Abstract
The rapid development of high-throughput technologies and computational frameworks enables the examination of biological systems in unprecedented detail. The ability to study biological phenomena at omics levels in turn is expected to lead to significant advances in personalized and precision medicine. Patients can be treated according to their own molecular characteristics. Individual omes as well as the integrated profiles of multiple omes, such as the genome, the epigenome, the transcriptome, the proteome, the metabolome, the antibodyome, and other omics information are expected to be valuable for health monitoring, preventative measures, and precision medicine. Moreover, omics technologies have the potential to transform medicine from traditional symptom-oriented diagnosis and treatment of diseases towards disease prevention and early diagnostics. We discuss here the advances and challenges in systems biology-powered personalized medicine at its current stage, as well as a prospective view of future personalized health care at the end of this review.
Personalized or precision medicine is expected to become the paradigm of future health care, owing to the substantial improvement of high-throughput technologies and systems approaches in the past two decades1, 2. Conventional symptoms-oriented disease diagnosis and treatment has a number of significant limitations: for example, it focuses on only late/terminal symptoms and generally neglects preclinical pathophenotypes or risk factors; it generally disregards the underlying mechanisms of the symptoms; the disease descriptions are often quite broad so that they may actually include multiple diseases with shared symptoms; the reductionist approach to identify therapeutic targets in traditional medicine may over-simplify the complex nature of most diseases3. Advances in the ability to perform large-scale genetic and molecular profiling are expected overcome these limitations by addressing individualized differences in diagnosis and treatment in unprecedented detail.
The rapid development of high-throughput technologies also drives modern biological and medical researches from traditional hypothesis-driven designs toward data-driven studies. Modern high-throughput technologies, such as high-throughout DNA sequencing and mass spectrometry, have enabled the facile monitoring of thousands of molecules simultaneously instead of just a few components that have been analysed in traditional research, thus generating a huge amount of data to document the real-time molecular details of a given biological system. Ultimately, when enough knowledge is gained, these molecular signatures, as well as the biological networks they form, may be associated with the physiological state/phenotype of the biological system at the very moment when the sample is taken.
Future personalized health care is expected to benefit from the combined personal omics data, which should include genomic information as well as longitudinal documentation of all possible molecular components. This combined information not only determines the genetic susceptibility of the person, but also monitors his/her real-time physiological states, as our integrated Personal Omics Profile (iPOP) study exemplified4. In this review we will cover recent advances in systems biology and personalized medicine. We will also discuss limitations and concerns in applying omics approaches to individualized, precision health care.
GENOMICS IN DISEASE-ORIENTED MEDICINE
The revolution of omics profiling technologies significantly benefited disease-oriented studies and health care, especially in disease mechanism elucidation, molecular diagnosis and personalized treatment. These new technologies greatly facilitated the development of genomics, transcriptomics, proteomics and metabolomics, which have become powerful tools for disease studies. Today, molecular disease analyses using large-scale approaches is pursued by an increasing number of physicians and pathologists5, 6.
Initially, genome-wide association studies (http://gwas.nih.gov/) were launched in search of association of common genetic variants to certain phenotypes of interest, which typically assayed more than 500,000 Single Nucleotide Polymorphisms (SNPs) and/or Copy Number Variations (CNVs) with DNA microarrays in thousands to hundred thousands of participants7. To date, 1,355 publications are listed in the NHGRI (National Human Genome Research Institute) GWAS Catalog reporting the association of 7,226 SNPs with 710 complex traits7. The studied complex traits vary vastly, from cancers (e.g. prostate cancer and breast cancer) and complex diseases (e.g. Type 1 and Type 2 Diabetes, Crohn’s Disease) to common traits (e.g. height and body mass index). These findings greatly broadened our knowledge on disease loci, and can potentially benefit disease risk prediction and drug treatments (as discussed in the section INTEGRATIVE OMICS IN PREVENTATIVE MEDICINE). Although powerful, GWAS studies have proven difficult for most complex diseases as typically a large number of loci are identified, each contributing to a small fraction of the genetic risk. These studies have many limitations including the small fraction of the genome that is analysed, and failure to account for gene-gene interactions, epistasis and environmental factors8.
Whole genome sequencing (WGS) and whole exome sequencing (WES) have become more and more affordable for genomic studies and are rapidly replacing DNA microarrays. Single-base analysis of a genome/exome is achieved, which allows scientists to investigate the genetic basis of health and disease in unprecedented detail. Assigning variants to paternal and maternal chromosomes i.e. “phasing” can be obtained through the analysis of families9 or other methods1, 10, 11. With the generation of massive amount of whole genome and exome data from diseased and healthy populations, understanding of both human population variation and genetic diseases, especially complex diseases, has been brought to a new level1, 12.
One field that significantly benefited from WGS technologies is cancer-related research. A large number of cancer genomes have been sequenced through individual or collaborative efforts, such as the International Cancer Genome Consortium (http://www.icgc.org/) and the Cancer Genome Atlas (http://cancergenome.nih.gov/). The DNA from many types of cancer have been sequenced, including breast cancer13-15, chronic lymphocytic leukaemia16, hepatocellular carcinoma17, pediatric glioblastoma13, melanoma18, ovarian cancer19, small-cell lung cancer20, and Sonic-Hedgehog medulloblastoma21, and databases are established, such as the Cancer Cell Line Encyclopedia22. In addition, single-cell level cancer genome has also been investigated by WES for clear cell renal cell carcinoma23 and JAK2-negative myeloproliferative neoplasm24. Somatic mutations and subtyping molecular markers were identified from these genomes. These different studies have revealed that nearly every tumour is different with distinct types of potential “driver” mutations. Importantly, cancer genome sequencing often reveals potential targets that may suggest precision cancer treatment for the specific patients. As an example, a novel spontaneous germline mutation in the p53 gene was identified by WGS in a female patient, which accounted for the 3 types of cancers she developed in merely 5 years25. An attempt has been made recently to treat a female patient with T Cell Lymphoma based on the target gene, CTLA4, identified by whole genome sequencing26. The patient’s cancer was suppressed for two months with the anti-CTLA4 drug ipilimumab, although she died of recurrence soon after.
Whole genome and exome sequencing can also facilitate the identification of possible causal genes for hereditary genetic diseases, and is increasingly used for trying to understand the basis of these “mystery diseases” once obvious candidates are ruled out. In one successful example, whole genome sequencing of a fraternal twin pair with dopa (3,4-dihydroxyphenylalanine)-responsive dystonia helped the identification of one pair of personalized compound heterozygous mutations in the gene SPR, which accounted for the disease in both individuals27. Importantly, based on the genome information the authors supplemented the L-dopa therapy with 5-hydroxytryptophan (SPR-dependent serotonin precursor) and significantly improved the health of both patients. In another example, Roach et al. sequenced the whole genomes of a family quartet and identified rare mutations in the genes DHODH and DNAH5 responsible for the two recessive disorders in both children – Miller syndrome and primary ciliary dyskinesia28.
Pharmacogenomics is another important application of genomic sequencing. It is known that the same drug may have different effect on different individuals due to their personal genomic background and living habits8, 29. Genetic information can be used to assign drug doses and reduce side effects. For example, genetic variants are known to affect patients’ response to antipsychotic drugs30. Based on pharmacogenomic trials, genetic tests for four drugs are required by the FDA (US Food and Drug Administration) before the administration of these drugs to patients, including the anti-cancer drugs cetuximab, trastuzumab, and dasatinib, and the anti-HIV drug maraviroc, and more are recommended such as the anticoagulant drug Warfarin and the anti-HIV drug Abacavir8.
OTHER OMICS TECHNOLOGIES AND MEDICINE
Other omics technologies are also likely to impact medicine. High throughput sequencing technologies have enabled whole transcriptome (cDNA) sequencing, or abbreviated as RNA-Seq31. RNA-Seq has become a powerful tool for disease-related studies, as it has great accuracy and sensitivity relative to microarray technology and it can also detect splicing isoforms32. As RNA profiles reflect actual gene activity, it is closer to the real phenotype compared to genomic sequence. With RNA-Seq, Shah et al. discovered varied clonal preference and allelic abundance in 104 cases of primary triple-negative breast cancers, and observed that ~36% of the genomic mutations were actually expressed33. Combining such information with genomic information may be valuable in treatment of cancer and other diseases. Moreover, RNA-Seq also captures more complex aspects of the transcriptome, such as splicing isoforms34 and editing events35, which are generally overlooked by hybridization-based methods. Splicing variants have now been associated with several distinct types of cancer and cancer prognosis36-40.
Although proteins have long been deemed as the executors of most biological functions, clinical proteomics is still a relatively young field due to technological limitations to profile the complexity of the proteome with high sensitivity and accuracy. Since the development of new soft desorption methods that enabled the analysis of biological macromolecules with mass spectrometry, proteomics advanced significantly in the past decade41, 42. With current mass spectrometry technology, one can now quantify thousands of proteins in a single sample. For example, we were able to reliably detect 6,280 proteins in the human Peripheral Blood Mononuclear Cell proteome4. Mass spectrometry also allows the detection of expressed mutations, allele-specific sequences and editing events in the human proteome4, 43, as well as profiling of the phosphoproteome44. Also of note is the MALDI-TOF (matrix-assisted laser desorption/ionization-time of flight) mass spectrometry-based imaging technology (MALDI-MSI) developed by Cornett et al., which allows the spatial proteome profiling in defined 2-dimentional laser-shot areas using tissue sections45. Using MALDI-MSI, Kang et al. identified immunoglobulin heavy constant alpha 2 as a novel potential marker for breast cancer metastasis46.
The field of metabolomics has also advanced significantly with the improvement of mass spectrometry. Both hydrophilic and hydrophobic metabolites can be profiled in specific samples4, 47. As the metabolome reflects the real-time energy status as well as metabolism of the living organism, it is expected that certain metabolome profiles may associate with different diseases48. Therefore, metabolomic profiles become an important aspect for personalized medicine49, 50. Jamshidi et al. profiled the metabolome of a female patient with Hereditary Hemorrhagic Telangiectasia (HHT) along with 4 healthy controls, and identified differences which highlighted the nitric oxide synthase pathway51. The authors then treated the patient with bevacizumab and shifted her metabolomic profile towards those of the healthy controls and improved the patient’s health. In addition, branched-chain amino acids such as isoleucine have been associated with Type 2 diabetes and may ultimately prove to be valuable biomarkers52. Finally, since some metabolites bind and direct regulate the activity of other biomolecules (e.g. kinases)53, there is significant potential to modulate cellular pathways using diet and metabolic analogs that serve as agonist or antagonist of protein function.
INTEGRATIVE OMICS IN PREVENTATIVE MEDICINE
The concept of personalized medicine emphasizes not only personalized diagnosis and treatment, but also personalized disease susceptibility assessment, health monitoring and preventative medicine. Because disease is easier to manage prior to it onset or when a disease is at its early stages, risk assessment and early detection will be transformative in personalized medicine. Systems biology has the potential to capture real-time molecular phenotypes of a biological system, which enables the detection of subtle network perturbations preluding the actual development of clinical symptoms.
Disease susceptibility and drug response can be assessed with a person’s genomic information8. This information may serve as a guideline for monitoring the health of a particular patient to achieve personalized health care, as showcased by Ashley et al.54. Whole genome sequence revealed variants for both high-penetrance Mendelian disorders, such as HTT (Huntington’s Disease55) and PAH (Phenylketonuria56), as well as common, complex diseases, such as the disease-associated genetic variants reported in GWAS studies57. Disease risks can be evaluated for a given person and an increase or decrease in disease risk compared with the population risk (of the same ethnicity, age and gender) can be estimated (Figure 1). In the study of Ashley et al., the genome of a patient was analysed and increased post-test probability risks for myocardial infarction and coronary artery disease were estimated54. Their estimation matched the fact that the patient, although generally healthy, had a family history of vascular disease as well as early sudden death58. Genetic variants associated with heart-related morbidities as well as drug response were identified in the patient’s genome, the information of which, as the authors stated, may direct the future health care for this particular patient. Similarly, Dewey et al. further extended this work by analysing a family quartet using a major allele reference sequence, and identified high-risk genes for familial thrombophilia, obesity and psoriasis59.
Figure 1.
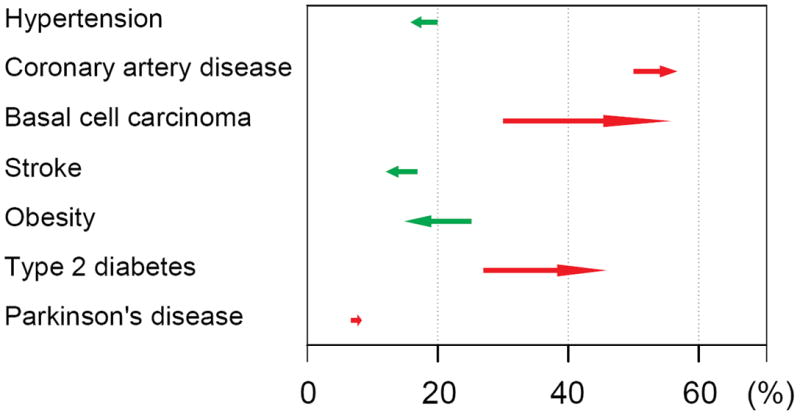
Example personalized RiskGraph. Each horizontal line symbolizes genetic risk of one disease tested for a specific individual. The tail of each arrow shows the pretest probability of a disease in a population of certain ethnicity, age and gender. The frond end of each arrow displays the posttest probability with consideration of the person’s genomic information. Red arrow, increased risk; green arrow, decreased risk.
To further explore variation and power of the full human genome, projects and databases (such as the Personal Genome Project60) are being launched to help advance this field. However, genomic information alone usually is not adequate to predict disease onset, and other factors such as environment are expected to play a critical role in this process61, 62. The predictive capability of whole genome sequence was assessed by Roberts et al. through modelling 24 disease risks in monozygotic twins63. For each disease, the authors modelled the genotype distribution in the twin population according to the observed concordance/discordance, and discovered that for most individuals and most diseases, the relative risk would be tested negative compared to the population, and in the best-case scenario, only one disease or more could be forewarned for any individual. The results of Roberts et al. are not surprising, as disease manifestation is probabilistic and not deterministic. Nonetheless, whole genome information by itself is expected to have partial value in disease prediction for complex diseases. In addition, from a systems point of view, peripheral components of the biological network would be more likely to contribute to complex diseases, as perturbation of the main nodes, which are usually essential genes, would be lethal64. Therefore it is more difficult to identify the exact contributors of complex diseases. Moreover, as stated above, non-genomic factors may also exist and further complicate the situation. As an example of this, multiple sclerosis is known to have genetic components however, Baranzini et al. failed to identify genomic, epigenomic or transcriptomic contributors in discordant monozygotic twins, which may indicate the existence of other factors, such as the environment65.
Current technologies, especially high-throughput sequencing and mass spectrometry, enable the monitoring of at least 105 molecular components, including DNA, RNA, protein and metabolites in the human body. Therefore it is now feasible to identify the profiles of these components that correlate with various physiological states of the body, and profile alterations as a result of physiological state changes and diseases. Compared with genomic sequences alone, the profiles of transcriptome, proteome and metabolome are closer indicators to the real-time phenotype, therefore collecting these omics information in a longitudinal manner would allow monitoring an individual’s physiological state. To test this concept, we implemented a study by following a generally healthy participant for 14 (now 28) months with integrated Personal Omics Profile (iPOP) analysis, incorporating information of the participant’s genome with longitudinal data from the person’s transcriptome, proteome, metabolome and autoantibodyome4. As blood constantly circulates the human body and exchanges biological matters with local tissues and is presently analysed in medical tests, we chose to monitor the participant’s physiological states by profiling the blood components (PBMCs, serum and plasma) with iPOP analysis. The genome of this individual was sequenced with 2 WGS (Illumina and Complete Genomics) and 3 WES (Agilent, Roche Nimblegen and Illumina) platforms to achieve high accuracy, which was further analysed for disease risk and drug efficiency. The identified elevated risks included coronary artery disease, basal-cell carcinoma, hypertriglyceridemia and Type 2 Diabetes (T2D), and the participant was estimated to have favourable response to rosiglitazone and metformin, both are anti-diabetic medications. Although the participant has a known family history for some of the high-risk diseases (but not T2D), he was free from most of them (except for hypertriglyceridemia, for which he used medication) and had a normal Body Mass Index at the start of our study. Nonetheless, these elevated disease risks served as a guideline to monitor his personal health with iPOP analysis. We profiled the transcriptome, proteome and metabolome from 20 time points in the 14 months, and monitored molecular profile changes for physiological state change events during our study, including 2 viral infections. The subject also acquired T2D during the study, immediately after one of the viral (respiratory syncytial virus) infection. Two types of changes were observed from the iPOP data: the autocorrelated trends that reflect chronic changes, and the spikes which include significantly up/down-regulated genes and pathways especially at the onset of each event. With our iPOP approach, we acquired a comprehensive picture of detailed molecular differences between different physiological states, as well as during disease onset. In particular, interesting changes in glucose and insulin signalling pathways were observed during the onset of T2D. We also obtained other important information from our omics data, such as dynamic changes in allele-specific expression and RNA-editing events, as well as personalized autoantibody profiles. Overall, this study revealed an important application of the use of genomics and other omics profiling for personalized disease risk estimation and precision medicine, as we discovered the increased T2D risk, monitored its early onset, and helped the participant effectively control and eventually reverse the phenotype by proactive interventions (diet change and physical exercise).
Another important feature of our study is that samples are collected in a longitudinal fashion so that aberrant/disease states can be compared to healthy states of the same individual. Another advantage of our iPOP approach is its modularity, as other omics and quantifiable information can also be included in the iPOP profile, which can be readily tailored to monitor any biological or pathological event of interest (Figure 2). Examples of other information are: epigenome66, gut microbiome67, microRNA profiles68 and immune receptor repertoire69. Moreover, quantifiable behavioural parameters such as nutrition, exercise, stress control and sleep may also be added to the profile70.
Figure 2.
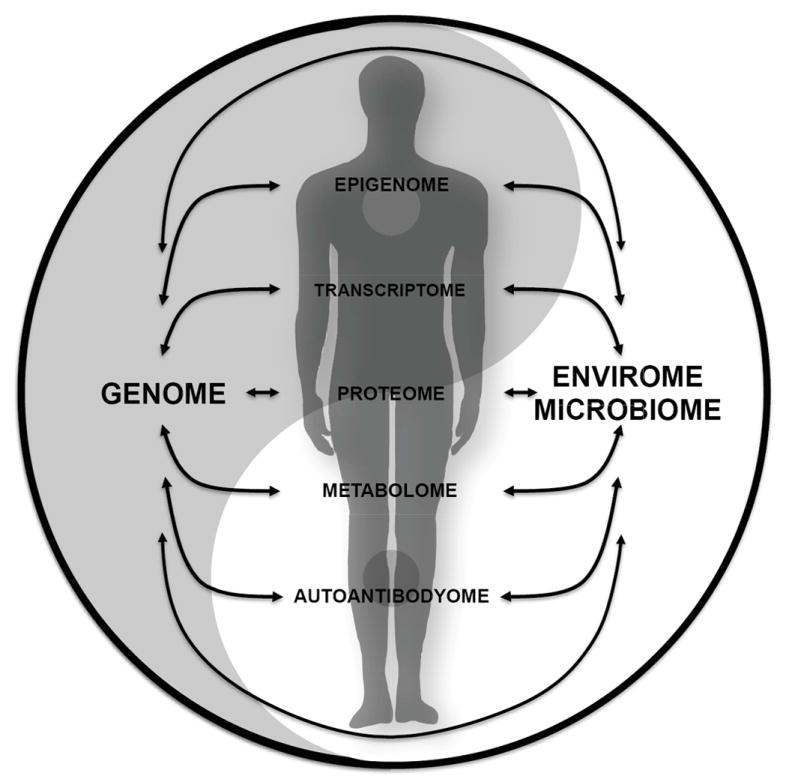
The concept of integrative Personal Omics Profile (iPOP). Physiological status of the body can be reflected by the integrated profiles of different omic profiles, as well as the interactions amongst them.
THE IMPORTANCE OF DATA MINING AND RE-MINING
One important aspect of systems biology is data mining. Data management and access can become a daunting task given the tremendous amount of data generated with current high-throughput technologies, and the data size is constantly increasing with time71. Challenges exist computationally in each step to handle, process and annotate high-throughput data, integrate data from different sources and platforms, and pursue clinical interpretation of the data72. These steps can be quite computationally intensive and require significant computational hardware; for example, to map short reads to achieve 30 X coverage of the human genome, 13 CPU days is typically required72 although these times are rapidly decreasing. Moreover, as biological systems act more than just the sum of its individual parts, knowledge from multiple levels (such as epistasis, interaction, localization and activation status) should be considered to capture the underlying highly organized networks for functional annotations73. Ultimately it will be important to have a comprehensive database that contains Electronic Health records (including treatment information), genome sequences with variant calls and as much molecular information as possible. In principle with appropriate algorithms such a database could be mined by physicians to make data-driven medical decisions.
Currently many high-throughput datasets of similar types (for example, expression and genome-wide association data collected from different populations of the same disease) were created as smaller, separate studies. Thus combining these publicly available datasets bioinformatically may provide more statistical power and lead to a clearer conclusion that could not be achieved in the individual studies. The work by Roberts et al. mentioned above serves as one example63. In order to test the capacity of whole genome information, the authors combined monozygotic twin pair data from a total of 5 sources in 13 publications to obtain a much large dataset for their test. Similarly, Butte and colleagues combined the results of 130 functional microarray experiments for T2D and remined the data for repeatedly appeared candidate genes74. They identified CD44 as the top candidate gene associated with T2D. In a related effort, by analysing curated data of 2510 individuals from 74 populations, the group led by Butte also discovered that T2D risk alleles were unevenly distributed across different human populations, with the risk higher in African and lower in Asian populations75.
CONCERNS AND LIMITATIONS
Personalized health monitoring and precision medicine is just accelerating at a rapid pace due to development of systems biology. As noted above, multiple efforts in both technology development and biological application have occurred, and an increasing number of researchers and physicians alike are sharing this vision. Hood et al. termed this approach as “P4 Medicine” for predictive, preventive, personalized and participatory medicine12.
Nevertheless, many concerns also exist, and guidelines on translational omics research have been recommended by the Institute of Medicine76. Khoury et al. suggested “a fifth P”, i.e. the population perspective be added to personalized medicine77 and population validation of systems results with strong evidence should be achieved before its clinical application. Many disease-associated genetic variants discovered in GWAS still need to be functionally validated78. In addition, Khoury et al. raised concerns that restricted health care resources might be wasted if unneeded disease screening/subclassification with systems approaches were conducted rather than lowering health care costs. However, with the rapid drop in technology costs and carefully designed pilot studies, the optimal screening frequencies/levels of subclassification necessary for precision medicine could be determined and costs maintained at affordable levels. It is worth noting that generating personalized omics data with appropriate interpretation can greatly benefit our understanding of physiological events for health and disease, and precision health care as we gain more knowledge in this field. In addition to personalized diagnosis and treatment, the future of precision medicine with omics approaches should emphasize personalized health monitoring, molecular symptom early detection and preventative medicine, a paradigm shift from traditional health care.
As the human body is a highly organized, complex system with multiple organs and tissues, it is important to select the correct sample type for understanding a specific biological problem. However, as many sample types are unavailable (e.g. brain tissue) or not regularly accessible (e.g. biopsy samples from internal organs) from living individuals, our scope for personalized health monitoring is thus restricted. Therefore systems biology results, especially iPOP results, should not be over-interpreted. Although iPOP data from blood components may indicate changes in the other parts of the human body, the actual profiles for the tissue of interest might be underrepresented in blood or delayed in phase.
It is still not clear who is to develop and deliver personalized treatments for personalized medicine if they are not available as conventional medication. The cost for developing personalized drugs may become prohibitive to accurately address personal specificity, and may face other difficulties such as Food and Drug Administration approval. However, advances in high-throughput drug discovery will help accelerate this field.
In addition, personalized medicine using omics approaches relies heavily on technology development for biological research. This includes advances in both research instrumentation and computational framework. For example, it is still not possible to accurately determine the entire sequence of a genome due to limitations of current WGS/WES methods79, 80, even after computational improvement of signal-to-noise ratio81, 82. A low sequencing error rate was claimed by both the Illumina HiSeq (for 2 × 100 bp reads, more than 80% of the bases have a quality score above Q30, or 99.9% accuracy, http://www.illumina.com/documents//products/datasheets/datasheet_hiseq_systems.pdf) and the Complete Genomics platform (1 × 10-5 at the time of our study80 and 2 × 10-6 as of October 8th, 2012, www.completegenomics.com); however, per variant error rate is still high (15.50% and 9.08% for Illumina and Complete Genomics respectively with no filter, and 1.01% and 1.12% post multiple filters) as reported by Reumers et al.81, which agreed with our observation that only 88.1% of the SNP calls overlapped when the same genome was sequenced with the two platforms80. Thus possible disease-associated variants in these regions would be overlooked or misinterpreted. Another issue lies in storage and processing of the omics data, as petabytes of data can easily be generated for a small iPOP study of 200 participants and demanding computing resources will be needed for data analysis. Therefore, interdisciplinary efforts from biologists, computer scientists and hardware engineers should be organized to ensure the continued improvement of this field.
CONCLUSION
The era of personalized precision medicine is about to emerge. The steady improvement of high-throughput technologies greatly facilitates this process by enabling omics profiling such as whole genome, epigenome, transcriptome, proteome and metabolome, which convey detailed information of the human body. Integrated profiles of these omes should reflect the physiological status of the host at the time the samples are collected. Personalized omics approach catalyzes precision medicine at two levels: for diseases and biological processes whose mechanisms are still unclear, omics approach will facilitate researches that would greatly advance our understanding; and when the mechanisms are clarified, individualized health care can be provided through health monitoring, preventative medicine, and personalized treatment. This would be especially helpful for complex diseases such as autism83 and Alzheimer’s disease84, where multiple factors are responsible for the phenotypes. Furthermore, omics approach also facilitates the development of other less-stressed but important health-related fields, such as nutritional systems biology, which studies personalized diet and its relationship to health in systems point of view85. With the rapid decrease in the cost of omics profiling, we anticipate an increased number of personalized medicine applications in many aspects of health care besides our proof-of-principle study. This will significantly improve the health and cut down health care costs of the general public. Scientists, governments, pharmaceutical companies and patients should work closely together to ensure the success of this transformation86.
Acknowledgments
This work is supported by funding from the Stanford University Department of Genetics and the National Institutes of Health. We thank Drs. George I. Mias and Hogune Im for their help in proof-reading the manuscript and the insightful discussions.
Biography
Michael Snyder M.S. serves as founder and consultant for Personalis, a member of the scientific advisory board of GenapSys, and a consultant for Illumina.
Contributor Information
Rui Chen, Department of Genetics, Stanford University School of Medicine, 300 Pasteur Drive, Stanford, CA 94305-5120, USA.
Michael Snyder, Department of Genetics, Stanford University School of Medicine, 300 Pasteur Drive, Stanford, CA 94305-5120, USA.
References
- 1.Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes & development. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Molecular systems biology. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loscalzo J, Barabasi AL. Systems biology and the future of medicine. Wiley interdisciplinary reviews Systems biology and medicine. 2011;3:619–627. doi: 10.1002/wsbm.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moch H, Blank PR, Dietel M, Elmberger G, Kerr KM, Palacios J, Penault-Llorca F, Rossi G, Szucs TD. Personalized cancer medicine and the future of pathology. Virchows Archiv : an international journal of pathology. 2012;460:3–8. doi: 10.1007/s00428-011-1179-6. [DOI] [PubMed] [Google Scholar]
- 6.Berman DM, Bosenberg MW, Orwant RL, Thurberg BL, Draetta GF, Fletcher CD, Loda M. Investigative pathology: leading the post-genomic revolution. Laboratory investigation; a journal of technical methods and pathology. 2012;92:4–8. doi: 10.1038/labinvest.2011.147. [DOI] [PubMed] [Google Scholar]
- 7.Hindorff LA, MacArthur J, Wise A, Junkins HA, Hall PN, Klemm AK, Manolio TA. [August 30th, 2012];A Catalog of Published Genome-Wide Association Studies. Available at: http://www.genome.gov/gwastudies.
- 8.Antman E, Weiss S, Loscalzo J. Systems pharmacology, pharmacogenetics, and clinical trial design in network medicine. Wiley interdisciplinary reviews Systems biology and medicine. 2012;4:367–383. doi: 10.1002/wsbm.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roach JC, Glusman G, Hubley R, Montsaroff SZ, Holloway AK, Mauldin DE, Srivastava D, Garg V, Pollard KS, Galas DJ, et al. Chromosomal haplotypes by genetic phasing of human families. American journal of human genetics. 2011;89:382–397. doi: 10.1016/j.ajhg.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters BA, Kermani BG, Sparks AB, Alferov O, Hong P, Alexeev A, Jiang Y, Dahl F, Tang YT, Haas J, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nature biotechnology. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. New biotechnology. 2012 doi: 10.1016/j.nbt.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 14.Natrajan R, Mackay A, Lambros MB, Weigelt B, Wilkerson PM, Manie E, Grigoriadis A, A’Hern R, van der Groep P, Kozarewa I, et al. A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers. The Journal of pathology. 2012;227:29–41. doi: 10.1002/path.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yost SE, Smith EN, Schwab RB, Bao L, Jung H, Wang X, Voest E, Pierce JP, Messer K, Parker BA, et al. Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic acids research. 2012 doi: 10.1093/nar/gks299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, et al. High-resolution characterization of a hepatocellular carcinoma genome. Nature genetics. 2011;43:464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 18.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Link DC, Schuettpelz LG, Shen D, Wang J, Walter MJ, Kulkarni S, Payton JE, Ivanovich J, Goodfellow PJ, Le Beau M, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA : the journal of the American Medical Association. 2011;305:1568–1576. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conventional Cancer Therapy and Whole Genome Sequencing. Available at: http://www.nytimes.com/interactive/2012/07/08/health/conventional-cancer-therapy-and-whole-genome-sequencing.html.
- 27.Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, et al. Whole-genome sequencing for optimized patient management. Science translational medicine. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Q, Lu AY. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacological reviews. 2011;63:437–459. doi: 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S, Jain S, Brahmachari SK, Kukreti R. Pharmacogenomics: a path to predictive medicine for schizophrenia. Pharmacogenomics. 2006;7:31–47. doi: 10.2217/14622416.7.1.31. [DOI] [PubMed] [Google Scholar]
- 31.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delahaye NF, Rusakiewicz S, Martins I, Menard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro M, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nature medicine. 2011;17:700–707. doi: 10.1038/nm.2366. [DOI] [PubMed] [Google Scholar]
- 38.Rajan P, Elliott DJ, Robson CN, Leung HY. Alternative splicing and biological heterogeneity in prostate cancer. Nature reviews Urology. 2009;6:454–460. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 39.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6004–6009. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 42.Wu L, Han DK. Overcoming the dynamic range problem in mass spectrometry-based shotgun proteomics. Expert review of proteomics. 2006;3:611–619. doi: 10.1586/14789450.3.6.611. [DOI] [PubMed] [Google Scholar]
- 43.Ruppen-Canas I, Lopez-Casas PP, Garcia F, Ximenez-Embun P, Munoz M, Morelli MP, Real FX, Serna A, Hidalgo M, Ashman K. An improved quantitative mass spectrometry analysis of tumor specific mutant proteins at high sensitivity. Proteomics. 2012;12:1319–1327. doi: 10.1002/pmic.201100611. [DOI] [PubMed] [Google Scholar]
- 44.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature protocols. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, Caprioli RM. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Molecular & cellular proteomics : MCP. 2006;5:1975–1983. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Kang S, Maeng H, Kim BG, Qing GM, Choi YP, Kim HY, Kim PS, Kim Y, Kim YH, Choi YD, et al. In situ Identification and Localization of IGHA2 in the Breast Tumor Microenvironment by Mass Spectrometry. Journal of proteome research. 2012 doi: 10.1021/pr3003672. [DOI] [PubMed] [Google Scholar]
- 47.Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273–295. doi: 10.1007/978-1-60327-811-9_20. [DOI] [PubMed] [Google Scholar]
- 48.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 49.van der Greef J, Hankemeier T, McBurney RN. Metabolomics-based systems biology and personalized medicine: moving towards n = 1 clinical trials? Pharmacogenomics. 2006;7:1087–1094. doi: 10.2217/14622416.7.7.1087. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson JK, Wilson ID, Lindon JC. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics. 2011;12:103–111. doi: 10.2217/pgs.10.157. [DOI] [PubMed] [Google Scholar]
- 51.Jamshidi N, Miller FJ, Mandel J, Evans T, Kuo MD. Individualized therapy of HHT driven by network analysis of metabolomic profiles. BMC systems biology. 2011;5:200. doi: 10.1186/1752-0509-5-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell metabolism. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 56.Kaufman S. A model of human phenylalanine metabolism in normal subjects and in phenylketonuric patients. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3160–3164. doi: 10.1073/pnas.96.6.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Human molecular genetics. 2011;20:R182–188. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nature biotechnology. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dewey FE, Chen R, Cordero SP, Ormond KE, Caleshu C, Karczewski KJ, Whirl-Carrillo M, Wheeler MT, Dudley JT, Byrnes JK, et al. Phased whole-genome genetic risk in a family quartet using a major allele reference sequence. PLoS genetics. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ball MP, Thakuria JV, Zaranek AW, Clegg T, Rosenbaum AM, Wu X, Angrist M, Bhak J, Bobe J, Callow MJ, et al. A public resource facilitating clinical use of genomes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11920–11927. doi: 10.1073/pnas.1201904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su MW, Tung KY, Liang PH, Tsai CH, Kuo NW, Lee YL. Gene-gene and gene-environmental interactions of childhood asthma: a multifactor dimension reduction approach. PloS one. 2012;7:e30694. doi: 10.1371/journal.pone.0030694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakravarti A. Genomics is not enough. Science. 2011;334:15. doi: 10.1126/science.1214458. [DOI] [PubMed] [Google Scholar]
- 63.Roberts NJ, Vogelstein JT, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. The predictive capacity of personal genome sequencing. Science translational medicine. 2012;4:133ra158. doi: 10.1126/scitranslmed.3003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome medicine. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutation research. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135:183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smarr L. Quantifying your body: A how-to guide from a systems biology perspective. Biotechnology journal. 2012;7:980–991. doi: 10.1002/biot.201100495. [DOI] [PubMed] [Google Scholar]
- 71.Nekrutenko A, Taylor J. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nature reviews Genetics. 2012;13:667–672. doi: 10.1038/nrg3305. [DOI] [PubMed] [Google Scholar]
- 72.Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazzocchi F. Complexity and the reductionism-holism debate in systems biology. Wiley interdisciplinary reviews Systems biology and medicine. 2012;4:413–427. doi: 10.1002/wsbm.1181. [DOI] [PubMed] [Google Scholar]
- 74.Kodama K, Horikoshi M, Toda K, Yamada S, Hara K, Irie J, Sirota M, Morgan AA, Chen R, Ohtsu H, et al. Expression-based genome-wide association study links the receptor CD44 in adipose tissue with type 2 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7049–7054. doi: 10.1073/pnas.1114513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen R, Corona E, Sikora M, Dudley JT, Morgan AA, Moreno-Estrada A, Nilsen GB, Ruau D, Lincoln SE, Bustamante CD, et al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLoS genetics. 2012;8:e1002621. doi: 10.1371/journal.pgen.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Institute of Medicine. [June 11th 2012];Evolution of Translational Omics: Lessons Learned and the Path Forward. URL: http://www.iom.edu/Reports/2012/Evolution-of-Translational-Omics.aspx. [PubMed]
- 77.Khoury MJ, Gwinn ML, Glasgow RE, Kramer BS. A population approach to precision medicine. American journal of preventive medicine. 2012;42:639–645. doi: 10.1016/j.amepre.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.On beyond GWAS. Nature genetics. 2010;42:551. doi: 10.1038/ng0710-551. [DOI] [PubMed] [Google Scholar]
- 79.Clark MJ, Chen R, Lam HY, Karczewski KJ, Euskirchen G, Butte AJ, Snyder M. Performance comparison of exome DNA sequencing technologies. Nature biotechnology. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lam HY, Clark MJ, Chen R, Natsoulis G, O’Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, et al. Performance comparison of whole-genome sequencing platforms. Nature biotechnology. 2012;30:562. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, Van Loo P, Van Den Bossche M, Catthoor K, Sabbe B, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nature biotechnology. 2012;30:61–68. doi: 10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- 82.Ideker T, Dutkowski J, Hood L. Boosting signal-to-noise in complex biology: prior knowledge is power. Cell. 2011;144:860–863. doi: 10.1016/j.cell.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu VW. A systems approach towards an understanding, diagnosis and personalized treatment of autism spectrum disorders. Pharmacogenomics. 2011;12:1235–1238. doi: 10.2217/pgs.11.94. [DOI] [PubMed] [Google Scholar]
- 84.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panagiotou G, Nielsen J. Nutritional systems biology: definitions and approaches. Annual review of nutrition. 2009;29:329–339. doi: 10.1146/annurev-nutr-080508-141138. [DOI] [PubMed] [Google Scholar]
- 86.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. The New England journal of medicine. 2012;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]


