Abstract
Malaria parasite transmission depends on the successful transition of Plasmodium through discrete developmental stages in the lumen of the mosquito midgut. Like the human intestinal tract, the mosquito midgut contains a diverse microbial flora, which may compromise the ability of Plasmodium to establish infection. We have identified an Enterobacter bacterium isolated from wild mosquito populations in Zambia that renders the mosquito 99% resistant to infection with the human malaria parasite Plasmodium falciparum by interfering with parasite development prior to invasion of the midgut epithelium. Phenotypic analyses showed that the anti-Plasmodium mechanism requires small populations of replicating bacteria and is mediated through a mosquito-independent interaction with the malaria parasite. We show that this anti-Plasmodium effect is largely caused by bacterial generation of reactive oxygen species.
Plasmodium parasites suffer considerable losses during their development in the mosquito midgut (1, 2), where they encounter a hostile environment of human blood-derived factors, mosquito innate immune responses, and resident microbiota. However, escape of only a few parasites is sufficient to ensure onward transmission. Midgut bacteria play a key role in modulating Plasmodium infection of the Anopheles mosquito vector (3–7), but the mechanism(s) of bacterially mediated parasite inhibition has not been described, although the mosquito’s innate immune responses to the microbiota and parasite challenge have been implicated in this phenomenon (6–10).
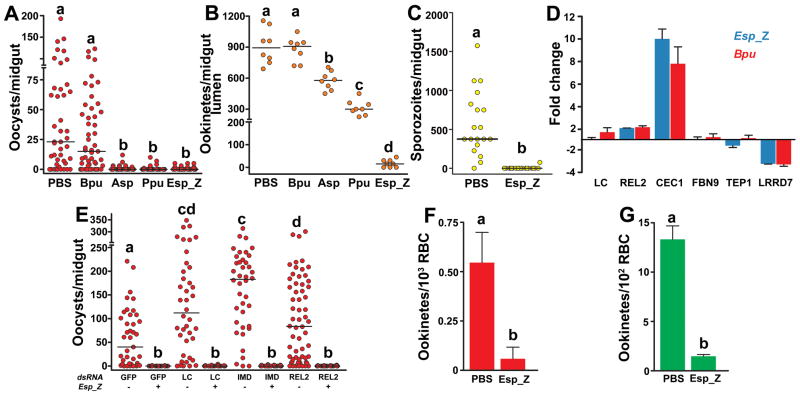
In the present study, we isolated bacteria from southern Zambian populations of A. arabiensis, an important malaria vector, during two collecting trips in two consecutive years (11). A majority of the captured mosquito’s midguts contained cultured bacteria, with an average concentration of 104 bacteria per non-bloodfed gut, similar to that observed for laboratory-reared Anopheles mosquitoes (6). Sixteen distinct bacterial strains were identified based on 16S ribosomal DNA sequence, with similar genera isolated from both collections (Table S1). Of these, some Gram-negative (G−) isolates reduced P. falciparum prevalence and intensity in the mosquito, while a Gram-positive (G+) isolate had no detectable effect on infection (Fig 1A), in accordance with previous reports involving other Plasmodium-Anopheles models (3–5). The G− bacteria inhibited P. falciparum oocyst formation in the main African and Asian malaria vectors, A. gambiae and A. stephensi, respectively (Fig. 1A, S1). To investigate the temporal specificity of parasite inhibition by G− bacteria, we monitored the development of the Plasmodium ookinetes in the presence of the field isolated bacteria. Significantly fewer ookinetes developed with G− bacteria present, although the level of inhibition was bacterial species-dependent (Fig 1B). The Enterobacter sp. (Esp_Z) bacterium inhibited ookinete, oocyst, and sporozoite development of a highly virulent laboratory Plasmodium strain by 98%, 99% and 99%, respectively (Fig 1A–C), leading us to further investigate the mechanism of this inhibition.
Figure 1. Field bacteria-mediated inhibition of Plasmodium development.
P. falciparum oocyst (A) and ookinete (B) loads in midguts of A. gambiae mosquitoes co-fed with parasites and field-isolated bacteria. Bpu, Bacillus pumilus; Asp, Acinetobacter sp.; Ppu, Pseudomonas putida; Esp_Z, Enterobacter sp. (C) P. falciparum sporozoite loads in salivary glands of A. gambiae mosquitoes co-fed with parasites and Esp_Z. For (A–C), circles represent the number of parasites from an individual mosquito and horizontal lines indicate the median number of parasites per tissue. (D) Midgut-specific transcript abundance of select genes at 8 h after bloodfeeding with equal quantities of either the Bpu or Esp_Z isolate. Each column and error bar represents the fold-change ± standard deviation in transcript abundance when compared to PBS-fed controls. LC= PGRP-LC; CEC1=cecropin1; FBN9=fibrinogen immunolection 9; TEP1=thioester-containing protein 1; LRRD7=leucine-rich repeat-containing protein 7. (E) Oocyst loads in mosquitoes depleted of transcripts for IMD pathway molecules and co-challenged with Esp_Z and P. falciparum. The dsRNA and absence (−) or presence (+) of Esp_Z are indicated below each column. Circles represent the same as in (A). (F–G) In vitro development of P. falciparum (E) and P. berghei (F) ookinetes co-cultured with Esp_Z bacteria. Bars represent the mean ± standard deviation in ookinetes. For all figures, statistical significance is represented by letters above each column, with different letters signifying distinct statistical groups (p<0.05; Mann-Whitney test for (A–C, E); p<0.05; unpaired t-test for (F–G)).
The immune deficiency (IMD) innate immune pathway defends mosquitoes against P. falciparum in the gut tissue, and the microbiota has been shown to activate this pathway through the receptor protein peptidoglycan recognition protein-LC (PGRP-LC) (6–8); thus, we hypothesized that the refractory phenotype could be caused by Esp_Z activation of the IMD pathway. Two independent approaches showed that the mechanism of refractoriness is independent of the IMD pathway. First, although a general antibacterial response is mounted through the increased transcription of the antimicrobial peptide cecropin1 (CEC1), regulation of IMD pathway controlled genes, including several potent anti-Plasmodium effector genes [fibrinogen immunolectin 9 (FBN9), leucine-rich repeat protein LRRD7, and thioester-containing protein 1 (TEP1) (12)] were similar in the midguts of mosquitoes challenged with Esp_Z or the non-inhibitory Bacillus bacterium (Bpu) (Fig 1D). Second, RNA interference-mediated depletion of the key pathway molecules, PGRP-LC, Imd, and REL2, did not rescue P. falciparum oocyst development in the presence of Esp_Z (Fig. 1E).
Hence, we investigated the potential for parasite inhibition exclusive of the mosquito. Although P. falciparum ookinete development was inefficient in vitro, co-culture of Esp_Z with gametocytes inhibited ookinete formation by 89% (Fig 1F). We observed a similarly strong in vitro inhibition of ookinete formation in the more-robust rodent malaria parasite P. berghei experimental model (Fig 1G).
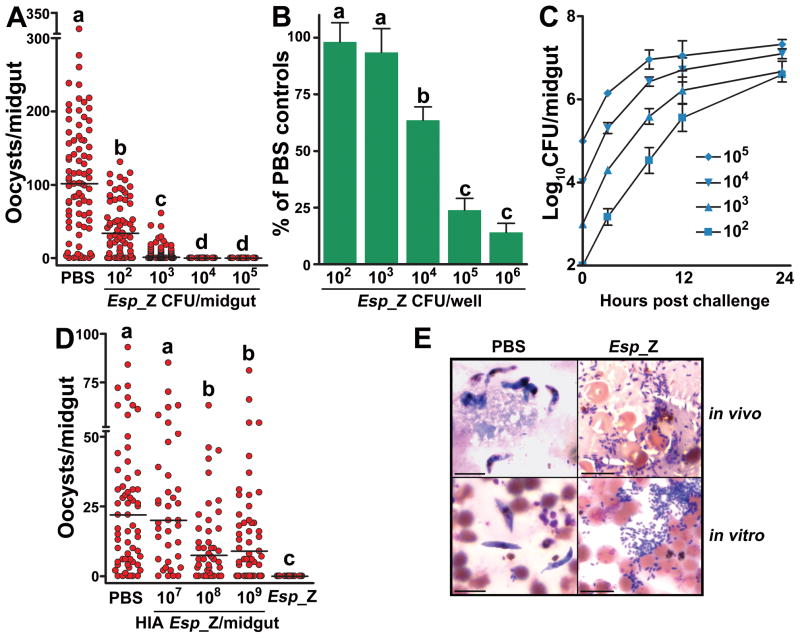
Inhibition of P. falciparum by Esp_Z in the mosquito was dose-dependent, with a threshold of 104 ingested bacteria providing near complete protection against parasite infection (Fig 2A). A remarkably low density of only 100 ingested bacteria was still able to significantly decrease oocyst intensity by 67% (Fig 2A). In vitro ookinete development of P. berghei was also inhibited in a dose-dependent manner (Fig. 2B). Esp_Z populations in the midgut expanded by 100–1,000 fold (Fig 2C) which is within the range of microbial proliferation that normally occurs in the midgut lumen after a blood meal (5, 6). The bacterial growth during the 24hr period immediately following blood ingestion correlates with the time period of parasite inhibition prior to ookinete formation (3–30hr after ingestion) (13). Negative correlations were also observed with the timing of bacterial replication and inhibition of P. berghei ookinete development in vitro (correlation coefficient= −0.95 for 106 Esp_Z and −0.94 for 107 Esp_Z) (Fig S2–3), and taken together, these results indicated that active replication of the bacteria was required for parasite inhibition. Mosquito exposure to heat-inactivated (HIA) Esp_Z upon feeding on a P. falciparum gametocyte culture did not result in the same level of refractoriness that was observed with exposure to live bacteria (Fig. 2D). Because of an overlap in antibacterial and anti-Plasmodium immune defenses in mosquitoes (6, 7, 9, 14), the observed decrease in oocyst numbers with high concentrations of HIA bacteria (Fig 2D) could be caused by the induction of an antibacterial response in the mosquito gut, while physical inhibition of parasite infection by the killed bacteria is also possible.
Figure 2. Phenotypic analyses of Esp_Z modulation of Plasmodium development.
(A–B) Effect of Esp_Z dosage on P. falciparum oocyst formation in mosquitoes (A) and P. berghei ookinete formation in vitro (B). Circles in (A) represent the number of oocysts in an individual mosquito midgut, and the horizontal lines indicate the median number of parasites per midgut. Bars in (B) represent the mean ± the standard deviation in percentage of the number of ookinetes formed in bacteria-treated groups as compared to PBS-treated controls. (C) Temporal replication of Esp_Z in mosquito midguts following bloodmeal administration of different inoculating doses of bacteria. CFU, colony-forming unit. Bars represent the mean ± the standard deviation. (D) Effect of heat-inactivated Esp_Z bacteria on P. falciparum oocyst formation. Circles represent the same as in (A). For (A), (B) and (D), statistical significance is represented by letters above each column, with different letters signifying distinct statistical groups (p<0.05; Mann-Whitney test for (A) and (D), unpaired t-test for (B)). (E) In vivo and vitro P. falciparum development in the presence of Esp_Z. Scale bar= 10μm.
These observations, along with microscopy (Figure 2E), indicated that Esp_Z inhibition of Plasmodium did not involve direct association between the bacteria and parasite and was mediated by diffusible bacterial factors produced during replication, or bacterial sequestration of mosquito factors that are essential for Plasmodium development. In subsequent assays, we observed that Plasmodium inhibition was independent of bacterial fatty acid metabolism (Figure S4) (15), xanthurenic acid (XA) (Fig S5) and iron (Fig S6) utilization by the parasite.
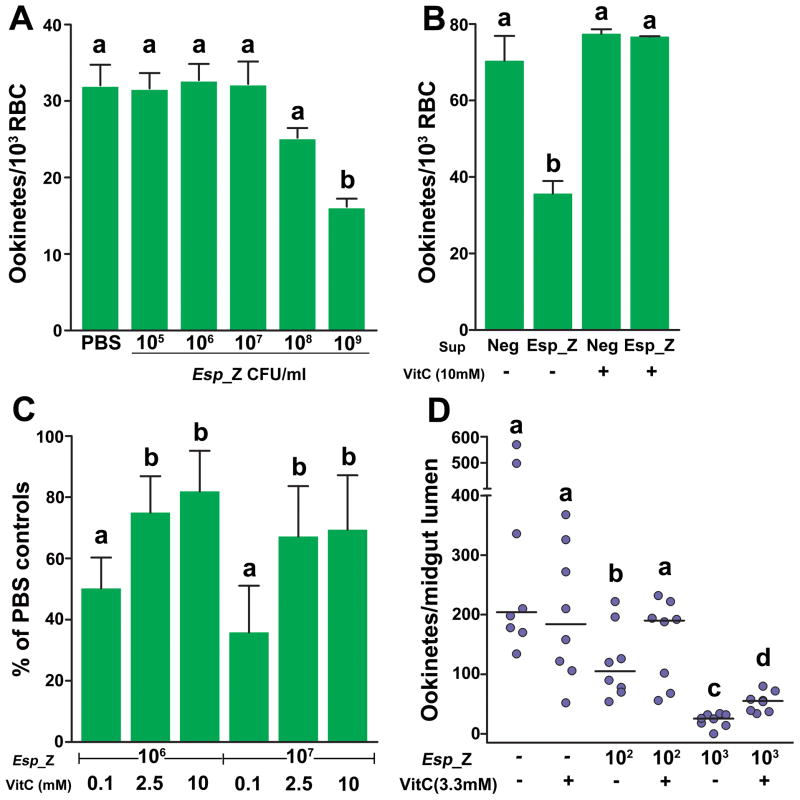
Because the inhibitory effect did not appear to be dependent on the sequestration or acquisition of a molecule necessary for the parasite, we hypothesized that the bacteria were producing an anti-Plasmodium molecule. We tested this by examining the in vitro development of P. berghei first in co-culture with physically separated bacteria and second in filtered fresh supernatant of a bacterial culture. Separation of bacteria and parasites abolished bacteria-mediated inhibition of ookinete formation, except at very high bacterial concentrations (Fig 3A). When parasites were cultured in filtered fresh supernatant from an Esp_Z culture, there was a two-fold reduction in numbers of ookinetes formed (Fig 3B). Together, these data indicated that the inhibitory activity was mediated by a short-lived molecule in a concentration-dependent manner. Reactive oxygen species (ROS) have a short half-life, kill Plasmodium (16–19), and can be generated by bacteria (20), so we tested the hypothesis that bacteria were inhibiting parasite development by producing ROS.
Figure 3. Involvement of ROS generation by Esp_Z in inhibition of Plasmodium development.
(A) Effect of physical separation of Esp_Z and parasites on P. berghei ookinete formation. (B) Effect of filtered culture supernatant and addition of vitamin C on P. berghei ookinete formation. Sup=supernatant; Neg=supernatant from a bacteria-negative culture; VitC=vitamin C. For (A–B), bars represent the mean ± the standard deviation. (C–D) Effect of vitamin C addition on P. berghei ookinete formation in vitro (C) and P. falciparum ookinete formation in A. gambiae midguts (D). For (C), bars represent the mean ± the standard deviation in percentage of the number of ookinetes formed in bacteria-treated groups as compared to PBS-treated controls. For (D), circles represent the number of ookinetes from an individual mosquito and horizontal lines indicate the median number of parasites per midgut. For all figures, statistical significance is represented by letters above each column, with different letters signifying distinct statistical groups (p<0.05; unpaired t-test for (A–C); Mann-Whitney test for (D)).
Among the field mosquito –derived bacteria, those lacking ookinete inhibitory activity (Fig 1B) did not produce detectable levels of ROS, whereas cultures of the inhibitory Esp_Z did (Table S2). To determine whether ROS was involved in the parasite inhibition, we supplemented the P. berghei culture with an antioxidant to neutralize free radicals formed. The addition of vitamin C (vitC) to in vitro cultures of P. berghei gametocytes rescued development of ookinetes to untreated control levels when grown in filtered Esp_Z culture medium (Fig 3B), and parasite development in the presence of replicating Esp_Z was rescued by vitC in a dose-dependent fashion (Fig 3C). Furthermore, reduced glutathione, another potent antioxidant, also rescued in vitro ookinete formation in the presence of Esp_Z (Fig S7). We also showed that vertebrate leukocytes were not responsible for the observed in vitro ookinete inhibition (Fig S8). More importantly, supplementing an infectious bloodmeal with vitC did not impact parasite numbers in the absence of Esp_Z but rescued P. falciparum ookinete development two-fold in the lumen of A. gambiae midguts upon co-feeding with Esp_Z (Fig 3D). The significant, yet incomplete, rescue of ookinete development with higher bacterial concentrations could be attributed to a variety of factors such as insufficient concentrations of antioxidant to neutralize the higher amount of bacteria produced ROS, excretion of significant amounts of antioxidant through mosquito diuresis, the intimate association between bacteria and parasites that may not enable detoxification of ROS prior to parasite inhibition, or the loss of antioxidant activity from prolonged exposure in the digestive environment of the midgut. Antioxidant concentrations higher than 10mM in the bloodmeal interfered with mosquito feeding propensity.
Genotypic analyses of laboratory and wild mosquito populations have suggested that a dominant refractory phenotype is associated with innate immunity and that Plasmodium infection is a result of immune failure (21–23). Our studies show a mechanism of Plasmodium inhibition that does not involve the mosquito-derived innate immune response, and they support the idea that the native microflora of Anopheles mosquitoes plays a crucial role in refractoriness to Plasmodium infection, and will therefore influence transmission success to humans.
Bacteria of the genus Enterobacter have been isolated from many anopheline mosquito species in diverse geographic regions (3, 5, 24). We show that mosquitoes do not become infected with Plasmodium parasites when exposed to an Enterobacter bacterium isolated from wild mosquito populations in southern Zambia, and we show that inhibition of parasite development can be mediated by bacterial generation of ROS. While Esp_Z was isolated from 25% of mosquitoes collected during one rainy season, it may be possible to manipulate the composition of the midgut microbial flora in wild mosquitoes to increase the prevalence of Esp_Z or other naturally inhibitory bacteria as part of an integrated malaria control strategy.
Supplementary Material
Acknowledgments
This work has been supported by the National Institutes of Health/National Institute of Allergy and Infectious Disease R01AI061576 and a Johns Hopkins Malaria Research Institute Pilot Grant (to G.D.), the Calvin A. and Helen H. Lang fellowship (to C.M.C.), a JHMRI postdoctoral fellowship (to J.S.-N.), and a fellowship from the National Science Foundation (to A.M.C.). The authors thank the mosquito collection team at the Malaria Institute at Macha, Zambia, the Johns Hopkins Malaria Research Institute (JHMRI) Parasitology and Insectary Core facilities, Sanaria Inc., Dr. Eric Nelson (Cornell University) for providing mutant bacteria strains, and Dr. Deborah McClellan for editorial services. GenBank accession numbers generated for bacterial 16S rDNA sequences are listed in Table S1, SOM.
Footnotes
Materials and Methods
References and notes
References and Notes
- 1.Alavi Y, et al. Int J Parasitol. 2003;33:933. doi: 10.1016/s0020-7519(03)00112-7. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan JA, Noden BH, Beier JC. Am J Trop Med Hyg. 1994;51:233. doi: 10.4269/ajtmh.1994.51.233. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Ceron L, Santillan F, Rodriguez MH, Mendez D, Hernandez-Avila JE. J Med Entomol. 2003;40:371. doi: 10.1603/0022-2585-40.3.371. [DOI] [PubMed] [Google Scholar]
- 4.Pumpuni CB, Beier MS, Nataro JP, Guers LD, Davis JR. Exp Parasitol. 1993;77:195. doi: 10.1006/expr.1993.1076. [DOI] [PubMed] [Google Scholar]
- 5.Pumpuni CB, Demaio J, Kent M, Davis JR, Beier JC. Am J Trop Med Hyg. 1996;54:214. doi: 10.4269/ajtmh.1996.54.214. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Manfredini F, Dimopoulos G. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meister S, et al. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garver LS, Dong Y, Dimopoulos G. PLoS Pathog. 2009;5:e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Y, et al. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. Science. 2010;327:1644. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science online.
- 12.Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Dev Comp Immunol. 2010;34:387. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan JA, Noden BH, Beier JC. J Parasitol. 1992;78:716. [PubMed] [Google Scholar]
- 14.Meister S, et al. Proc Natl Acad Sci USA. 2005;102:11420. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk K, Nelson EB. Appl Environ Microbiol. 2000;66:5340. doi: 10.1128/aem.66.12.5340-5347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, et al. Proc Natl Acad Sci USA. 2003;100:14139. doi: 10.1073/pnas.2036262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luckhart S, Vodovotz Y, Cui L, Rosenberg R. Proc Natl Acad Sci USA. 1998;95:5700. doi: 10.1073/pnas.95.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina-Cruz A, et al. J Biol Chem. 2008;283:3217. doi: 10.1074/jbc.M705873200. [DOI] [PubMed] [Google Scholar]
- 19.Peterson TML, Gow AJ, Luckhart S. Free Radic Biol Med. 2007;42:132. doi: 10.1016/j.freeradbiomed.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai-Prochnow A, et al. J Bacteriol. 2008;190:5493. doi: 10.1128/JB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blandin S, et al. Science. 2009;326:147. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niaré O, et al. Science. 2002;298:213. doi: 10.1126/science.1073420. [DOI] [PubMed] [Google Scholar]
- 23.Riehle MM, et al. Science. 2006;312:577. doi: 10.1126/science.1124153. [DOI] [PubMed] [Google Scholar]
- 24.Terenius O, et al. J Med Entomol. 2008;45:172. doi: 10.1603/0022-2585(2008)45[172:srgsfb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.