. coli upon antibiotic treatment.
Table 1.
MIC of enrofloxacin and target mutation profiles of rat GI tract E. coli.
| Animal group | Origin of E. coli (enrofloxacin selective plates) | Number of isolates | MIC range of enrofloxacin (mg/L) | Target mutations |
|
|---|---|---|---|---|---|
| GyrA (No. of E. coli) | ParC | ||||
| A* | 0.125 mg/L | 20 | 0.015∼0.06 | Wt (20/20) | Wt |
| 0.5 mg/L | 20 | 0.25∼1 | S83L (20/20) | Wt | |
| B | 0.125 mg/L | 20 | 0.015∼0.06 | Wt (20/20) | Wt |
| 0.5 mg/L | 20 | 0.25∼1 | S83L (20/20) | Wt | |
| C | 0.125 mg/L | 20 | 0.015∼0.5 | Wt (17/20), S83L (3/20) | Wt |
| 0.5 mg/L | 20 | 0.25∼1 | S83L (20/20) | Wt | |
| D | 0.125 mg/L | 20 | 0.06∼0.5 | Wt (15/20), S83L (5/20) | Wt |
| 0.5 mg/L | 20 | 0.125∼1 | S83L (20/20) | Wt | |
| E | 0.125 mg/L | 20 | 0.06∼1 | Wt (11/20), S83L (9/20) | Wt |
| 0.5 mg/L | 20 | 0.125∼1 | S83L (20/20) | Wt | |
*A, high dose (10-fold therapeutic dose) enrofloxacin treatment; B, therapeutic dose of enrofloxacin treatment; C, sub-therapeutic dose (1/10 of therapeutic dose) of enrofloxacin treatment; D, low dose (1/100 of therapeutic dose) enrofloxacin treatment; E, very low dose (1/1000 of therapeutic dose) of enrofloxacin treatment.
EFFECT OF THERAPEUTIC DOSE OF ANTIBIOTIC TREATMENT ON RAT GI TRACT E. coli
For the therapeutic treatment group, the GI tract E. coli exhibited slightly less susceptible E. coli background. Hence the therapeutic treatment dose of enrofloxacin did not have any effect on the numbers of E. coli in rat GI tract (Figure 1Bi). However, after antibiotic treatment, the number of less susceptible E. coli decreased slightly and then increased to high level throughout the first course of antibiotic treatment and antibiotic cessation periods. During the second course of treatment, the numbers of less susceptible E. coli reduced gradually during treatment and remained at a lower level during the second antibiotic withdrawal period. During the third course of antibiotic treatment, the number of less susceptible E. coli increased to a high level and remained so until the end of the experiment (Figure 1Bii).
The numbers of E. coli that grew on MacConkey agar with 0.5 mg/L enrofloxacin (mutation rate) increased to around half of the total E. coli at the second day upon the treatment of enrofloxacin and remained at this level during the first course of the treatment. E. coli strains with mutation disappeared during the antibiotic withdrawal period and reappeared during the second course of treatment albeit at a lower level. E. coli strains with mutation increased to a high level at the second day of the third course of treatment, then decreased and reappeared again during the antibiotic cessation period (Figure 1Biii). The data showed that mutation in E. coli might mainly triggered by antibiotic treatment and release of the antibiotic pressure can reduce the numbers of E. coli with mutation, which suggested that without antibiotic pressure, the E. coli with mutation may be less competitive than other normal E. coli in animal GI tract. The long term antibiotic treatment may make E. coli with mutation to adapt to animal GI tract, which can be seen from the increased number of E. coli with mutation at the end of third course of antibiotic treatment and the following cessation period.
EFFECT OF SUB-THERAPEUTIC AND LOWER DOSES OF ANTIBIOTIC TREATMENT ON RAT GI TRACT E. coli
Similar to the effect of the therapeutic dose of antibiotic treatment, sub-therapeutic dose of enrofloxacin did not have any impact on the overall numbers of E. coli in rat GI tract (Figure 1Ci). The less susceptible E. coli strains emerged upon antibiotic treatment and remained at a stable level throughout the course of the experiment (Figure 1Cii). The numbers of GI tract E. coli with target mutation slightly increased upon antibiotic treatment and disappeared during the antibiotic cessation period for the first two courses of treatments. The mutation rate was significantly increased in E. coli upon the third course of antibiotic treatment (Figure 1Ciii).
For the lower-level antibiotic treatment groups, namely 1/100 and 1/1000 of the therapeutic doses, no change in the number of GI tract E. coli could be observed throughout the experiment (Figures 1Di,Ei). The less susceptible E. coli could be selected during the second and third courses of antibiotic treatment and the E. coli with mutation could also be selected during second and third courses of the antibiotic treatments, with a higher rate at the end of the third course of the treatment, suggesting long term antibiotic treatment is the major trigger for the development of resistance in GI tract E. coli (Figures 1Dii,Eii). For the control group, neither less susceptible E. coli nor E. coli with target mutation could be selected (Figure 1F). Throughout the whole experiment, no E. coli that can grow on 2 mg/L of enrofloxacin could be obtained (data not shown).
Each of 20 E. coli isolates that grew on MacConkey with 0.125 mg/L and 0.5 mg/L enrofloxacin from different treatment experiments were selected to check their MIC and target mutation profiles. Different from the results obtained from the high dose treatment group, E. coli that grew on MacConkey with 0.125 mg/L enrofloxacin showed MIC range of enrofloxacin of 0.015~0.5, a little bit higher MIC than E. coli from high dose treatment group, with no mutation on any of the four target genes gyrA, gyrB, parC, and parE for most of the strains, but not all strains; on the other hand, E. coli that grew on MacConkey with 0.5 mg/L enrofloxacin exhibited MIC of enrofloxacin of 0.25~1 mg/L with a single mutation on GyrA (S83L) and no mutation at other target genes for all test strains, similar to the results from high dose treatment group (Table 1). The results further confirmed the use of MacConkey plate with 0.5 mg/L enrofloxacin as a tool to check for the mutation rate of GI tract E. coli upon antibiotic treatment.
DISCUSSION
Improper uses of antibiotics from clinical applications and promotion of animal growth are the main causes for high prevalence of antimicrobial resistance in bacterial pathogens (Kelly et al., 2004; Marshall and Levy, 2011; Looft et al., 2012). Evidences have shown that in-feed use of antibiotics could dramatically lead to an increase in the number of antimicrobial resistant genes and the size of microbial flora pool in anima GI tract (Looft et al., 2012). Oral usage of antibiotics may be the direct route that facilitates the selection of antimicrobial resistant gene and bacteria pool in animal GI tract (Zhang et al., 2013). These studies reinforced the concept that the antibiotic pressure in animal GI tract favored the amplification of antibiotic-resistant bacterial pool and cause it to become dominant in the animal GI tract, therefore increasing both antibiotic-resistant gene and bacteria pool. This study focuses mainly on the understanding of how GI tract bacteria, in particular E. coli, develop resistance upon encountering different levels of antibiotic pressure.
To obtain meaningful interpretation of the data, all rats were checked for the initial load of GI tract E. coli and their background level of susceptibility to enrofloxacin. Five rats with a predominant background of low level resistance to enrofloxacin (grow at MacConkey with 0.5 mg/L, but not with 2 mg/L) were arranged into one group and treated with high dose of enrofloxacin to check whether high dose of antibiotic could eradicate organisms with intermediate enrofloxacin resistance, while other groups of mice with lower or no background of less resistant E. coli were treated with different doses of antibiotic. This study has come up with several conclusions that may be useful for the understanding of bacterial resistance development in animal GI tract. Firstly, high dose of antibiotic treatment can eradicate less susceptible E. coli (with target mutation) and prevent mutation development in E.coli in the early course of enrofloxacin treatment, while became less effective for long term treatment; therapeutic dose of enrofloxacin could select for the less susceptible E. coli and those containing target mutations. It is commonly accepted that intake of therapeutic dose of antibiotics for the whole course of treatment could prevent the development of resistance in bacterial pathogens. Our data showed that only high dose of antibiotic could effectively eliminate E. coli with target mutation, which are present in rat GI tract before treatment. However, therapeutic dose, 1/10 and 1/100 of therapeutic doses of enrofloxacin could select for less susceptible E. coli without target mutation and E. coli with de novo target mutation. Extremely low dose of enrofloxacin treatment, such as 1/1000 of therapeutic dose, can also select for less susceptible E. coli without target mutation and E. coli with de novo target mutation, but with lower efficiency. From the data obtained, we can also see that most of the target mutations in E. coli were selected during the later course of antibiotic treatment and antibiotic cessation periods. Long term antibiotic treatment seems to be the major trigger for the development of target mutations in GI tract E. coli. Another interesting finding for this study is that antibiotic treatment, except for those with high dosage, did not affect the total number of GI tract E. coli. Even under high dosage antibiotic treatment, the number of GI tract E. coli decreased upon treatment and then became normal even during the second treatment.
Recent studies have reported the selection of resistant bacteria under very low concentration of antibiotics in vitro (Gullberg et al., 2011). However, these studies mainly focus on determining the concentration of antibiotics that allow the resistant bacteria to compete with their drug-susceptible counterparts favorably. In another word, how concentration of antibiotics lower than the MIC of susceptible cells, namely sub-MIC levels, promotes the enrichment of resistant bacteria. One previous study also showed how low concentration of antibiotic selects for de novo resistant mutants (Gullberg et al., 2011). The selection process in such study was antibiotic dependent. At 1 μg/ml concentration of streptomycin, Salmonella could develop resistance to up to 128 μg/ml of streptomycin after 700 generations, whereas at 2.3 ng/ml of ciprofloxacin, E. coli could be selected at 184 ng/ml, but not resistant to ciprofloxacin after 600 generations (Gullberg et al., 2011). Similar to in vitro studies, our data showed that regardless of the dosages of the enrofloxacin within three courses of treatment and cessation regimen, no enrofloxacin-resistant (MIC ≥4 mg/L) E. coli could be selected. In addition, no double target mutations in gyrA or parC could be detected in E. coli that could grow on MacConkey with 0.5 mg/L enrofloxacin. Our data is consistent with an early study in chicken in which treatment with enrofloxacin at doses routinely prescribed (50 ppm) rapidly reduced the fecal counts of colonized E. coli below the detection limit and did not induce resistance, whereas high frequencies of fluoroquinolone-resistant colonized C. jejuni were selected due to the de novo mutation at the target genes (van Boven et al., 2003). E. coli with S83, D87, or both mutations have commonly been reported in in vitro selection experiments and contributed to fluoroquinolone resistance in E. coli (Vashist et al., 2009). However, in the GI tract, no E. coli with resistant to enrofloxacin could be selected and only E. coli with reduced susceptibility to enrofloxacin and S83F target mutation could be selected, which is probably due to the unique GI environment. These data suggested that the generation of double target mutations in GyrA and/or ParC, which is the major mechanism of fluoroquinolone resistance in E. coli, may be associated with a fitness cost that hampers its survival in animal GI tract (Komp Lindgren et al., 2005; Gualco et al., 2007). In our current research design, we were not able to select enrofloxacin-resistant E. coli in mice GI tract. The fact that the high prevalence of fluoroquinolone-resistant E. coli in animal GI tract may be due to the long term selection under antibiotic selective pressure in animal gut or colonization of fluoroquinolone-resistant E. coli that has been selected outside the animal gut. Lastly, the development of enrofloxacin-resistant E. coli in animal GI tract may possibly require the acquisition of plasmid mediated resistance determinant, which may further facilitate the development of target mutation and therefore development of enrofloxacin resistance in E. coli. Recent study has shown the high carriage of different PMQR genes in animal E. coli isolates, which contribute to the development of enrofloxacin resistance in E. coli (Zhao et al., 2010; Yang et al., 2014). Different hypotheses of how GI tact E. coli development resistance to fluoroquinolone require further investigations.
In conclusion, this study demonstrated that long term antibiotic treatment seems to be the major trigger for the development of target mutations in GI tract E. coli, which provided insights into the rational use of antibiotics in animal husbandry.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the critical reading of the manuscript by Dr. Edward Chan and useful discussion with members from Sheng’s lab. This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127201).
SUPPLEMENTARY MATERIAL
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00468/abstract
REFERENCES
- Bernier S. P., Lebeaux D., DeFrancesco A. S., Valomon A., Soubigou G., Coppee J. Y., et al. (2013). Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet. 9:e1003144 10.1371/journal.pgen.1003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau J. M. (2009). New concepts in antimicrobial susceptibility testing: the mutant prevention concentration and mutant selection window approach. Vet. Dermatol. 20 383–396 10.1111/j.1365-3164.2009.00856.x [DOI] [PubMed] [Google Scholar]
- Brazier J. S. (2008). Clostridium difficile: from obscurity to superbug. Br. J. Biomed. Sci. 65 39–44 [DOI] [PubMed] [Google Scholar]
- CLSI. (2013). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-third Informational Supplement. CLSI document M100-S23 Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- Duval V., Lister I. M. (2013). MarA, SoxS and Rob of – Global regulators of multidrug resistance, virulence and stress response. Int. J. Biotechnol. Wellness Ind. 2 101–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett M. J., Jin Y. F., Ricci V., Piddock L. J. (1996). Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40 2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber D. (2010). Infectious disease. From pigs to people: the emergence of a new superbug. Science 329 1010–1011 10.1126/science.329.5995.1010 [DOI] [PubMed] [Google Scholar]
- Firsov A. A., Strukova E. N., Shlykova D. S., Portnoy Y. A., Kozyreva V. K., Edelstein M. V., et al. (2013). Bacterial resistance studies using in vitro dynamic models: the predictive power of the mutant prevention and minimum inhibitory antibiotic concentrations. Antimicrob. Agents Chemother. 57 4956–4962 10.1128/AAC.00578-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firsov A. A., Vostrov S. N., Lubenko I. Y., Zinner S. H., Portnoy Y. A. (2004). Concentration-dependent changes in the susceptibility and killing of Staphylococcus aureus in an in vitro dynamic model that simulates normal and impaired gatifloxacin elimination. Int. J. Antimicrob. Agents 23 60–66 10.1016/j.ijantimicag.2003.06.001 [DOI] [PubMed] [Google Scholar]
- Ghafur A. (2013). Call for global action to halt the superbug. Med. J. Aust. 198 251 10.5694/mja13.10099 [DOI] [PubMed] [Google Scholar]
- Gualco L., Schito A. M., Schito G. C., Marchese A. (2007). In vitro activity of prulifloxacin against Escherichia coli isolated from urinary tract infections and the biological cost of prulifloxacin resistance. Int. J. Antimicrob. Agents 29 679–687 10.1016/j.ijantimicag.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Gullberg E., Cao S., Berg O. G., Ilback C., Sandegren L., Hughes D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes M., Barton M., Conly J., Nicolle L., Barry C., Ford-Jones E. L. (2007). Community-associated MRSA: superbug at our doorstep. CMAJ 176 54–56 10.1503/cmaj.061370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. X., Lu D. H., Chen Z. L., Wang X. M., Chen J. R., Liu Y. H., et al. (2011). High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet. J. 187 99–103 10.1016/j.tvjl.2009.10.017 [DOI] [PubMed] [Google Scholar]
- Kelly L., Smith D. L., Snary E. L., Johnson J. A., Harris A. D., Wooldridge M., et al. (2004). Animal growth promoters: to ban or not to ban? A risk assessment approach. Int. J. Antimicrob. Agents 24 205–212 10.1016/j.ijantimicag.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Komp Lindgren P., Marcusson L. L., Sandvang D., Frimodt-Moller N., Hughes D. (2005). Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob. Agents Chemother. 49 2343–2351 10.1128/AAC.49.6.2343-2351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T., Johnson T. A., Allen H. K., Bayles D. O., Alt D. P., Stedtfeld R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U.S.A. 109 1691–1696 10.1073/pnas.1120238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. M., Levy S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24 718–733 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334 982–986 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2012). Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 20 227–234 10.1016/j.tim.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Srinivasan V. B., Vaidyanathan V., Mondal A., Rajamohan G. (2012). Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 7:e33777 10.1371/journal.pone.0033777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R. K., Ichhpujani R. I., Khare S., Rai A., Chauhan L. S. (2011). Superbug – the so-called NDM-1. Indian J. Med. Res. 133 458–460 [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C. (2006). Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 34(5 Suppl. 1) S3–S10 10.1016/j.ajic.2006.05.219 [DOI] [PubMed] [Google Scholar]
- van Boven M., Veldman K. T., de Jong M. C., Mevius D. J. (2003). Rapid selection of quinolone resistance in Campylobacter jejuni but not in Escherichia coli in individually housed broilers. J. Antimicrob. Chemother. 52 719–723 10.1093/jac/dkg402 [DOI] [PubMed] [Google Scholar]
- Vashist J., Vishvanath Kapoor R., Kapil A., Yennamalli R., Subbarao N., Rajeswari M. R. (2009). Interaction of nalidixic acid and ciprofloxacin with wild type and mutated quinolone-resistance-determining region of DNA gyrase A. Indian J. Biochem. Biophys. 46 147–153 [PubMed] [Google Scholar]
- Yang T., Zeng Z., Rao L., Chen X., He D., Lv L., et al. (2014). The association between occurrence of plasmid-mediated quinolone resistance and ciprofloxacin resistance in Escherichia coli isolates of different origins. Vet. Microbiol. 170 89–96 10.1016/j.vetmic.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Zhang L., Huang Y., Zhou Y., Buckley T., Wang H. H. (2013). Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 57 3659–3666 10.1128/AAC.00670-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Chen Z., Chen S., Deng Y., Liu Y., Tian W., et al. (2010). Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 54 4219–4224 10.1128/AAC.00139-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Drlica K. (2002). Restricting the selection of antibiotic-resistant mutant bacteria: measurement and potential use of the mutant selection window. J. Infect. Dis. 185 561–565 10.1086/338571 [DOI] [PubMed] [Google Scholar]
- Zheng H., Zeng Z., Chen S., Liu Y., Yao Q., Deng Y., et al. (2012). Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int. J. Antimicrob. Agents 39 305–310 10.1016/j.ijantimicag.2011.12.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.