Abstract
Asian cultivated rice Oryza sativa L. was domesticated from its wild ancestor, O. rufipogon. During domestication, the cultivated rice lost its seed-shattering behaviour. Previous studies have shown that two major quantitative trait loci (QTLs; qSH1 and sh4) are responsible for the seed-shattering degree. Here, we produced introgression lines carrying non-functional alleles from O. sativa ‘Nipponbare’ at the two major QTLs in the genetic background of wild rice O. rufipogon W630, and examined the effects of the two QTLs on seed shattering and abscission layer formation. The introgression lines, with Nipponbare alleles at either or both loci, showed complete or partial abscission layer formation, respectively, indicating that other unknown loci might be involved in enhancing seed shattering in wild rice. We detected a single QTL named qSH3 regulating seed-shattering degree using an F2 population between Nipponbare and the introgression line carrying Nipponbare alleles at the two QTLs. Although we generated an introgression line for qSH3 alone, no effects on seed shattering were observed. However, a significant effect on seed-shattering degree was observed for the introgression line carrying Nipponbare alleles at qSH3 and the two QTLs, suggesting an important role of qSH3 on seed shattering in coordination with the two QTLs.
Keywords: wild rice, Oryza rufipogon, seed shattering, abscission layer, QTL
Introduction
Seed shattering is one of the most important characteristics enabling the efficient propagation in wild plants. In many monocot plants, seed shattering is achieved by degradation of the abscission layer formed at the basal part of grains. After fertilisation, plants transfer nutrients to seeds, but as soon as the seed matures, the abscission layer begins to degrade. Since seed shattering causes reduction in the yield, hunter-gatherers and early agriculturists may have selected non-shattering plants to harvest the seeds more efficiently. Therefore, loss of seed shattering is proposed to be one of the most important traits selected for in early crop domestication (Fuller 2007, Harlan 1975).
Asian cultivated rice Oryza sativa L. is known to have been domesticated from its wild ancestor O. rufipogon about 10,000 years ago (Fuller 2007, Oka 1988). During domestication, rice plants with weak or no seed shattering were preferably selected and an increase in yield was proposed to have contributed human civilisation in Asia. Among rice cultivars, a wide variation in seed-shattering degree has been observed (Konishi et al. 2006), suggesting that seed shattering is a quantitatively regulated trait. Genomic loci involved in seed shattering have been identified based on quantitative trait locus (QTL) analyses using segregating populations between natural variations (Cai and Morishima 2000, Ishikawa et al. 2010, Lee et al. 2005, Li et al. 2006a, Onishi et al. 2007a, Thomson et al. 2003, Xiong et al. 1999). A major QTL for seed shattering between O. rufipogon and O. sativa Indica was sh4, which explained 69% of the total phenotypic variance in their segregating populations. A gene responsible for sh4 was identified and shown to encode a Myb-type transcription factor. A cultivated allele with a non-functional mutation was found to inhibit the normal development of the abscission layer (Li et al. 2006b). Another major QTL is qSH1, which was identified using a segregating population between O. sativa Indica cv. Kasalath and Japonica cv. Nipponbare explaining 68.6% of the phenotypic variance. A causative mutation at qSH1 was shown to be an SNP located upstream of the rice homolog of the Arabidopsis REPLUMLESS (RPL) gene, that is involved in the formation of a dehiscence zone in the Arabidopsis silique. The Nipponbare allele causes the reduction of rice RPL gene expression, resulting in the complete absence of abscission layer formation (Konishi et al. 2006). Since the non-functional mutation at sh4 is commonly observed in cultivated rice, it is widely accepted that the mutation played an important role in rice domestication (Lin et al. 2007, Onishi et al. 2007b, Zhang et al. 2009). A reverse genetic approach was also used to identify genes involved in seed shattering. SHAT1 encoding an APETALA2 transcription factor was identified by the suppressor mutagenesis of cultivated rice cv. Guangluai 4 with strong seed shattering due to a wild sh4 allele (Zhou et al. 2012). Mutant analysis of seed-shattering cultivars identified OsCPL1, encoding a nuclear phosphatase, as a repressor of abscission layer formation (Ji et al. 2010).
We previously evaluated the two major seed-shattering loci, qSH1 and sh4, using a backcross population between cultivated rice, O. sativa Japonica cv. ‘Nipponbare’, as a donor parent, and wild rice, O. rufipogon acc. W630, as a recurrent parent, aiming to elucidate the effects of the Nipponbare alleles with non-functional mutations (Htun et al. 2011, Ishikawa et al. 2010). Interestingly, non-shattering behaviour was not observed, even though the Nipponbare non-functional allele at either the qSH1 or sh4 locus was introgressed into the wild genetic background. Further, we found that the introgression line that had the Nipponbare non-functional alleles at both loci showed weak shattering. These results suggest that wild rice still had additional effective gene(s) controlling seed shattering. In this study, we carried out a histological analysis to understand the role of the non-functional alleles at seed-shattering loci on abscission layer formation using these introgression lines. Subsequently, QTL analysis was performed to explore novel loci involved in seed shattering in wild rice.
Materials and Methods
Plant materials
A Japonica rice cultivar, O. sativa cv. Nipponbare, and an annual wild accession, O. rufipogon acc. W630, originating from Myanmar, were used in this study. O. sativa cv. Nipponbare has non-shattering behaviour while O. rufipogon W630 has strong shattering behaviour. Introgression lines of wild rice carrying the Nipponbare alleles at either the sh4 or qSH1 locus have previously been produced (Ishikawa et al. 2010). We also produced the introgression line containing the Nipponbare alleles at both the qSH1 and sh4 loci in the genetic background of wild rice by backcrossing (Htun et al. 2011). This introgression line was further crossed with Nipponbare and their F2 plants were grown in pots in the greenhouse at Kobe University. To minimise the variation in heading date, F2 individuals were genotyped for Hd5 (chr. 8) and Hd6 (chr. 3) using dCAPS and CAPS markers, respectively (Table 1), and short day treatment was given to the plants with late-flowering genotypes. Further, introgression lines of the newly identified shattering locus were generated in the genetic background of wild rice by using two flanking simple sequence repeat (SSR) markers (Table 1). Chromosomal information of the Nipponbare segment covering qSH1, sh4, and qSH3 loci in the five introgression lines was also analysed (Supplemental Table 1).
Table 1.
List of dCAPS, CAPS, and flanking SSR markers used in this study
| Chr. | Locus | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Restriction Enzyme | Reaction condition |
|---|---|---|---|---|---|
| 1 | qSH1 | TGATGGTATTGATGTATACTGGACAaTa | TCGAAATGTGGAGACAGCTC | EcoT22I (AvaIII) | 37°C |
| 4 | sh4 | AACGCCGGCGCGGCGGTCGTCGTCCAGtCGa | ACGGGCACCTGACTGCTAC | TaqI | 60°C |
| 3 | Hd6 | ACCTGGCAGCATGTTATGAC | CTACAGATCCACAGAACAGG | Hind III | 37°C |
| 8 | Hd5 | GCGCCGATGGCGTCGTCATCGACGGGaATTa | ACGCCTTGTTCCCTGACG | EcoRI | 37°C |
| 3 | qSH3 | ||||
| RM16 | CGCTAGGGCAGCATCTAAA | AACACAGCAGGTACGCGC | |||
| RM3513 | TACTCCTATCCTGCCATGGC | TGTAGTAGACGAGAGGCCGG |
Nucleotide with a small letter indicates the primer mismatch creating restriction endonuclease-sensitive polymorphism.
Histological analysis of abscission layer formation
Histological samples for abscission layer analysis were collected from pedicel tissue of rice grains just after flowering. Samples were fixed in an FAA solution (5% v/v formaldehyde, 5% v/v acetic acid and 63% ethanol) with vacuum infiltration. These samples were kept at 4°C and then were dehydrated through an ethanol series (50%, 70%, 80%, 90%, 95%, 99.5% and 100%) for 1 h each. Then, they were embedded in Technovit 7100 resin (Heraeus Kulzer, Germany) according to the manufacturer’s instruction. Samples were cut into 3-μm-thick sections by using a rotary microtome, RM2125RT (Leica Biosystems) and stained with toluidine blue O solution. These sections were observed under a microscope.
Evaluation of seed shattering degree
To precisely evaluate the shattering degree in plants in an F2 segregating population, a digital force gauge (FGP 0.5, Nidec-Shimpo Co., Japan) was employed. The seed-shattering degree was evaluated by measuring the breaking tensile strength (BTS) required to detach the seeds from the pedicels. At the maturation stage, approximately 35 days after flowering, shattering degree was measured using 125 spikelets (25 randomly chosen spikelets from 5 panicles of each plant), and their average BTS values were calculated.
SSR marker genotyping and QTL analysis
To determine the genotypes at 179 polymorphic marker loci covering the whole rice genome (Thanh et al. 2010), genomic DNA was extracted from 90 F2 plants and PCR was conducted as previously described (Ishikawa et al. 2010). The amplified products were electrophoresed in 4.0% polyacrylamide gels and banding patterns were visualised by the silver staining method of Panaud et al. (1996). QTL analysis for seed shattering was carried out with phenotypic data (BTS values) by composite interval mapping (CIM) using WinQTL cartographer ver. 2.5 (Wang et al. 2007). The optimal log of odds (LOD) threshold values (with the permutation value set at 1000) were used to determine the presence of a putative QTL, and the percentage of variation explained by the QTL was estimated. Information on the candidate genes at qSH3 was obtained from RAP-DB (IRGSP-1.0, Rice Annotation Project, http://rapdb.dna.affrc.go.jp/) (Sakai et al. 2013).
Results
Effects of two major quantitative trait loci (QTLs) for seed shattering on abscission layer formation in wild rice
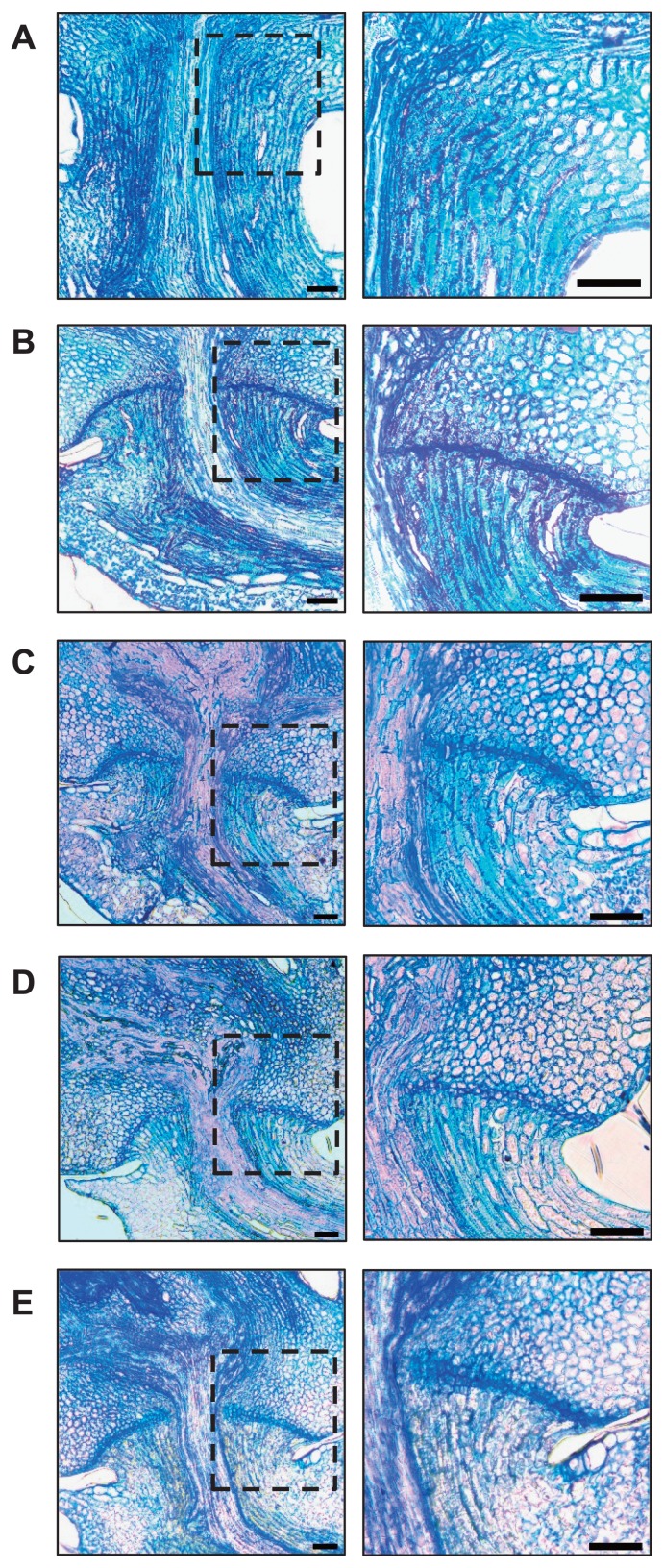
In our previous study, we generated and characterised backcross plants individually having the Nipponbare non-functional alleles at qSH1 or sh4 in the genetic background of wild rice. We found that these two introgression lines showed strong seed-shattering habits similar to those of wild rice, indicating that a single mutation at either the qSH1 or sh4 locus is not sufficient to confer the non-shattering phenotype (Ishikawa et al. 2010). We also observed partial seed shattering in the introgression line having the Nipponbare alleles at both the qSH1 and sh4 loci in the genetic background of wild rice. This line could keep mature seeds on the panicles, giving small BTS values to detach the seeds (Htun et al. 2011, Ishikawa et al. 2010). To better characterise the shattering habits in these plants, we observed their abscission layer formation at the flowering stage (Fig. 1). In the parental lines, non-shattering cultivated rice (O. sativa cv. Nipponbare) formed no abscission layer, while wild rice (O. rufipogon acc. W630) formed a complete abscission layer across the pedicel region (Fig. 1A, 1B). In both introgression lines having the Nipponbare alleles at qSH1 or sh4, complete abscission layers across the pedicel, as in wild rice, were observed (Fig. 1C, 1D). Partial inhibition of the abscission layer was observed for the introgression line carrying both the Nipponbare alleles at qSH1 and sh4 (Fig. 1E), although the complete loss of the abscission layer was observed for Nipponbare having non-shattering alleles at qSH1 and sh4. Taken together, these results agree with previous observations of seed shattering and indicate that some other minor gene(s) are still involved in promoting abscission layer formation in wild rice.
Fig. 1.
Abscission layer formation of the introgression lines carrying non-functional alleles from O. sativa cv. Nipponbare at one or both major QTLs for seed shattering in the genetic background of wild rice O. rufipogon W630. (A) A Japonica rice cultivar O. sativa cv. Nipponbare. (B) Wild rice O. rufipogon W630. (C, D) Introgression lines carrying Nipponbare alleles at qSH1 (C) or sh4 (D). (E) The introgression line carrying Nipponbare alleles at both qSH1 and sh4 loci. Magnification of the boxed abscission layer on the left is shown on the right-hand side. Bar = 50 μm.
Distribution of seed-shattering degree in an F2 population produced between Nipponbare and the introgression line carrying both the Nipponbare alleles at qSH1 and sh4
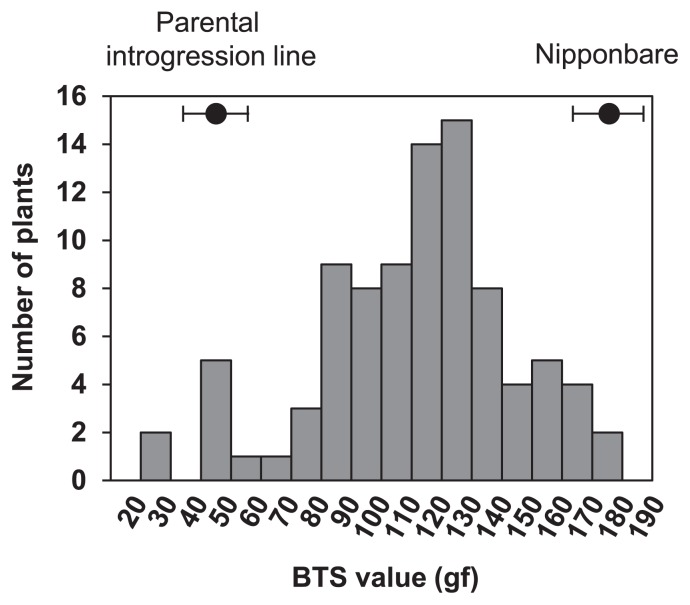
To identify novel gene(s) controlling seed shattering in wild rice, we crossed Nipponbare and the introgression line carrying both the Nipponbare alleles at qSH1 and sh4. Since these two loci are fixed with the Nipponbare alleles, variation of seed shattering-degree in the F2 population was not affected by them. QTL analysis of seed shattering was carried out to search for the additional loci. We used 90 F2 individuals, however, the heading date was expected to segregate depending on the genotypes at Hd5 and Hd6, as these two loci were previously shown to influence the flowering time of Nipponbare and O. rufipogon W630 (Thanh et al. 2010). To minimise the environmental effects on seed-shattering degree, these F2 lines were required to flower in a similar period. Therefore, we screened the F2 plants for late flowering by PCR genotyping at two loci, Hd5 and Hd6 (Table 1). We performed short-day treatment for plants with late-flowering genotypes and induced their flowering in the period from Aug. 9 to 16, 2011. We measured BTS values for all the plants at approximately 35 days after flowering. A continuous frequency distribution for BTS values was observed ranging from 28 to 180 gf, and a few plants showed transgressive segregation (Fig. 2). These observations indicate that seed shattering in wild rice is still regulated by QTLs apart from qSH1 and sh4.
Fig. 2.
Frequency distribution of breaking tensile strength (BTS) values for 90 F2 individuals between cultivated rice O. sativa cv. Nipponbare, and wild introgression line carrying Nipponbare alleles at both qSH1 and sh4 loci. BTS values for parental lines are shown with black dots with s.d. (n = 125).
Identification of additional QTL involved in the regulation of seed shattering in wild rice
QTL analysis for shattering degree was carried out with 90 F2 plants using 179 polymorphic markers in the linkage map previously constructed between O. sativa Nipponbare and O. rufipogon W630 (Thanh et al. 2010). A QTL was detected near RM16 on chr. 3 with an LOD score of 5.8. It explained 16.0% of the total phenotypic variance with the Nipponbare allele enhancing BTS value and the dominant effect was 4.9 (Table 2). We named this QTL, ‘qSH3’, a name previously given for the QTL close to RM16 (Onishi et al. 2007b). No additional loci were detected over threshold score (LOD = 3.8) in this QTL analysis.
Table 2.
Characteristics of the QTL for seed-shattering degree detected in this study
| Chr. | QTL region | Nearest marker | Sourcea | LOD | PVb (%) | Additive effect (gf) | Dominant effect (gf) |
|---|---|---|---|---|---|---|---|
| 3 | RM16-RM3513 | RM16 | Nipponbare | 5.8 | 16.0 | 19.0 | 4.9 |
Allele source increasing trait values.
Percentage of total phenotypic variance explained by the QTL.
Evaluation of qSH3 in the genetic background of wild rice
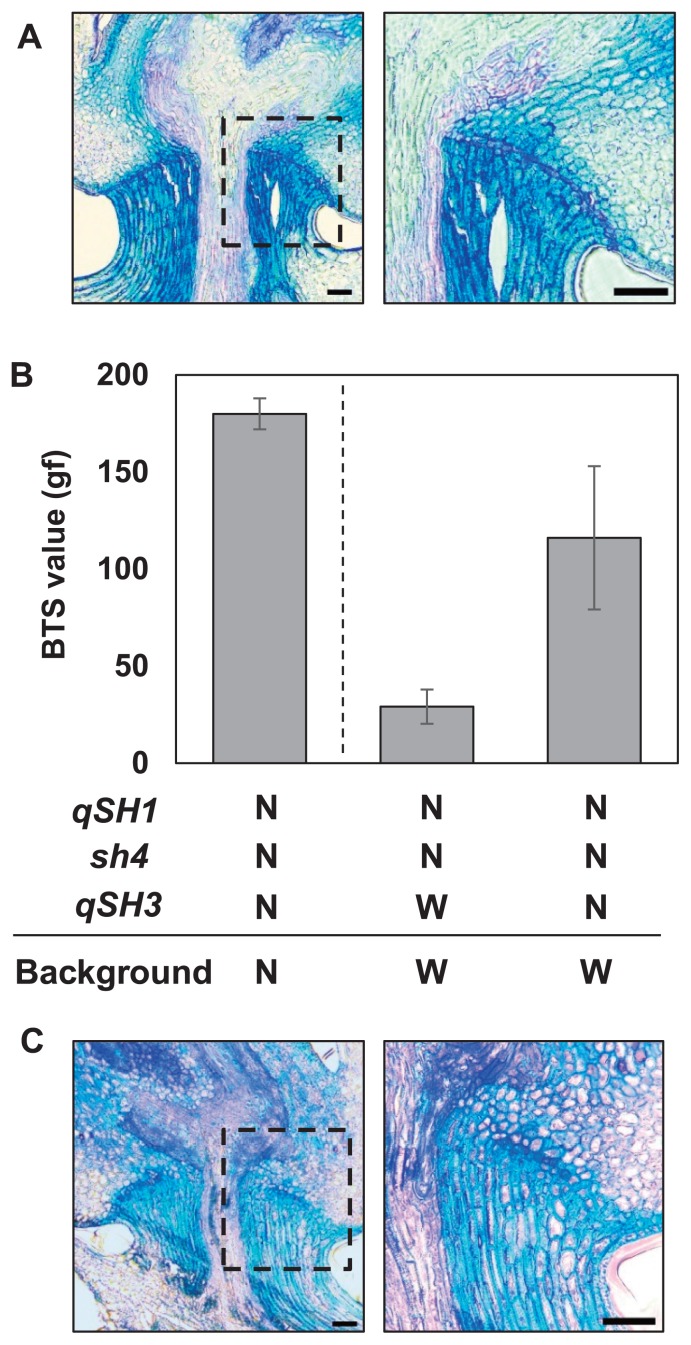
On chr. 3, QTLs for seed shattering were reported in several studies (Cai and Morishima 2000, Onishi et al. 2007a, 2007b). However, as mentioned in Onishi et al. (2007b), the effect of the allele at qSH3 from wild rice on seed shattering is small in the genetic background of cultivated rice. This could be due to the interactions among seed-shattering loci in rice. To evaluate the effect of the allele at qSH3 on seed shattering, we generated an introgression line with the Nipponbare allele at qSH3 in the genetic background of wild rice. This line showed strong seed-shattering behaviour with complete abscission layer formation, as observed in wild rice (Fig. 3A). This indicates that the Nipponbare allele at qSH3 alone did not contribute at all to abscission layer formation in the genetic background of wild rice. We next evaluated shattering degree of introgression lines carrying the Nipponbare alleles at two (qSH1 and sh4) and three loci (qSH1, sh4, and qSH3). A significant difference was observed for the BTS value between them, namely, the latter showed approximately 4 times higher value (116 ± 37 gf) than the former (29 ± 9 gf), although the latter is still lower than Nipponbare (180 ± 8 gf) (Fig. 3B). We further carried out a histological analysis with these introgression lines, and found that abscission layer formation is partially blocked outside the region of pedicel due to the additional Nipponbare allele at qSH3 (Fig. 3C). Inhibition of abscission layer formation outside the region of pedicel might contribute to the significant increase in the BTS value (Fig. 3B), suggesting an important role of the Nipponbare allele at qSH3. Taken together, our results indicate that the Nipponbare allele at qSH3 alone does not show significant effects on abscission layer formation in the genetic background of wild rice, however, its additive effect was observed in coordination with the Nipponbare non-functional alleles at qSH1 and sh4.
Fig. 3.
Evaluation of qSH3 locus in the genetic background of wild rice. (A) Abscission layer formation of introgression line carrying Nipponbare allele at qSH3. (B) Breaking tensile strength (BTS) values of introgression lines carrying Nipponbare alleles at two (qSH1 and sh4) and three loci (qSH1, sh4, and qSH3). Genotypes at qSH1, sh4, and qSH3 and background information are as follows: N, O. sativa cv. Nipponbare; W, O. rufipogon W630. BTS value for Nipponbare is shown as control on the left. Data are mean ± s.d. (n = 75). (C) Abscission layer formation for the introgression line carrying Nipponbare alleles at qSH1, sh4, and qSH3. Bar = 50 μm.
Discussion
Loci contributed to loss of seed shattering in cultivated rice
Wild rice has strong seed-shattering habits while cultivated rice has non-shattering habits. A wide range of natural variation in seed-shattering degree is observed for cultivated rice, suggesting that several genes or alleles might be involved in their quantitative regulation. Using these variations, seed shattering has been extensively studied and two major QTLs have been identified as the qSH1 and sh4 loci in rice (Konishi et al. 2006, Li et al. 2006b). In most cases, these loci were evaluated in cultivated rice (Ishikawa et al. 2010, Konishi et al. 2006, Onishi et al. 2007b). A qSH1 mutation was detected in the variation between O. sativa Indica and Japonica cultivars, and the causative mutation was proposed to be selected among rice cultivars. Since introgression or transformation of a functional allele at sh4 into cultivated rice causes the shattering phenotype and a non-functional mutation at sh4 is widely conserved in cultivated rice (Lin et al. 2007, Onishi et al. 2007b), it is widely accepted that the mutation at sh4 plays an important role in changing seed-shattering degree and was selected in rice domestication. However, we do not know how many genes or mutations were involved in the complete loss of seed shattering as observed in Japonica cultivars. To precisely study seed shattering in rice, we evaluated the effects of the Nipponbare non-functional alleles in the genetic background of wild rice. We did not find either of the Nipponbare alleles at qSH1 and sh4 to be sufficient in conferring loss of the abscission layer (Fig. 1C, 1D). This finding confirms that other loci are involved in promoting complete seed shattering in wild rice. In this study, qSH3 was found to be necessary for establishment of non-shattering in coordination with qSH1 and sh4. However, the BTS value of the introgression line carrying the Nipponbare alleles at qSH1, sh4, and qSH3 (116 ± 37 gf) is still lower than that of Nipponbare (180 ± 8 gf), implying these three loci are not enough to explain the complete non-shattering habit. It is likely that some additional minor loci might be involved in the complete loss of shattering as observed in Nipponbare. In our recent study, we found that the closed panicle trait regulated by OsLG1 was selected for in the early phase of rice domestication to increase gathering efficiency (Ishii et al. 2013). Interestingly, a closed panicle trait was also shown to promote self-pollination, suggesting that recessive mutations may have easily been fixed. Therefore, several mutations at seed-shattering loci might have accumulated under artificial selection during rice domestication.
We have successfully identified a novel QTL for seed shattering by fixing two major loci previously detected in the segregating population. A similar approach was previously tested for identifying two new QTLs regulating internode elongation in deepwater rice (Nagai et al. 2012). Therefore, an approach that involves fixing the major QTL(s) in the segregating population is very effective to explore and uncover the additional QTL(s) involved.
Role of qSH3 on abscission layer formation
Previously, interaction at three seed-shattering loci from wild rice was studied in the genetic background of cultivated rice, O. sativa Japonica cv. A58 (Onishi et al. 2007b). The results showed that qSH1 and qSH4 (same as sh4) played significant roles in seed shattering. In contrast, the effect of qSH3 on seed shattering was relatively small compared to those of qSH1 and qSH4. In the present study, we detected a strong effect of the Nipponbare allele at qSH3 on abscission layer formation with the Nipponbare alleles at qSH1 and sh4 in the genetic background of wild rice. The difference in the effects of qSH3 can be due to the unidentified mutations at seed-shattering loci potentially harboured in cultivated rice. Thus, evaluation of non-functional alleles for seed shattering in the genetic background of wild rice may reveal the role of qSH3 more clearly. Complete abscission layer formation was observed for introgression lines carrying any one of the Nipponbare alleles at qSH1, sh4, or qSH3 (Figs. 1C, 1D, 3A), suggesting that abscission layer formation in wild rice is redundantly regulated by these loci. We found that abscission layer formation was disturbed around vascular bundles in the introgression line carrying the Nipponbare alleles at qSH1 and sh4 (Fig. 1E). Moreover, the introgression line carrying the Nipponbare alleles at qSH1, sh4, and qSH3 showed additional loss of abscission cells outside of the pedicel, resulting in a large increase in the BTS value (Fig. 3B, 3C). These observations suggest that redundancy and gradient regulation may exist in the regulation of abscission layer formation in rice. Therefore, tissue-specific analysis of the abscission cells and expression domain of the genes controlling seed shattering will be an important scope of future study. It would be of interest to analyse genetic interaction between qSH3 and two major loci of qSH1 and sh4 in the genetic background of wild rice. These genetic studies will shed light on how non-shattering cultivated rice was established during rice domestication.
Candidate genes related to qSH3
In our QTL analysis, qSH3 was identified on chr. 3 between two SSR markers, RM16 and RM3513 (Table 2). We surveyed candidate genes involved in seed shattering or abscission layer formation reported around this genomic region. One candidate gene is OsSh1 (Os03g0650000), a rice homolog of the sorghum Shattering 1 (Sh1) gene, encoding YABBY transcription factor and controlling cell numbers at the abscission layer (Lin et al. 2012). Interestingly, the genomic region harbouring OsSh1 was shown to be under strong artificial selection (He et al. 2011, Xu et al. 2012), suggesting that OsSh1 may have been involved in the loss of seed shattering during rice domestication (Lin et al. 2012). However, studies have not shown any of the selected causative mutations of OsSh1, because the phenotypic effect of OsSh1 on seed shattering was examined only by using an artificially induced non-shattering mutant SR-5 (Lin et al. 2012). To explore the involvement of OsSh1 in this study, we compared nucleotide sequences of the OsSh1 gene between O. sativa Nipponbare and O. rufipogon W630. Among several mutations detected in the genic region, one SNP was found located on exon 1 of OsSh1. This SNP contributes to an amino acid change from leucine (L) in W630 to phenylalanine (F) in Nipponbare (Supplemental Fig. 1A). Recently, another gene, SpWRKY, located approximately 300 kb away from Sh1 in sorghum, was also shown to be involved in seed shattering (Tang et al. 2013). We also found the homologue of the rice WRKY gene (Os03g0657400), that is located approximately 450 kb away from the OsSh1 gene. Sequence analysis between O. sativa Nipponbare and O. rufipogon W630 identified two amino acid changes; one amino acid (glycine: G) deletion in Nipponbare and one amino acid change from alanine (A) in W630 to threonine (T) in Nipponbare (Supplemental Fig. 1B). We do not know whether these changes are associated with the seed-shattering degree between wild and cultivated rice. Further fine mapping analysis and transformation study are required to identify the causative mutation at qSH3. Moreover, the additional effect of qSH3 identified in this study may be important in the breeding process, used to fine-tune the shattering degree for efficient harvesting of cultivated rice in coordination with the two major QTLs at qSH1 and sh4.
Supplemental Table 1. The Nipponbare chromosomal segments covering qSH1, sh4, and qSH3 loci in the five introgression lines in the genetic background of wild rice
Supplemental Fig. 1. Amino acid sequences of OsSh1 and OsWRKY proteins. Alignment of amino acid sequences of OsSh1 (A) andOsWRKY (B) proteins between cultivated rice O. sativa cv. Nipponbare and wild rice O. rufipogon acc. W630.
Acknowledgements
The wild rice accession used in this study was kindly provided from the National Institute of Genetics (Japan) supported by the National BioResource Project (MEXT, Japan). This work was supported in part by a Grant-in-Aid (No. 23580006 to T.I. and No. 23580473 to R.I.) from Japanese Society for Promotion of Science.
Literature Cited
- Cai, H.W. and Morishima, H. (2000) Genomic regions affecting seed shattering and seed dormancy in rice. Theor. Appl. Genet. 100: 840–846 [Google Scholar]
- Fuller, D.Q. (2007) Contrasting patterns in crop domestication and domestication rates, recent archaeobotanical insights from the Old World. Ann. Bot. 100: 903–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan, J.R. (1975) Crops and Man. American Society of Agronomy, Madison [Google Scholar]
- He, Z., Zhai, W., Wen, H., Tang, T., Wang, Y., Lu, X., Greenberg, A.J., Hudson, R.R., Wu, C.I. and Shi, S. (2011) Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7: e1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun, T.M., Ishii, T. and Ishikawa, R. (2011) Temporal changes of seed shattering degree of substitution lines having non-shattering alleles from cultivated rice (Oryza sativa) in the genetic background of wild rice (O. rufipogon). J. Crop Res. 56: 39–44 [Google Scholar]
- Ishii, T., Numaguchi, K., Miura, K., Yoshida, K., Thanh, P.T., Htun, T.M., Yamasaki, M., Komeda, N., Matsumoto, T., Terauchi, R.et al. (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45: 462–465 [DOI] [PubMed] [Google Scholar]
- Ishikawa, R., Thanh, P.T., Nimura, N., Htun, T.M., Yamasaki, M. and Ishii, T. (2010) Allelic interaction at seed-shattering loci in the genetic backgrounds of wild and cultivated rice species. Genes Genet. Syst. 85: 265–271 [DOI] [PubMed] [Google Scholar]
- Ji, H., Kim, S.R., Kim, Y.H., Kim, H., Eun, M.Y., Jin, I.D., Cha, Y.S., Yun, D.W., Ahn, B.O., Lee, M.C.et al. (2010) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 61: 96–106 [DOI] [PubMed] [Google Scholar]
- Konishi, S., Izawa, T., Lin, S.Y., Ebana, K., Fukuta, Y., Sasaki, T. and Yano, M. (2006) An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Lee, S.J., Oh, C.S., Suh, J.P., McCouch, S.R. and Ahn, S.N. (2005) Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed. 124: 209–219 [Google Scholar]
- Li, C., Zhou, A. and Sang, T. (2006a) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 170: 185–194 [DOI] [PubMed] [Google Scholar]
- Li, C., Zhou, A. and Sang, T. (2006b) Rice domestication by reducing shattering. Science 311: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Lin, Z., Griffith, M.E., Li, X., Zhu, Z., Tan, L., Fu, Y., Zhang, W., Wang, X., Xie, D. and Sun, C. (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226: 11–20 [DOI] [PubMed] [Google Scholar]
- Lin, Z., Li, X., Shannon, L.M., Yeh, C.-T., Wang, M.L., Bai, G., Peng, Z., Li, J., Trick, H.N., Clemente, T.E.et al. (2012) Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, K., Kuroha, T., Ayano, M., Kurokawa, Y., Angeles-Shim, R.B., Shim, J.-H., Yasui, H., Yoshimura, A. and Ashikari, M. (2012) Two novel QTLs regulate internode elongation in deepwater rice during the early vegetative stage. Breed. Sci. 62: 178–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, H.I. (1988) Origin of Cultivated Rice. Elsevier, Tokyo, p. 254 [Google Scholar]
- Onishi, K., Horiuchi, Y., Ishigoh-Oka, N., Takagi, K., Ichikawa, N., Maruoka, M. and Sano, Y. (2007a) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 57: 7–16 [Google Scholar]
- Onishi, K., Takagi, K., Kontani, M., Tanaka, T. and Sano, Y. (2007b) Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50: 757–766 [DOI] [PubMed] [Google Scholar]
- Panaud, O., Chen, X. and McCouch, S.R. (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597–607 [DOI] [PubMed] [Google Scholar]
- Sakai, H., Lee, S.S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., Wakimoto, H., Yang, C.C., Iwamoto, M., Abe, T.et al. (2013) Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6 (1–11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H., Cuevas, H.E., Das, S., Sezen, U.U., Zhou, C., Guo, H., Goff, V.H., Ge, Z., Clemente, T.E. and Paterson, A.H. (2013) Seed shattering in a wild sorghum is conferred by a locus unrelated to domestication. Proc. Natl. Acad. Sci. USA 110: 15824–15829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh, P.T., Phan, P.D.T., Ishikawa, R. and Ishii, T. (2010) QTL analysis for flowering time using backcross population between Oryza sativa Nipponbare and O. rufipogon. Genes Genet. Syst. 85: 273– 279 [DOI] [PubMed] [Google Scholar]
- Thomson, M.J., Tai, T.H., McClung, A.M., Lai, X.H., Hinga, M.E., Lobos, K.B., Xu, Y., Martinez, C.P. and McCouch, S.R. (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493 [DOI] [PubMed] [Google Scholar]
- Wang, S., Basten, C.J. and Zeng, Z.B. (2007) Windows QTL cartographer 2.5. (http://statgen.ncsu.edu/qtlcart/WQTLCart.htmZ). Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Xiong, L.Z., Liu, K.D., Dai, X.K., Xu, C.G. and Zhang, Q. (1999) Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251 [Google Scholar]
- Xu, X., Liu, X., Ge, S., Jensen, J.D., Hu, F., Li, X., Dong, Y., Gutenkunst, R.N., Fang, L., Huang, L.et al. (2012) Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111 [DOI] [PubMed] [Google Scholar]
- Zhang, L.B., Zhu, Q., Wu, Z.Q., Ross-Ibarra, J., Gaut, B.S., Ge, S. and Sang, T. (2009) Selection on grain shattering genes and rates of rice domestication. New Phytol. 184: 708–720 [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Lu, D., Li, C., Luo, J., Zhu, B.F., Zhu, J., Shangguan, Y., Wang, Z., Sang, T. and Zhou, B. (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor SHATTERING ABORTION1. Plant Cell 24: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. The Nipponbare chromosomal segments covering qSH1, sh4, and qSH3 loci in the five introgression lines in the genetic background of wild rice
Supplemental Fig. 1. Amino acid sequences of OsSh1 and OsWRKY proteins. Alignment of amino acid sequences of OsSh1 (A) andOsWRKY (B) proteins between cultivated rice O. sativa cv. Nipponbare and wild rice O. rufipogon acc. W630.