Abstract
The health of an individual depends upon their DNA as well as environmental factors (environome or exposome). It is expected that although the genome is the blueprint of an individual, its analysis with that of the other omes, such as the DNA methylome, the transcriptome, proteome, as well as metabolome will further provide a dynamic assessment of the physiology and health state of an individual. This review will help to categorize the current progress of omics analyses, and how omics integration can be used for medical research. We believe that integrative Personal Omics Profiling (iPOP) is a stepping stone to a new road to personalized health care and may improve 1) Disease risk assessment, 2) Accuracy of diagnosis, 3) Disease monitoring, 4) Targeted treatments and 5) Understanding the biological processes of disease states for their prevention.
Introduction
Health care has always been personal in that patients are treated according to their individual symptoms, exposures and family history. Current medical exams measure a limited number of components in healthy individuals and slightly more in those with suspected diseases. However, the revolution in new technologies allows measurements of DNA and other molecules at an unprecedented level for more precise diagnostics and treatment at a personalized level.
A personal genome, the ~6 billion nucleotides that encode each individual, can now be deduced at a reasonable cost. The effects of environmental exposures (pathogens, food, and other contacts) are also likely to shape ones physiology and their effects on other omes such as epigenome (DNA methylation), transcriptome (RNA), proteome, metabolome, auto-antibodyome, and microbiome (summary in Table 1), can be measured by many new technologies (e.g. DNA sequencing, mass spectrometry, protein microarrays). These measurements, along with standard medical tests, are expected to capture the additional in-progress omic dimensions of an individual’s biological condition. The integration of omics information, referred to as integrated Personal –Omic Profiling (iPOP), is expected to better assist in health care in many ways including 1) disease risk assessment, 2) early and accurate diagnosis, 3) monitoring disease progression, 4) targeted therapeutic treatments and 5) understanding the biological basis of disease states and 6) disease prevention.
Table 1.
Selected topics in the expanding world of omics
| -ome | Level | Description | Selected resources |
|---|---|---|---|
| Genome | DNA | Complete/whole DNA sequence, chromosomes |
http://www.personalgenomes.org/ http://www.1000genomes.org/ dbSNP: http://www.ncbi.nlm.nih.gov/snp |
| Exome | DNA | DNA sequence assoc. to coding regions |
http://www.nhlbi.nih.gov/resources/exome.htm http://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/mendelian.htm |
| Epigenome | DNA/RNA | DNA methylation and histone modification, can affect chromatin and gene expression |
NIH Roadmap Epigenomics Mapping Consortium http://www.roadmapepigenomics.org/ |
| Methylome | DNA | DNA methylation | - |
| Regulome | DNA binding regions |
Regulation factors that affect gene expression |
ENCODE: ENCyclopedia Of DNA Elements http://www.genome.gov/ENCODE/ |
| Transcriptome | RNA | Gene expression, isoforms, miRNA, allelic specific expression |
http://www.h-invitational.jp/ http://www.ncbi.nlm.nih.gov/refseq/ |
| Splice-ome | RNA | Alternative splicing (not the spliceosome complex) |
http://jbirc.jbic.or.jp/h-dbas/ |
| Editome | RNA | RNA edits, variants not present in DNA |
- |
| miRNome | RNA | miRNAs | http://genetrail.bioinf.uni-sb.de/wholemirnomeproject/ |
| Proteome | protein | Protein expression, isoforms |
http://www.humanproteinpedia.org/ http://www.hprd.org/ |
| Autoantibodyome | protein | Antibody targeted against one’s own protein(s) |
- |
| Metabolome | metabolites | Small molecules of metabolism (eg. nucleotides, amino acids, vitamins) |
Human Metabolome Database: http://www.hmdb.ca/ |
| Metagenome | DNA | Genomes of multiple organisms |
https://www.ebi.ac.uk/metagenomics/ |
| Microbiome | DNA/RNA/ protein |
Microbial characterization at multiple human body sites |
http://www.human-microbiome.org/ NIH: http://www.hmpdacc.org/ |
| Interactome | All | Networks of all –omic interactions |
http://interactome.dfci.harvard.edu/ |
| Pharmacogenome | DNA/RNA/ protein |
-omic variants reflecting an individual’s response to drugs |
http://hapmap.ncbi.nlm.nih.gov/ http://www.pharmgkb.org/ |
Omics information can in principle be collected from a variety of tissues and cell types, although for medical purposes typically only accessible fluids such as blood or urine are analyzed in healthy individuals. Of particular importance in such studies is the collection of longitudinal information so that changes can be identified during the onset of disease states. However, for individuals with diseases, biopsies and surgically removed samples can be analyzed and subjected to detailed analyses.
Below we summarize the different omes and their application to personalized medicine.
The use of ‘–ome’ or ‘-omics’ as a suffix to represent an encompassing new field of study is rapidly growing, whereby in 2010 over 200 omic types were annotated in Medline (McDonald et al., 2012). Bolded –omes are those used in an initial iPOP study by Chen et al., 2012. In this case, iPOP collection was performed at relatively-healthy time points which was compared to un-healthy states (viral infections, green figure in Figure 1D). iPOP can be implemented over a lifetime; particularly informative with individuals with high disease risk assessment (from genome), and/or family histories of illness. This will allow for analysis of patterns in disease manifestation, progression and ideally identification of gene/pathway targets for treatment; many of which are hypothesized to overlap in large-scale population iPOP studies (personalized ↔ population).
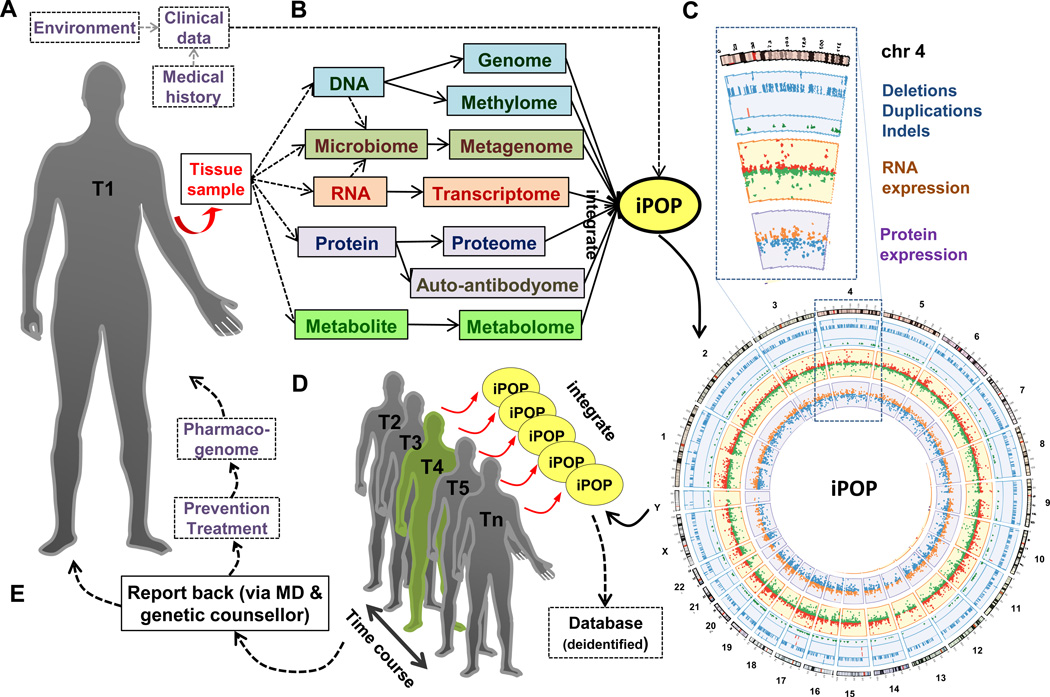
Figure 1. Schematic representing the implementation of iPOP for personalized medicine.
(A) Participant tissue sample (e.g. PBMC) is collected, while environment (incl. diet, exercise, etc.), medical history and clinical data are recorded. T1 is the first time point.
(B) Selected omic analysis involved in a sample iPOP study (Chen et al., 2012).
(C) Sample Circos plot (Krzywinski et al., 2009) of DNA (outer ring), RNA (middle ring) and protein (inner ring) data matching to chromosomes.
(D) iPOP performed and integrated at multiple time points: T2, T3, T4 (viral-infected), T5 up to Tn states, including disease-state(s). Grey and green forms represent relative-healthy individual and a disease-state, respectively.
(E) Report data back to genetic counsellor and medical practitioner with better informed choices for prevention and/or treatment (matched with pharmacogenetic data), if needed.
More than meets the eye: The complexity of personalized –omics
The 1941 article communicating the ‘one-gene/one-enzyme/one-function’ hypothesis by Beadle and Tatum, discussed the first insights that this was “ranging from simple one-to-one relations to relations of great complexity” (Beadle and Tatum, 1941). Crick in 1970 re-assessed his ‘central dogma of molecular biology’, and discussed the intricate inter- and intra- relationships between DNA ↔ RNA ↔ proteins (Crick, 1958). A myriad of different omes can currently be measured, each with its own contributions and challenges to detect disease states. Current research continues to reveal the multi-faceted mechanisms first within each omic level, as well as the crosstalk between the omes.
Genome (DNA)
The reference human genome was ’completed‘ in 2003 (Genomics, 2004) and is a haploid composite of multiple individuals. It has been revised over the years but still contains several gaps and errors. Since then the sequences of a large number of human genomes have been determined including with the Personal Genomes Project and the International Hapmap Project. Many of these have been determined at low coverage as part of the 1000 Genomes Project (Abecasis et al., 2010). In personal genome sequencing, genomes are typically sequenced using short read technologies (currently ~100 bp) and variants are called relative to the reference genome (Flicek et al., 2011; Snyder, Du, & Gerstein, 2010). The variants fall into three classes: 1) single nucleotide variations (SNVs), 2) small insertion and deletions (indels) typically 1-100 bp and 3) structural variants which are large rearrangements that include deletions, duplications, inversions, and translocations. To assign variants to each chromosome (i.e. DNA phasing) several technologies can be used such as sequencing of family members and imputation using known haplotypes. DNA phasing is important in order to identify compound heterozygous mutations (different mutations that lie in the same gene but on different chromosomes), and to better interpret disease risk. Other key distinctive features of the genome are pseudogenes, transposons and repetitive regions, which collectively comprise about 45% of the genome. While much attention has been placed on coding sequences (which comprise only ~1.5% of the genome and can be targeted by exome sequencing); growing focus is on non-coding regions of DNA (Boyle et al., 2012; Chen et al., 2012). For example, new insights based on the ENCODE (ENCyclopedia Of DNA Elements) project have revealed millions of transcription factor-binding sites, as well as epigenetic modifications (histone marks, chromatin accessibility, DNA methylation) that ultimately effect the transcription landscape (Birney et al., 2007). The genome sequence can be used to predict disease risk using two approaches: 1) examination of rare variants in protein coding genes that are highly penetrant and associated with human disease and 2) examination of complex disease risk by integration of information over multiple variants, each of low penetrance (Ashley et al., 2010). Curation of such DNA variants relative to different forms of cancers has been ongoing since 2005 as part of The Cancer Genome Atlas (TCGA). In spite of the many advances in this area, interpretation of most variants remains a formidable challenge requiring considerable effort (we spend 100 manual hours per genomes) and in many cases the effects of personal variants remain unclear.
Transcriptome
Analysis of gene expression levels of mRNAs and their various spliced isoforms is typically the main focus of transcriptomics, although there is interest in other RNAs such as rRNA, tRNA, miRNA, lincRNA, snoRNAs (Table 1). Changes in an individual’s RNA expression are tissue specific and time-course dependent. Measurements of RNA transcripts by RNA sequencing affords a very large dynamic range (greater than five orders of magnitude) enabling the detection of transcripts expressed at a low level. Deep RNA sequencing allows for more accurate quantification of expression of the heterozygous variants namely, allelic specific expression (ASE). Similarly those RNA variants that are absent at the DNA level, suggest the occurrence of post-transcriptional RNA-editing (editome), typically in mammals either A→G or I and C→U (Chen et al., 2012; Li et al., 2009). Many of these RNA variants result in missense or nonsense changes that can also be identified at the proteomic level.
Proteome and metabolome
The proteome and metabolome are expected to be more closely connected to phenotype and thereby provide more precise measures of a physiological state. Liquid chromatography mass spectrometry (LC-MS/MS) analysis can currently identify up to ~5000 expressed proteins, typically the most abundant proteins in the sample. A personal proteome that detects the variants present in an individual can be generated using a personal reference genome or transcriptome enabling a better detection of protein variants. Like RNA, protein expression is time course- and tissue-dependent. Increasing evidence shows that the expression levels of RNAs are only partially correlated with those of their protein counterparts (De Sousa Abreu, Penalva, Marcotte, & Vogel, 2009). Proteins can undergo several modifications, their detection limited by current methods. Such modifications include: phosphorylation (via kinases), ubiquitination, methylation, acetylation, glycosylation, oxidation and nitrosylation.
Metabolome profiling is a considerable challenge due to the diverse chemical nature of metabolites (e.g. hydrophobic/hydrophilic, basic/acidic). It is typically either targeted (e.g. GC-MS) which allows the analysis of several hundred metabolites, or untargeted (LC-MS) which reveals >4000 mass spectrometry peaks, a fraction of which can be tentatively assigned based on column retention time and molecular mass. Nonetheless, key metabolites have been associated with important diseases such as Type II Diabetes (glucose and branched amino acids) and cancer.
Integrative Analyses of Omics information
Omics information has been combined to better understand and monitor healthy and disease states. Two recent examples include cancer and healthy person profiling.
Cancer Omics
Cancer omics began with genome sequencing, which revealed that each individual cancer contains unique somatic mutations. These mutations often fall into particular pathways, for example in ovarian cancer patients, mutations often occur in PIK3/Rb, BRCA1 and Notch pathways (Cancer & Atlas, 2011). More recently, efforts have been made to incorporate other types of information, notably RNA-expression, DNA methylation, and phosphoprotein studies (using reverse phase arrays). These studies have revealed common genes and pathways that can be affected. For example, although mutations in the EGFR gene are uncommon in colon cancer, the EGFR signalling pathway is often elevated from phosphosignaling analyses (Perkins et al., 2010). As such, integrative profiling is expected to reveal such changes and provide a better understanding of the disease state and its etiology. There is also growing value in longitudinal genome analysis (monitoring somatic changes), particularly in cases with early genome information and/or family history that are associated to diseases like cancer (Dawson et al., 2013).
Personal Omics Profiling
Chen et al., 2012 discussed the first example of integrated Personalized Omics Profiling (iPOP), where the complete genome was sequenced and overlaid with the corresponding exome, transcriptome, proteome, metabolome and auto-antibodyome (Figure 1). This was performed on a 54 year old, initially healthy male, at 20 time points over the time course of 726 days which included two viral infections and the onset of Type 2 Diabetes (T2D). Peripheral blood mononuclear cells (PBMC) and serum or plasma were isolated for nucleic acid, protein and metabolite analyses. A plethora of biologically relevant information was gleaned using iPOP, and several novel highlights are summarized below (Figures 2 and 3):
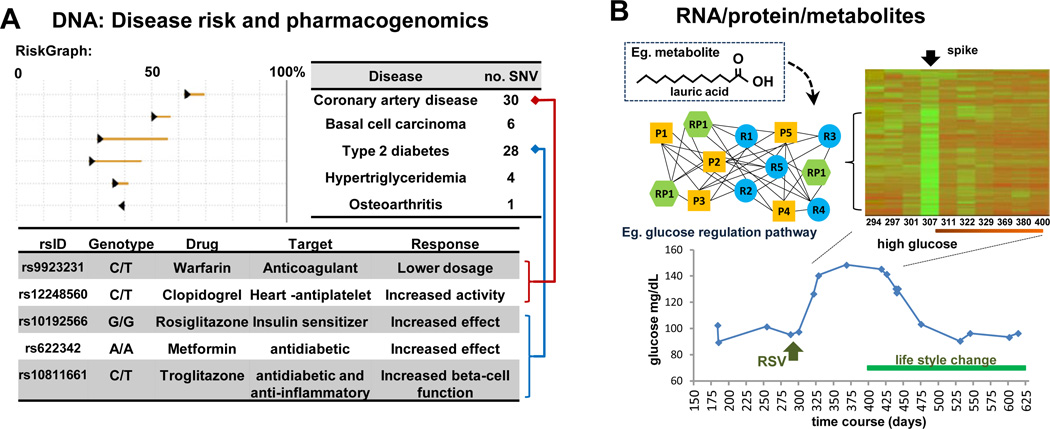
Figure 2. Highlights in iPOP.
(A) Integration of DNA variants to assess disease risk (RiskGraph, top panel) and a sample pharmacogenome (bottom panel). Arrow heads point in the direction of the change in post-test probability (%).
(B) Expression analysis (partial heatmap) of the transriptome and proteome over a time course spanning a respiratory syncytial viral (RSV) infection, with glucose monitoring (bottom, onset of T2D). Genes showing relative change in expression are clustered and represented as a network of inter- and intra-connected pathways: RNA (blue circle), protein (yellow square) and both RNA and protein (green hexagon). An example of a metabolite identified during the time course (inset panel).
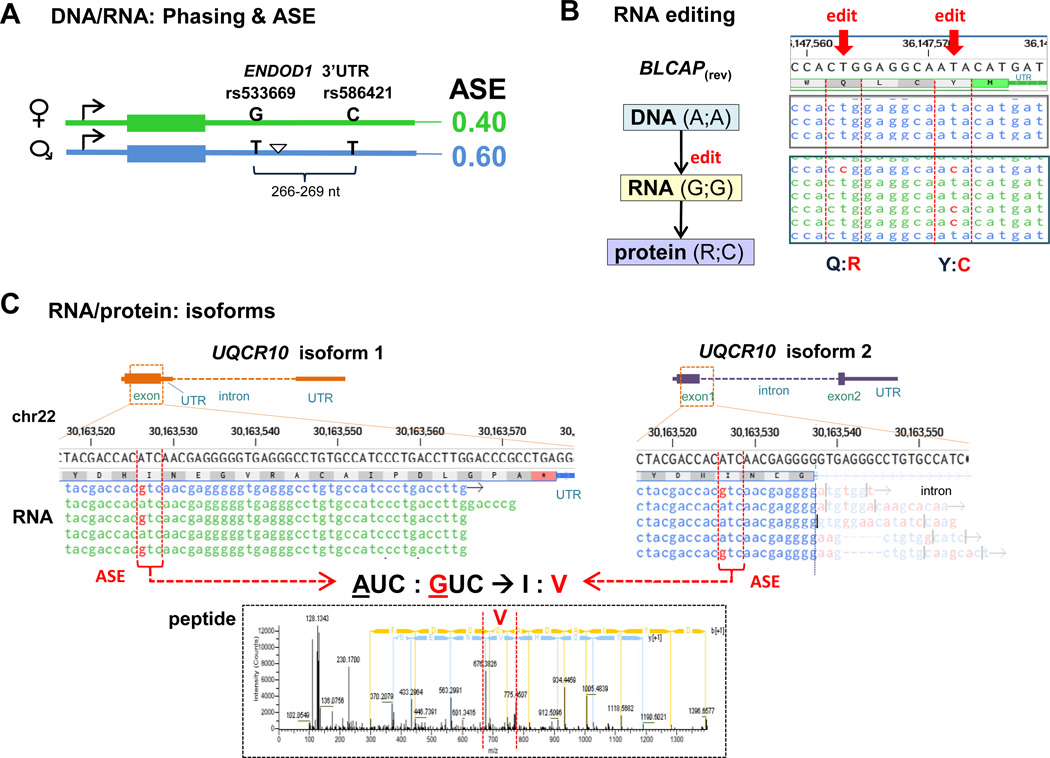
Figure 3. Highlights in iPOP.
(A) Sample phased DNA overlaid with RNA variant data corroborated with allelic specific expression (ASE) ratios for the ENDOD1 gene. Maternal transcript is expressed at 0.40, while the respective paternal transcript is expressed at 0.60. The triangle represents an indel in the paternally inherited transcript.
(B) RNA editing in BLCAP (red arrows) result in protein level amino acid changes. BLCAP is found on the reverse strand (rev), and A→G editing appear as T→C.
(C) An example of the diversity of isoforms observed in UQCR10 RNA and protein data. The two isoforms each contain the allelic specific variant A and G (rs76013375). Note isoform 2 spans into the intron position of DNA (faded); the true match is at the alternate spliced region located further downstream (not shown). RNA variant (A/G) results in amino acid change I:V, as identified in proteome mass spectrometric data (bottom).
For (B) and (C), DNAnexus was used as a genome browser, where red nucleic acid represents mis-matches to reference genome (top). Blue and green nucleic acid strands represent forward and reverse Illumina reads, respectively.
1) Disease-risk and pharmacogenetic assessment via DNA variants
A major component of iPOP analysis is the ability to accurately assess disease risk. This is challenging, given both the large number of genetic variants and the fact that many diseases likely involve combinations of both genetic and environmental factors. The RiskGraph (Ashley et al., 2010) takes into account age, gender, and ethnicity as well as multiple independent disease-associated genomic SNVs to calculate the subject’s posttest probability (%) of illness risk (Figure 2A, top). In this case, higher risks were shown for coronary artery disease and triglycemia (known family history) as well as T2D (unexpected). Importantly during the course of this study the subject experience high levels of glucose and Hemoglobin A1c (HgA1c) and was diagnosed as diabetic. Thus, the genome information indicated risk for a previously unsuspected disease which the subject acquired (and managed) during the study (Figure 2B, bottom).
A pharmacogenetic assessment for drug response to common pharmaceuticals was also ascertained and the response relevant to the subject potential disease treatments noted (Figure 2A, bottom). For example, sensitivity to particular statins (which were being taken by the subject) and metformin (which was not used but is pertinent to diabetes) was noted. This type of personalized matching of patients to medications has the potential to greatly reduce cost, treatment time and adverse side effects to medications. While genome analysis is predictive, complementary analysis can be used at the different –omic levels, including their overall crosstalk with environmental factors, to better gauge disease progression.
2) RNA, proteins and metabolites exhibit dynamic changes over disease-states and provide a more comprehensive view in monitoring disease progression
A key highlight to iPOP is monitoring changes in omics profiles over time. Molecules that are up- or down- regulated and linked to disease states are of particular interest as they can: A) be used for understanding mechanisms of disease progression and B) in turn, be targets for therapeutics. An example in this study showed that a number of RNAs, proteins and metabolites changed in abundance during both viral infections (human rhinovirus - HRV and respiratory syncytial virus -RSV) and the elevation of glucose levels. Both gradual changes as well as aberrant spikes in molecular profiles were systematically extracted from the profiles. Notably, of the biological pathways that showed spike maxima (Figure 2B, heatmap), these occurred post- RSV infection, at elevated glucose (Figure 2B, bottom) and HgA1c levels. Indeed, up-regulated RNA and/or the protein levels linked to genes involved in similar function were found to group together. Based on GO ontology, several biological pathways were enriched (network representation in Figure 2B, left). One pathway was involved in glucose regulation and insulin secretion which corroborated with clinical data (and the onset of T2D). Metabolites from overlapping time points also clustered in similar patterns to RNA and protein data. This included lauric acid (Figure 2B, inset), found to be associated to the sterol regulatory element binding transcription factor 1 (SREBF1) involved in glucose metabolism and lipid production (Nafikov et al., 2013). Thus, changes of RNA, protein and metabolic components were elucidated and provided a clearer picture of the biological changes that occurred during disease onset and progression.
3) DNA phasing to allele specific expression (ASE) and RNA editing
Though disease-risk can be estimated from DNA variants, the expression of genes and proteins is equally if not more, phenotypically relevant. In this study, additional variants were assigned to parental chromosomes (i.e. paternal and maternal), and heterozygous gene expression was monitored. The majority of allelic specific expression (ASE) was found at equal levels, although a number of RNAs exhibited differential allele expression. An example is represented in Figure 3A, where ASE variants on correspondent 3’ UTRs of the ENDOD1 (an endonuclease) transcript were each found to be differentially expressed, those on the maternal copy at 0.40 and paternal copy at 0.60. This data was integrated with Chip-Seq data (regulome) to further investigate the biology of selective transcript expression.
The editome is a new field of investigation whereby variants in RNA (undetectable in DNA) are observed. These are due to post-transcriptional modifications. Figure 3B illustrates two consecutive edit sites (red arrows) in BLCAP (bladder cancer-associated protein gene, on the reverse strand) resulting in amino acid changes at the protein level. A ratio of transcripts ~0.20 (A→G) were found to be edited in PMBCs, suggesting low levels of RNA editing are detectable (Chen et al., 2012). Edit sites are also commonly found in non-coding regions as well as non-genic regions, including Alus (Ramaswami et al., 2012).
Allele specific gene expression and RNA editing can be observed at the protein level. One example is shown in Figure 3C where the two known isoforms of UQCR10a mitochondrial ubiquinol-cytochrome c reductase, are both expressed, each containing an allelic specific variant A or G (rs76013375). This holistic approach of integration of the transcriptome with the proteome allowed for a full scope analysis of all the missense and nonsense changes detectable using mass spectrometry, whereby both amino acid types, I (AUC) and V (GUC), were expressed at the protein level (Figure 3C inset).
Overall, we speculate that allele specific gene expression and differential RNA editing is important for phenotypic differences among individuals in disease susceptibility and progression.
Progress and the future of iPOP and health care
Development of iPOP and its application to an individual’s health needs have focused on: 1) the advancement of high-throughput technology, 2) data storage and sharing, and 3) world-wide bioethics and discussion on health-states and the environment.
Current genome-wide methods, as Illumina sequencing, Complete Genomics, SOLiD (ABI), 454 Life Sciences, Ion Torrent (Life Technologies) and other platforms are improving in cost, speed, and quality. For genomic sequencing, the cost has greatly reduced from $500M-1B/genome in 2001 to <5K/genome (Green & Guyer, 2011). Still there are significant challenges such as sequencing and mapping errors that are present from using short reads. De novo and direct sequencing are technologies that are still under development, with limitations in quality and depth. Likewise, the analyses of large numbers of proteomes and metabolomes can be challenging, as can be the storage and dissemination of large data sets.
The use of diverse technologies and the generation of large data reflect the growing need for an amalgamation and cooperation of overlapping expertise in diverse fields. In our Chen et al., study, the least-squares spectral time-series analysis was implemented to compensate for unevenly sampled data when integrating the transcriptome, proteome and metabolome (Figure 2B). This method was based on methodology developed for astronomical observations by physicists Lomb and Scargle. Consortiums are being formed for optimization of resources and expertise, and omic methods as iPOP are being recognized as tangible study options in multi-disciplinary sciences.
The growing understanding of the multifaceted layers of omics, as the regulation of allele specific expression (ASE) resulting in diverse translated products (Figure 3C); reveal the importance of an integrated approach. Such studies will improve understanding of compound heterozygous mutations (inherited and private), as well as compensatory mutations, which may unravel the biology of complex diseases. Longitudinal studies, whereby healthy versus disease-states can be compared (Figures 1D, 2B), reveal clearer biological patterns matched to an individual and captured in a dynamic environment. Thus, more precise methods of monitoring, treatment and prevention can be applied and fine-tuned with each patient. In turn, iPOP studies overlapping large cohorts are underway for a better understanding of overall biochemical mechanisms in disease manifestation and progression.
The medical interpretation of iPOP is a balance of both nature (omics) and nurture (environment). Major concerns in data interpretation and re-distribution to the individual (and to the public) raise bioethical questions on information privacy (de-identification). The challenging path from the laboratory to the clinic is also being addressed; few biomarkers of the 150K described through research have made it to the clinic (Poste, 2011) and in our study it is yet undetermined which sets of markers are likely to be of highest clinical significance. Genetic counsellors partnered with medical staff are indispensable for a patient assessing their omics and medical choices, both biologically and psychologically. Particularly in cases of high disease risk, the individual can make a more informed decision on their medical course of action. In this light, iPOP has opened the way to a new perspective on the plasticity of biological processes that underlie health states in flux. Integrated personal omics profiling is facilitating the discovery of novel genes and pathways and their interplay with the environment, allowing for improved health monitoring and targeted therapeutics, and leading the road to an improved personalized health care.
ACKNOWLEDGMENTS
We thank G. Mias and R. Chen for insightful discussions. M.S. is a founder and member of the scientific advisory board for Personalis, a member of the scientific advisory board of GenapSys and a consultant for Illumina.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
J.L.P.T declares no conflict of interest.
REFERENCES
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs Ra, Hurles ME, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA. 1941;27(11):499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos Ja, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer T, Atlas G. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R*, Mias GI*, Li-Pook-Than J*, Jiang L*, Lam HYK, Chen R, Miriami E, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148(6):1293–1307. doi: 10.1016/j.cell.2012.02.009. *equal contribution
- Crick F. Central Dogma of Molecular Biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- De Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol bioSyst. 2009;5(12):1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, et al. Ensembl 2011. NAR. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomics C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, Leproust E, Zhang K, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324(5931):1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- McDonald D, Clemente JC, Kuczynski J, Rideout J, Stombaugh J, Wendel D, Wilke A, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience. 2012;1(1):7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafikov RA, Schoonmaker JP, Korn KT, Noack K, Garrick DJ, Koehler KJ, Minick-Bormann J, Reecy JM, Spurlock DE, Beitz DC. Sterol regulatory element binding transcription factor 1 (SREBF1) polymorphism and milk fatty acid composition. J Dairy Sci. 2013;96(4):2605–2616. doi: 10.3168/jds.2012-6075. [DOI] [PubMed] [Google Scholar]
- Pan S, Dewey FE, Perez MV, Knowles JW, Chen R, Butte AJ, Ashley Ea. Personalized Medicine and Cardiovascular Disease: From Genome to Bedside. Curr Cardiovasc Risk Rep. 2011;5(6):542–551. [Google Scholar]
- Perkins G, Lièvre A, Ramacci C, Méatchi T, De Reynies A, Emile J-F, Boige V, et al. Additional value of EGFR downstream signaling phosphoprotein expression to KRAS status for response to anti-EGFR antibodies in colorectal cancer. International journal of cancer. Journal international du cancer. 2010;127(6):1321–1331. doi: 10.1002/ijc.25152. [DOI] [PubMed] [Google Scholar]
- Poste G. Bring on the biomarkers. Nature. 2011;469(7329):156–157. doi: 10.1038/469156a. [DOI] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012;9(6):579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Du J, Gerstein M. Personal genome sequencing : current approaches and challenges. Genes Dev. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]